Significance
The giant panda and red panda are obligate bamboo-feeders that independently evolved from meat-eating ancestors and possess adaptive pseudothumbs, making them ideal models for studying convergent evolution. In this study, we identified genomic signatures of convergent evolution associated with bamboo eating. Comparative genomic analyses revealed adaptively convergent genes potentially involved with pseudothumb development and essential bamboo nutrient utilization. We also found that the umami taste receptor gene TAS1R1 has been pseudogenized in both pandas. These findings provide insights into genetic mechanisms underlying phenotypic convergence and adaptation to a specialized bamboo diet in both pandas and offer an example of genome-scale analyses for detecting convergent evolution.
Keywords: de novo genome, phenotype convergence, amino acid convergence, positive selection, pseudogenization
Abstract
Phenotypic convergence between distantly related taxa often mirrors adaptation to similar selective pressures and may be driven by genetic convergence. The giant panda (Ailuropoda melanoleuca) and red panda (Ailurus fulgens) belong to different families in the order Carnivora, but both have evolved a specialized bamboo diet and adaptive pseudothumb, representing a classic model of convergent evolution. However, the genetic bases of these morphological and physiological convergences remain unknown. Through de novo sequencing the red panda genome and improving the giant panda genome assembly with added data, we identified genomic signatures of convergent evolution. Limb development genes DYNC2H1 and PCNT have undergone adaptive convergence and may be important candidate genes for pseudothumb development. As evolutionary responses to a bamboo diet, adaptive convergence has occurred in genes involved in the digestion and utilization of bamboo nutrients such as essential amino acids, fatty acids, and vitamins. Similarly, the umami taste receptor gene TAS1R1 has been pseudogenized in both pandas. These findings offer insights into genetic convergence mechanisms underlying phenotypic convergence and adaptation to a specialized bamboo diet.
Similar selective pressures can lead to the parallel evolution of identical or similar traits in distantly related species, often referred to as adaptive phenotypic convergence (1–3). A critical mechanism underlying phenotypic convergence is genetic convergence, including the same metabolic and regulatory pathways, protein-coding genes, or even identical amino acid substitutions in the same gene (1–3). However, genome-wide surveys of convergent evolution are relatively rare (4–6), and more empirical genome-scale studies are needed to elucidate the genetic bases of phenotypic convergence.
The giant panda (Ailuropoda melanoleuca) and red panda (Ailurus fulgens), two endangered and sympatric species that diverged approximately 43 million years ago (Mya), have distinct phylogenetic positions in the order Carnivora (7). The giant panda belongs to the family Ursidae (8), whereas the red panda belongs to the family Ailuridae within the superfamily Musteloidea (9). Uniquely in the Carnivora, both pandas are specialized herbivores with an almost exclusive bamboo diet (>90%), although they still retain a typical Carnivore digestive tract. Bamboo is a low-nutrition, high-fiber food with only 13.2% protein, 3.4% fat, and 3.3% soluble carbohydrate (10). Therefore, efficient absorption of nutrients, especially essential amino acids, essential fatty acids, and vitamins, from the specialized bamboo diet is vital to growth, development, and reproduction in both species.
Remarkably, both pandas have evolved a pseudothumb, an enlarged radial sesamoid (Fig. 1) that significantly facilitates feeding dexterity by grasping bamboo (11–14), a phenotype of long-standing interest to evolutionary biologists. D. Dwight Davis, for example, noted that in the giant panda, “the highly specialized and obviously functional radial sesamoid has a specific, but probably very simple, genetic base” (13), and Stephen J. Gould later featured the new digit in the title of his popular 1980 book, “The Panda’s Thumb” (14). In the red panda, the pseudothumb also facilitates arboreal locomotion (15). Despite this widespread interest, its genetic basis has remained elusive. These shared characteristics between giant and red pandas represent a classic model of convergent evolution under presumably the same environmental pressures.
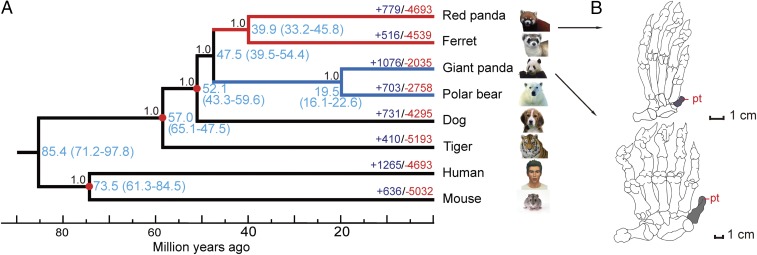
Fig. 1.
Genome-wide phylogenetic tree of giant and red pandas, and their convergent pseudothumbs. (A) Phylogenomic tree, divergence time (blue), and expansion (purple, +) and contraction (red, ‒) of gene family in red and giant pandas, and other six Eutherian species. Three divergence times (red node) were used as the calibration points for estimating divergence time. The estimated divergence times were showed with 95% confidence intervals (in parentheses). The posterior probabilities for each branch of the tree were showed. (B) Diagram of pseudothumb (pt) of red panda (Upper) and giant panda (Lower).
In this study, we identified genomic signatures of convergent evolution in both pandas by comparing two genome assemblies, a de novo sequenced red panda genome and a much improved giant panda genome assembly with added sequencing data. These findings yield rich insights into pseudothumb development and nutritional utilization of bamboo.
Results and Discussion
We sequenced the genome of a wild male red panda by using the Illumina Hiseq 2000 platform with a whole-genome shotgun sequencing strategy (SI Appendix, SI Materials and Methods). A total of 292 Gb of sequence data (121.7-fold genome coverage) was generated (SI Appendix, Table S1). The genome size of the final de novo assembly was 2.34 Gb, comparable to that of the ferret (2.41 Gb) and dog (2.44 Gb), with contig N50 of 98.98 Kb and scaffold N50 of 2.98 Mb (SI Appendix, Figs. S1–S5 and Table S2). Alignment of the genome scaffolds to BAC clone sequences indicated that scaffold coverage was in the range of 97.04‒99.85% without any scaffold inconsistencies (SI Appendix, Fig. S6 and Table S3), Core Eukaryotic Genes Mapping Approach (CEGMA) evaluation (16) found that 242 (97.58%) of 248 core eukaryotic genes were complete (SI Appendix, Table S4), and Benchmarking Universal Single-Copy Orthologs (BUSCO) assessment (17) on genome assembly showed that 2,632 (87.07%) of 3,023 conserved vertebrate genes were assembled to be complete (SI Appendix, Table S5). A total of 21,940 protein-coding genes were annotated by combining homology-based and ab initio gene prediction methods, with 90.77% of gene models supported by RNA-seq transcripts (SI Appendix, Fig. S7 and Table S6). BUSCO assessment on gene annotation showed that 2,533 (83.79%) of 3,023 conserved vertebrate genes were annotated to be complete (SI Appendix, Table S5). Repeat elements occupied 41.23% of the red panda genome, similar to that of the giant panda (41.29%) and dog (41.95%) (SI Appendix, Fig. S8 and Tables S7–S11). To make the assembly quality of the giant panda genome comparable to that of the red panda genome, we reassembled the giant panda genome by combination of ∼82 × new sequencing data and published mate-pair reads (SI Appendix, Table S12) (18). The reassembled genome was significantly improved compared with the reference (version ailMel1) (18), with contig N50 from 40 Kb to 126.71 Kb and scaffold N50 from 1.28 Mb to 9.9 Mb (SI Appendix, Table S13). Furthermore, extra contig sequences of 136 Mb were assembled, presumably from gap regions of the reference genome. Totally, 23,371 protein-coding genes were annotated in the improved genome, where 88.79% of gene models were verified by RNA-seq transcripts (SI Appendix, Fig. S7 and Table S14).
To reveal the genomic signatures of convergent evolution in both pandas, we first constructed a genome-wide phylogenetic tree combining the published genomes of the polar bear, ferret, dog, tiger, human, and mouse (SI Appendix, Table S15). A total of 171,041 protein-coding genes from these eight species were used for gene family analysis, and 14,534 gene families were identified (SI Appendix, Fig. S9), including 2,855 single-copy true orthologous genes across all eight species. After removing 326 genes with convergent amino acid substitutions and excluding the third codon positions of exons, the constructed genome-wide phylogenetic tree confirmed recent molecular conclusions (7–9) that the giant panda belongs to the family Ursidae together with the polar bear, whereas the red panda and ferret belong to the superfamily Musteloidea (Fig. 1). Based on the 133 genes evolving under the strict molecular clock, a divergence time of 47.5 Mya (95% confidence interval, 39.5∼54.4 Mya) between giant and red pandas was derived by using three calibration points (Fig. 1). This result was slightly higher than previous molecular-based estimate of 43 Mya (7).
Based on homologous gene annotations by syntenic alignment across the eight species (Fig. 1), 14,254 orthologous genes were used for genomic convergence and positive selection analyses. First, convergent amino acid substitutions between both pandas were identified based on Zhang and Kumar’s method (19). Considering noise resulting from chance amino acid substitutions (6, 20), we performed a statistical test to compare the observed number of convergent sites with random expectation under the JTT-fgene and JTT-fsite amino acid substitution models, respectively (21), and found that 1,066 and 645 genes with convergent amino acid substitutions contained significantly more sites than the random expectation (q < 0.05). Because the result under the JTT-fsite model has a relatively large bias when the number of species analyzed is small (eight species in our study), we used the result under the JTT-fgene model for the next analysis. Because of possible impact of gene tree discordance on the identification of amino acid convergence (3, 22), we further removed 15 genes whose gene trees supported the clustering of the giant and red panda lineages. Second, 434 positively selected genes were identified by a branch-site model in PAML (SI Appendix, Tables S16–S18) (23). Similar to the above, because gene tree discordance could also affect the identification of positive selection (24), we removed 18 genes whose positive selection signatures were lost under their respective gene trees. Finally, to obtain more conservative signatures of adaptive convergence, we focused on positively selected genes with nonrandom convergent amino acid substitutions (i.e., adaptively convergent genes) (5, 20). As a result, 70 adaptively convergent genes were identified (SI Appendix, Tables S19 and S20), and gene ontology and KEGG enrichment analyses discovered significant terms and pathways involved in limb development and nutrient utilization, including appendage and limb development (GO:0048736, P = 0.0321), cilium assembly (GO:0042384, P = 0.0376), protein digestion and absorption (ko04974, P = 0.0086), and retinol metabolism (ko00830, P = 0.0217) (SI Appendix, Tables S21 and S22).
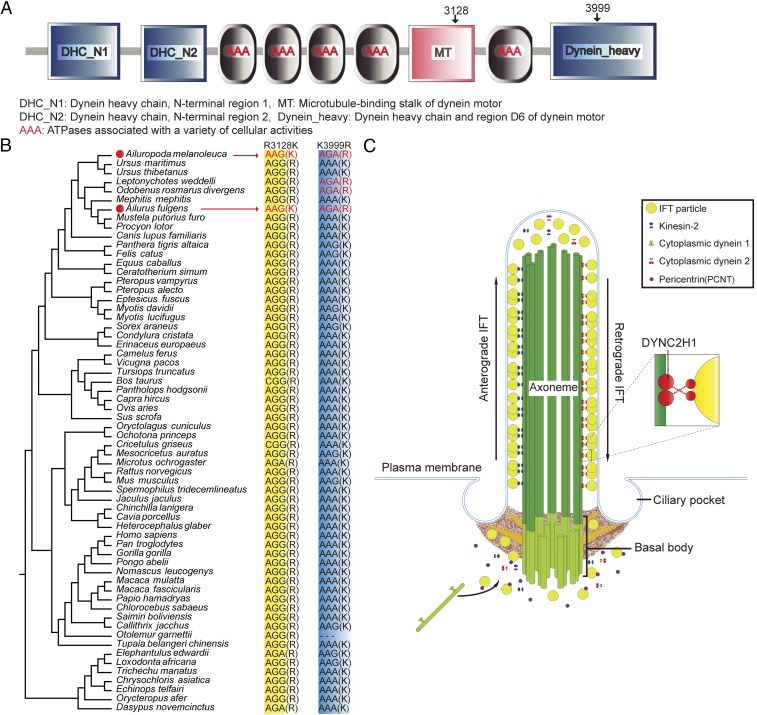
Among the 70 adaptively convergent genes, DYNC2H1 and PCNT are involved in limb development and their missense or null mutations result in a polydactyly phenotype and abnormal skeletogenesis in both mice and humans (25–28). These findings suggest that convergent amino acid substitutions in these genes may introduce subtle changes in the functional spectrum of focal proteins and consequently contribute to pseudothumb development in both pandas. The DYNC2H1 protein is a core component of the dynein complex, the motor of retrograde intraflagellar transport (IFT) during ciliogenesis (25, 26). Two convergent substitutions of R3128K and K3999R were identified (Fig. 2 A and B and Table 1). In particular, the R3128K substitution is located in a stalk domain between AAA domains 4 and 5 implicated in microtubule binding (Fig. 2A) (28). It has been reported that amino acid changes in the stalk domain of DYNC2H1 affect retrograde IFT and produce abnormal primary cilia (26). Primary cilia are signal transduction organelles of the Sonic Hedgehog (SHH) signaling pathway, and abnormal cilia would inhibit the functions of GLI3, a major downstream target of the SHH pathway (25). Dysfunction of GLI3 produces ectopic digits and polydactyly in mice (29). Therefore, structural or functional variation in cilia may well contribute to developmental novelty of bones and limbs through the SHH pathway and GLI3 blockade (25). Indeed, across 62 Eutherian species, the R3128K substitution occurs exclusively in giant and red pandas and is verified by more additional individuals (Fig. 2B and SI Appendix, Table S23–S25). Moreover, these mutations have not been reported in population-level SNP datasets or databases of polar bears, dogs, humans, and mice (SI Appendix, Table S25), highlighting the uniqueness of these variations and their potential functional importance in pseudothumb development. Similarly, the centrosomal coiled-coil protein PCNT is also involved in primary cilia assembly by forming a complex with IFT proteins (e.g., IFT20, IFT57, IFT88) (Fig. 2C, Table 1, and SI Appendix, Fig. S10). PCNT modification could affect basal body localization of IFT proteins, thus inhibiting primary cilia assembly (30). We propose that convergent amino acid substitutions in the two genes may work synergistically for pseudothumb development in both pandas, although their effects on other aspects of skeletal development remain elusive. This inference warrants further experimental validation in the future.
Fig. 2.
Adaptively convergent genes closely related with limb development. (A) Structural domains of DYNC2H1 protein and the locations of two convergent amino acid sites. The structural domain annotation derives from the SMART database. (B) Comparison of two convergent amino acid substitutions of DYNC2H1 gene among the genomes of 59 Eutherian species and the gene fragments of three other Eutherian species (Ursus thibetanus, Mephitis mephitis, and Procyon lotor). The first convergent site is R3128K and the amino acid K only occurs in giant and red pandas. The second convergent site is K3999R and the amino acid R only occurs in giant and red pandas, Weddell seal, and walrus. (C) Roles of DYNC2H1 and PCNT proteins in IFT and ciliogenesis. DYNC2H1 is a core component of cytoplasmic dynein 2 complex.
Table 1.
Positively selected genes with nonrandom convergent amino acid substitution between giant and red pandas, which are closely related with limb development and essential nutrient utilization
| Gene symbol | Full gene name | P value | FDR | Convergent amino acid substitution |
| DYNC2H1* | Cytoplasmic dynein 2, heavy chain 1 | 1.06e-04 | 0.0372 | R3128K, K3999R |
| PCNT | Pericentrin | 3.18e-05 | 0.0097 | S2327P, Q2458R |
| PRSS1* | Protease, serine, 1 | 3.47e-04 | 0.0362 | D119N, I140V |
| PRSS36 | Protease, serine, 36 | 7.73e-12 | 3.45e-09 | R57S |
| CPB1* | Carboxypeptidase B1 | 3.13e-06 | 4.73e-04 | V218F, T271I, M310L |
| GIF* | Gastric intrinsic factor | 2.42e-04 | 0.0363 | M212L, D406K, H407D |
| CYP4F2 | Cytochrome P450, family 4, subfamily F, polypeptide 2 | 9.99e-06 | 0.0022 | M92F |
| CYP3A5* | Cytochrome P450, family 3, subfamily A, polypeptide 5 | 2.58e-09 | 3.03e-06 | T363S, V393L, M395I, T400S |
| ADH1C* | Alcohol dehydrogenase 1C (class I), gamma Polypeptide | 1.75e-04 | 0.02 | V187I |
The gene passed the nonrandom convergence test not only under the JTT-fgene substitution model but also under the JTT-fsite model.
Bamboo is an almost exclusive source of essential amino acids, essential fatty acids, and vitamins for giant and red pandas. To meet this nutritional challenge, pandas need to improve efficiency of nutrient absorption and utilization and, therefore, signatures of adaptive convergence related to essential nutrient utilization might be observed. Three genes (PRSS1, PRSS36, and CPB1) involved in dietary protein digestion showed adaptive convergence (Table 1 and SI Appendix, Fig. S10). All of these proteins belong to serine proteases and are secreted by the pancreas into the small intestine. Interestingly, both PRSS1 and PRSS36 are endopeptidases for proteolytic cleavage of Lys or Arg residues from the carboxyl terminal, whereas CPB1 is an exopeptidase and acts on preferential release of Lys or Arg from C-terminal. Quantitative measurements indicate that bamboo has much lower Lys and Arg content than animal meats and plant leaves (SI Appendix, Table S26). Taken together, it seems that the adaptive convergence of these genes may in synergy result in elevated efficiency for releasing Lys and Arg from dietary proteins and amino acid recycling, thus offsetting the limited nutrient supply in bamboo.
Whereas vitamins A and B12 and arachidonic acid are essential nutrients, they are either absent or their content in bamboo is much lower than in meat, nuts, or green plants (SI Appendix, Tables S27 and S28). Four genes (ADH1C, CYP3A5, CYP4F2, and GIF) involved in utilization of these nutrients were identified to be under adaptive convergence (Table 1 and SI Appendix, Fig. S10). ADH1C and CYP3A5 are involved in the regulation of vitamin A metabolism that is essential to dark vision maintenance. ADH1C catalyzes conversion between retinal and retinol, whereas CYP3A5 is involved in the degradation of retinoic acids to prevent detrimental accumulation of excessive vitamin A (31). Vitamin A is needed by the retina of the eye in the form of retinal, which combines with protein opsin to form rhodopsin, the light-absorbing molecule necessary for dark vision (32). The giant and red pandas are active both in the daytime and at night, with slightly lower activity rate at night (10, 33). Because vitamin A exists only in meat foods, both pandas must absorb dietary β-carotene as the source of vitamin A, although the content in bamboo is low. Therefore, the adaptive convergence of the two genes may improve the utilization of vitamin A, which may function to meet the need of maintaining dark vision (SI Appendix, Fig. S11).
It has been suggested that a long-term vegetarian diet is prone to vitamin B12 deficiency because vitamin B12 cannot be supplied by plant foods and is synthesized exclusively by gut microbes (34, 35). Vitamin B12 deficiency is an important risk factor for cardiovascular disease such as angiosclerosis (34, 35), this nutrient challenge needs to be overcome by both obligate bamboo-eating pandas. GIF, a glycoprotein secreted by parietal cells of the gastric mucosa, is essential for adequate absorption of vitamin B12 and its adaptive convergence may improve vitamin B12 absorption efficiency (34). The cytochrome P450 enzyme CYP4F2 can catalyze arachidonic acid into 20-hydroxyeicosatetraenoic acid, an important bioactive substance for regulating vascular endothelial cells with antiangiosclerosis activity (36), thus potentially alleviating the physiological consequences of vitamin B12 deficiency (SI Appendix, Fig. S11).
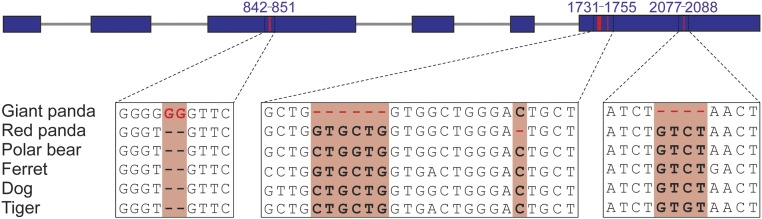
The selective relaxation of functional constraint on protein-coding genes may occur during the dietary shift and specialization of both pandas. Thus, we identified the signatures of pseudogenization in both pandas based on genome-wide comparison of these eight species. We identified 129 and 140 pseudogenes in giant and red pandas, respectively, among which 10 pseudogenes were shared (SI Appendix, Table S29). Interestingly, the umami taste receptor gene TAS1R1 has been pseudogenized in both pandas (Fig. 3), representing a remarkable scenario of genetic convergence. Umami is a critical taste sense for meat-eating animals that can perceive components of meat and other protein-rich foods through the umami taste receptor TAS1R1/TAS1R3 heterodimer (37). In the red panda, TAS1R1 has become a pseudogene because of one nucleotide deletion in the sixth exon, as confirmed by Sanger sequencing of three additional individuals (Fig. 3 and SI Appendix, Table S24), whereas the loss of function of TAS1R1 in the giant panda is due to three insertion/deletion mutations in the third and sixth exons, as reported (18, 38). In contrast, TAS1R3 is an intact gene rather than a pseudogene in both pandas. Based on the change in the rate ratio (ω) of nonsynonymous to synonymous substitutions (SI Appendix, SI Materials and Methods), we infer that the functional constraint on TAS1R1 in red pandas was relaxed approximately 1.58 Mya (95% confidence interval, 0.1 ∼4.36 Mya) (SI Appendix, Fig. S12). The fossil records from sister lineages of the red panda (Pristinailurus bristoli and the genus Parailurus) suggest that the ailurines were partially herbivorous from the late Miocene (7 ∼4.5 Mya) (33). Thus, pseudogenization of TAS1R1 in red pandas may have occurred after its diet was at least partially herbivorous, as in giant pandas (38). Convergent pseudogenization of TAS1R1 in both pandas may be an evolutionary response to the dietary shift from carnivory and omnivory to herbivory.
Fig. 3.
Convergent pseudogenization of umami receptor gene TAS1R1 between the giant and red pandas, as opposed to the polar bear, ferret, dog, and tiger. The giant panda has one 2-bp insertion on the third exon and two deletions (6-bp and 4-bp) on the sixth exon, and the red panda has one 1-bp deletion on the sixth exon.
Convergent evolution has long interested evolutionary biologists. Classic examples include the wings of bats and birds, echolocation in bats and dolphins, and adaptation of marine mammals to extreme marine environments (1–3). Although the functional nature of these convergent specializations is often obvious, the genetic basis underpinning particular examples of convergent evolution is far less clear. Charles Darwin suggested that convergent evolution stems from similarity in independent changes that underpin the same features in different organisms (39). Although there have been advances in understanding the molecular basis of such parallel and independent phenotype convergence in recent decades, insights at the genomic level are rare (4–6, 20). To survive in a novel environment, animals can drastically change their diet, such as from carnivory to omnivory, to herbivory, or even become dietary specialists (40), all of which have profound impacts on species ecology, behavior, physiology, and even morphology and genetics. The giant panda and red panda are obligate bamboo-feeders that evolved from meat-eating ancestors and are remarkable examples of dietary shift and specialization (41), representing an ideal model to study convergent evolution in these traits. In the present study, comparative genomics has revealed the signatures of adaptive protein convergence associated with digestive physiology and limb development, taking the first step toward confirming D. Dwight Davis’s genetic hypothesis for the pseudothumb (13). Furthermore, we have identified convergent pseudogenization related to dietary shift from the perspective of selective relaxation of functional constraints on protein-coding genes. Our findings provide rich insights into genetic convergence mechanisms underlying phenotypic convergence and adaptation to a specialized bamboo diet in both pandas. These findings demonstrate that genetic convergence occurred at multiple levels spanning metabolic pathways, amino acid convergence, and pseudogenization, providing a fascinating example for genome-scale convergent evolution analysis of dietary shift and specialization.
Materials and Methods
Additional detailed information is provided in SI Appendix, SI Materials and Methods, including the genome and transcriptome sequencing, de novo genome assembly, genome assembly assessment, and genome annotation. Animal care and experiments were conducted according to the guidelines established by the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, 2013) and were approved by the Committee for Animal Experiments of the Institute of Zoology, Chinese Academy of Sciences, China.
Phylogenomic Tree and Divergence Time Estimation.
Protein sequences for each single-copy gene family were concatenated and aligned by MUSCLE (42), and poorly aligned sites were trimmed by trimAl (43). Then protein sequence alignment was transformed back to coding DNA sequence alignment. Considering the potential impacts of convergent amino acid substitutions and the large GC-content variance in the third codon position of exons (44, 45), we removed the genes with convergent amino acid substitutions and excluded the third codon position of exons before constructing the phylogenomic tree. Additionally, we considered the partition of the first and second codon positions and separately determined their best-fitting nucleotide substitution models. The nucleotide substitution model was determined by ModelGenerator (46) and selected by Bayesian information criterion. MrBayes 3.2.6 (47) was then used to reconstruct the phylogenomic tree. The Markov chain Monte Carlo analyses were run for 2,500,000 generations, sampled every 1,000 generations, and the first 25% samples were burn-in. The posterior probability for each branch of the phylogenetic tree was determined from the remaining samples. To estimate divergence times between species, we performed strict molecular clock test in baseml program of PAML 4.8 (23) for those single-copy orthologous genes used to reconstruct phylogenomic tree. After removing the genes that significantly deviated from molecular clock model and excluding the third codon positions of exons, the mcmctree program of PAML 4.8 (23) was used for divergence time estimation, with three calibration points applied: the fossil-based divergence time of Caniformia and Feliformia (min = 43 Mya, max = 65 Mya) (7, 48–50), the fossil-based divergence time of Canidae and Arctoidea (min = 37 Mya) (50, 51), and the divergence time of Primatomorpha and Glires (min = 65 Mya) (50).
Homologous Gene Annotations by Syntenic Alignment Across Multiple Species.
The draft nature of the genome made detailed comparisons of protein-coding genes inefficient because gene annotation was frequently incomplete because of gaps or missing exons. To gain a better resolution for gene-level analysis, we took advantage of the prealigned 100 vertebrate genomes in the University of California, Santa Cruz genome browser [hgdownload.soe.ucsc.edu/goldenPath/hg19/multiz100way/, in multiple alignment format (MAF) file format], where the mouse, dog, ferret, and giant panda genomes have been aligned against the human genome (hg19, GRCh37). The corresponding human proteins were then used to blast against the red panda, tiger, and polar bear gene sets, and genomic sequences were retrieved from each species by extending 5 Kb from either side of the gene’s start and stop codon positions. Genome sequences of these eight species were then aligned by CLUSTALW 2.1 (52). Exons with Ns or internal stop codons were excluded, and intact exons of the same gene were concatenated for positive selection and genomic convergence analyses.
Identification of Positively Selected Genes.
Based on the MAF alignment sequences from six Carnivora species (red panda, giant panda, polar bear, ferret, dog, and tiger), positive selections in the red panda and giant panda were tested under the reconstructed phylogenomic tree (Fig. 1A) by using PAML 4.8 (23). The branch-site model of codon evolution (53) was used with model = 2 and NSsites = 2. We compared model A (allows sites to be under positive selection; fix_omega = 0) with the null model A1 (sites may evolve neutrally or under purifying selection; fix_omega = 1 and omega = 1), with likelihood ratio tests (LRT) performed by the Codeml program in PAML. Significance (P < 0.05) of the compared LRTs was evaluated by χ2 tests from PAML, assuming that the null distribution was a 50:50 mixture of a χ2 distribution with a point mass at zero, and QVALUE in R was used to correct for multiple testing (54) with a false discovery rate (FDR) cutoff of 0.1. To find positively selected genes related to the pseudothumb development and bamboo nutrient utilization, positive selections were analyzed through six different datasets by using the giant panda, red panda, or both pandas as foreground branches (SI Appendix, Table S16). Then the positively selected genes were combined together for further analysis. Because gene tree discordance resulting from incomplete lineage sorting or hybridization may affect the identification of positive selection signatures (24), we constructed a gene tree for each positively selected gene by using MrBayes 3.2.6, and then reperformed the positive selection test when the gene tree was different from the expected species tree. We removed those genes whose positive selection signature was lost under the gene tree. Finally, the functional enrichment of these positively selected genes in GO terms and KEGG pathways was tested by using the GeneTrail2 method (55), with the 14,254 orthologous genes as the reference gene set. Significantly enriched category included at least two genes, and the hypergeometric test was used to estimate significance (P < 0.05).
Genomic Convergence Analysis.
Convergent sites include both “parallel” and “convergent” sites as defined in ref. 19. In this study, ancestral protein sequences were reconstructed for 14,254 orthologs among eight Eutherian species (Fig. 1A) by using the Codeml program in PAML 4.7. We identified convergent amino acid sites between giant and red pandas with the following rules: (i) the amino acid residues of both the extant giant and red panda lineages were identical; (ii) amino acid change was inferred to have occurred between the extant giant panda lineage and its most recent common ancestor with polar bear lineage; and (iii) amino acid change was inferred to have occurred between the extant red panda lineage and its most recent common ancestor with ferret lineage. To filter out noise resulting from chance amino acid substitutions (6, 20), we then performed a Poisson test to verify whether the observed number of convergent sites of each gene was significantly more than the expected number caused by random substitution under the JTT-fgene and JTT-fsite amino acid substitution models, respectively, by following the method of ref. 21. The Benjamini–Hochberg method was used to correct for multiple comparisons (q < 0.05). We used the result under the JTT-fgene model rather than under the JTT-fsite model in our analysis, because the performance of JTT-fsite model depends on the number of species analyzed and the use of a few sequences may bias the estimation of equilibrium amino acid frequencies (21). Then, considering that gene tree discordance resulting from incomplete lineage sorting or hybridization may produce the false identification of amino acid convergence (22), we constructed a gene tree for each gene with nonrandom amino acid convergence, and then removed those genes whose gene trees supported the giant panda‒red panda clade. Finally, to obtain more conservative signatures of adaptive convergence, genes with nonrandom convergent amino acid sites were compared with genes under positive selection (5, 20), and overlapping genes between the two gene sets were inferred to have undergone adaptive convergence. The functional enrichment of these adaptively convergent genes in GO terms and KEGG pathways was tested by using the GeneTrail2 method, with the 14,254 orthologous genes as the reference gene set. The significantly enriched category included at least two genes, and the hypergeometric test was used to estimate significance (P < 0.05).
Genome-Wide Pseudogene Identification.
To identify common pseudogenes in the red and giant panda genomes, we work with the precompiled MAF alignment file across eight Eutherian species (Fig. 1A). The MAF alignments with (i) at least five species present including two pandas, (ii) minimal sequence identity of 80%, and (iii) corresponding to human coding exon, were retrieved to check the presence of SNPs or indels that introduce premature stop codon or frame-shift mutation, and genomic positions with known heterozygous SNP or indel were excluded.
Supplementary Material
Acknowledgments
We thank Yujun Zhang, Jiayan Wu, and Meili Chen for assistance with high-throughput sequencing. This project was supported by Microevolution Key Research Project of the National Natural Science Foundation of China Grant 91531302, the National Key Program of Research and Development, Ministry of Science and Technology Grant 2016YFC0503200, National Natural Science Foundation of China Grant 31470441, and Key Research Program of the Chinese Academy of Sciences Grant KJZD-EW-L07.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.M.P. is a Guest Editor invited by the Editorial Board.
Data deposition: Sequence data have been deposited in the Sequence Read Archive under accession nos. SRP064935 and SRP064940. The red panda whole genome shotgun project has been deposited at DDBJ/EMBL/GenBank under accession no. LNAC00000000. The version described in this paper is version LNAC01000000. The giant panda whole genome shotgun project has been deposited at DDBJ/EMBL/GenBank under accession no. LNAT00000000. The version described in this paper is version LNAT01000000.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613870114/-/DCSupplemental.
References
- 1.Christin PA, Weinreich DM, Besnard G. Causes and evolutionary significance of genetic convergence. Trends Genet. 2010;26(9):400–405. doi: 10.1016/j.tig.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Stern DL. The genetic causes of convergent evolution. Nat Rev Genet. 2013;14(11):751–764. doi: 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]
- 3.Storz JF. Causes of molecular convergence and parallelism in protein evolution. Nat Rev Genet. 2016;17(4):239–250. doi: 10.1038/nrg.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denoeud F, et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345(6201):1181–1184. doi: 10.1126/science.1255274. [DOI] [PubMed] [Google Scholar]
- 5.Foote AD, et al. Convergent evolution of the genomes of marine mammals. Nat Genet. 2015;47(3):272–275. doi: 10.1038/ng.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou Z, Zhang J. No genome-wide protein sequence convergence for echolocation. Mol Biol Evol. 2015;32(5):1237–1241. doi: 10.1093/molbev/msv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eizirik E, et al. Pattern and timing of diversification of the mammalian order Carnivora inferred from multiple nuclear gene sequences. Mol Phylogenet Evol. 2010;56(1):49–63. doi: 10.1016/j.ympev.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei F, et al. Black and white and read all over: The past, present and future of giant panda genetics. Mol Ecol. 2012;21(23):5660–5674. doi: 10.1111/mec.12096. [DOI] [PubMed] [Google Scholar]
- 9.Yu L, et al. Phylogenetic utility of nuclear introns in interfamilial relationships of Caniformia (order Carnivora) Syst Biol. 2011;60(2):175–187. doi: 10.1093/sysbio/syq090. [DOI] [PubMed] [Google Scholar]
- 10.Schaller GB, Hu J, Pan W, Zhu J. The Giant Pandas of Wolong. Univ of Chicago Press; Chicago: 1985. [Google Scholar]
- 11.Endo H, et al. Role of the giant panda’s ‘pseudo-thumb’. Nature. 1999;397(6717):309–310. doi: 10.1038/16830. [DOI] [PubMed] [Google Scholar]
- 12.Antón M, Salesa MJ, Pastor JF, Peigné S, Morales J. Implications of the functional anatomy of the hand and forearm of Ailurus fulgens (Carnivora, Ailuridae) for the evolution of the ‘false-thumb’ in pandas. J Anat. 2006;209(6):757–764. doi: 10.1111/j.1469-7580.2006.00649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis DD. The Giant Panda: A Morphological Study of Evolutionary Mechanisms. Chicago Nat Hist Museum; Chicago: 1964. [Google Scholar]
- 14.Gould SJ. The Panda’s Thumb: More Reflections in Natural History. WW Nortoni; New York: 1980. [Google Scholar]
- 15.Salesa MJ, Antón M, Peigné S, Morales J. Evidence of a false thumb in a fossil carnivore clarifies the evolution of pandas. Proc Natl Acad Sci USA. 2006;103(2):379–382. doi: 10.1073/pnas.0504899102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 17.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 18.Li R, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463(7279):311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Kumar S. Detection of convergent and parallel evolution at the amino acid sequence level. Mol Biol Evol. 1997;14(5):527–536. doi: 10.1093/oxfordjournals.molbev.a025789. [DOI] [PubMed] [Google Scholar]
- 20.Thomas GWC, Hahn MW. Determining the null model for detecting adaptive convergence from genomic data: A case study using echolocating mammals. Mol Biol Evol. 2015;32(5):1232–1236. doi: 10.1093/molbev/msv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou Z, Zhang J. Are convergent and parallel amino acid substitutions in protein evolution more prevalent than neutral expectations? Mol Biol Evol. 2015;32(8):2085–2096. doi: 10.1093/molbev/msv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendes FK, Hahn Y, Hahn MW. Gene tree discordance can generate patterns of diminishing convergence over time. Mol Biol Evol. 2016;33(12):3299–3307. doi: 10.1093/molbev/msw197. [DOI] [PubMed] [Google Scholar]
- 23.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 24.Mendes FK, Hahn MW. Gene tree discordance causes apparent substitution rate variation. Syst Biol. 2016;65(4):711–721. doi: 10.1093/sysbio/syw018. [DOI] [PubMed] [Google Scholar]
- 25.May SR, et al. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287(2):378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 26.Merrill AE, et al. Ciliary abnormalities due to defects in the retrograde transport protein DYNC2H1 in short-rib polydactyly syndrome. Am J Hum Genet. 2009;84(4):542–549. doi: 10.1016/j.ajhg.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Endoh-Yamagami S, et al. A mutation in the pericentrin gene causes abnormal interneuron migration to the olfactory bulb in mice. Dev Biol. 2010;340(1):41–53. doi: 10.1016/j.ydbio.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 28.Dagoneau N, et al. DYNC2H1 mutations cause asphyxiating thoracic dystrophy and short rib-polydactyly syndrome, type III. Am J Hum Genet. 2009;84(5):706–711. doi: 10.1016/j.ajhg.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mo R, et al. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124(1):113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- 30.Jurczyk A, et al. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J Cell Biol. 2004;166(5):637–643. doi: 10.1083/jcb.200405023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molotkov A, Ghyselinck NB, Chambon P, Duester G. Opposing actions of cellular retinol-binding protein and alcohol dehydrogenase control the balance between retinol storage and degradation. Biochem J. 2004;383(Pt 2):295–302. doi: 10.1042/BJ20040621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blomhoff R, Blomhoff HK. Overview of retinoid metabolism and function. J Neurobiol. 2006;66(7):606–630. doi: 10.1002/neu.20242. [DOI] [PubMed] [Google Scholar]
- 33.Glatston AR. Red Panda: Biology and Conservation of the First Panda. Academic Press; London: 2011. [Google Scholar]
- 34.Herrmann W, Geisel J. Vegetarian lifestyle and monitoring of vitamin B-12 status. Clin Chim Acta. 2002;326(1-2):47–59. doi: 10.1016/s0009-8981(02)00307-8. [DOI] [PubMed] [Google Scholar]
- 35.O’Leary F, Samman S. Vitamin B12 in health and disease. Nutrients. 2010;2(3):299–316. doi: 10.3390/nu2030299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulu A, et al. Soluble epoxide hydrolase inhibitors reduce the development of atherosclerosis in apolipoprotein e-knockout mouse model. J Cardiovasc Pharmacol. 2008;52(4):314–323. doi: 10.1097/FJC.0b013e318185fa3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444(7117):288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- 38.Zhao H, Yang JR, Xu H, Zhang J. Pseudogenization of the umami taste receptor gene Tas1r1 in the giant panda coincided with its dietary switch to bamboo. Mol Biol Evol. 2010;27(12):2669–2673. doi: 10.1093/molbev/msq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darwin CR. The Origin of Species. John Murray; London: 1859. [Google Scholar]
- 40.Price SA, Hopkins SSB, Smith KK, Roth VL. Tempo of trophic evolution and its impact on mammalian diversification. Proc Natl Acad Sci USA. 2012;109(18):7008–7012. doi: 10.1073/pnas.1117133109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gittleman JL. Are the pandas successful specialists or evolutionary failures? Bioscience. 1994;44(7):456–464. [Google Scholar]
- 42.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nabholz B, Künstner A, Wang R, Jarvis ED, Ellegren H. Dynamic evolution of base composition: Causes and consequences in avian phylogenomics. Mol Biol Evol. 2011;28(8):2197–2210. doi: 10.1093/molbev/msr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jarvis ED, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346(6215):1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keane TM, Naughton TJ, McInerney JO. ModelGenerator: Amino Acid and Nucleotide Substitution Model Selection. Natl Univ of Ireland; Maynooth, Ireland: 2004. [Google Scholar]
- 47.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flynn JJ. Carnivoran phylogeny and rates of evolution: Morphological, taxonomic, and molecular. In: Gittleman JL, editor. Carnivore Behavior, Ecology, and Evolution. Vol 2. Cornell Univ Press; Ithaca: 1996. pp. 542–581. [Google Scholar]
- 49.Wesley-Hunt GD, Flynn JJ. Phylogeny of the carnivora: Basal relationships among the carnivoramorphans, and assessment of the position of ‘miacoidea’ relative to carnivore. J Syst Palaeontol. 2005;3(1):1–28. [Google Scholar]
- 50.Meredith RW, et al. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334(6055):521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Tedford RH. Canidae. In: Prothero DR, Emry RJ, editors. The Terrestrial Eocene-Oligocene Transition in North America. Cambridge Univ Press; Cambridge: 1996. pp. 433–452. [Google Scholar]
- 52.Labarga A, Valentin F, Anderson M, Lopez R. Web services at the European bioinformatics institute. Nucleic Acids Res. 2007;35(Web Server issue) suppl 2:W6–W11. doi: 10.1093/nar/gkm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22(12):2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 54.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stöckel D, et al. Multi-omics enrichment analysis using the GeneTrail2 web service. Bioinformatics. 2016;32(10):1502–1508. doi: 10.1093/bioinformatics/btv770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.