Significance
Diterpenes play important roles in plant biology and serve as industrial bioproducts and therapeutics, including the anticancer drug Taxol. Enzymes of the diterpene synthase family produce the many core structural scaffolds that form the foundation of the large diversity of biologically active diterpenes. This paper describes the identification and the mechanism of a distinct diterpene synthase, pseudolaratriene synthase, from the golden larch tree, Pseudolarix amabilis. The enzyme catalyzes the first committed reaction in the biosynthesis of pseudolaric acids, complex diterpenes with potential anticancer activity.
Keywords: diterpene biosynthesis, Pseudolarix amabilis, plant natural products, pseudolaric acid, chemotherapeutic drug
Abstract
The diversity of small molecules formed via plant diterpene metabolism offers a rich source of known and potentially new biopharmaceuticals. Among these, the microtubule-destabilizing activity of pseudolaric acid B (PAB) holds promise for new anticancer agents. PAB is found, perhaps uniquely, in the coniferous tree golden larch (Pseudolarix amabilis, Pxa). Here we describe the discovery and mechanistic analysis of golden larch terpene synthase 8 (PxaTPS8), an unusual diterpene synthase (diTPS) that catalyzes the first committed step in PAB biosynthesis. Mining of the golden larch root transcriptome revealed a large TPS family, including the monofunctional class I diTPS PxaTPS8, which converts geranylgeranyl diphosphate into a previously unknown 5,7-fused bicyclic diterpene, coined “pseudolaratriene.” Combined NMR and quantum chemical analysis verified the structure of pseudolaratriene, and co-occurrence with PxaTPS8 and PAB in P. amabilis tissues supports the intermediacy of pseudolaratriene in PAB metabolism. Although PxaTPS8 adopts the typical three-domain structure of diTPSs, sequence phylogeny places the enzyme with two-domain TPSs of mono- and sesqui-terpene biosynthesis. Site-directed mutagenesis of PxaTPS8 revealed several catalytic residues that, together with quantum chemical calculations, suggested a substantial divergence of PxaTPS8 from other TPSs leading to a distinct carbocation-driven reaction mechanism en route to the 5,7-trans-fused bicyclic pseudolaratriene scaffold. PxaTPS8 expression in microbial and plant hosts provided proof of concept for metabolic engineering of pseudolaratriene.
Drawing on centuries of knowledge of ethnomedicinal plants from around the world, plant natural products, also known as “secondary metabolites” or “specialized metabolites,” remain a valuable but largely untapped source for drug discovery (1). Diterpenes form a diverse class of plant secondary metabolites, including some with functions known to be critical for plant vigor and basic survival (2). The various biological activities of diterpenes also form the foundation of their use as biopharmaceuticals, such as the anticancer drug Taxol (paclitaxel) and the cAMP-activating compound forskolin (3, 4).
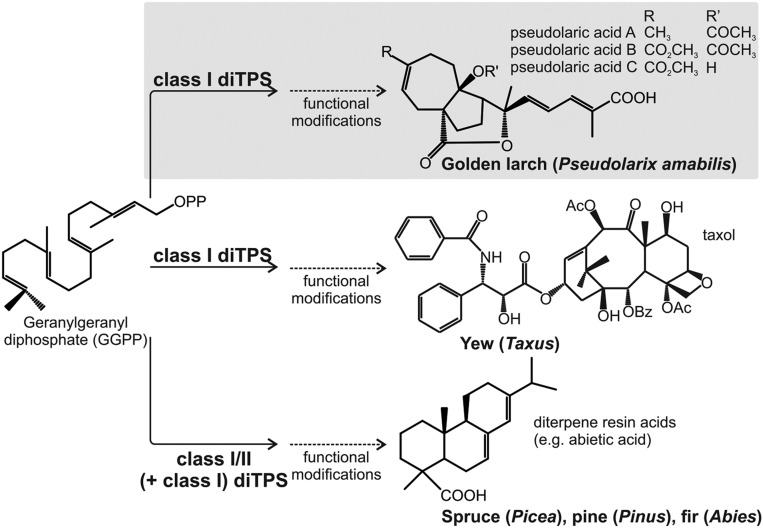
Golden larch (Pseudolarix amabilis, Pinaceae) is a deciduous gymnosperm tree renowned as one of the 50 fundamental herbs in traditional Chinese medicine. Golden larch forms a set of diterpenes, the pseudolaric acids, that are distinct from other diterpenes (Fig. S1) (5). The major bioactive component, pseudolaric acid B (PAB), has demonstrated antitumor properties against several cancer types (6, 7). Similar to the widely used chemotherapeutics Taxol and vinblastine, PAB exhibits antiproliferative activity by binding to microtubules (8). Specifically, PAB inhibits microtubule polymerization (9) and has the capacity to circumvent multidrug resistance (6, 7). Development of PAB as an anticancer drug is limited by supply, which depends on its isolation from golden larch roots or may be achieved through multistep chemical synthesis (5, 9). Knowledge of the genes and enzymes of PAB biosynthesis in golden larch would provide the resources needed to develop enzymatic biomanufacturing systems. On this premise, we recently established a genomics-enabled gene-discovery strategy for diterpene metabolism in nonmodel plants (10). Target genes include the family of diterpene synthases (diTPSs), which catalyze the carbocation-driven cyclization and rearrangement of the central precursor geranylgeranyl diphosphate (GGPP) into various diterpene scaffolds that form the bedrock for diterpene structural diversity (4, 11). The large family of diTPSs and their distinctive biological functions result from evolutionary divergence through repeated gene duplication and neofunctionalization events (4, 12). Given their shared ancestry, known plant diTPSs of different origin and functions are structurally conserved with variations of three α-helical domains and two distinct active sites (class II and class I). The domain structure and the presence and contour of these active sites define diTPS catalytic specificity (13–15). In gymnosperms, such as P. amabilis, most diTPSs of secondary metabolism are bifunctional class I/II enzymes that combine both functional active sites in one protein and form various labdane diterpenes (Fig. S1) (4, 16). Select gymnosperms also evolved monofunctional class I diTPSs with roles in secondary metabolism, including enzymes of labdane biosynthesis in pines (Pinus) (17) and taxadiene synthases, which form the precursor for Taxol and other taxoids in yew (Taxus) (18). Because all known gymnosperm class II/I diTPSs and monofunctional class II diTPSs are known to form labdane-related diterpenes exclusively, and pseudolaric acids (albeit distinct) more closely resemble the taxane scaffold (Fig. S1), we hypothesized that a monofunctional class I diTPS may catalyze the first committed step in pseudolaric acid biosynthesis.
Fig. S1.
Natural diterpene scaffolds in gymnosperms.
Here we report the functional and mechanistic characterization of the class I diTPS, golden larch terpene synthase 8 (PxaTPS8), that yields pseudolaratriene as the characteristic 5,7-bicyclic scaffold in pseudolaric acid biosynthesis. Structural–functional and quantum chemical analysis of the proposed carbocation cyclization and rearrangement reactions provide insight into the enzyme’s catalytic mechanism and may offer future opportunity for metabolic pathway engineering.
Results
TPS in the Golden Larch Root Transcriptome.
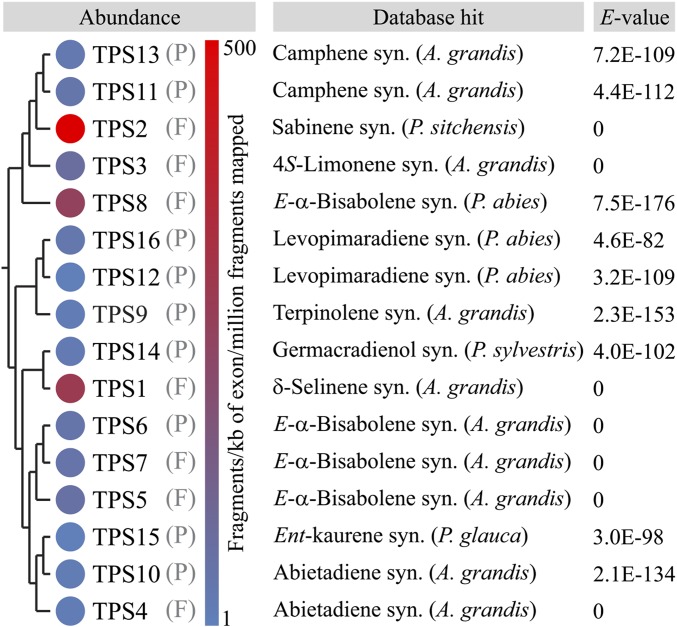
Pseudolaric acids have traditionally been isolated from the roots of golden larch for use as a Chinese materia medica (5). To investigate the underlying biosynthetic pathway, we previously developed a root-specific transcriptome resource (10), which revealed 16 candidate terpene synthase (TPS) genes (Fig. S2). Of the five candidates with highest database matches to known diTPSs (PxaTPS4, 10, 12, 15, and 16), four represented partial sequences putatively annotated as ent-kaurene synthase or class I/II diTPSs. The full-length class I/II diTPS PxaTPS4 was characterized as a levopimaradiene/abietadiene synthase (10). All identified diTPS candidates most closely matched enzymes of labdane diterpene biosynthesis, leaving a possible monofunctional class I diTPS predicted to form the distinct nonlabdane pseudolaric acid scaffold elusive. We therefore investigated the remaining TPSs, whose top database matches were either mono- or sesqui-TPSs. Four genes (PxaTPS5–8) were of particular interest because they most closely resembled three-domain gymnosperm E-α-bisabolene synthases (BISs), sesqui-TPSs. Of these, PxaTPS8 was given priority for further characterization because of its higher transcript abundance in roots as identified by RNA-sequencing–based transcript mapping (Fig. S2).
Fig. S2.
TPS gene annotation in the P. amabilis root transcriptome. TPS candidate genes were identified with an E-value threshold of 1E-50. Relative transcript abundance is based on fragments per kilobase of exon per million fragments mapped (FPKM) values obtained via mapping of Illumina reads against assembled TPS reads. F, full-length transcript; P, partial transcript.
Sequence Phylogeny Places the Three-Domain PxaTPS8 Distant from Known Gymnosperm TPSs.
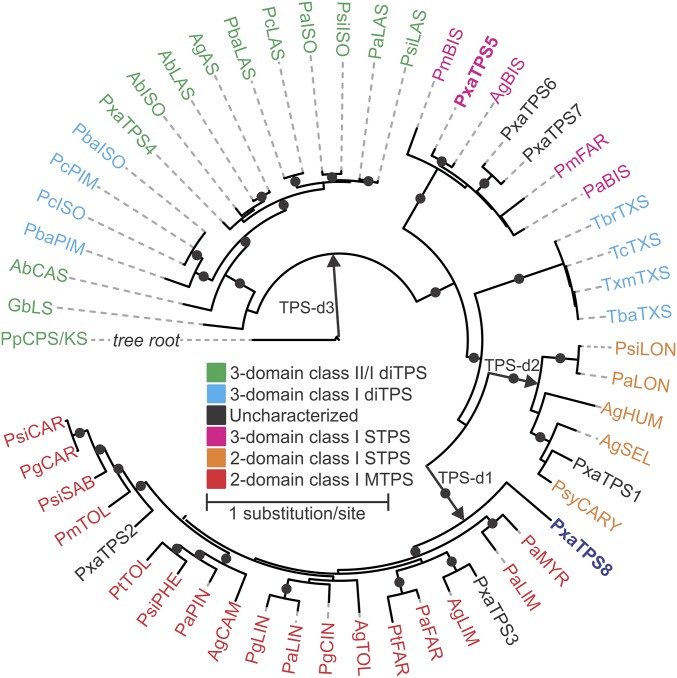
The four BIS-like candidates identified in the golden larch root transcriptome resembled three-domain class I TPSs, lacking a class II active site and featuring the DDxxD and Asn-Ser-Glu/Asp-Thr-Glu (NSE/DTE) class I catalytic motifs (19). A γβα-domain structure is conserved among enzymes of the TPS-d3 clade. As predicted, our sequence phylogeny placed golden larch PxaTPS5, -6, and -7 closely with known BISs from grand fir (Abies grandis), Douglas fir (Pseudotsuga menziesii), and Norway spruce (Picea abies) within the gymnosperm TPS-d3 clade (Fig. 1). Surprisingly, despite its three-domain structure, PxaTPS8 emerged outside the TPS-d3 clade as a distant branch at the base of the TPS-d1 clade. This clade contains known two-domain mono- and sesqui-TPSs, including PXaTPS1–3, that cluster with known terpinolene, limonene, and caryophyllene synthases. The phylogenetic position of a three-domain TPS with the TPS-d1 clade is unprecedented and suggested a substantial evolutionary divergence leading to PxaTPS8 in golden larch. The unusual pairing of a three-domain structure and TPS-d1 clade association prohibited a functional prediction of PxaTPS8 based on similarity with any known gymnosperm TPS. However, these features also made it a prime candidate for a potentially unusual function.
Fig. 1.
Maximum likelihood tree illustrating the phylogenetic relationship of PxaTPS8 within the gymnosperm TPS-d clade. Dots represent bootstrap support of >80% (1,000 repetitions). The tree is rooted with Physcomitrella patens ent-kaurene synthase. For sequence abbreviations Table S1.
Table S1.
Abbreviations and GenBank/European Bioinformatics Institute Data Bank accession numbers of diterpene synthases used for phylogenetic analyses
| Abbreviation | Protein | Accession no. |
| AbCAS | Cis-abienol synthase, Abies balsamea | JN254808 |
| AbISO | Isopimaradiene synthase, Abies balsamea | JN254806 |
| AbLAS | Levopimaradiene/abietadiene synthase, Abies balsamea | JN254805 |
| AgAS | Abietadiene synthase, Abies grandis | AAK83563 |
| AgBIS | E-α-Bisabolene synthase, Abies grandis | AF006195 |
| AgCAM | Camphene synthase, Abies grandis | AAB70707 |
| AgHUM | γ-Humulene synthase, Abies grandis | AAC05728 |
| AgLIM | 4S-Limonene synthase, Abies grandis | AAB70907 |
| AgSEL | δ-Selinene synthase, Abies grandis | AAC05727 |
| AgTOL | Terpinolene synthase, Abies grandis | AAF61454 |
| GbLS | Levopimaradiene synthase, Ginkgo biloba | AAS89668 |
| PaBIS | E-α-bisabolene synthase, Picea abies | AY473619 |
| PaFAR | α-Farnesene synthase, Picea abies | AAS47697 |
| PaISO | Isopimaradiene synthase, Picea abies | AY473620 |
| PaLAS | Levopimaradiene/abietadiene synthase, Picea abies | AAS47691 |
| PaLIM | Limonene synthase, Picea abies | AAS47694 |
| PaLIN | (-)-Linalool synthase, Picea abies | AAS47693 |
| PaLON | Longifolene synthase, Picea abies | AAS47695 |
| PaMYR | Myrcene synthase, Picea abies | AAS47696 |
| PaPIN | α/β-Pinene synthase, Picea abies | AAS47692 |
| PbaISO | Isopimaradiene synthase, Pinus banksiana | M4HXW5 |
| PbaLAS | Levopimaradiene/abietadiene synthase, Pinus banksiana | M4HXU6 |
| PbaPIM | Pimaradiene synthase, Pinus banksiana | M4HY08 |
| PcISO | Isopimaradiene synthase, Pinus contorta | JQ240314 |
| PcLAS | Levopimaradiene/abietadiene synthase, Pinus contorta | JQ240310 |
| PcPIM | Pimaradiene synthase, Pinus contorta | JQ240315 |
| PgCAR | 3-Carene synthase, Picea glauca | ADZ45510 |
| PgCIN | 1,8-Cineole synthase, Picea glauca | ADZ45498 |
| PgLIN | (-)-Linalool synthase, Picea glauca | ADZ45500 |
| PmBIS | E-γ-Bisabolene synthase, Pseudotsuga menziesii | AAX07266 |
| PmFAR | β-Farnesene synthase, Pseudotsuga menziesii | AAX07265 |
| PmTOL | Terpinolene synthase, Pseudotsuga menziesii | AAX07264 |
| PpCPSKS | Ent-kaurene/kaurenol synthase, Physcomitrella patens | BAF61135 |
| PsiCAR | 3-Carene synthase, Picea sitchensis | ADU85924 |
| PsiISO | Isopimaradiene synthase, Picea sitchensis | ADZ45512 |
| PsiLAS | Levopimaradiene/abietadiene synthase, Picea sitchensis | ADZ45517 |
| PsiLON | Longipinene synthase, Picea sitchensis | ADZ45516 |
| PsiPHE | β-Phellandrene synthase, Picea sitchensis | ADZ45506 |
| PsiSAB | Sabinene synthase, Picea sitchensis | ADU85929 |
| PsyCARY | β-Caryophyllene synthase, Pinus sylvestris | B4XAK4 |
| PtFAR | α-Farnesene synthase, Pinus taeda | AAO61226 |
| PtTOL | α-Terpineol synthase, Pinus taeda | Q84KL4 |
| PxaTPS4 | Levopimaradiene/abietadiene synthase, Pseudolarix amabilis | AGN70885 |
| TbaTXS | Taxadiene synthase, Taxus baccata | AAR02861 |
| TbrTXS | Taxadiene synthase, Taxus brevifolia | U48796 |
| TcTXS | Taxadiene synthase, Taxus cuspidata | DQ305407 |
| TxmTXS | Taxadiene synthase, Taxus x media | AAS18603 |
PxaTPS8 Produces a Nonlabdane Diterpene.
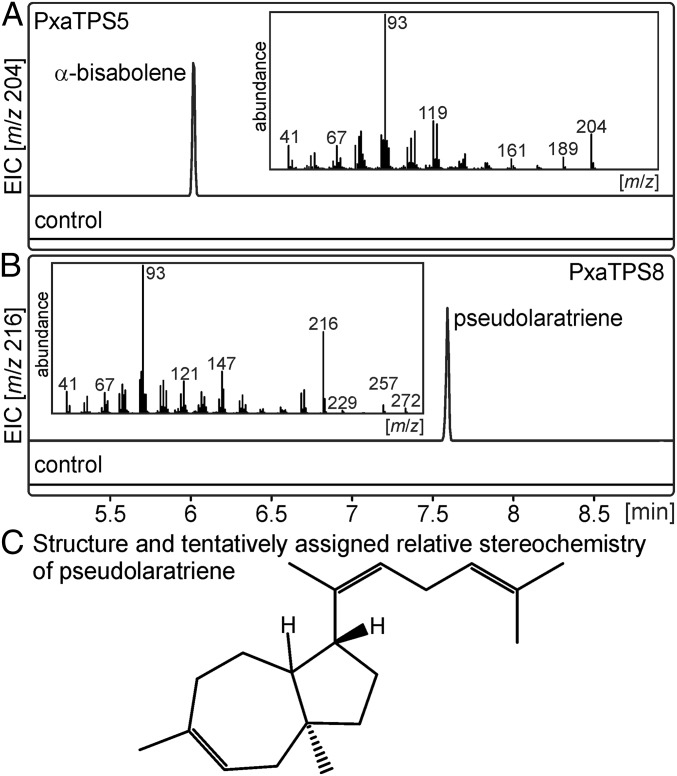
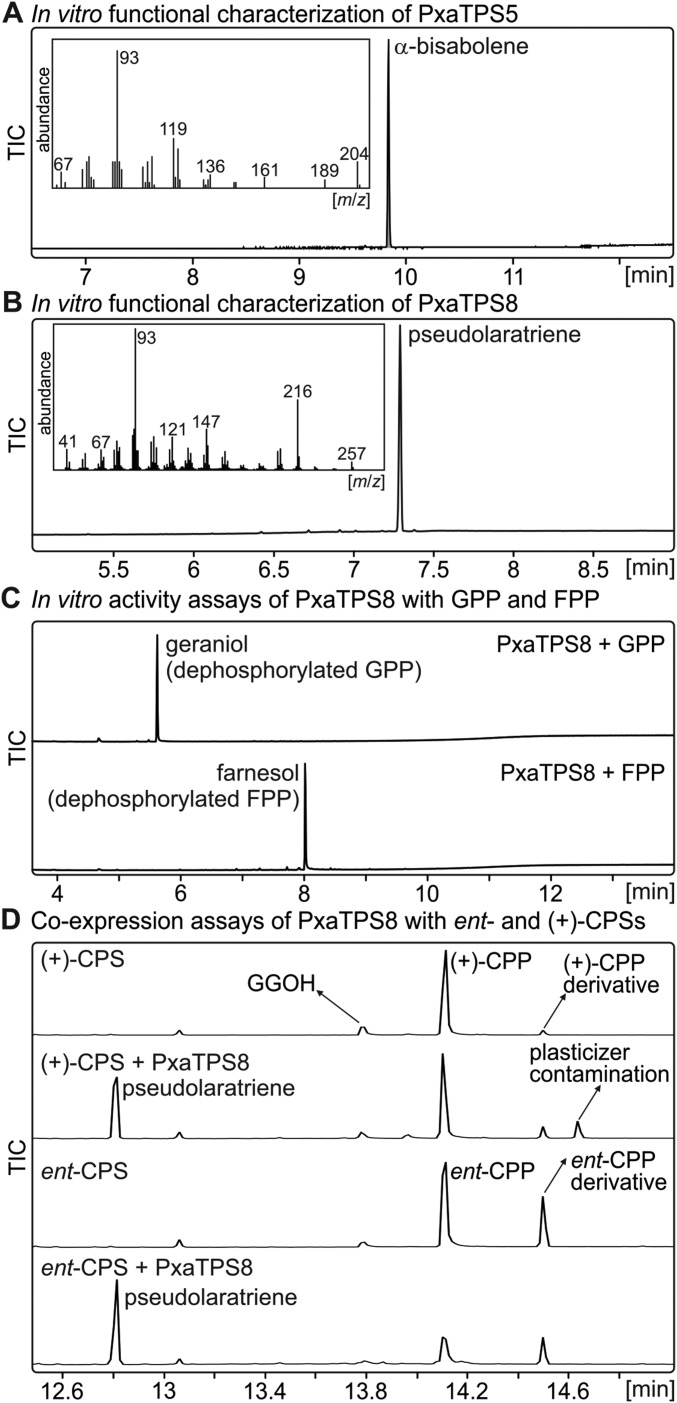
We performed in vitro enzyme assays with PxaTPS8 expressed in Escherichia coli and in vivo assays using transient Agrobacterium-mediated expression in Nicotiana benthamiana. For comparison we also characterized PxaTPS5, which represents a TPS-d3 BIS-like enzyme. Transient expression assays verified PxaTPS5 as an α-BIS by comparison with product reference mass spectra of known α-BISs from grand fir and Norway spruce (Fig. 2) (20, 21). By contrast, in vivo assays of PxaTPS8 revealed as a single product, a previously unknown diterpene with a fragmentation pattern featuring signature ions of m/z 93 (100), 121 (25), 147 (31), 216 (57) (Fig. 2). In vitro assays using affinity-purified proteins confirmed the activity of PxaTPS5 and PxaTPS8 (Fig. S3). Furthermore, PxaTPS8 was active only with GGPP as a substrate; no products were detected with geranyl diphosphate (GPP) or farnesyl diphosphate (FPP) or in assays with class II diTPSs forming copalyl diphosphate (CPP) of ent- or (+)-stereochemistry (Fig. S3).
Fig. 2.
Functional analysis of PxaTPS5 and PxaTPS8. (A and B) GC-MS analysis of products resulting from the transient expression of PxaTPS5 (A) and PxaTPS8 (B) in N. benthamiana shown as extracted ion chromatograms (EICs). Expression of the silencing suppressor protein p19 alone was used as a control. (C) Validation of the PxaTPS8 product as pseudolaratriene via NMR analysis and quantum chemical shift prediction.
Fig. S3.
Products of in vitro assays of PxaTPS5 and PxaTPS8. (A and B) GC-MS analysis of reaction products obtained from in vitro enzyme assays of recombinant PxaTPS5 with FPP as a substrate (A) and with PxaTPS8 with GGPP as a substrate (B). (C) Activity assays of PxaTPS8 with GPP or FPP as substrate resulted only in the corresponding dephosphorylated substrates, demonstrating no conversion by PxaTPS8. (D) E. coli coexpression assays of PxaTPS8 with a (+)-CPP synthase from Marrubium vulgare or an ent-CPP synthase from Zea mays. No additional products were observed compared with the individual CPP synthase reaction products. GC/MS traces are illustrated as TICs.
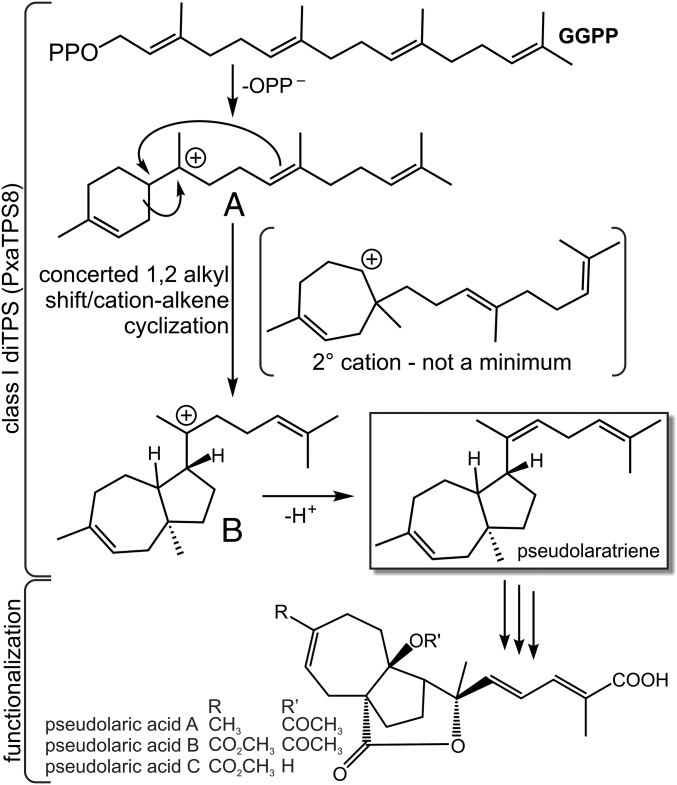
Using an engineered E. coli expression system (22) yielded sufficient amounts and purity of the PxaTPS8 product to enable 1D and 2D NMR analysis, which identified a distinct bicyclic diterpene, termed “pseudolaratriene” (Fig. 2C). Additional quantum chemical calculations (23, 24) of 1H and 13C NMR chemical shifts for several pseudolaratriene isomers and diastereomers of each verified the structure shown in Fig. 2C (Dataset S1) and were consistent with all available NMR data (chemical shifts, coupling constants, and NOE experiments). The trans configuration of the bridgehead methyl group and hydrogen atom at the connection point of the “tail” was assigned based on the known relative configuration of pseudolaric acids. Although the relative configuration of the bridgehead proton could not be assigned conclusively, coupling constants favor the structure in which the two hydrogen atoms are trans with respect to each other. These two hydrogen atoms have unusual chemical shifts (1.82 and 3.43 ppm for the bridgehead and allylic protons, respectively) that are ∼1 ppm further downfield than expected. The possibility that a hydroxyl group causes this discrepancy was excluded based on the IR spectrum of pseudolaratriene that showed no evidence for such a group (Dataset S1). Also, no other conceivable carbon skeleton matched the experimental NMR data. Therefore, we propose that the unusual chemical shifts result from the participation of these hydrogens in CH–π interactions [i.e., close contacts between the hydrogen atoms and π-bonds from elsewhere in the molecule (25)]. Preliminary quantum chemical calculations on model systems confirm that chemical shift changes of ∼1 ppm are possible for such structures. For this interaction to occur in pseudolaratriene, it appears that the internal alkene of the tail would need to be Z rather than E. Formation of a C = C π-bond with this stereochemistry is not precluded by the cyclization and rearrangement mechanism described below and is consistent with the known relative configurations of pseudolaric acids. Indeed, consistent with a Z alkene, an NOE between the hydrogen atom and the methyl group on this alkene was observed (Dataset S1). The bridgehead hydrogen may approach the same alkene if it is cis with respect to the allylic hydrogen or the π-bond in the seven-membered ring if it is trans with respect to the allylic hydrogen. Collectively, these studies show that PxaTPS8 is a distinct diTPS that adds an additional catalyst to the diversity of the enzyme family that transforms GGPP into a 5,7-trans–fused bicyclic scaffold, the characteristic core structure of PAB (5).
Occurrence of Pseudolaratriene, PAB, and PxaTPS8 in P. amabilis Tissues.
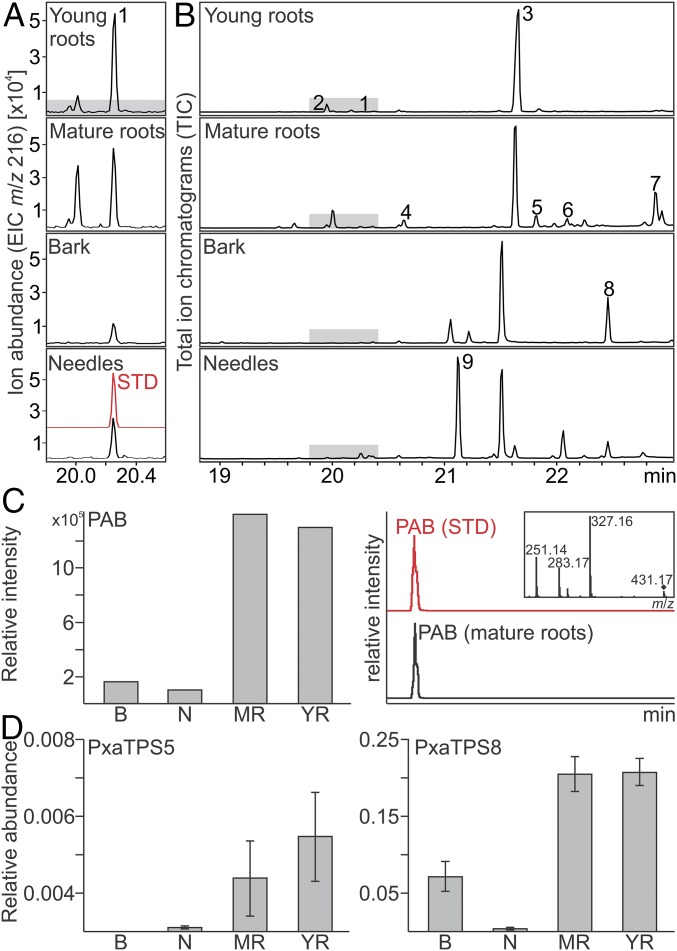
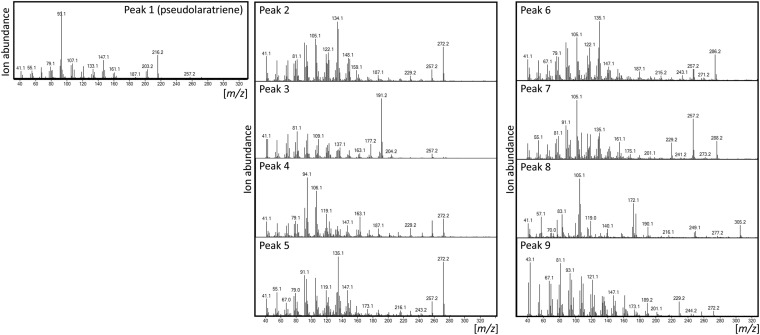
To test if pseudolaratriene is present in planta, we analyzed metabolite extracts from young and mature roots, bark, and needle tissue of golden larch via GC-MS compared with the enzyme product. In tandem, PAB abundance in planta was measured against an authentic standard using ultra performance LC (UPLC)-MS/MS analysis. Although present only at trace levels, pseudolaratriene was detected in all tissues and was most abundant in young and mature roots (Fig. 3). Notably, several compounds resembling olefin or oxygenated labdane diterpenes were detected also (Fig. 3 and Fig. S4, peaks 2–9), consistent with the presence of bifunctional class II/I diTPSs in the P. amabilis transcriptome (Fig. S2). However, the abundance of these metabolites did not enable structural verification. Despite its historical association with roots for their medicinal use (5), PAB was observed in all tested tissues; however, its abundance was highest in roots (Fig. 3). Next, we performed gene-expression studies of PxaTPS5 and PxaTPS8 by quantitative real-time PCR (qPCR) analysis. Consistent with the obtained metabolite profiles, the PxaTPS8 transcript was detected predominantly in roots, in bark, and near the limit of detection in needle tissue (Fig. 3). Similarly, PxaTPS5 showed highest expression in roots but was present only at low levels in needles and was absent in bark tissue.
Fig. 3.
Abundance of PAB biosynthetic components in planta. (A and B) GC-MS analysis of pseudolaratriene (peak 1) in young and mature roots, bark, and needles of P. amabilis depicted as EIC m/z 216 (A) and total ion chromatograms (TICs) (B) spectra, respectively. Purified PxaTPS8 product served as a standard (STD). Other detected compounds resembling putative diterpenes are labeled (peaks 2–9), and corresponding mass spectra are given in Fig. S4. (C) UPLC-MS/MS analysis of PAB in the same tissues. (D) Relative transcript abundance of PxaTPS5 and PxaTPS8 in P. amabilis tissues based on qPCR analysis (n = 3). B, bark; M, mature roots; N, needles; Y, young roots.
Fig. S4.
Mass spectra of compounds identified in P. amabilis. Shown are mass spectra of select compounds identified in P. amabilis root, bark, and needle tissue that may represent endogenous diterpenes as based on signature mass ions.
Active Site Determinants of PxaTPS8 Function.
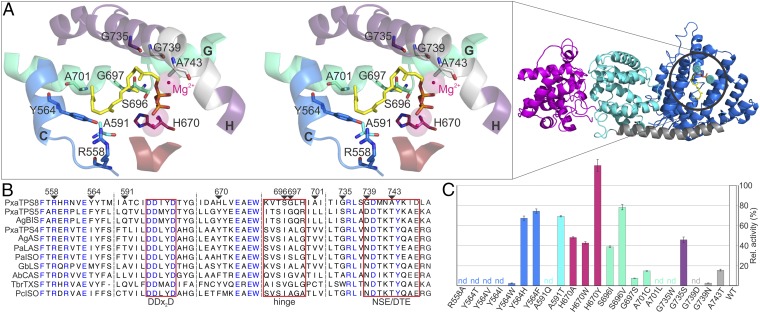
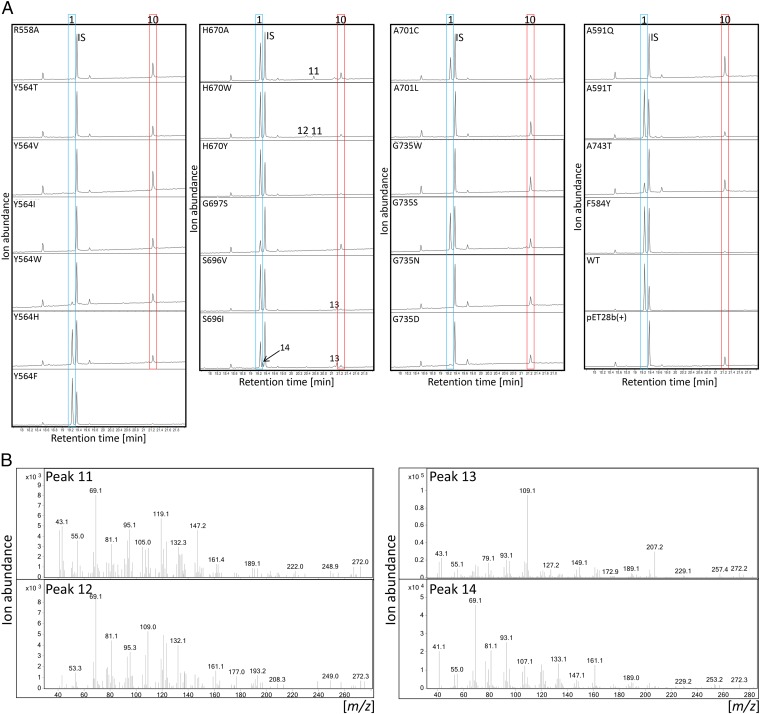
We conducted homology modeling of PxaTPS8 based on the structure of grand fir α-BIS (AgBIS) (19) and molecular docking of GGPP into the active site to probe residues that contribute to the distinct activity of PxaTPS8 (Fig. 4). Among common features, the Mg2+-coordinating DDxxD motif and three arginines (R558, R560, and R736) shown to be critical for catalysis in AgBIS (19) and other class I TPSs (26) are conserved in PxaTPS8. Concurrently, alanine substitution of R558 as a representative of the catalytic arginine triad abolished activity in E. coli in vivo assays (Fig. 4 and Fig. S5). However, a protein sequence identity of only 32–38% between the class I active sites of PxaTPS8 and known gymnosperm TPSs showed a large degree of evolutionary divergence. Within an 8-Å radius around the docked ligand in the PxaTPS8 active site cavity, several residues with possible catalytic impact were identified, including positions known to direct product outcome in class I TPSs, such as the hinge region on helix G (Fig. 4) (19, 26, 27). Substitution of PxaTPS8 S696, G697, and A701 for different functional residues common in other gymnosperm TPSs led to a decrease (S696I/V, G697S, and A701C) or loss (A701L) of pseudolaratriene formation. Likewise, substituting G735, G739, and A743 (the latter two unique to PxaTPS8) located at the catalytic NSE/DTE motif on helix H resulted in reduced (G735S and A743T) or abolished (G735W and G739D/N) activity. In addition, residues A591, H670, and Y564 were positioned proximal to the docked GGPP (Fig. 4). Mutagenesis of A591 for threonine and glutamine resulted in reduced and abolished activity, respectively (Fig. 4). Variants H670A/W reduced product formation, whereas H670Y showed wild-type activity, suggesting that residue size and functionality impact catalysis at this position. Similarly, substitution of Y564 (unique to PxaTPS8) for threonine, isoleucine, valine, and tryptophan abolished or severely decreased activity, whereas exchange for histidine or phenylalanine had only minor catalytic impact. These results suggest the relevance of an aromatic side chain to control pseudolaratriene formation, likely via carbocation stabilizing to enable alkyl migration before deprotonation. Surprisingly, of all tested mutations, altered product outcome was detected only for the variants S696I/V and H670A/W, which formed minor amounts of diterpene olefins as based on the corresponding mass spectra (Fig. S5, peaks 11–14). To test if more expansive changes to the active site composition would alter product outcome, we conducted further domain swaps by substituting helix G or H with the corresponding helical segments of the sesqui-TPS AgBIS. Both protein variants caused a loss of function with no detectable alternate products (Fig. S5).
Fig. 4.
(A, Right) Homology model of PxaTPS8 based on the structure of A. grandis α-BIS (PDB-ID: 3EAS) (19) showing the typical diTPS three-domain structure including the γ-, β-, and α-domains (magenta, cyan, and blue, respectively). (Left) Stereoview of the PxaTPS8 active site with docked GGPP (yellow), DDxxD motif (red), and NSE/DTE motif (gray). Analyzed residues and helices H and G (containing the nonhelical hinge region) are color coded. (B) Alignment of select PxaTPS8 amino acids with known gymnosperm TPSs (for abbreviations Table S1). (C) Pseudolaratriene formation by PxaTPS8 variants relative to the wild-type enzyme quantified with eicosene as an internal standard. Color coding is as in A.
Fig. S5.
(A) Total ion chromatograms of products resulting from E. coli coexpression assays of PxaTPS8 protein variants (Fig. 4) compared with the wild-type enzyme and the empty vector control pET28b(+). Peak IS, internal standard 1-eicosene; peak 1, pseudolaratriene; peak 10, dephosphorylated GGPP; peaks 11–14, unidentified putative diterpene olefins. (B) Mass spectra corresponding to peaks 11–14 in A.
Quantum Chemical Calculations Indicate an Unusual Reaction Mechanism for PxaTPS8.
Quantum chemical calculations [mPW1PW91/6-31+G(d,p)//B3LYP/6-31+G(d,p)] (24, 28) were used to assess the viability of carbocation cyclization/rearrangement mechanisms for pseudolaratriene formation (Dataset S1). According to these calculations, the first carbocation intermediate, A, results from initial 1,6-cyclization (Fig. 5). The subsequent 1,2-alkyl shift and 6,10-cyclization occur in a single step (29) to form a carbocation, B, with the characteristic 5,7-fused bicyclic scaffold. Carbocations A and B are predicted to be similar in energy, and the A-to-B reaction is predicted to have an activation barrier of less than 15 kcal/mol. Deprotonation of B would form the final product (pseudolaratriene).
Fig. 5.
Proposed PxaTPS8-catalyzed mechanism based on quantum chemical calculations. After cleavage of the diphosphate group of GGPP, the initial carbocation, A, forms via 1,6-cyclization. Single-step 1,2-alkyl shift and 6,10-cyclization afford a second carbocation, B, with the characteristic 5,7-fused bicyclic structure. Deprotonation of B yields pseudolaratriene.
Formation of Pseudolaratriene in Engineered Yeast.
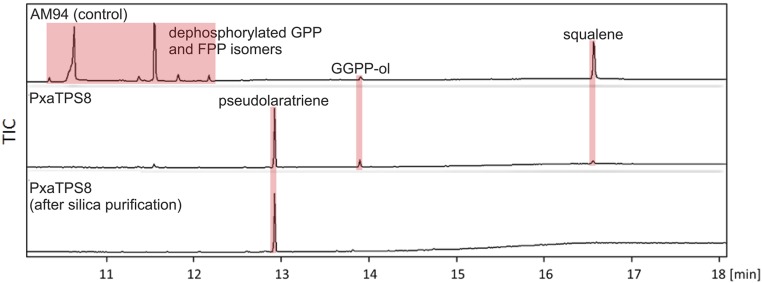
To develop a proof-of-concept production platform for pseudolaratriene, we cotransformed PxaTPS8 with the yeast (Saccharomyces cerevisiae) GGPP synthase BTS1 (30) in the engineered yeast strain AM94 that provides elevated terpene precursor yield (31). Pseudolaratriene was abundant solely in cell pellets after induction with galactose for 41 h, yielding a product amount of 1 mg/L culture (Fig. S6). Dephosphorylated GGPP and squalene as major by-products after diethyl ether extraction could be removed readily by simple chromatography on silica matrix to afford pseudolaratriene in greater than 90% purity. These findings outline a promising foundation for a microbial production system for pseudolaratriene to enable efficient discovery of downstream pathway components and related diterpene structures.
Fig. S6.
Pseudolaratriene formation in engineered yeast. Shown are GC/MS TICs of the PxaTPS8 production after coexpression with the yeast GGPP synthase (BTS1) in the engineered yeast (S. cerevisiae) strain AM94. Pseudolaratriene was purified on silica matrix. GGPP-ol, geranylgeraniol (dephosphorylated GGPP).
Discussion
The diversity of diTPSs and their ability to function in modular biosynthetic networks produces a vast chemical space of diterpene metabolites (4, 11). Exploring this space has the potential to contribute to devising therapeutic lead compounds in the face of drug discovery over recent decades having fallen short of meeting the demand for new and improved therapies (32). Currently, only a few plant-derived diterpene pharmaceuticals, including Taxol and forskolin, are available at industrial scale (4, 33, 34) because of the often low availability of diterpenes from natural sources or uneconomic chemical synthesis.
Pseudolaric acids, especially PAB, from the traditional Chinese medicinal plant golden larch have been recognized for their chemotherapeutic potential and prevention of multidrug resistance (6, 7). The discovery of golden larch PxaTPS8, which forms a 5,7-fused bicyclic diterpene, here coined pseudolaratriene, adds a hitherto unique catalyst to the diTPS family with possible uses for PAB biomanufacture. The concordant occurrence of PxaTPS8, its product pseudolaratriene, and PAB in P. amabilis root, bark, and needle tissue supports the intermediacy of pseudolaratriene in PAB biosynthesis. Biochemical and quantum chemical mechanistic insights into the PxaTPS8-catalyzed conversion of GGPP into pseudolaratriene exemplifies an efficient and short mechanism of carbocation rearrangement that underscores nature’s ability to use the inherent carbocation reactivity to generate a structurally complex product without much concern about diversion to other products that may arise if more discrete intermediates were involved (Fig. 5) (28, 29, 35). This conclusion is supported by the lack of product alteration when replacing select PxaTPS8 active site positions (including Y654 and G697) that were shown to impact product specificity in other gymnosperm diTPSs (27, 36, 37). Only mutation of S696 and H670 afforded small amounts of alternative putative diterpene olefin products. This finding is consistent with the prevalence of isoleucine and valine residues in this position in labdane-forming gymnosperm diTPSs (Fig. 4). Although our mutagenic studies identified several active site determinants (A591, S696, G697, A701, G735, G739, A743, and Y564) with apparent roles in PxaTPS8 catalysis, their possible contributions to product specificity remain elusive. However, the need for an aromatic ring at position 564 offers some insight into the steric control of the intermediary carbocations.
Before this study, knowledge of secondary diterpene metabolism in gymnosperms included the bifunctional class I/II diTPSs involved in the biosynthesis of labdane diterpenes, derived diterpene resin acids of conifer chemical defense (16, 38), and a few monofunctional class I diTPSs involved in diterpene resin acid biosynthesis in pine and taxane formation in species of yew (Fig. S1) (17, 18). All represent three-domain enzymes of the TPS-d3 clade (Fig. 1). Surprisingly, the mechanistically distinct PxaTPS8 was identified as the first three-domain diTPS at the base of the TPS-d1 clade. The three-domain architecture of PxaTPS8 suggests a possible common origin with BIS enzymes. It appears that BIS-like genes in golden larch, of which there are at least four different members, may indeed have undergone more substantial evolution with regard to gene number and functions than the corresponding, seemingly single copy BIS genes in other gymnosperm species (20, 21, 39, 40). The route of pseudolaric acid biosynthesis may have arisen from this diversification in the golden larch tree as the sole species of its genus. The pseudolaratriene scaffold is perhaps unique to golden larch, but structurally similar sphenolobane and tormesane diterpenes have been described in liverworts of the genus Anastrophylum (41) and the eudicot Halimium viscosum (Cristaceae) (42). However, diTPSs of these species are presently not known.
Given the low abundance of PAB in tissues of golden larch (Fig. 3), metabolic engineering of PxaTPS8 in microbial or plant hosts could pave the way for pseudolaratriene production for semisynthesis of PAB and related compounds. Such a recombinant system also would provide a platform for accelerating the discovery of downstream enzymes of PAB biosynthesis.
Materials and Methods
Plant Material.
One-year-old P. amabilis saplings were obtained from the Camellia Forest nursery (www.camforest.com). N. benthamiana plants were grown in Conviron TCR120 growth chambers (www.conviron.com) under a photoperiod of 16 h, 60% relative humidity, 100 μmol⋅m−2⋅s−1 light intensity, and a day/night temperature cycle of 21/18 °C.
Gene Discovery and cDNA Cloning.
TPS candidate genes were identified by querying a previously reported P. amabilis root transcriptome against a curated TPS database followed by phylogenetic analysis (10). Transcript abundance was calculated by mapping adapter-trimmed Illumina reads against the assembled transcripts using BWA version 0.5.9-r16. Reads were mapped as paired with an insert size ≤350 bp. Selected cDNAs were amplified from total RNA with gene-specific oligonucleotides (Table S2) and ligated into the pJET vector (www.Clontech.com) for sequence verification.
Table S2.
Oligonucleotides used in this study
| Designation | Gene | Sequence (5′–3′) |
| pET28b(+) | PxaTPS5-forward | TATCATATGGCTGGCGTTTCTGTTG |
| PxaTPS5-reverse | TATGTCGACTTAAAGTGGGAGCGGATCA | |
| PxaTPS8-forward | TATCATATGATACGGTCAAATACTTCG | |
| PxaTPS8-reverse | TGAGAGCTCTCATAAAGACACAGCTTG | |
| pET28b(+) PxaTPS8 variants | R558A-sense | CCACGTTTCTATGTGCAGTGAACCTC |
| R558A-antisense | GAGGTTCACTGCACATAGAAACGTGG | |
| S696I-sense | ATGTAAACCAATCGTGACCTTGCTGTTC | |
| S696I-antisense | GAACAGCAAGGTCACGATTGGTTTACAT | |
| S696V-sense | CTATATGTAAACCAACCGTGACCTTGCTGT | |
| S696V-antisense | ACAGCAAGGTCACGGTTGGTTTACATATAG | |
| G697S-sense | ATCGCTATATGTAAACTACTCGTGACCTTGCTG | |
| G697S-antisense | CAGCAAGGTCACGAGTAGTTTACATATAGCGAT | |
| Y564T-sense | CCATAGTGTAAGTTTCCACGTTTC | |
| Y564T-antisense | GAAACGTGGAAACTTACACTATGG | |
| Y564V-sense | CCATAGTGTAAACTTCCACGTTTC | |
| Y564V-antisense | GAAACGTGGAAGTTTACACTATGG | |
| Y564H-sense | ATAGAAACGTGGAACATTACACTATGGC | |
| Y564H-antisense | GCCATAGTGTAATGTTCCACGTTTCTAT | |
| Y564F-sense | CCATAGTGTAAAATTCCACGTTTC | |
| Y564F-antisense | GAAACGTGGAATTTTACACTATGG | |
| Y564W-sense | TAGAAACGTGGAATGGTACACTATGGCAT | |
| Y564W-antisense | ATGCCATAGTGTACCATTCCACGTTTCTA | |
| A591T-sense | AATTGTGTGGAATCACTACTTGTATCGATG | |
| A591T-antisense | CATCGATACAAGTAGTGATTCCACACAATT | |
| A591Q-sense | ATTGTGTGGAATCCCAACTTGTATCGAT | |
| A591Q-antisense | ATCGATACAAGTTGGGATTCCACACAAT | |
| H670A-sense | TATAATTGATGCGGCTCTGGTAGAAGC | |
| H670A-antisense | GCTTCTACCAGAGCCGCATCAATTATA | |
| H670Y-sense | TATAATTGATGCGTATCTGGTAGAAGCG | |
| H670Y-antisense | CGCTTCTACCAGATACGCATCAATTATA | |
| H670W-sense | TATAATTGATGCGTGGCTGGTAGAAG | |
| H670W-antisense | CTTCTACCAGCCACGCATCAATTATA | |
| A701C-sense | GTGAGGATGGGCAATATACATATATGTAAACCAC | |
| A701C-antisense | GTGGTTTACATATATGTATATTGCCCATCCTCAC | |
| A701L-sense | GATGGGCAATATCAGTATATGTAAACCACTCG | |
| A701L-antisense | CGAGTGGTTTACATATACTGATATTGCCCATC | |
| G735S-sense | GCCTCCACCATTAGTCGACTAAGTGGT | |
| G735S-antisense | ACCACTTAGTCGACTAATGGTGGAGGC | |
| G735W-sense | ACTTGGCCTCCACCATTTGGCGACTAAGT | |
| G735W-antisense | ACTTAGTCGCCAAATGGTGGAGGCCAAGT | |
| G739N-sense | TTGGTCGACTAAGTAATGACATGAACG | |
| G739N-antisense | CGTTCATGTCATTACTTAGTCGACCAA | |
| G739D-sense | GGTCGACTAAGTGATGACATGAACGCC | |
| G739D-antisense | GGCGTTCATGTCATCACTTAGTCGACC | |
| A743T-sense | GGTGACATGAACACCTACAAGATTGACC | |
| A743T-antisense | GGTCAATCTTGTAGGTGTTCATGTCACC | |
| PxaTPS8:helix H chimera 5′-forward | CAAATGGGTCGCGGATCCATGTCAAATACTTCGAAATATG | |
| PxaTPS8:helix H chimera 3′-reverse | GGTGGTGGTGCTCGAGTTATAAAGACACAGCTTGAACG | |
| PxaTPS8:helix H chimera internal-forward | GGTCTTTGTGTCGTCTGCCAGGCGGGAAATGAGGCTGTTAAGCTCTGTGAGAACACGCCTAGACAGAGTATCAATTTCCTGCA | |
| PxaTPS8:helix H chimera internal-reverse | GCCTCATTTCCCGCCTGGCAGACGACACAAAGACCTACAAAGCTGAGAAGGCTCGTGGAGAAGAATCTTCATGCATATCGTGTTATATG | |
| pLIFE33 | PxaTPS5-forward | GGCTTAAUAATGGCTGGCGTTTCTGTTG |
| PxaTPS5-reverse | GGTTTAAUTTAAAGTGGGAGCGGATCAATTAGAC | |
| PxaTPS8-forward | GGCTTAAUAATGTCAAGATTTACATCTGCTGC | |
| PxaTPS8-reverse | GGTTTAAUTTATAAAGACACAGCTTGAACGAG | |
| pESC-His | PxaTPS8-forward | GAATTCAACCCTCACTAAAGGGCGGCCGCATGACGAAAAGGAGTGAAGCAGATG |
| PxaTPS8-reverse | CTTGTAATCCATCGATACTAGTGCGGCCGCTTATAAAGACACAGCTTGAACGAGAGTTC | |
| qRT-PCR | PxaTPS5-forward | CTTGATTCGAAGAACGGCCAA |
| PxaTPS5-reverse | TCGTTGATTCTCCATCTCCTGC | |
| PxaTPS8-forward | TGCTAGAAACGCTTGGGAGG | |
| PxaTPS8-reverse | ATGGTGGAGGCCAAGTGATG | |
| EF-1α-forward | CCTGGCCATCGTGACTTCAT | |
| EF-1α-reverse | AAGTGAACGCCAATAGGGCA |
Enzyme Assays.
For in vitro enzyme assays, a truncated form of PxaTPS8 (lacking the plastidial transit peptide, Δ26) and the full-length (FL) PxaTPS5 cDNA in the pET28b vector (www.emdmillipore.com) were expressed in E. coli BL21DE3-C41, Ni2+-affinity purified, and assayed as described elsewhere (10), with 15 µM of GPP, FPP, or GGPP (www.sigmaaldrich.com/) as substrates. For expression in N. benthamiana, FL PxaTPS5 and PxaTPS8 constructs in the pLIFE33 vector were transformed into Agrobacterium tumefaciens strain GV3101 as reported earlier (10). Cultures containing the TPS constructs were mixed with one culture volume of the RNA-silencing suppressor construct p19 and were infiltrated into the abaxial side of the leaves of 6-wk-old plants and incubated for 5 d (10). Expression of only p19 served as a control.
Metabolite Analysis.
Terpenes were isolated directly from E. coli cultures or by grinding plant tissues in liquid N2, extraction with 1 mL of hexane (10), and GC-MS analysis on an Agilent 7890B gas chromatograph with a 5977 Extractor XL mass selective detector (Agilent Technologies) at 70 eV and 1.2 mL/min He flow using a HP5-ms column (30 m, 250 µm i.d., 0.25 µm film). The GC parameters were 40–50 °C for 1–2 min, 10–20 °C/min to 300 °C, hold for 3 min; pulsed splitless injection at 250 °C. For UPLC-MS/MS analysis, hexane extracts were dried under a N2 stream, resuspended in 1 mL MeOH, and purified through 0.2-μm GH Polypro filters (www.pall.com/main/home.page) before analysis on an Agilent 6560 quadrupole time-of-flight system coupled with an Agilent 1290 Infinity II LC system, using a Waters Acquity UPLC CSH C18 column (100 × 2.1 mm; 1.7 μm) in electrospray ionization (ESI) in negative mode. For NMR analysis, pseudolaratriene was produced in an E. coli system engineered for terpene formation (22) using protein expression in 2 L culture for 72 h at 16 °C. Enzyme product was extracted with 1 L hexane and purified on silica matrix (70-230 mesh size) using hexane. 1H, 13C, COSY, HSQC, NOESY, and selective 1D NOE NMR spectra were acquired in deuterated CHCl3 on a Bruker 800 MHz Avance III spectrometer equipped with a Bruker 5 mm CPTCI cryoprobe.
Pseudolaratriene Formation in Yeast.
The PxaTPS8Δ26 construct was cloned into the pESC-HIS:BTS1 plasmid for coexpression with the yeast GGPP synthase BTS1 (30). After transformation into the S. cerevisiae strain AM94 (31), cells were grown in 1 L selective dropout medium [−His, −Leu, 2% (mass/vol) dextrose] at 30 °C to an OD600 of ∼0.6. Cells then were transferred into 1 L of yeast extract peptone (YEP) medium with 2% (mass/vol) galactose for induction. After 41 h, pseudolaratriene was extracted by vortexing the cells with glass beads in 5 mL of diethyl ether and separation on a silica matrix with 95:5% (vol/vol) hexane:ethyl acetate.
Quantum Chemical Calculations.
Computational NMR and quantum chemical calculations on the carbocation cyclization/rearrangement mechanism were carried out as previously described for theoretical studies on diterpene-forming carbocation rearrangements (28, 43). Experimental details are given in Dataset S1.
qPCR Analysis.
Total RNA was isolated as described previously (44). After cDNA amplification with the SuperScript III reverse transcriptase (Invitrogen), qRT-PCR was performed on a Bio-Rad CFX96 Real-Time system using iTaq SYBR Green Supermix (www.bio-rad.com) and gene-specific primers (Table S2). Transcript abundance was calculated using efficiency-corrected ΔCT values based on elongation factor 1α (EF-1α) as the reference gene and triplicate measurements with three biological replicates. Sequence verification of amplicons confirmed target specificity.
Phylogenetic Analysis.
Amino acid alignments were performed using clustalW2 and curated with gBlocks (molevol.cmima.csic.es/castresana/Gblocks_server.html). A maximum likelihood tree was generated in PhyML (atgc.lirmm.fr/phyml/) with 1,000 bootstrap repetitions.
Homology Modeling and Site-Directed Mutagenesis.
Homology models of PxaTPS5 and PxaTPS8 were generated using SWISS-MODEL based on the structure of A. grandis α-bisabolene synthase (PDB-ID: 3EAS) (19) with stereochemical validation using Ramachandran plots. GGPP was docked in the active site using Molegro Virtual Docker (45). Protein variants were generated using site-specific oligonucleotides (Table S2) and the pET28b:PxaTPS8 construct as template. DpnI treatment removed template plasmids. PxaTPS8 chimeric variants containing the helix G or H sequences of PxaTPS5 were generated by gene synthesis and overlap extension PCR, respectively (46). All protein variants were sequence verified.
Supplementary Material
Acknowledgments
We thank Dr. Bennett Addison, Carla Sanders, and Nhu Nguyen for assistance with structural analyses, Dr. Reuben Peters for sharing the pGG and pIRS plasmids, and Dr. David Baulcombe for the p19 plasmid. This work was supported by a University of California, Davis Academic Senate Faculty Research Grant (to P.Z.), the Natural Sciences and Engineering Research Council of Canada (J.B.), National Science Foundation Grants CHE-1361807 and CHE030089 [Extreme Science and Engineering Discovery Environment (XSEDE) program] (to D.J.T.), and National Institutes of Health Grants U24-DK097154 and S10-RR031630 (to O.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The nucleotide sequences described in this study have been deposited in the National Center for Biotechnology Information GenBank/European Bioinformatics Institute Data Bank [accession nos. KU685114 (PxaTPS8) and KU685113 (PxaTPS5)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612901114/-/DCSupplemental.
References
- 1.Wurtzel ET, Kutchan TM. Plant metabolism, the diverse chemistry set of the future. Science. 2016;353(6305):1232–1236. doi: 10.1126/science.aad2062. [DOI] [PubMed] [Google Scholar]
- 2.Tholl D. Biosynthesis and biological functions of terpenoids in plants. Adv Biochem Eng Biotechnol. 2015;148:63–106. doi: 10.1007/10_2014_295. [DOI] [PubMed] [Google Scholar]
- 3.Bohlmann J, Keeling CI. Terpenoid biomaterials. Plant J. 2008;54(4):656–669. doi: 10.1111/j.1365-313X.2008.03449.x. [DOI] [PubMed] [Google Scholar]
- 4.Zerbe P, Bohlmann J. Plant diterpene synthases: Exploring modularity and metabolic diversity for bioengineering. Trends Biotechnol. 2015;33(7):419–428. doi: 10.1016/j.tibtech.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Chiu P, Leung LT, Ko BCB. Pseudolaric acids: Isolation, bioactivity and synthetic studies. Nat Prod Rep. 2010;27(7):1066–1083. doi: 10.1039/b906520m. [DOI] [PubMed] [Google Scholar]
- 6.Wong VKW, et al. Pseudolaric acid B, a novel microtubule-destabilizing agent that circumvents multidrug resistance phenotype and exhibits antitumor activity in vivo. Clin Cancer Res. 2005;11(16):6002–6011. doi: 10.1158/1078-0432.CCR-05-0209. [DOI] [PubMed] [Google Scholar]
- 7.Sun Q, Li Y. The inhibitory effect of pseudolaric acid B on gastric cancer and multidrug resistance via Cox-2/PKC-α/P-gp pathway. PLoS One. 2014;9(9):e107830. doi: 10.1371/journal.pone.0107830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarkar T, et al. Interaction of pseudolaric acid B with the colchicine site of tubulin. Biochem Pharmacol. 2012;84(4):444–450. doi: 10.1016/j.bcp.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trost BM, Waser J, Meyer A. Total synthesis of (-)-pseudolaric acid B. J Am Chem Soc. 2008;130(48):16424–16434. doi: 10.1021/ja806724x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zerbe P, et al. Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol. 2013;162(2):1073–1091. doi: 10.1104/pp.113.218347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters RJ. Two rings in them all: The labdane-related diterpenoids. Nat Prod Rep. 2010;27(11):1521–1530. doi: 10.1039/c0np00019a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen F, Tholl D, Bohlmann J, Pichersky E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011;66(1):212–229. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhou K, et al. Insights into diterpene cyclization from structure of bifunctional abietadiene synthase from Abies grandis. J Biol Chem. 2012;287(9):6840–6850. doi: 10.1074/jbc.M111.337592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Köksal M, Jin Y, Coates RM, Croteau R, Christianson DW. Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature. 2011;469(7328):116–120. doi: 10.1038/nature09628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Y, Honzatko RB, Peters RJ. Terpenoid synthase structures: A so far incomplete view of complex catalysis. Nat Prod Rep. 2012;29(10):1153–1175. doi: 10.1039/c2np20059g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keeling CI, Bohlmann J. Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol. 2006;170(4):657–675. doi: 10.1111/j.1469-8137.2006.01716.x. [DOI] [PubMed] [Google Scholar]
- 17.Hall DE, et al. Evolution of conifer diterpene synthases: Diterpene resin acid biosynthesis in lodgepole pine and jack pine involves monofunctional and bifunctional diterpene synthases. Plant Physiol. 2013;161(2):600–616. doi: 10.1104/pp.112.208546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams DC, et al. Heterologous expression and characterization of a “Pseudomature” form of taxadiene synthase involved in paclitaxel (Taxol) biosynthesis and evaluation of a potential intermediate and inhibitors of the multistep diterpene cyclization reaction. Arch Biochem Biophys. 2000;379(1):137–146. doi: 10.1006/abbi.2000.1865. [DOI] [PubMed] [Google Scholar]
- 19.McAndrew RP, et al. Structure of a three-domain sesquiterpene synthase: A prospective target for advanced biofuels production. Structure. 2011;19(12):1876–1884. doi: 10.1016/j.str.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 20.Martin DM, Fäldt J, Bohlmann J. Functional characterization of nine Norway Spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol. 2004;135(4):1908–1927. doi: 10.1104/pp.104.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bohlmann J, Crock J, Jetter R, Croteau R. Terpenoid-based defenses in conifers: cDNA cloning, characterization, and functional expression of wound-inducible (E)-α-bisabolene synthase from grand fir (Abies grandis) Proc Natl Acad Sci USA. 1998;95(12):6756–6761. doi: 10.1073/pnas.95.12.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrone D, et al. Increasing diterpene yield with a modular metabolic engineering system in E. coli: Comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl Microbiol Biotechnol. 2010;85(6):1893–1906. doi: 10.1007/s00253-009-2219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaughan MM, et al. Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. Plant Cell. 2013;25(3):1108–1125. doi: 10.1105/tpc.112.100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lodewyk MW, Siebert MR, Tantillo DJ. Computational prediction of 1H and 13C chemical shifts: A useful tool for natural product, mechanistic, and synthetic organic chemistry. Chem Rev. 2012;112(3):1839–1862. doi: 10.1021/cr200106v. [DOI] [PubMed] [Google Scholar]
- 25.Marchand AP, Rose JE. On the question of bridge-proton absorptions in the nuclear magnetic resonance spectra of norbornene and related systems. J Am Chem Soc. 1968;90(14):3724–3731. [Google Scholar]
- 26.Peters RJ, Croteau RB. Abietadiene synthase catalysis: Mutational analysis of a prenyl diphosphate ionization-initiated cyclization and rearrangement. Proc Natl Acad Sci USA. 2002;99(2):580–584. doi: 10.1073/pnas.022627099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeling CI, Weisshaar S, Lin RPC, Bohlmann J. Functional plasticity of paralogous diterpene synthases involved in conifer defense. Proc Natl Acad Sci USA. 2008;105(3):1085–1090. doi: 10.1073/pnas.0709466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tantillo DJ. Walking in the woods with quantum chemistry--applications of quantum chemical calculations in natural products research. Nat Prod Rep. 2013;30(8):1079–1086. doi: 10.1039/c3np70028c. [DOI] [PubMed] [Google Scholar]
- 29.Tantillo DJ. The carbocation continuum in terpene biosynthesis--where are the secondary cations? Chem Soc Rev. 2010;39(8):2847–2854. doi: 10.1039/b917107j. [DOI] [PubMed] [Google Scholar]
- 30.Ro DK, Bohlmann J. Diterpene resin acid biosynthesis in loblolly pine (Pinus taeda): Functional characterization of abietadiene/levopimaradiene synthase (PtTPS-LAS) cDNA and subcellular targeting of PtTPS-LAS and abietadienol/abietadienal oxidase (PtAO, CYP720B1) Phytochemistry. 2006;67(15):1572–1578. doi: 10.1016/j.phytochem.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Ignea C, et al. Efficient diterpene production in yeast by engineering Erg20p into a geranylgeranyl diphosphate synthase. Metab Eng. 2015;27:65–75. doi: 10.1016/j.ymben.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 32.Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11(3):191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- 33.Pateraki I, et al. Manoyl oxide (13R), the biosynthetic precursor of forskolin, is synthesized in specialized root cork cells in Coleus forskohlii. Plant Physiol. 2014;164(3):1222–1236. doi: 10.1104/pp.113.228429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts SC. Production and engineering of terpenoids in plant cell culture. Nat Chem Biol. 2007;3(7):387–395. doi: 10.1038/nchembio.2007.8. [DOI] [PubMed] [Google Scholar]
- 35.Tantillo DJ. Biosynthesis via carbocations: Theoretical studies on terpene formation. Nat Prod Rep. 2011;28(6):1035–1053. doi: 10.1039/c1np00006c. [DOI] [PubMed] [Google Scholar]
- 36.Zerbe P, Chiang A, Bohlmann J. Mutational analysis of white spruce (Picea glauca) ent-kaurene synthase (PgKS) reveals common and distinct mechanisms of conifer diterpene synthases of general and specialized metabolism. Phytochemistry. 2012;74:30–39. doi: 10.1016/j.phytochem.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 37.Leonard E, et al. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc Natl Acad Sci USA. 2010;107(31):13654–13659. doi: 10.1073/pnas.1006138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zerbe P, et al. Bifunctional cis-abienol synthase from Abies balsamea discovered by transcriptome sequencing and its implications for diterpenoid fragrance production. J Biol Chem. 2012;287(15):12121–12131. doi: 10.1074/jbc.M111.317669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parveen I, et al. Investigating sesquiterpene biosynthesis in Ginkgo biloba: Molecular cloning and functional characterization of (E,E)-farnesol and α-bisabolene synthases. Plant Mol Biol. 2015;89(4-5):451–462. doi: 10.1007/s11103-015-0381-3. [DOI] [PubMed] [Google Scholar]
- 40.Huber DP, Philippe RN, Godard KA, Sturrock RN, Bohlmann J. Characterization of four terpene synthase cDNAs from methyl jasmonate-induced Douglas-fir, Pseudotsuga menziesii. Phytochemistry. 2005;66(12):1427–1439. doi: 10.1016/j.phytochem.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 41.Buchanan MS, Connolly JD, Rycroft DS. Sphenolobane diterpenoids of the liverwort Anastrophyllum donnianum. Phytochemistry. 1996;43(6):1297–1301. [Google Scholar]
- 42.Urones JG, Marcos IS, Garrido MD. Tormesane derivatives of Halimium viscosum. Phytochemistry. 1990;29(10):3243–3246. [Google Scholar]
- 43.Potter KC, et al. Blocking Deprotonation with retention of aromaticity in a plant ent-copalyl diphosphate synthase leads to product rearrangement. Angew Chem Int Ed Engl. 2016;55(2):634–638. doi: 10.1002/anie.201509060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolosova N, et al. Isolation of high-quality RNA from gymnosperm and angiosperm trees. Biotechniques. 2004;36(5):821–824. doi: 10.2144/04365ST06. [DOI] [PubMed] [Google Scholar]
- 45.Thomsen R, Christensen MH. MolDock: A new technique for high-accuracy molecular docking. J Med Chem. 2006;49(11):3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 46.Horton RM, Cai Z, Ho SM, Pease LR. Gene splicing by overlap extension: Tailor-made genes using the polymerase chain reaction. Biotechniques. 2013;8(5):528–535. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.