Significance
The mitochondrial F1Fo ATP synthase is an essential membrane protein machine that supplies all eukaryotic cells with ATP. The proton-driven rotation of the rotor assembly in the membrane transmits energy to the catalytic F1 head, where ATP is generated by rotary catalysis. We determined the in situ structures of ATP synthase dimers from the lethal human parasite Trypanosoma brucei and its free-living relative Euglena gracilis. In both ATP synthases, the catalytic subunits form a threefold pyramid rather than the usual near-sixfold ring. This unexpected finding indicates that the structure of the F1 head, and therefore its catalytic action, is less highly conserved than previously thought, and provides insight into the fundamental mechanism of ATP production in higher organisms.
Keywords: mitochondrial ATP synthase, electron cryotomography, subtomogram averaging, trypanosome, rotary catalysis
Abstract
We used electron cryotomography and subtomogram averaging to determine the in situ structures of mitochondrial ATP synthase dimers from two organisms belonging to the phylum euglenozoa: Trypanosoma brucei, a lethal human parasite, and Euglena gracilis, a photosynthetic protist. At a resolution of 32.5 Å and 27.5 Å, respectively, the two structures clearly exhibit a noncanonical F1 head, in which the catalytic (αβ)3 assembly forms a triangular pyramid rather than the pseudo-sixfold ring arrangement typical of all other ATP synthases investigated so far. Fitting of known X-ray structures reveals that this unusual geometry results from a phylum-specific cleavage of the α subunit, in which the C-terminal αC fragments are displaced by ∼20 Å and rotated by ∼30° from their expected positions. In this location, the αC fragment is unable to form the conserved catalytic interface that was thought to be essential for ATP synthesis, and cannot convert γ-subunit rotation into the conformational changes implicit in rotary catalysis. The new arrangement of catalytic subunits suggests that the mechanism of ATP generation by rotary ATPases is less strictly conserved than has been generally assumed. The ATP synthases of these organisms present a unique model system for discerning the individual contributions of the α and β subunits to the fundamental process of ATP synthesis.
F1Fo (F-type) ATP synthases are ancient, energy-converting nanomachines that generate ATP by rotary catalysis (1). In mitochondria, the ATP synthases form rows of dimers along the highly curved ridges of the inner membrane cristae (2, 3). All known F1Fo ATP synthases consist of a catalytic hydrophilic (αβ)3 hexamer, a hydrophobic membrane-embedded region containing a rotor ring driven like a turbine by the proton gradient, and a pair of stalks connecting the two. The central stalk transmits the torque of the rotor ring to the (αβ)3 hexamer to power catalysis, whereas the peripheral stalk acts as a stator to prevent idle rotation of the catalytic subunits with the rotor assembly. The (αβ)3 subunits and the central stalk make up the membrane-extrinsic F1 subcomplex. The membrane-intrinsic subunits, including the rotor ring and the peripheral stalk, form the Fo subcomplex.
The F1 subcomplex has three catalytic sites, each located at an interface of the α and β subunits. The two subunits alternate in the (αβ)3 hexamer, which forms a ring of near-sixfold symmetry (4). ATP synthesis is powered by the proton-driven rotation of the central stalk, which induces conformational changes in the α and β subunits (5, 6). In the course of these changes, a conserved α subunit arginine inserts into the nucleotide binding pocket of the β subunit (7). Substitution of the arginine residue inhibits ATP production, highlighting the essential role of the α subunit in catalysis (8, 9).
Trypanosomes belong to a group of parasitic protists, which cause severe and widespread insect-borne human and animal diseases. Biochemistry and mass spectrometry have indicated that the α subunit in the mitochondrial ATP synthase of euglenozoa (which include trypanosomes) exists as two separate fragments: an N-terminal 14-kDa fragment (αN) and a C-terminal 44-kDa fragment (αc) (10–14). Even though the α subunit is cleaved, the complex still hydrolyzes and synthesizes ATP, which is critical for the survival of Trypanosoma brucei, the sleeping sickness parasite (10, 11, 14–19). How this cleavage affects the structure and molecular mechanism of the enzyme is unknown.
Here we report the in situ structure of the mitochondrial ATP synthase from the “procyclic” insect stage of the lethal human parasite T. brucei and the related Euglena gracilis, a photosynthetic protist. The structures were determined by electron cryotomography and subtomogram averaging to a resolution of 32.5 Å and 27.5 Å, respectively. Both structures reveal an unexpected arrangement of the catalytic subunits, which form a triangular pyramid rather than a hexameric ring of near-sixfold symmetry, as is seen in all other known rotary ATPases. Rigid body fitting of X-ray structures showed that the unusual arrangement of catalytic subunits in the F1 head results from the phylum-specific cleavage of the α subunit. The subtomogram averages from both organisms indicate that the αc fragment is displaced from the central stalk by ∼2 nm and forms a link between neighboring β subunits. Consequently, the typical catalytic interfaces of rotary ATPases cannot form, and therefore the mechanism of ATP synthesis must be different.
Results
Structure of Euglenozoan ATP Synthase Dimers in Situ.
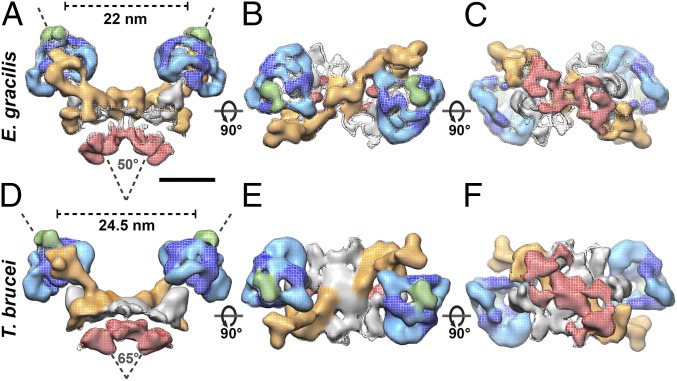
Mitochondrial membranes were isolated from procyclic (insect stage) T. brucei or light-grown E. gracilis cells and imaged by electron cryotomography. Tomographic volumes revealed that cristae were discs decorated with double rows of 10-nm particles. The structure of the particles obtained by subtomogram averaging confirmed that they were ATP synthase dimers. A total of 1,294 dimers from E. gracilis and 925 dimers from T. brucei were averaged to generate maps at a resolution of 27.5 Å and 32.5 Å, respectively (Fig. 1, Fig. S1, and Movie S1). The ATP synthase dimers of both species are twofold-symmetric V-shaped structures, with distances between the catalytic F1 heads of 24.5 nm in T. brucei and 22 nm in E. gracilis (Fig. 1 A and D) and an angle between the long axes of the monomers of 65° in T. brucei and 50° in E. gracilis. The peripheral stalks are positioned on opposite sides of the dimer and are structurally unrelated to other peripheral stalk subcomplexes (Fig. 1 B and E) (3, 20). On the lumenal side of the membrane, the monomers are connected by several interlinked protein densities (Fig. 1 C and F). In the complex from E. gracilis, two protein domains near the dimer axis tie the monomers together on the matrix side.
Fig. 1.
In situ structure of mitochondrial ATP synthase dimers in E. gracilis and T. brucei. Subtomogram average of ATP synthase dimers from E. gracilis (Upper) and T. brucei (Lower). (A and D) Side view. (B and E) Matrix view. (C and F) Lumenal view. Dark and light blue indicate the catalytic region; orange, peripheral stalk; red, lumenal density; gray, central stalk; green, OSCP. See also Movie S1 and Fig. S1. (Scale bar: 10 nm.)
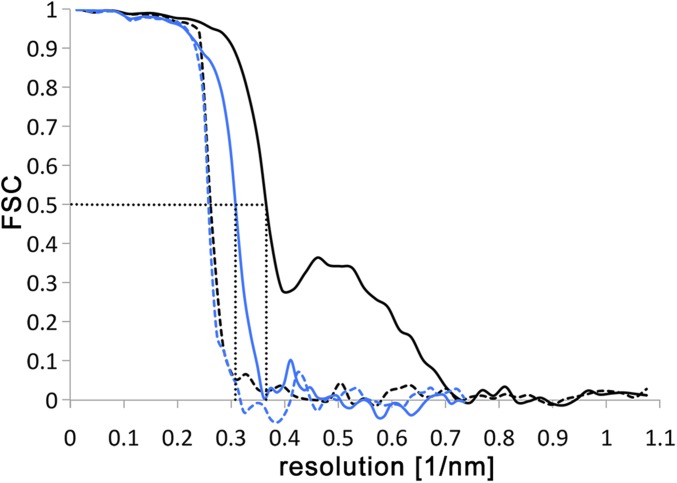
Fig. S1.
Resolution estimates of subtomogram averages. FSC curves of the ATP synthase dimer averages from E. gracilis (black) and T. brucei (blue). To assess mask bias, FSC was performed on half-maps created from tomograms with phases randomized beyond 40 Å (dashed lines). The correlation drops at the resolution above which the phases were randomized, indicating that the mask does not affect the resolution estimate. FSC curves for the E. gracilis and T. brucei maps indicate resolutions of 27.5 Å and 32.5 Å for the two maps using the 0.5 FSC criterion (59) (dotted lines).
Cleavage of the α Subunit Results in an Unusual Arrangement of Catalytic Subunits.
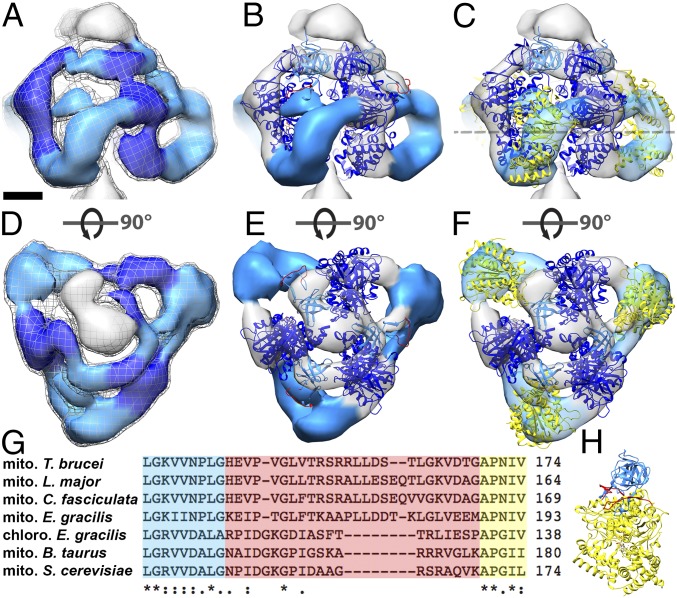
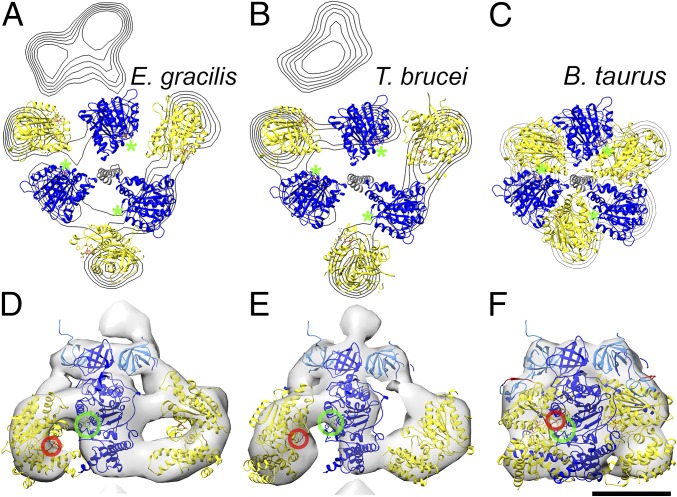
The most conspicuous feature of the two new ATP synthase dimer maps is the catalytic F1 head (Fig. 2 and Fig. S2). In all known rotary ATPases, the F1 head is a hexagonal prism of near-sixfold symmetry, consisting of alternating α and β subunits or their equivalents (4, 21–24). In the T. brucei and E. gracilis ATP synthases, the F1 head resembles a triangular pyramid (Fig. 2 A and D and Fig. S2 A and E). A comparison with the bovine F1 reveals that the subtomogram average map accommodates only every other subunit of the (αβ)3 hexamer (Fig. 2 B and E and Fig. S2 B and C and S3). The densities of the three other intervening subunits are arc-shaped and look strikingly different from those in the bovine or yeast F1 heads.
Fig. 2.
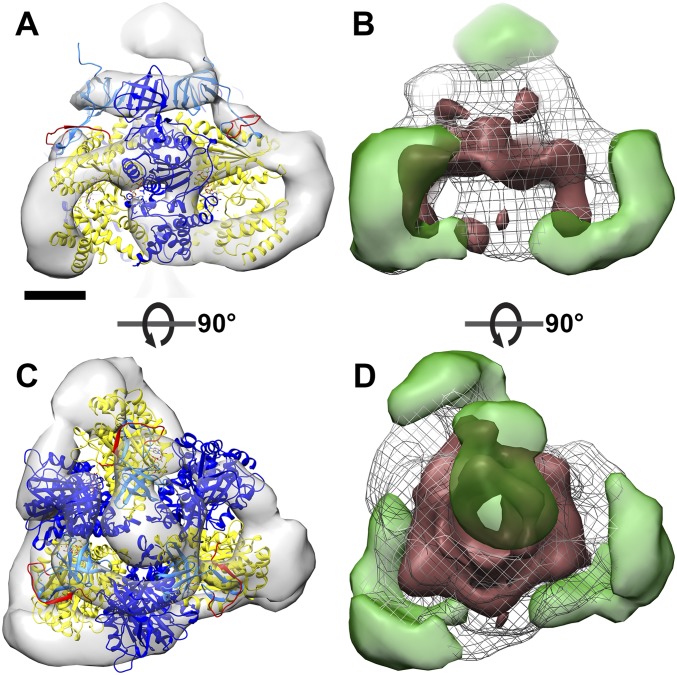
Fit of the atomic model to the catalytic region of mitochondrial ATP synthase dimers from E. gracilis. Side views (A–C) and matrix views (D–F) of the F1 head region in the subtomogram average of E. gracilis ATP synthase dimer. (A and D) Subtomogram average. (B and E) The same as A and D, with a fitted atomic model of the bovine (αβ)3 hexamer (PDB ID code 1BMF) (4) without αc. (C and F) The same as B and E, but with αc fragments fitted as a rigid body. The dashed line indicates a cross-sectional plane for Fig. 4 A–C. (G) Sequence alignment of the mitochondrial (mito) α subunit of T. brucei, Leishmania major, C. fasciculata, E. gracilis, Bos taurus, and Saccharomyces cerevisiae, and the chloroplast (chloro) α subunit of E. gracilis. Blue, C-terminal sequence of αN; yellow, N-terminal sequence of αc; red, nonconserved loop region and region of protease cleavage site in mitochondrial α subunit of euglenozoa. (H) α subunit of bovine heart ATP synthase color-coded as in G. Light blue, αN; yellow, αc; red, loop with protease cleavage site; dark blue, β subunit. (Scale bar in A–F: 2.5 nm.) See also Movie S2, Fig. S2, and Fig. S3.
Fig. S2.
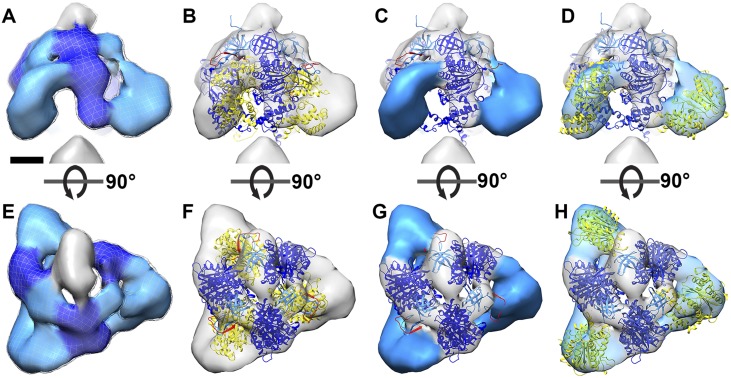
Catalytic region of the ATP synthase from T. brucei. Side view (A–D) and matrix view (E–H) of catalytic regions from subtomogram averages of T. brucei ATP synthase dimers. (A and E) Map density. (B and F) The same as A and E, with a fitted atomic model of the bovine (αβ)3 hexamer (PDB ID code 1BMF) (4). Dark blue, β subunit; light blue, αN fragment; yellow, αC fragment; red, loop with cleavage site. (C and G) Atomic model of the fitted bovine (αβ)3 hexamer with central and C-terminal domain removed. (D and H) The same as C and G, but with the αc fragment fitted to the arc-shaped density region as a rigid body. (Scale bar: 2.5 nm.) See also Movie S2.
Fig. S3.
Fit of the bovine (αβ)3 hexamer to the F1 region of E. gracilis ATP synthase dimer. Side view (A) and matrix view (C) of the F1 region with a fitted bovine (αβ)3 hexamer (PDB ID code 1BMF) (4). Dark blue, β subunit; light blue, αN fragments; yellow, αC fragment; red, loop with proteolytic cleavage site. (B and D) Difference map between the E. gracilis F1 region and bovine (αβ)3 hexamer (PDB ID code 1BMF) (4). Green indicates density present in E. gracilis F1 but not in the bovine (αβ)3 hexamer; dark red indicates density present in the bovine (αβ)3 hexamer but not in E. gracilis. Densities not present in the E. gracilis structure correspond mainly to the central and C-terminal domains of the α subunit as positioned in the bovine structure. (Scale bar: 2.5 nm.)
Previous mass spectrometry and biochemical studies indicated that the α subunit of euglenozoan ATP synthases exists as two separate fragments, αN and αC (10–13). To investigate whether the cleavage of the α subunit might account for the unusual structure of the F1 head, we fitted a cleaved model of the bovine F1 structure into the subtomogram average densities (Fig. 2 B and E and Fig. S2 C and G). The α subunit is cleaved by an unidentified protease in a sequence found only in euglenozoa (residues 145–169 in T. brucei) (10, 13). This cleavage site maps to a loop that connects the N-terminal β barrel to the central domain of the bovine α subunit (Fig. 2 G and H). We severed the α subunit of the bovine (αβ)3 atomic model (4) at this location and positioned the resulting subcomplex (β + αN subunits) into the EM maps as a rigid body. In both subtomogram averages, the atomic model closely matched the density, with the N-terminal β barrels from both the αN fragment and the β subunit occupying a narrow ring of density below the oligomycin sensitivity-conferring protein (OSCP), known as the crown region. The remainder of the β subunits occupied the three densities adjacent to the central stalk (Fig. 2 B and E and Fig. S2 C and G).
The fit of the αN and β subunits did not occupy the volumes of the arc-shaped densities that form the vertices of the triangular pyramid (blue densities in Fig. 2 B and E and Fig. S2 C and G). These densities, which span the gap between neighboring β subunits, are 10 nm long and ∼2 nm wide and enclose a volume of ∼49 × 103 Å3 in E. gracilis and 58 × 103 Å3 in T. brucei. Based on an average partial specific volume of 1.2 Å3/Da (25), each arc-shaped density should accommodate a protein with a molecular mass of 41–48 kDa. This is very similar to the size of the αC fragment (44 kDa), which in T. brucei has been shown to remain bound to the F1 subcomplex, as demonstrated by αC-specific antisera (26) and mass spectrometry (10).
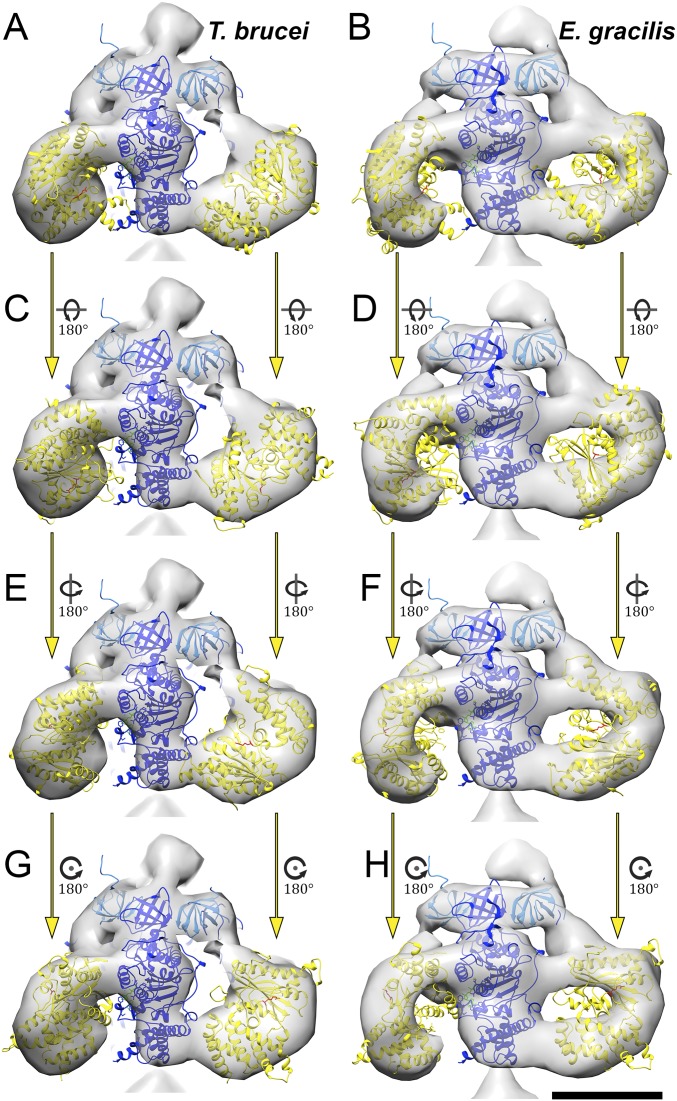
To assess whether the cleaved αC fragments could be located in the arc-shaped densities, we performed rigid body fitting (Fig. 2 C and F, Fig. S2 D and H, and Movie S2). The αC fragments fitted the profiles of the arc-shaped density well, indicating that they do indeed constitute the vertices of the triangular F1. Despite the good overall match, rigid body fitting permits several possible orientations of the αC fragment (Fig. S4). We consider the αC fragment position that involves an outward displacement of ∼20 Å and a rotation by 30° to be the most likely orientation (Fig. 2 C and F and Fig. S2 D and H), because it requires the minimal offset from the canonical structure. The accuracy of the fit is limited by the map resolution. Nevertheless, based on our difference map (Fig. S3 B and D), the rigid body fits (Fig. 2 C and F and Fig. S2 D and H), and the fact that the αC fragment remains attached to the F1 subcomplex after cleavage (10, 26), we can unambiguously assign the αC fragments to the arc-shaped densities. Thus, our results show that in euglenozoa, cleavage of the mitochondrial α subunit results in an unusual arrangement of catalytic subunits in a triangular F1 pyramid. We conclude that the structure of the catalytic F1 head in euglenozoa is substantially different from that of any other known rotary ATPase.
Fig. S4.
Alternative orientation of the αC fragments in the F1 subcomplex. Shown are F1 subcomplexes from T. brucei (A, C, E, and G) and E. gracilis (B, D, F, and H) with a fitted bovine atomic model (PDB ID code 1BMF) (4) with β subunits (dark blue), αN fragments (light blue), and αC fragments (yellow). Rigid body fits of different orientations of the αC fragments to the arc-shaped densities are shown in each row. Whereas the αC fragment orientations shown in A and B are related by a 20-Å outward shift and a 30° rotation to the canonical structure, all alternative models (C–H) require additional rotations of ∼180° around the long or short axis of the αC fragment and are thus unlikely. (Scale bars: 5 nm.)
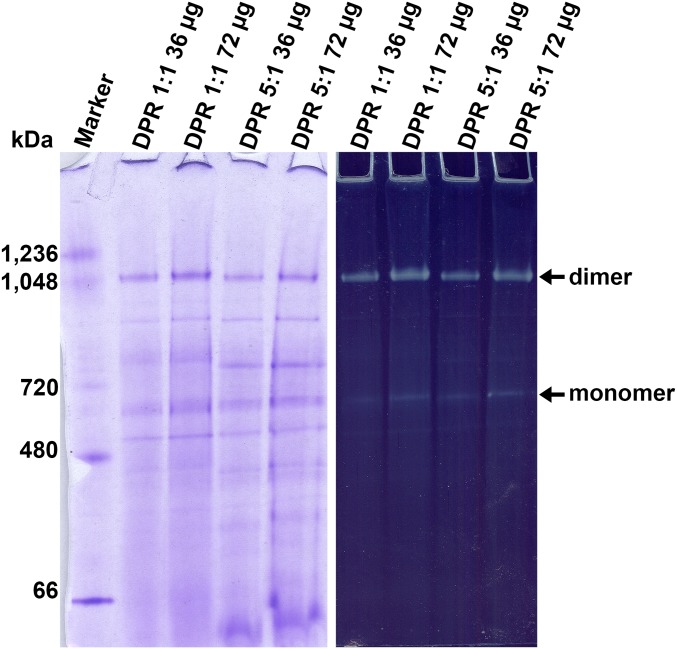
To assess whether the in situ structures obtained by subtomogram averaging are of an active complex, we solubilized samples of the E. gracilis mitochondrial membranes used for electron cryotomography with n-Dodecyl β-d-maltoside (DDM) and performed blue native gel electrophoresis (BN-PAGE). Two bands reacted to an in-gel ATPase activity assay, one in the megadalton range, which is the ATP synthase dimer, and the other at ∼700 kDa, which is the ATP synthase monomer (Fig. S5). These results indicate that the ATP synthases averaged in the tomograms of isolated mitochondrial membranes from E. gracilis are catalytically active.
Fig. S5.
Solubilized mitochondrial membrane complexes from E. gracilis exhibit ATPase activity. (Left) BN-PAGE of mitochondrial membranes from E. gracilis solubilized in DDM (DPRs of 1:1 and 5:1). (Right) In-gel ATPase activity assay performed on the same gel shown on the left. The upper band is the ATP synthase dimer. The weaker activity of the ∼700-kDa band is the ATP synthase monomer.
Dimer Rows in Euglena and Trypanosomes Indicate an Unexpected Arrangement of ATP Synthase Monomers.
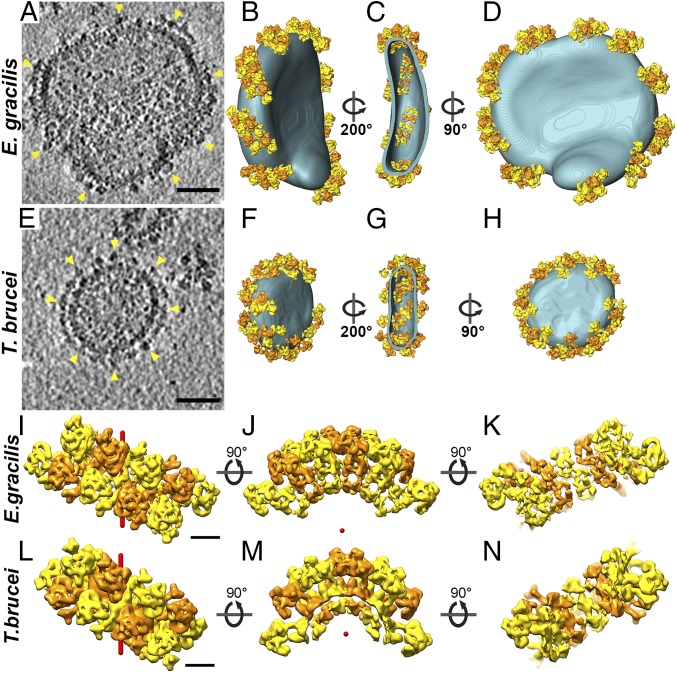
To determine the macromolecular arrangement of ATP synthase dimers from E. gracilis and T. brucei in the membrane, the subtomogram averages were placed into the original tomographic volumes in the positions and orientations determined in the averaging process. In both species, the isolated mitochondrial membranes were decorated with multiple ATP synthase dimer rows that are arranged in short left-handed helix segments around the edges of discoid vesicles (Fig. 3 A–H and Movie S3). Each row contained between three and six dimers. The short dimer rows encompass the entire circumference of the disk-shaped membranes (Fig. 3 D and H).
Fig. 3.
Macromolecular arrangement of mitochondrial ATP synthase dimers from E. gracilis and T. brucei in situ. Tomographic slice through discoid mitochondrial membrane vesicles from E. gracilis (A) and T. brucei (E) containing ATP synthases (arrowheads). (B and F) Surface representations of A and (E) highlighting discoid membranes (blue) and ATP synthase dimer ribbons (yellow and orange). (C and G) Section through the vesicles in B and F showing the arrangement of ATP synthase dimers viewed from cristae lumen. (D and H) Same as in B and F, but rotated 90°. (Scale bar: 50 nm.) See also Movie S3. (I–N) Row arrangement of ATP synthase dimers as determined from subtomogram averages. (I and L) Matrix view. (J and M) Side view. (K and N) Lumenal view. (I–K) E. gracilis. (L–N) T. brucei. Alternating dimers are colored orange and yellow, and form rows with left-handed helicity (helix axes shown in red). Adjacent ATP synthase monomers in the row belong to different dimers; thus, the dimers are interdigitated. (Scale bar: 10 nm.) See also Movie S3.
In each row, the two F1 monomers from one dimer interdigitate with monomers from the next dimers along the row. As a result, the nearest-neighbor monomers, both across and along the row, belong to different dimers (Fig. 3 I and L). This contrasts with the arrangement of ATP synthase dimers in metazoans and fungi, where the nearest monomers across the row belong to the same dimer (3). When viewed from the lumen, the dimers form a ladder-like assembly (Fig. 3 C, G, K, and N). Examination of each row in isolation shows that the principal curvature is caused by the association of dimers into rows, rather than by the dimers themselves (Fig. 3 J and M). The resulting membrane curvature is stronger in T. brucei than in E. gracilis (Fig. 3 J and M), most likely due to the different dimer angles in the two species (Fig. 1 A and D).
Discussion
Structural Variation of Mitochondrial ATP Synthase Dimers and Dimer Rows.
The assembly of the mitochondrial ATP synthase into dimers, and of dimers into rows, is a feature common to mitochondria of all species examined to date (2, 3, 27–29). The principal role of dimer rows appears to be in the formation of mitochondrial inner membrane cristae. Tomographic analysis has revealed two different row architectures that bend the membrane in different ways. The first type of row architecture, which we call the metazoan type, is found in metazoans, plants, and fungi. In this group, the ATP synthases form V-shaped dimers with a dimer angle of ∼90° between monomers (3). Dimers of the metazoan type assemble into straight rows along the edges of lamellar cristae, without any lateral offset between adjacent dimers (3). The V-shape of individual dimers induces a sharp, local 90°-membrane curvature that is responsible for driving the self-association of dimers into rows and shaping cristae membranes (30, 31).
In ciliates (29), euglenozoa, and green algae (20), the ATP synthase dimers form a left- or right-handed helix along the curved ridges of the cristae or along the outer perimeter of helical tubular cristae. In these organisms, the principal membrane curvature is caused by the macromolecular association of dimers into helical arrays, rather than by the individual dimers as in metazoans (Fig. 3 J and M). It will be interesting to investigate the architecture of ATP synthase complexes in the bloodstream stage of T. brucei, which lacks oxidative phosphorylation and contains sparse, if any, mitochondrial cristae (32) and uses the enzyme as an ATP-powered proton pump to generate the mitochondrial membrane potential (16, 18).
A striking difference between the metazoan-type ATP synthase dimers and ATP synthase dimers of protozoans or unicellular algae, which we refer to as the protozoan type, is the structural diversity of the peripheral stalk and dimer interfaces (33). The architectures of all metazoan-type ATP synthase dimers studied so far are virtually identical (3), and sequence analysis and proteomics have identified homologous subunits for nearly all of the subunits that constitute the mitochondrial ATP synthase in these organism (34, 35). In contrast, there seem to be no subunits in the protozoan-type ATP synthases that correspond to the peripheral stalk or dimer interface in the metazoan type. Instead, proteomics and biochemical analysis have identified several unique subunits, none of which are homologous among the various phyla (10, 11, 36, 37). This variation in subunit composition correlates with the observed diversity in structure, which may reflect adaptations to different energetic requirements or environmental conditions. Apparently, these adaptations occurred before trypanosomes adopted their parasitic lifestyle.
Unusual Arrangement of Catalytic Subunits on the F1 Head of Euglenozoan ATP Synthase Dimers.
Despite the diversity of ATP synthase dimer architectures observed in different species, the catalytic subunits that are involved in ATP synthesis and form the F1 subcomplex are structurally conserved in all previously reported structures (36, 38, 39). Here we show that the catalytic F1 head of the mitochondrial ATP synthase dimers from E. gracilis and T. brucei exhibits an unprecedented pyramidal structure (Fig. 2 and Fig. S2). This unusual arrangement of catalytic subunits is surprising, given the otherwise strictly conserved pseudo-sixfold symmetry and catalytic mechanism of the (αβ)3 hexamer in all other known F-type ATPases, including those of bacteria and chloroplasts (4, 24, 40). The observed alterations in the T. brucei and E. gracilis F1 region are in line with previous biochemical studies reporting a complete cleavage of the α subunit into two fragments, both of which remain bound to the ATP synthase (10–14). Furthermore, it has been shown that despite the α-subunit fragmentation, the T. brucei ATP synthase remains catalytically active throughout the life cycle and is essential for parasite survival in both the procyclic and bloodstream forms (10, 16, 18, 19); as mentioned above, it is the reverse function as a proton-pumping ATPase that is essential for survival in the mammalian host. In addition, the isolated mitochondrial membranes from E. gracilis isolated in this study contained catalytically active ATPase, as demonstrated by in-gel activity (Fig. S5).
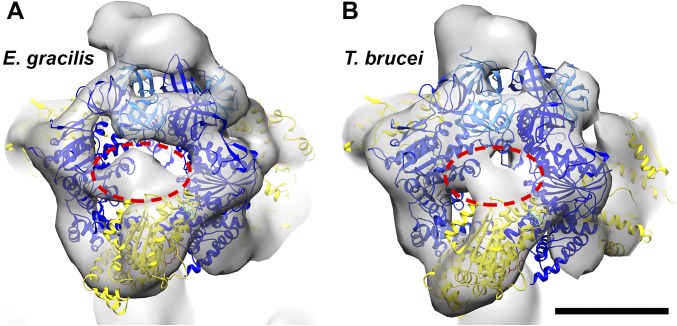
In the bovine ATP synthase, the catalytic sites are located at the interface of the α- and β subunits (Fig. 4C). The nucleotide-binding sites are formed largely by the central domain of the β subunits, whereas the α subunits contribute a conserved arginine (the arginine finger) that is essential for catalysis (8) (Fig. 4 C and F). In euglenozoa, the nucleotide-binding site in the β subunit is conserved (13), but the α/β interface is not (Fig. 2 B and E and Fig. S2 C and G); however, in the unusual arrangement of catalytic subunits, it appears unlikely that the arginine in the α subunit, which is present in T. brucei and E. gracilis, can reach the bound nucleotide from its remote position in the central domain of the αc fragment (Fig. 4 D and E). Because the arginine finger is critical for catalysis in both the ATP synthase (8) and in many other ATPase domains in general (e.g., AAA+ ATPases) (41), it must be located at the active site and thus must be contributed by a different part of the structure. Apart from a suitably positioned residue in the αc fragment, a prime candidate is the euglenozoa-specific F1 subunit p18 (10–13) (systematic ID in T. brucei Tb927.5.1710). Knockdown of p18 in procyclic T. brucei by RNAi has been shown to result in slowed growth and disappearance of F1, indicating that p18 is required for F1 integrity (42). Although functional data on the role of p18 in other trypanosomatids has not been reported to date, p18 has been identified as a subunit of mitochondrial ATP synthase in other trypanosomatids, including Leishmania tarentolae (13) and Crithidia fasciculata (12), as well as in the euglenoid E. gracilis (11), strongly suggesting that this subunit is generally conserved in euglenozoa. The location of p18 in the unusual F1 structure is currently unknown; however, an unassigned density region between αN and αC and the likely presence of more than one copy of p18 per F1 molecule (10) suggest that this region may contain p18 (Fig. S6). Resolving the question of how the euglenozoan ATP synthase dimers produce ATP will require a higher-resolution structure.
Fig. 4.
Domain arrangement in the catalytic F1 region. (A and B) Cross-section of catalytic domain of E. gracilis and T. brucei with fitted atomic models as indicated in Fig. 2C, contoured in steps of 0.8 σ. (C) Cross-section of the conserved bovine structure (PDB code: 1BMF) (4) for comparison. Blue, β subunits; yellow αC fragment; gray, central stalk. Green asterisks indicate nucleotide-binding sites in the β subunits. (D–F) Enlarged view of the catalytic interface from E. gracilis (D), T. brucei (E), and B. taurus (F), with the arginine finger circled in red and the nucleotide- binding site in green. (Scale bar: 5 nm.)
Fig. S6.
Unaccounted-for density region in the euglenozoan F1 subcomplex. Shown are the F1 regions of the subtomogram averages from E. gracilis (A) and T. brucei (B) fitted with atomic models from the bovine (αβ)3 hexamer (PDB ID code 1BMF) (4). Whereas the positions of the β subunits (dark blue) and αN fragments (light blue) are unchanged relative to the conserved F1 structure, the αC fragments (yellow) shift to the arc-shaped densities that appear to be characteristic of euglenozoan ATP synthases. A small unassigned density between the αN and αC fragments (red dashed ellipses) may contain the euglenozoan-specific p18 subunit (10, 11, 42). (Scale bar: 5 nm.)
The in Situ Structure of Euglenozoan ATP Synthase Dimers Provides Insight into Rotary Catalysis.
ATP synthesis by rotary catalysis involves three key stages: (i) proton movement across the membrane-embedded Fo sector drives the rotation of the central stalk; (ii) rotation of the central stalk causes conformational changes at the catalytic α/β interface, which triggers phosphate bond formation; and (iii) conformational changes are cooperatively transmitted between catalytic sites. The exact molecular mechanism of this process is not known. A central question is the extent to which each of the α and β subunits contributes to this process. In the euglenozoan ATP synthase, the contributions of each subunit to rotary catalysis can be dissected. The displacement of the αC fragments away from the central stalk means that the γ subunit is unable to interact with the α subunits directly (Fig. 4 A and B). Thus, at least in the euglenozoan ATP synthase, but possibly also in the conserved F1 structure, interactions between the β and γ subunits are sufficient for the conversion of γ-subunit rotation into conformational changes in the process of mechanochemical coupling.
The different position of the αC fragments in the euglenozoan ATP synthase also rules out involvement of the γ subunit in the transmission of conformational changes between catalytic sites, because this subunit can interact with only one β subunit at a time. This observation is consistent with results of high-speed atomic force microscopy demonstrating that in Bacillus PS3, conformational transmission between catalytic sites does not require the central stalk, but the loss of a single α subunit stops the propagation of conformational changes (43). In the T. brucei and E. gracilis structures, the central and C-terminal domains of the α subunit are displaced from their canonical position but remain bound to the complex, indicating an essential role of the fragment in enzymatic activity. In these complexes, the αC fragment connects the C-terminal domain of one β subunit to the central domain of the neighboring subunit (Fig. 2C and Fig. S2D). In the bovine complex, the distance between these two contact points in one β subunit changes significantly during nucleotide binding and release (4). Therefore, the arc-shaped αc fragments in the euglenozoan complexes likely transmit conformational changes from one β subunit to the next, ensuring cooperativity among the three catalytic sites in the F1 head.
Conclusion
Electron cryotomography of mitochondria from E. gracilis and insect-stage T. brucei shows that the ATP synthases of both organisms form dimers, which assemble into short left-handed helical arcs around the edge of disk-shaped cristae. Subtomogram averaging of the ATP synthase dimers in these species reveals a noncanonical F1 head structure. This unusual structure is the result of a proteolytic cleavage of the α subunit, which causes a displacement of the central and C-terminal domains of the α subunit from their canonical position. The structure of the euglenozoan F1 heads provides insight into the mechanism of rotary catalysis, in particular how rotation of the central stalk induces conformational changes and how these changes are transmitted between catalytic sites. The unique structure of the F1 head of the euglenozoan ATP synthases represents a potential model system for establishing future in-depth studies on the mechanism of ATP synthesis, a fundamental and essential life process.
Materials and Methods
Culture Conditions and Sample Preparation.
T. brucei procyclic form cells [strain 29.13 (44)] were grown at 27 °C and 5% (vol/vol) CO2 in SDM-79 medium (45) containing hemin (7.5 mg/mL) and 10% (vol/vol) FBS (Gibco). Cells were counted with a Z2 Cell Counter (Beckman Coulter) and harvested by centrifugation at 1,300 × g for 5 min. Then 600 × 106 cells were resuspended in 1.5 mL of homogenization buffer (20 mM Tris pH 7.4, 250 mM sucrose, and 5 mM MgCl2) and homogenized by 350 passages in a ball-bearing cell homogenizer with 4-µm clearance (Isobiotec). The cell lysate was centrifuged at 500 × g for 10 min at 4 °C. Mitochondrial membranes were pelleted by centrifugation at 16,000 × g for 10 min at 4 °C.
E. gracilis (Lebendkulturen Helbig) cultures were grown in Euglena medium (46) under a cycle of 10 h of light at 10,000 lux and 14 h of darkness at 21 °C. Then 100-mL cultures were harvested by centrifugation at 800 × g for 10 min at 4 °C, followed by resuspension in 10 mL of homogenization buffer (20 mM Tris pH 7.4, 250 mM sucrose, and 5 mM MgCl2). Cells were homogenized by 30 passages in a cell homogenizer with 8-µm clearance, and the homogenate was centrifuged at 1,500 × g for 5 min at 4 °C. The supernatant was collected and centrifuged at 6,000 × g for 10 min at 4 °C. The resulting pellet contained an upper green layer, which was carefully removed and discarded, and a brownish pellet containing the mitochondrial membranes.
BN-PAGE of Solubilized Membrane Complexes and In-Gel ATPase Activity Assay.
E. gracilis mitochondrial membranes were resuspended in solubilization buffer (final concentration, 50 mM Tris⋅HCl pH 8.0, 1 mM MgCl2 with Roche Complete Protease Inhibitor Mixture added), mixed with DDM in a final detergent-to-protein ratio (DPR) of 1:1 (wt/wt) or 5:1 (wt/wt), and incubated for 30 min on ice. Samples were centrifuged at 15,000 × g for 15 min at 4 °C, after which solubilized protein complexes were collected from the supernatant. Solubilized E. gracilis mitochondrial membrane protein complexes were separated by BN-PAGE as described by Wittig et al. (47), using 3–12% NativePAGE Novex Bis-Tris gradient gels (Life Technologies), which were subsequently stained with Coomassie blue R-250. In-gel ATPase activity assays were performed as described previously (48), with slight modifications. After gel electrophoresis, native gels were rinsed twice in ddH2O and once in 1 M Tris⋅HCl pH 7.8, followed by incubation in activity buffer (35 mM Tris⋅HCl pH 7.8 and 14 mM MgSO4). ATP, lead (II) nitrate, and N,N-Dimethyl-n-dodecylamine N-oxide were added in trace amounts, whereupon white lead phosphate precipitate formed inside the protein bands with ATPase activity during overnight incubation.
Electron Cryotomography and Tomogram Processing.
Mitochondrial membrane pellets were resuspended in 50 µL of freezing buffer (20 mM Tris pH 7.4 and 250 mM trehalose) and mixed 1:1 with 6-nm colloidal gold fiducial markers (Aurion). Then 3 µL was applied to a glow-discharged R 2/2 Cu 300-mesh holey carbon-coated support grid (Quantifoil). Excess liquid was removed by blotting (#4 Whatman paper; Sigma-Aldrich), and the grid was rapidly frozen in liquid ethane using a home-built guillotine. Frozen-hydrated specimens of T. brucei mitochondrial membranes were imaged in a Titan Krios electron microscope (FEI) at a nominal magnification of 42,000× (specimen pixel size, 3.35 Å). Tomographic tilt series were collected at 4-µm defocus to ±60° in 2° increments, starting at +24°.
E. gracilis mitochondrial membranes were imaged with an FEI Tecnai Polara transmission electron microscope at a nominal magnification of 90,000× (specimen pixel size, 2.28 Å), and tilt series were collected at 3 µm defocus from -50° to +60° in 2° increments, starting at +24°. In both cases, tilt series were collected using the program LATITUDE (Digital Micrograph; Gatan). The cumulative dose of a tilt series was ∼100 electrons/Å2. Both microscopes were operated at 300 kV and equipped with a Quantum Energy filter and a K2 summit direct electron detector (Gatan) operated in counting mode. Tomographic volumes were reconstructed using IMOD (49). The contrast transfer function was estimated and corrected in IMOD (50). The handedness of tomographic volumes was determined by evaluating the tilt axis rotation angle in tilt series of a sample of known handedness. For 3D visualization, tomographic volumes were contrast-enhanced by nonlinear anisotropic diffusion filtering (51) and segmented manually with AMIRA (FEI). Subtomogram averages were placed into original tomographic volumes using the EMPackage plugin (52) for AMIRA.
Subtomogram Averaging.
ATP synthase dimers were averaged as described previously (3). In brief, particle pairs were manually identified in tomographic volumes, and initial rotation angles were assigned based on the F1 head positions relative to the membrane in SPIDER (53). Particles were extracted, rotated, and averaged to create an initial reference. Particle alignment was optimized iteratively in PEET (54), using a restricted search range. Resolution of final maps was estimated by Fourier shell correlation (FSC). Particle sets were randomly split into two halves after alignment. The resulting half-maps were multiplied with a Gaussian mask before FSC. To prevent mask bias, tomographic volumes were phase-randomized beyond 40 Å using PEET, and two half-maps were generated by extracting, aligning, and averaging the same particle subsets. The FSC curves of the phase-randomized half-sets indicated a drop at the randomization frequency. 3D visualization, rigid body fitting, and difference map generation were performed in UCSF Chimera (55).
Sequence Alignment.
Sequence alignment was performed with ClustalW2 (56). Sequences were obtained from TriTrypDB (57), UniProt (58), and Perez et al. (11).
Supplementary Material
Acknowledgments
We thank Deryck Mills for maintaining the EM facility and Özkan Yildiz and Juan Francisco Castillo Hernandez for maintaining the computer system. This work was funded by the Max Planck Society (W.K.); the German Research Foundation-funded Cluster of Excellence Frankfurt, “Macromolecular Complexes” (K.M.D.); and UK Medical Research Council Grant G0600129 (to A.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structures of the mitochondrial ATP synthase dimers from E. gracilis and T. brucei have been deposited in the Electron Microscope Data Bank, emdatabank.org (EMDB ID codes EMD-3559 and EMD-3560, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612386114/-/DCSupplemental.
References
- 1.Boyer PD. The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim Biophys Acta. 1993;1140(3):215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- 2.Strauss M, Hofhaus G, Schröder RR, Kühlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27(7):1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies KM, et al. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci USA. 2011;108(34):14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8-Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370(6491):621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 5.Duncan TM, Bulygin VV, Zhou Y, Hutcheon ML, Cross RL. Rotation of subunits during catalysis by Escherichia coli F1-ATPase. Proc Natl Acad Sci USA. 1995;92(24):10964–10968. doi: 10.1073/pnas.92.24.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabbert D, Engelbrecht S, Junge W. Intersubunit rotation in active F-ATPase. Nature. 1996;381(6583):623–625. doi: 10.1038/381623a0. [DOI] [PubMed] [Google Scholar]
- 7.Menz RI, Walker JE, Leslie AGW. Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: Implications for the mechanism of rotary catalysis. Cell. 2001;106(3):331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- 8.Komoriya Y, et al. Principal role of the arginine finger in rotary catalysis of F1-ATPase. J Biol Chem. 2012;287(18):15134–15142. doi: 10.1074/jbc.M111.328153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yukawa A, Iino R, Watanabe R, Hayashi S, Noji H. Key chemical factors of arginine finger catalysis of F1-ATPase clarified by an unnatural amino acid mutation. Biochemistry. 2015;54(2):472–480. doi: 10.1021/bi501138b. [DOI] [PubMed] [Google Scholar]
- 10.Zíková A, Schnaufer A, Dalley RA, Panigrahi AK, Stuart KD. The F(0)F(1)-ATP synthase complex contains novel subunits and is essential for procyclic Trypanosoma brucei. PLoS Pathog. 2009;5(5):e1000436. doi: 10.1371/journal.ppat.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez E, et al. The mitochondrial respiratory chain of the secondary green alga Euglena gracilis shares many additional subunits with parasitic Trypanosomatidae. Mitochondrion. 2014;19(Part B):338–349. doi: 10.1016/j.mito.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Speijer D, et al. Characterization of the respiratory chain from cultured Crithidia fasciculata. Mol Biochem Parasitol. 1997;85(2):171–186. doi: 10.1016/s0166-6851(96)02823-x. [DOI] [PubMed] [Google Scholar]
- 13.Nelson RE, Aphasizheva I, Falick AM, Nebohacova M, Simpson L. The I-complex in Leishmania tarentolae is a uniquely structured F(1)-ATPase. Mol Biochem Parasitol. 2004;135(2):221–224. doi: 10.1016/j.molbiopara.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Dean S, Gould MK, Dewar CE, Schnaufer AC. Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proc Natl Acad Sci USA. 2013;110(36):14741–14746. doi: 10.1073/pnas.1305404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bochud-Allemann N, Schneider A. Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J Biol Chem. 2002;277(36):32849–32854. doi: 10.1074/jbc.M205776200. [DOI] [PubMed] [Google Scholar]
- 16.Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K. The F1-ATP synthase complex in bloodstream-stage trypanosomes has an unusual and essential function. EMBO J. 2005;24(23):4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams N, Frank PH. The mitochondrial ATP synthase of Trypanosoma brucei: Isolation and characterization of the intact F1 moiety. Mol Biochem Parasitol. 1990;43(1):125–132. doi: 10.1016/0166-6851(90)90137-b. [DOI] [PubMed] [Google Scholar]
- 18.Brown SV, Hosking P, Li J, Williams N. ATP synthase is responsible for maintaining mitochondrial membrane potential in bloodstream form Trypanosoma brucei. Eukaryot Cell. 2006;5(1):45–53. doi: 10.1128/EC.5.1.45-53.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Šubrtová K, Panicucci B, Zíková A. ATPaseTb2, a unique membrane-bound FoF1-ATPase component, is essential in bloodstream and dyskinetoplastic trypanosomes. PLoS Pathog. 2015;11(2):e1004660. doi: 10.1371/journal.ppat.1004660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudkina NV, Sunderhaus S, Braun HP, Boekema EJ. Characterization of dimeric ATP synthase and cristae membrane ultrastructure from Saccharomyces and Polytomella mitochondria. FEBS Lett. 2006;580(14):3427–3432. doi: 10.1016/j.febslet.2006.04.097. [DOI] [PubMed] [Google Scholar]
- 21.Numoto N, Hasegawa Y, Takeda K, Miki K. Inter-subunit interaction and quaternary rearrangement defined by the central stalk of prokaryotic V1-ATPase. EMBO Rep. 2009;10(11):1228–1234. doi: 10.1038/embor.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau WCY, Rubinstein JL. Subnanometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature. 2011;481(7380):214–218. doi: 10.1038/nature10699. [DOI] [PubMed] [Google Scholar]
- 23.Benlekbir S, Bueler SA, Rubinstein JL. Structure of the vacuolar-type ATPase from Saccharomyces cerevisiae at 11-Å resolution. Nat Struct Mol Biol. 2012;19(12):1356–1362. doi: 10.1038/nsmb.2422. [DOI] [PubMed] [Google Scholar]
- 24.Shirakihara Y, et al. The crystal structure of the nucleotide-free α3β3 subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer. Structure. 1997;5(6):825–836. doi: 10.1016/s0969-2126(97)00236-0. [DOI] [PubMed] [Google Scholar]
- 25.Harpaz Y, Gerstein M, Chothia C. Volume changes on protein folding. Structure. 1994;2(7):641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 26.Brown B SV, Chi TB, Williams N. The Trypanosoma brucei mitochondrial ATP synthase is developmentally regulated at the level of transcript stability. Mol Biochem Parasitol. 2001;115(2):177–187. doi: 10.1016/s0166-6851(01)00282-1. [DOI] [PubMed] [Google Scholar]
- 27.Dudkina NV, Oostergetel GT, Lewejohann D, Braun HP, Boekema EJ. Row-like organization of ATP synthase in intact mitochondria determined by cryo-electron tomography. Biochim Biophys Acta. 2010;1797(2):272–277. doi: 10.1016/j.bbabio.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Daum B, Walter A, Horst A, Osiewacz HD, Kühlbrandt W. Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc Natl Acad Sci USA. 2013;110(38):15301–15306. doi: 10.1073/pnas.1305462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mühleip AW, et al. Helical arrays of U-shaped ATP synthase dimers form tubular cristae in ciliate mitochondria. Proc Natl Acad Sci USA. 2016;113(30):8442–7. doi: 10.1073/pnas.1525430113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies KM, Anselmi C, Wittig I, Faraldo-Gómez JD, Kühlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci USA. 2012;109(34):13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paumard P, et al. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21(3):221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown RC, Evans DA, Vickerman K. Changes in oxidative metabolism and ultrastructure accompanying differentiation of the mitochondrion in Trypanosoma brucei. Int J Parasitol. 1973;3(5):691–704. doi: 10.1016/0020-7519(73)90095-7. [DOI] [PubMed] [Google Scholar]
- 33.Lapaille M, et al. Atypical subunit composition of the chlorophycean mitochondrial F1FO-ATP synthase and role of Asa7 protein in stability and oligomycin resistance of the enzyme. Mol Biol Evol. 2010;27(7):1630–1644. doi: 10.1093/molbev/msq049. [DOI] [PubMed] [Google Scholar]
- 34.Wittig I, Schägger H. Structural organization of mitochondrial ATP synthase. Biochim Biophys Acta. 2008;1777(7-8):592–598. doi: 10.1016/j.bbabio.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 35.Eubel H, Jänsch L, Braun HP. New insights into the respiratory chain of plant mitochondria: Supercomplexes and a unique composition of complex II. Plant Physiol. 2003;133(1):274–286. doi: 10.1104/pp.103.024620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balabaskaran Nina P, et al. Highly divergent mitochondrial ATP synthase complexes in Tetrahymena thermophila. PLoS Biol. 2010;8(7):e1000418. doi: 10.1371/journal.pbio.1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Lis R, Mendoza-Hernández G, Groth G, Atteia A. New insights into the unique structure of the F0F1-ATP synthase from the chlamydomonad algae Polytomella sp. and Chlamydomonas reinhardtii. Plant Physiol. 2007;144(2):1190–1199. doi: 10.1104/pp.106.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubinstein JL, Walker JE, Henderson R. Structure of the mitochondrial ATP synthase by electron cryomicroscopy. EMBO J. 2003;22(23):6182–6192. doi: 10.1093/emboj/cdg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allegretti M, et al. Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase. Nature. 2015;521(7551):237–240. doi: 10.1038/nature14185. [DOI] [PubMed] [Google Scholar]
- 40.Böttcher B, Gräber P. The structure of the H(+)-ATP synthase from chloroplasts and its subcomplexes as revealed by electron microscopy. Biochim Biophys Acta. 2000;1458(2-3):404–416. doi: 10.1016/s0005-2728(00)00090-6. [DOI] [PubMed] [Google Scholar]
- 41.Ogura T, Whiteheart SW, Wilkinson AJ. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J Struct Biol. 2004;146(1-2):106–112. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 42.Hashimi H, et al. The assembly of F(1)F(O)-ATP synthase is disrupted upon interference of RNA editing in Trypanosoma brucei. Int J Parasitol. 2010;40(1):45–54. doi: 10.1016/j.ijpara.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 43.Uchihashi T, Iino R, Ando T, Noji H. High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science. 2011;333(6043):755–758. doi: 10.1126/science.1205510. [DOI] [PubMed] [Google Scholar]
- 44.Wirtz E, Leal S, Ochatt C, Cross GAM. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99(1):89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 45.Brun R, Schönenberger Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 1979;36(3):289–292. [PubMed] [Google Scholar]
- 46.Starr RC. The culture collection of algae at Indiana University. Am J Bot. 1964;51(9):1013–1044. [Google Scholar]
- 47.Wittig I, Braun HP, Schägger H. Blue native PAGE. Nat Protoc. 2006;1(1):418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 48.Zerbetto E, Vergani L, Dabbeni-Sala F. Quantification of muscle mitochondrial oxidative phosphorylation enzymes via histochemical staining of blue native polyacrylamide gels. Electrophoresis. 1997;18(11):2059–2064. doi: 10.1002/elps.1150181131. [DOI] [PubMed] [Google Scholar]
- 49.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 50.Xiong Q, Morphew MK, Schwartz CL, Hoenger AH, Mastronarde DN. CTF determination and correction for low-dose tomographic tilt series. J Struct Biol. 2009;168(3):378–387. doi: 10.1016/j.jsb.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135(3):239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 52.Pruggnaller S, Mayr M, Frangakis AS. A visualization and segmentation toolbox for electron microscopy. J Struct Biol. 2008;164(1):161–165. doi: 10.1016/j.jsb.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 53.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 54.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313(5789):944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 55.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 56.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 57.Aslett M, et al. TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38(Database issue):D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bateman A, et al. UniProt Consortium UniProt: A hub for protein information. Nucleic Acids Res. 2015;43(Database issue) D1:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386(6620):88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 60.Dautant A, Velours J, Giraud MF. Crystal structure of the Mg·ADP-inhibited state of the yeast F1c10-ATP synthase. J Biol Chem. 2010;285(38):29502–29510. doi: 10.1074/jbc.M110.124529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.