Significance
Hepatitis E virus (HEV) is responsible for an estimated 20 million enterically transmitted cases of viral hepatitis globally. Here, we demonstrate that one of HEV’s three major gene products, ORF3, is an ion channel. Deletion of ORF3 abrogates release of infectious virions, and we show that viral egress can be rescued by expressing the influenza A virus (IAV) matrix-2 protein in trans. Expression of ORF3 facilitates ion flux across the plasma membrane, providing direct evidence for its viroporin activity. We identify regions within ORF3 abrogating both ion channel activity and particle release, thereby linking these two processes for a quasienveloped human virus and providing an attractive potential target for antiviral drug development.
Keywords: hepatitis E virus, hepatitis E, viroporin, virion release, drug development
Abstract
Hepatitis E virus (HEV) is the leading cause of enterically transmitted viral hepatitis globally. Of HEV’s three ORFs, the function of ORF3 has remained elusive. Here, we demonstrate that via homophilic interactions ORF3 forms multimeric complexes associated with intracellular endoplasmic reticulum (ER)-derived membranes. HEV ORF3 shares several structural features with class I viroporins, and the function of HEV ORF3 can be maintained by replacing it with the well-characterized viroporin influenza A virus (IAV) matrix-2 protein. ORF3’s ion channel function is further evidenced by its ability to mediate ionic currents when expressed in Xenopus laevis oocytes. Furthermore, we identified several positions in ORF3 critical for its formation of multimeric complexes, ion channel activity, and, ultimately, release of infectious particles. Collectively, our data demonstrate a previously undescribed function of HEV ORF3 as a viroporin, which may serve as an attractive target in developing direct-acting antivirals.
In the general population, hepatitis E virus (HEV)-associated mortality is roughly 1%. However, the risk to pregnant women is significantly higher, reaching as high as 25% among women with HEV infections during their third trimester. This risk ultimately results in 70,000 deaths and 3,000 stillbirths every year. Whereas most HEV infections occur in developing countries, recent epidemiological studies have found a high seroprevalence of anti-HEV antibodies in industrialized countries (1), suggesting exposure to the virus from travel to HEV endemic areas or from contact with pigs, a major reservoir of HEV. In a majority of cases, HEV causes an acute infection, but among immunocompromised patients—notably organ transplant recipients (2, 3) and individuals coinfected with HIV (4–6)—HEV can progress to chronicity. An effective vaccine preventing HEV infection has been developed, but it is only licensed in China (7). Pegylated IFN (peg-IFN) and the nucleoside analog ribavirin (RBV) have been successfully used to treat HEV infection (8, 9), but the use of these drugs is not recommended in certain patient groups, including pregnant women and organ transplant recipients. Therefore, novel antiviral compounds are still needed, especially because HEV isolates resistant to RBV have been identified (10).
HEV is a quasienveloped, positive-sense RNA virus with three ORFs. HEV’s three gene products and their associated functions could serve as druggable targets. However, only ORF1 and ORF2 are the most fully characterized. ORF1 encodes for a nonstructural polyprotein composed of a methyltransferase, papain-like cysteine protease, RNA helicase, and RNA-dependent RNA polymerase (11). ORF2 encodes for the viral capsid protein and is involved in virion assembly, interaction with the host cell, and immunogenicity. It contains three glycosylation sites necessary for formation of infectious particles (12). In contrast, ORF3 and its corresponding function(s) have been largely elusive. The smallest ORF of the HEV genome, ORF3 is translated from a subgenomic RNA into a protein of 113–115 amino acids. Previous studies showed that ORF3 is bound to viral particles found in patient sera (13) and produced in cell culture (13, 14). Although in cultured cells ORF3 has not appeared essential for HEV RNA replication, viral assembly, or infection, it is required for particle release (14–16).
In this study, we aimed to further elucidate the function of ORF3. Building on a previously established cell culture system (17), we developed a transcomplementation system to uncouple HEV RNA replication from the assembly and release of infectious virions. This platform enabled us to discover and characterize a previously undescribed function of ORF3 as a viroporin—a virally encoded ion channel. Because HEV ORF3 shares multiple features with class IA viroporins, we tested whether its function could be substituted by another well-characterized viroporin, influenza A virus (IAV) matrix-2 (M2). Expression of IAV M2 can indeed partially rescue release of infectious virions. Furthermore, voltage-clamp experiments directly demonstrate that expression of HEV ORF3 can facilitate the flux of ions across the plasma membrane of Xenopus laevis oocytes. To identify residues within ORF3 critical for its viroporin function and the release of infectious virions, we performed a comprehensive alanine scanning mutagenesis. Two regions outside of the PXXP motifs previously shown to be critical for HEV egress (15, 18) were particularly sensitive to amino acid substitutions and abrogated ORF3’s ability to facilitate HEV release and to flux ions. Our work ascribes a function to one of the three gene products of a major human pathogen. Whereas viroporins have been implicated in the release of other viruses, to our knowledge this is an example of a quasienveloped virus requiring an ion channel to release infectious particles.
Results
ORF3 Is a Transmembrane Protein Localized at ER Membranes and Forms Multimeric Complexes.
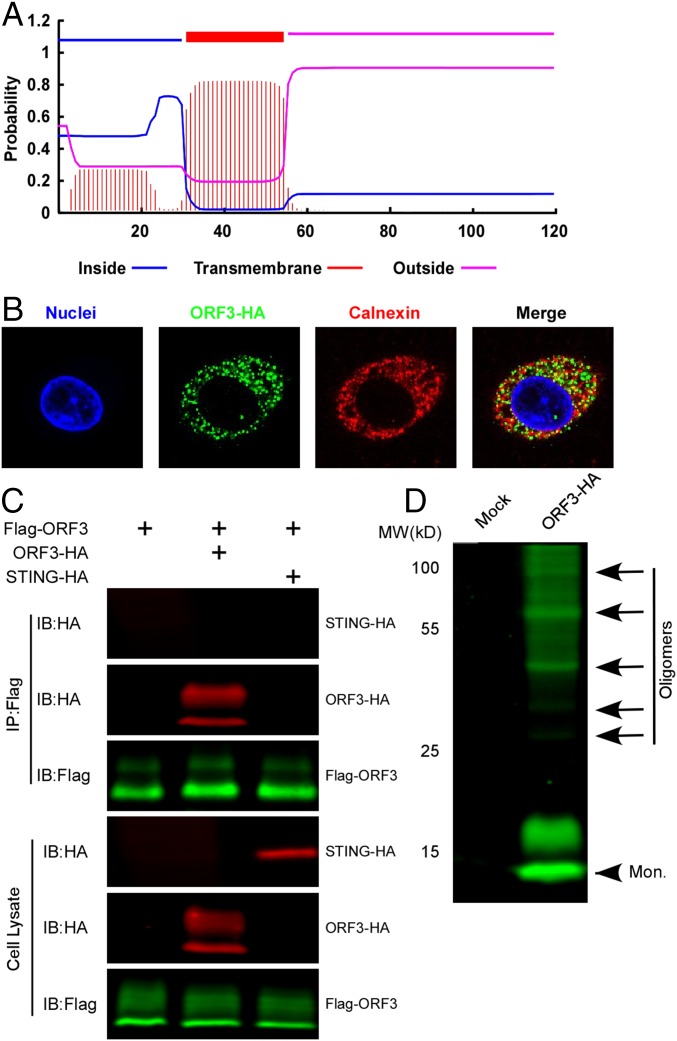
Our initial bioinformatic analysis indicated that HEV ORF3 contained a putative transmembrane domain (Fig. 1A). This prediction was corroborated following expression of HEV ORF3 in HepG2C3A cells, a commonly used human hepatoma cell line permissive to HEV infection. In these cells, we observed colocalization of ORF3 with the ER-associated protein calnexin (Fig. 1B), suggesting ORF3 associates with intracellular membranes presumably derived from the ER. The punctate pattern in which ORF3 was expressed motivated us to determine whether the protein forms larger complexes. FLAG-tagged ORF3 immunoprecipitated with an anti-HA antibody in lysates derived from HepG2C3A cells coexpressing FLAG- and HA-tagged ORF3 (Fig. 1C). In contrast, coexpression of HA-tagged STING, a well-characterized ER-resident protein (19), did not pull down ORF3, demonstrating that the interactions between different ORF3 molecules are specific. Additionally, larger protein complexes were observed in Western blots of HepG2C3A lysates even under denaturing conditions, suggesting multimerization (Fig. 1D). Collectively, these data confirm that HEV ORF3 is a transmembrane protein localized at ER membranes (20) and forms multimeric complexes, presumably through homophilic interactions.
Fig. 1.
ORF3 is a predicted transmembrane protein that associates with ER membranes and can form homooligomers. (A) Schematic representation of the predicted transmembrane topology [TMHMM (transmembrane helices in proteins) Server v. 2.0] of HEV ORF3 protein (Kernow C1/p6). (B) HEV ORF3 protein associates with the ER membrane. HepG2C3A cells lentivirally transduced with HEV ORF3-HA were stained with anti-HA and anti-calnexin antibodies. Nuclei were stained with Hoechst dye. Colocalization was analyzed by ImageJ, and Pearson's r = 0.44 ± 0.063. Shown are representative images of at least triplicate experiments. (C) Coimmunoprecipitation assay of FLAG-ORF3 and ORF3-HA or STING-HA in HepG2C3A cells. Cells were transduced with FLAG-ORF3 and/or ORF3-HA or HA-STING lentivirus and lysed 72 h posttransduction. Cell lysates were subjected to immunoprecipitation with an anti-FLAG antibody. IP, immunoprecipitation; IB, immunoblotting. (D) The cell lysates from cells lentivirally transduced to express ORF3-HA or mock transduced were analyzed by Western blotting with an anti-HA antibody. The monomer and oligomers are indicated with arrowhead and arrows, respectively.
HEV ORF3 Resembles a Class IA Viroporin.
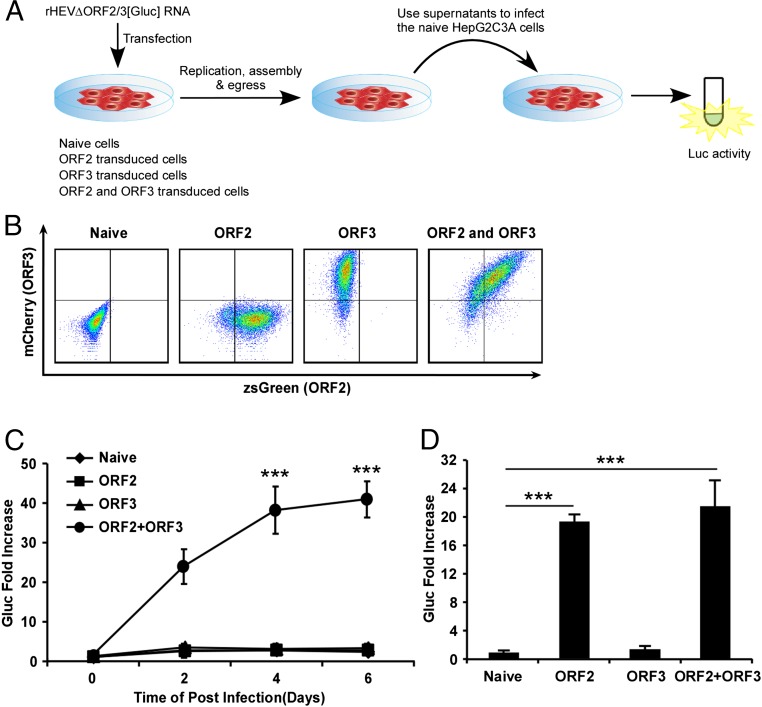
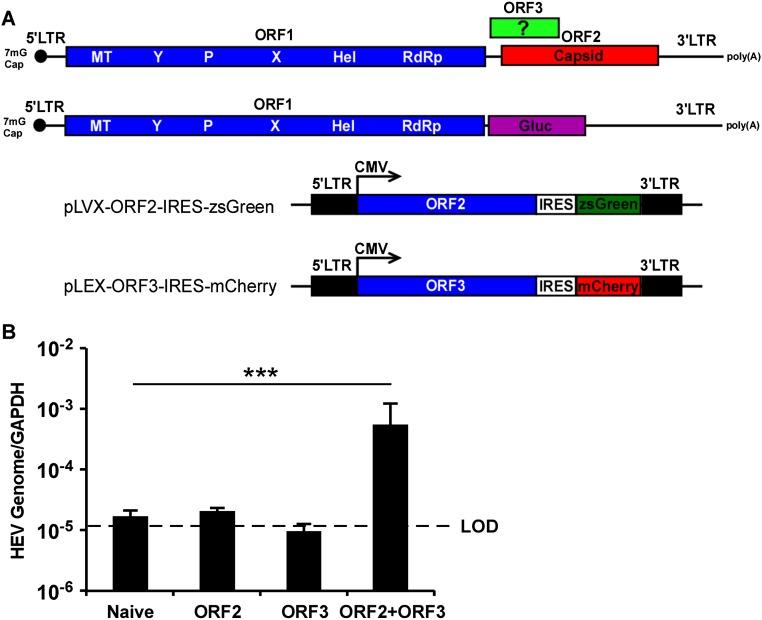
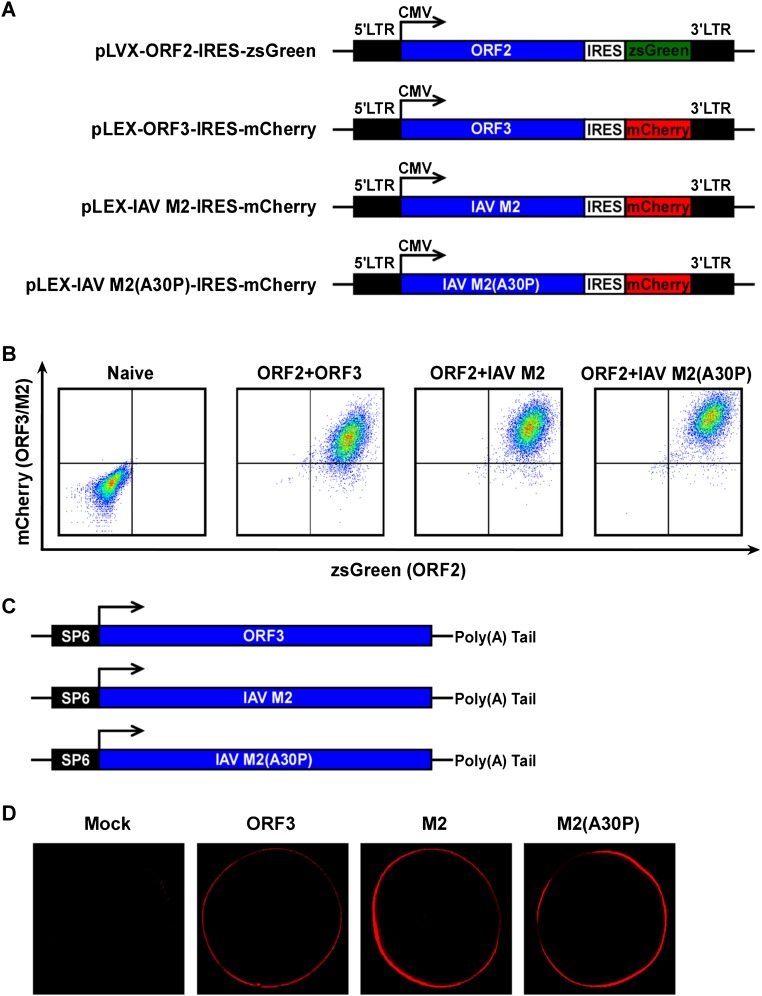
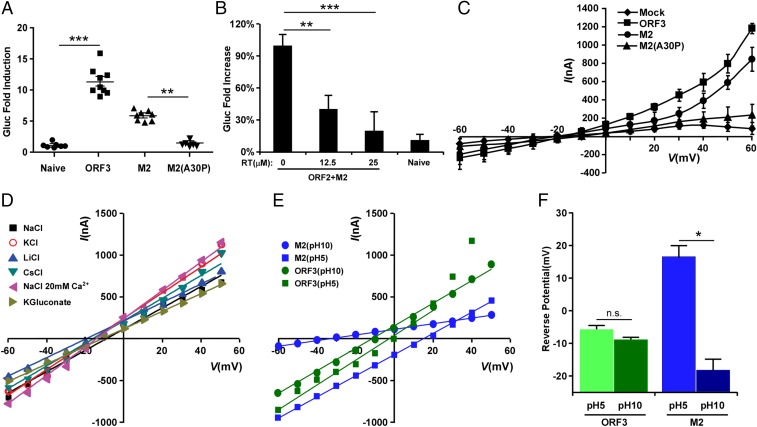
When we examined the characteristics of ORF3 more closely, we noted its similarity to known viroporins, i.e., virally encoded protein complexes that serve as functional ion channels. Like other viroporins, ORF3 is a small hydrophobic protein that tends to oligomerize in ER-derived membranes. Similar to class IA viroporins such as IAV M2 (21), HIV-1 Vpu (22), or the coronavirus E protein (23), ORF3 has a short tail at the N terminus that resides in the ER lumen and a long cytosolic tail at the C terminus that is prone to phosphorylation at a serine in position 70 (24, 25). To enable direct analysis of ORF3’s function, we expressed ORF2 and/or ORF3 lentivirally in HepG2C3A cells (Fig. 2 A and B and Fig. S1A). These cells were subsequently transfected with in vitro transcribed RNA from a recombinant HEV genome derived from the KernowC1/p6 genome (17) in which ORF2 and ORF3 are replaced by a secreted version of Gaussia luciferase (Gluc), termed rHEVΔORF2/3[Gluc] (Fig. 2A). Supernatants collected from these cultures at day 5 posttransfection were then used to infect naïve HepG2C3A cells. The recombinant HEV subgenome replicated equally efficiently in all cells, irrespective of ORF2/3 expression. In cells expressing ORF2 only, infectious virions assembled but were retained intracellularly (Fig. 2D). Only supernatants collected from HepG2C3A cells expressing HEV ORF2/3 together, and not separately, led to robust reinfection, as indicated by an ∼35- to 45-fold increase in luciferase activity over background (Fig. 2C). Consistent with these luciferase data HEV RNA was also only significantly increased in HepG2C3A cells that had been exposed to supernatants from rHEVΔORF2/3[Gluc] transfected ORF2/3 producer cells (Fig. S1B). These data are consistent with previous reports showing that ORF2 is essential for packaging and ORF3 for release of infectious particles (16, 25). Next, we aimed to determine whether ORF3’s essential function in HEV release could be replaced by IAV M2, a well-characterized class IA viroporin (21). Notably, infection of HepG2C3A cells with supernatants from HepG2C3A cells in which transfection of rHEVΔORF2/3[Gluc] was complemented in trans with HEV ORF2 and IAV M2 (Fig. S2 A and B) resulted in a 5- to 8-fold increase in Gluc activity (Fig. 3A). This signal was approximately twofold lower than the 8- to 15-fold increase in Gluc activity, resulting from infection with particles packaged in cells expressing HEV ORF2 and ORF3 (Fig. 3A). Furthermore, IAV M2-mediated HEV particle release was dependent on M2’s ion channel activity, as the IAV M2(A30P) mutant, which abolishes its ion channel activity (26), did not support HEV particle egress. These data are consistent with our observation that addition of rimantadine, a known inhibitor of the M2, during transcomplementation of the reporter subgenome inhibited particle release (Fig. 3B), providing further evidence that the viroporin activity of M2 is required. Taken together, these data strongly suggest that HEV ORF3 functions as an ion channel.
Fig. 2.
ORF2 and ORF3 are required for releasing viral particles to infect naïve HepG2C3A cells. (A) Schematic representation of the transcomplementation system for packaging HEV virions. (B) Representative flow cytometry plots demonstrating efficient ORF2 and ORF3 expression. HepG2C3A cells were transduced with pLVX-ORF2-IRES-zsGreen and/or pLEX-ORF3-IRES-mCherry or not transduced. Flow cytometric analysis was performed 4 d following transduction to quantify the frequencies of ORF2 and/or ORF3 expressing cells. (C) Infection kinetics of transcomplemented HEV in HepG2C3A cells. Cell culture supernatants from naïve HepG2C3A, or HepG2C3A cells transduced with HEV ORF2 or/and ORF3, were collected 5 d posttransfection with rHEVΔORF2/3[Gluc] RNA. Naïve HepG2C3A cells were incubated with these supernatants. After 12 h, cells were washed and Gaussia luciferase activity quantified in the cell culture supernatants at the indicated time points. (D) Five days following transfection of rHEVΔORF2/3[Gluc] RNA into HepG2C3A, or HepG2C3A cells transduced with HEV ORF2 or/and ORF3, lysates were used to infect naïve HepG2C3A cells. Gluc activity was measured in the supernatants 4 d postinfection. Shown are averages and SDs of triplicate measurements of three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001.
Fig. S1.
(A) Schematic representation of the wild-type HEV and rHEVΔORF2/3[Gluc] genomes and the bicistronic lentiviral constructs for expressing ORF2 and ORF3. (B) Cell culture supernatants from naïve HepG2C3A, or HepG2C3A cells transduced with HEV ORF2 and ORF3, M2, or M2(A30P), were collected 5 d posttransfection with rHEVΔORF2/3[Gluc] RNA. Naïve HepG2C3A cells were incubated with these supernatants. After 12 h, cells were washed and intracellular total RNA was extracted 4 d later for RT-qPCR assay to quantify HEV RNA. HEV RNA was normalized to the housekeeping gene GAPDH. Data represent the mean ± SD (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001. LOD, limit of detection.
Fig. S2.
(A) Schematic representation of the bicistronic lentiviral constructs for expressing HEV ORF2, ORF3, IAV M2, and IAV M2(A30P). (B) Representative flow cytometry plots demonstrating efficient ORF2 and ORF3, IAV M2, or M2(A30P) expression. HepG2C3A cells were transduced with LVX-ORF2-IRES-zsGreen and/or LEX-[ORF3 or M2 or M2(A30P)]-IRES-mCherry or not transduced. Flow cytometric analysis was performed 4 d following transduction to quantify the frequencies of ORF2- and/or ORF3- and M2-expressing cells. (C) Schematic representation of SP6-driven constructs used for in vitro transcription of HEV ORF3, IAV M2, and IAV M2(A30P) mRNAs. (D) HEV ORF3, IAV M2, or M2(A30P) expressed in X. laevis oocytes localizes to the plasma membrane. Water-injected and HEV ORF3, IAV M2, or M2(A30P) mRNA-injected oocytes were immunolabeled with polyclonal ORF3 or M2 antibodies and analyzed by confocal microscopy.
Fig. 3.
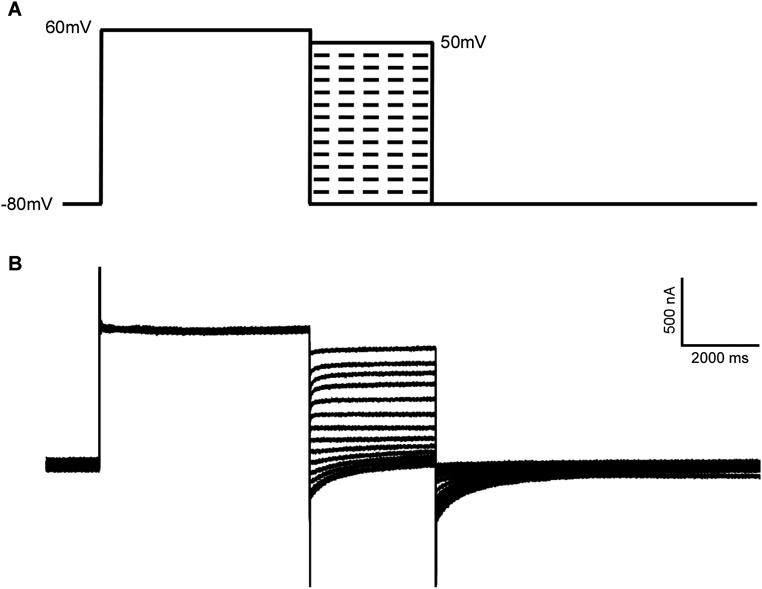
The HEV ORF3 protein displays ion channel activity. (A) IAV M2 can substitute ORF3 function. Cell culture supernatants from naïve HepG2C3A, or HepG2C3A cells transduced with HEV ORF2 and ORF3, M2 or M2(A30P), were collected 5 d posttransfection with rHEVΔORF2/3[Gluc] RNA. Naïve HepG2C3A cells were incubated with these supernatants. After 12 h, cells were washed and Gaussia luciferase activity quantified in the cell culture supernatants 4 d later. Data represent the mean ± SD (n = 6–9). *P < 0.05, **P < 0.01, and ***P < 0.001. (B) Inhibition of IAV M2 function with rimantadine (Sigma-Aldrich) suppresses release of HEV particles. Cell supernatants from naïve HepG2C3A or HepG2C3A cells transduced with HEV ORF2 and IAV M2 in the presence of indicated concentrations of rimantadine, were collected 5 d posttransfection with rHEVΔORF2/3[Gluc] RNA. Naïve HepG2C3A cells were incubated with these supernatants. After 12 h, cells were washed and Gaussia luciferase activity quantified in the cell culture supernatants 4 d later. Results are presented as percentage relative to the carrier-treated control. Data represent the mean ± SD *P < 0.05, **P < 0.01, and ***P < 0.001. (C) Current–voltage relationship of X. laevis oocytes expressing HEV ORF3, IAV M2, IAV M2(A30P) or control oocytes. During the current recording, the oocytes were bathed in Ringer solution. The standard voltage-clamp protocol consisted of rectangular voltage step pulses from −90 to +60 mV in 10-mV increments applied from a holding voltage of −60 mV. Each point represents the steady-state current (average current between 4,000–5,000 ms) at the corresponding voltage step. Data represent the mean ± SD (n = 5). (D) ORF3 has little to no ion selectivity. Shown is the current–voltage relationship of the peak tail currents of HEV ORF3 at different ionic conditions. To assess monovalent cation selectivity, K+, Cs+, or Li+ replaced Na+ at equimolar concentration. These substitutions did not alter the reversal potential of the HEV ORF3 currents. In addition, no significant change in reversal potential was observed when 20 mM CaCl2 was added to the solution or when Cl− was replaced by gluconate. (E) ORF3 channels, unlike M2, are nonselective to protons. Current–voltage relationship of the instantaneous current of HEV ORF3 (green) and M2 (blue). To assess for proton permeability oocytes expressing M2 or ORF3 channels were exposed to bath solutions at pH 10 and 5 (Experimental Procedures). Oocytes were clamped at −80 mV, followed by a 1-s depolarization to +20 mV. The voltage was then stepped to test voltages ranging from −80 to +50 mV in 10-mV increments. (F) Graph shows average reversal potential (Erev) for HEV ORF3 (green) and M2 (blue) channels. Error bars show SEM (n = 7). A one-way ANOVA confirmed difference between the different groups (F(3, 27) = 34.187, P < 0.005). Tukey’s honest significant difference (HSD) tests revealed significant differences between the M2 at pH 5 and M2 at pH 10 (P < 0.004) groups. This shift of Erev for M2 at different pHs support proton selectivity; in contrast, no difference was found for HEV ORF3.
HEV ORF3 Has Ion Channel Activity.
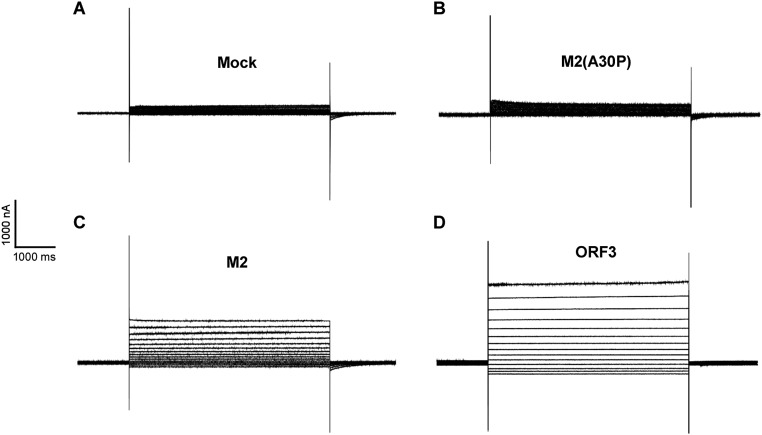
To directly test the ability of ORF3 to facilitate ion fluxes across the plasma membrane, we used the two-electrode voltage-clamp procedure in X. laevis oocytes injected with in vitro transcribed wild-type ORF3, wild-type M2, or mutant M2(A30P) mRNAs (Fig. S2C). Ionic currents were measured from oocytes that were injected with mRNAs 2 d before the recordings. Immunofluorescence imaging confirmed expression of ORF3 and the M2 proteins on the outer oocyte membrane (Fig. S2D). The currents of oocytes expressing the wild-type ORF3, M2, or M2 mutant proteins were studied by holding the membrane voltage of the oocytes at −60 mV and then applying voltage step pulses every 10 mV from −90 to +60 mV. Both hyperpolarization and depolarizing pulses induced large currents with minimal time dependence that increased to a steady value immediately after the voltage pulse was applied (Fig. S3). This current was significantly larger than the endogenous current evoked by identical changes of membrane voltage in control oocytes injected with M2 mutant (A30P) mRNA or mock injected but was very similar to that observed in oocytes injected with wild-type M2 mRNA (Fig. 3C). Whole-cell instantaneous currents were examined under different ionic conditions to estimate the selectivity properties of the HEV ORF3 channel. Little difference in the reversal potential for each of the tested solutions suggested that HEV ORF3 formed nonselective ion channels (Fig. 3D and Fig. S4). Likewise, and in contrast to M2 channels that are selective to protons and display a shift in the reversal potential at acidic (5) and basic (10) pH, ORF3’s do not show significant changes in the reversal potential as expected for nonselective channels (Fig. 3 E and F). Together, these data demonstrate that HEV ORF3 serves as a functional ion channel but does not have selectivity for specific ions.
Fig. S3.
Current traces demonstrating the viroporin function of HEV ORF3. Representative current traces showing baseline current in mock (A) and IAV M2(A30P)-injected oocytes (B). Traces of burst activity in X. laevis oocytes injected with IAV M2 (C) or HEV ORF3 (D). Scales for time (horizontal axis) and current (vertical axis) are shown.
Fig. S4.
(A) Voltage protocol to determine the instantaneous current–voltage relations. Oocytes were clamped at −80 mV, followed by a 5-s depolarization to +60 mV. The voltage was then stepped to test voltages ranging from −80 to +50 mV in 10-mV increments. This was followed by a step to −80 for 7 s to restore initial conditions. (B) Representative current traces from X. laevis oocyte expressing HEV ORF3 wild type using the protocol in A. The bathing solution contained 117 mM NaCl, 1.8 mM CaCl2, and 10 mM Hepes. Scales for time (horizontal axis) and current (vertical axis) are shown.
Identification of Domains Within ORF3, Which Are Required for Viroporin Function and Particle Release.
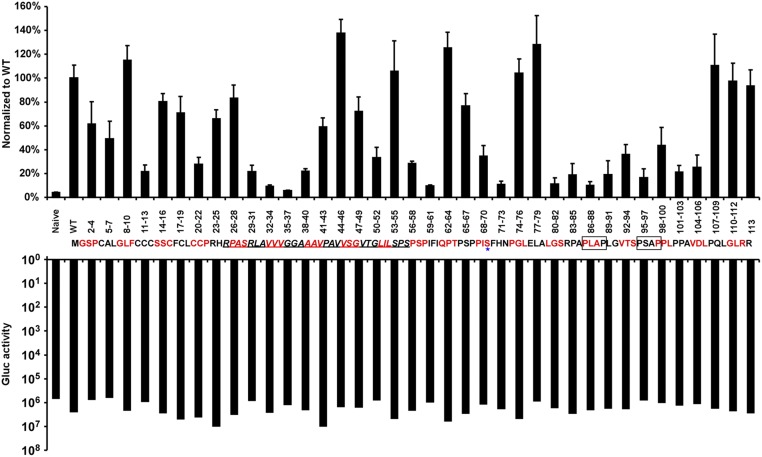
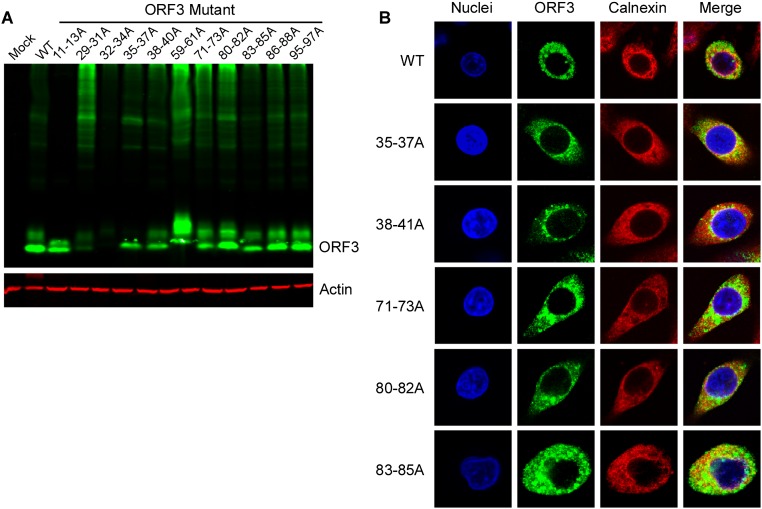
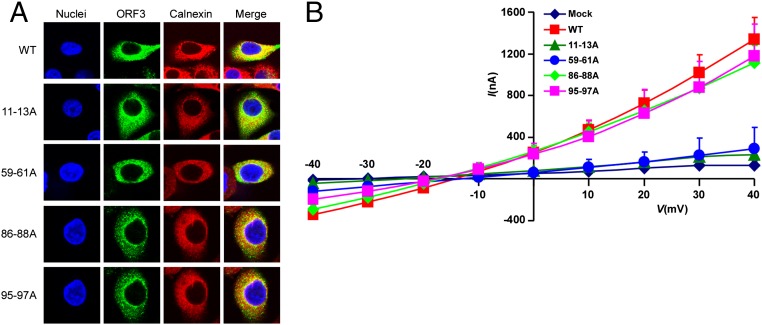
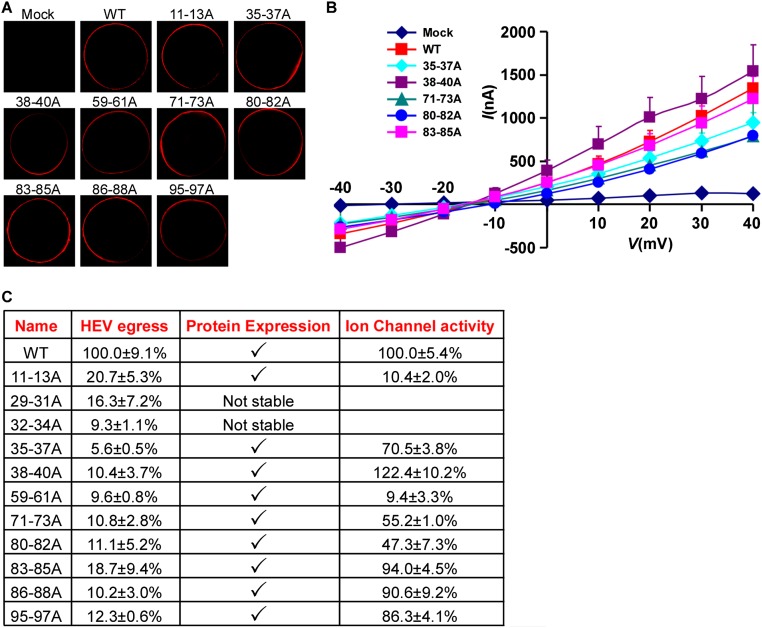
Next, we aimed to systematically identify regions within HEV ORF3, important for its ion channel function and release of infectious HEV particles. Thus, we performed alanine scanning mutagenesis to change triplets of amino acids to alanine across the entire ORF3 protein. Lentiviral delivery of individual ORF3 mutants, along with unmodified ORF2, led to high expression of the proteins in the majority of HepG2C3A cells. Dually transduced cells were subsequently transfected with rHEVΔORF2/3[Gluc] RNA and supernatants collected 5 d thereafter. To assess whether any of the ORF3 mutants affected release of HEV, supernatants were used to infect naïve HepG2C3A cells. Several positions led to ≥90% reduction in particle release compared with HEV released from cells expressing wild-type ORF2 and ORF3 (Fig. 4, Top). In particular, we confirmed that ORF3 residues 86–89 and 95–98, each containing a PXXP motif previously shown to be necessary for HEV release (14, 15, 18, 27), serve essential functions. In addition, mutations in positions 11–13, 29–40, 59–61, 71–73, and 80–85 reduced virion release by 80–95%. Gluc levels in the transfected cells were equivalent across all experimental conditions, demonstrating that the observed differences cannot be simply attributed to differences in RNA transfection and/or HEV replication efficiency in the different producer cells (Fig. 4, Bottom). Next, we eliminated from the analysis ORF3 mutants that simply affected protein stability. Western blots of lysates from HepG2C3A cells expressing HA-tagged ORF3 showed that all mutants except ORF3[RLA29-31AAA] and ORF3[VVV32-34AAA] could readily be detected with an anti-HA antibody (Fig. S5A) and those that did express well, with the exception of ORF3[CCC11-13AAA], still formed higher molecular weight complexes indicative of multimerized ORF3. All stable mutants were then subjected to voltage-clamp experiments. Notably, ORF3[CCC11-13AAA] and ORF3[IFI59-61AAA] exhibited a significant decrease in ion flux across the membrane compared with oocytes expressing wild-type ORF3 (Fig. 5B and Fig. S6 A–C). ORF3[PLA86-88AAA] and ORF3[PSA95-97AAA], which had alanine triplets disrupting the PXXP motifs critical for interactions with components of the ESCRT machinery, and thereby interfere with the release of infectious particles through the vacuolar protein secretion pathway (14–16, 18, 28), did not diminish ion channel activity. Of note, none of these mutants, including ORF3[CCC11-13AAA] and ORF3[IFI59-61AAA], displayed vastly different subcellular localization compared with wild-type ORF3 (Fig. 5A and Fig. S5 A and B). Thus, residues 11–13 and 59–61 likely fall in regions important for ORF3’s ion channel function.
Fig. 4.
Identification via alanine scanning mutagenesis of essential positions within HEV ORF3 for release of infectious virions. HEV ORF3 mutants generated by changing triplets of amino acids to alanines across the entire ORF3 protein were lentivirally delivered to HepG2C3A cells expressing ORF2. Dually transduced cells were subsequently transfected with rHEVΔORF2/3[Gluc] RNA. Five days posttransfection, supernatants were collected and used to infect naïve HepG2C3A cells. Gaussia luciferase activity was quantified in the producer cells (Bottom) and in the cell culture supernatant 4 d postinfection (Top). The putative transmembrane regions (TM) are underlined. The asterisk marks the serine residue of ORF3, which can be phosphorylated. The two boxes mark the PXXP motifs within ORF3. Shown are averages and SDs of triplicate measurements of three independent experiments.
Fig. S5.
Expression of ORF3 mutant constructs. (A) The cell lysates from HepG2C3A cells lentivirally transduced to express HA-tagged wild-type ORF3, the indicated alanine mutants of ORF3 or mock transduced were analyzed by Western blot with anti-HA or anti-actin antibodies. (B) HepG2C3A cells lentivirally transduced with HA-tagged wild type or the indicated ORF3 mutants were stained with anti-HA and anti-calnexin antibodies. Nuclei were stained with Hoechst dye. Shown are representative images of at least triplicate experiments.
Fig. 5.
Identification of amino acid residues critical for HEV ORF3’s ion channel activity. (A) HepG2C3A cells lentivirally transduced with HA-tagged wild type or the indicated ORF3 mutant were stained with anti-HA and anti-calnexin antibodies. Nuclei were stained with Hoechst dye. Shown are representative images of triplicate experiments. (B) Current–voltage relationship of X. laevis oocytes expressing HEV wild type or the indicated ORF3 mutants. Experiments were conducted as detailed in Fig. 3. Data represent the mean + SD (n = 5).
Fig. S6.
Ion channel activity of different ORF3 mutants. (A) Wild type or alanine mutants of HEV ORF3 expressed in X. laevis oocytes localize to the plasma membrane. Water-injected and HEV ORF3 mRNA-injected oocytes were immunolabeled with polyclonal ORF3 antibody and analyzed by confocal microscopy. (B) Current–voltage relationship of X. laevis oocytes expressing HEV wild type or the indicated ORF3 mutants. Experiments were conducted as detailed in Fig. 3. Data represent the mean + SD (n = 5). (C) Summary of the features of triple alanine ORF3 mutants that do not support HEV release.
Altogether, our data demonstrate that ORF3’s ion channel activity is important for particle release, which is an additional, distinct function to the previously described, essential interactions of HEV ORF3 with components of the ESCRT pathway.
Discussion
In the present study, we provide evidence that HEV ORF3 is a functional ion channel required for the release of infectious virions from host cells. To date, viroporins have been identified in nine different viruses, including six enveloped viruses (HCV, HIV, IAV, rotavirus, alphavirus/Sindbis virus, and coronaviruses) but only three nonenveloped viruses [simian virus 40 (SV40), coxsackie B virus (CBV), and polio virus] (reviewed in refs. 29, 30). Viroporins play a central role in promoting viral pathogenesis. For both enveloped and nonenveloped viruses, viroporin function can influence cell entry and genome replication but is most frequently linked to virus release. In line with our observations of HEV ORF3, viroporin-defective viruses are often impaired in their ability to properly assemble and release infectious particles (31–36). The mechanisms underlying viroporin-mediated viral release remain opaque. Cell lysis and the subsequent release of infectious virions can be triggered by increased membrane permeability as a result of viroporin accumulation (36). It has also been proposed that viroporin insertion in cellular membranes may disrupt the electrochemical gradient by facilitating ion fluxes across membranes, thus dissipating the membrane potential of internal vesicles or the plasma membrane and thus stimulating viral budding (30).
HEV is a particularly intriguing example of a virus apparently reliant on viroporin function for viral release. Although classically defined as a nonenveloped virus, recent work has shown it can be shed from infected cells as a quasienveloped virus (37) similar to hepatitis A virus (HAV) (38). HAV and presumably other nonenveloped viruses, such as HEV, hijack intracellular membranes to shield from neutralizing antibody epitopes contained in the viral capsid. Quasienvelopment is also thought to promote HAV spread within the liver but is mechanistically incompletely understood (39). For HAV, it was demonstrated that the biogenesis of the membranes surrounding virions is dependent on host proteins associated with the ESCRT pathway, specifically VPS4B and ALIX (38). HEV also relies on components of the ESCRT pathway for virus budding through the interaction of PXXP motifs contained within ORF3 with TSG101 (15, 18). However, the important role of ORF3 viroporin activity was not known and thus adds a critical piece to forming a more complete picture of HEV release. Our present study lays the foundation for also gaining a deeper understanding of the budding of nonenveloped and other quasienveloped viruses. Future studies will focus on elucidating the structural details of how the ORF3 ion channel may function and possibly regulate the release of infectious virions.
Because viroporins are essential for the release of a variety of viruses, they are attractive targets for antiviral therapy. For example, it was previously shown that amantadine and rimantadine can inhibit the ion channel activity of M2 (40) and thereby interfere with the uncoating of IAV. Likewise, long alkyl iminosugar derivatives block HCV p7 function and decrease the release of infectious virions (41–43), and amiloride derivatives impair the ion channel activity of HIV-1 Vpu and severe acute respiratory syndrome coronavirus (SARS-CoV) E (44, 45). Besides pegylated IFN and ribavirin, both of which have substantial side effects, there are currently no specific drugs approved for the treatment of hepatitis E that directly interfere with the viral life cycle. HEV ORF3’s function as a viroporin may serve as a potential “druggable” target and could pave the way for the discovery of therapeutics that block this ion channel and thus disrupt HEV infectivity. This would address the important need for developing direct-acting antivirals to combat HEV in particularly vulnerable populations, such as pregnant women and immunocompromised individuals, who cannot be easily treated by current drug regimens.
Experimental Procedures
Additional procedures are described in detail in SI Materials and Methods.
Cell Lines and Animals.
HEK293T cells (ATCC) and HepG2C3A cells (ATCC) were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% (vol/vol) FBS, 50 IU/mL penicillin and streptomycin, in a humidified 5% (vol/vol) CO2 incubator at 37 °C. All experiments involving oocytes derived from X. laevis were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee at Rutgers University.
Coimmunoprecipitation and Western Blot Assay.
Cell lysates were prepared from 5 × 106 cells in a 10-mM Tris buffer (pH 7.5) containing 0.1% Nonidet P-40 and 1 mM EDTA, along with a mixture of protease and phosphatase inhibitors. Lysates were then incubated with Flag (M2, Sigma-Aldrich) antibody (1 μg, at 4 °C for 6 h), after which the immune complexes were precipitated with protein A sepharose. These immunoprecipitates were resolved on a 12% (wt/vol) SDS-polyacrylamide gel, transferred onto a nitrocellulose membrane, and then analyzed by Western blot using anti-Flag (M2, Sigma-Aldrich) or anti-HA (clone HA-7, Sigma-Aldrich) antibodies. Membranes were then washed three times with TBS-T for 15 min total. Membranes were incubated with goat anti-mouse DyLight800-conjugated antibody (Thermo Fisher Scientific) diluted 1:5,000 for 30 min and washed with TBS-T three times for 5 min each. Membranes were visualized using the Odyssey CLx Imaging System and images were processed using Image Studio Lite Ver5.0.
Voltage-Clamp Experiments.
The ORF3 or M2 cDNA was cloned into pSP64-polyA vector with restriction sites for HindlII (5′ end) and BamHI (3′ end) enzymes. The plasmid was linearized by EcoRI digestion and transcribed in vitro to synthesized mRNA using the mMESSAGE mMACHINE high-yield capped RNA transcription SP6 kit (Ambion). Healthy X. laevis oocytes in stage V to VI were injected with 20 ng of mRNA per oocyte and incubated at 16 °C in an ND-96 solution. Two-electrode voltage clamp (OC-725C, Warner Instruments) was used to record the currents at 48 h postinjection. The oocytes were first bathed in standard Ringer solution (115 mM NaCl, 2 mM KCl, 1.8 mM CaCl2 and 5 mM HEPS, pH 7.4) at room temperature and impaled with microelectrodes filled with 3 M KCl. Currents were generated by applying a 5-s voltage step protocol from −90 to +60 mV in 10-mV increments with a holding voltage of −60 mV. As for the ion selectivity, oocytes were clamped at −80 mV, followed by a 5-s depolarization to +60 mV. The voltage was then stepped to test pulses ranging from −80 to +50 mV in 10-mV increments. This was followed by a step to −80 for 7 s to restore initial conditions. The bathing solutions contained 117 mM NaCl, 1.8 mM CaCl2, and 10 mM Hepes, pH 7.4. Na+ was replaced at equimolar concentrations by K+, Cs+, or Li+. In addition, a 20 mM CaCl2 solution and a gluconate (in replacement of Cl−) solution was also used. To assess for proton permeability oocytes expressing M2 or ORF3 channels were exposed to bath solutions at pH 10 and 5. Bathing solution contained 100 mM NaCl, 1.8 mM CaCl2, 5 mM glycine (pH 10, adjusted with 1 M NMG) or 5 mM citric acid/Na-citrate (pH 5, adjusted with NMG). Oocytes were clamped at −80 mV, followed by a 1-s depolarization to +20 mV. The voltage was then stepped to test voltages ranging from −80 to +50 mV in 10- mV increments. Current recording and analysis were performed using pClamp 10.3 and Clampfix softwares (Axon Instruments), respectively.
SI Materials and Methods
Plasmid Construction.
To construct a lentiviral construct that encodes Kernow C1/p6 ORF2 or Kernow C1/p6 ORF3, the Kernow C1/p6 ORF2 or Kernow C1/p6 ORF3 cDNA was amplified by PCR from the HEV Kernow C1/p6 construct (a kind gift from Suzanne Emerson, NIH, Bethesda, MD) and then cloned into pLVX-IRES-zsGreen1 or pLEX-IRES-mCherry vectors using the In-Fusion HD Cloning Kit (Clontech). To construct the pLEX-IAV M2-IRES-mCherry vector, cDNA encoding Influenza A virus M2 (A/Puerto Rico/8/34/Mount Sinai/Wi(H1N1)) was synthesized by IDT with gBlock, and then cloned into pLEX-IRES-mCherry vectors using In-Fusion HD Cloning Kit (Clontech). IAV M2 mutants were generated by Quikchange (Stratagene) site-directed mutagenesis. All of the constructs were verified by DNA sequencing analysis.
Generation of ORF3 Alanine Mutants.
Kernow C1/p6 ORF3 alanine triplet mutants were synthesized by GenScript or by PCR amplification with primers containing the desired changes. Primary PCR products containing the engineered mutations were assembled and cloned into the pLEX-IRES-mCherry vector using In-Fusion HD Cloning Kit (Clontech) according to the instructions.
In Vitro Transcription and Viral RNA Transfection.
HEV Kernow-C1 p6/Gluc plasmid (genotype 3 subgenomic replicon expressing Gaussia luciferase; a gift from Suzanne Emerson, NIH) was linearized by MluI and the viral-capped RNAs were transcribed in vitro from linearized plasmid using mMESSAGE mMACHINE T7 Ultra Kit (Ambion). The in vitro transcription (IVT) reaction mixture of 20 μL was assembled by adding DNA template (1 μg), T7 reaction buffer, T7 NTP/ARCA, GTP, and T7 enzyme mix. The IVT reaction mixture was incubated at 37 °C for 3 h. To remove the template DNA, 1 μL TURBO DNase (from MEGAscript T7 Kit) was added to the IVT reaction mixture and incubated for 15 min at 37 °C. Then, the viral RNA was purified using RNeasy Mini Kit (Qiagen). Viral RNA was transfected into HepG2C3A cells using TransIT-mRNA Transfection reagent (Mirus Bio) according to the instructions.
Gaussia Luciferase Assay.
Gaussia luciferase activity was measured with the Renilla Luciferase Assay System (Promega). Ten microliters of harvested cell culture medium was added per well of a 96-well black, flat-bottom microplate (Corning), followed by the addition of Renilla luciferase assay substrate and the detection of luminescence using a Tristart2 Multimode Reader LB942 (Berthold Technologies).
Lentiviral Particles Production and Infection.
VSV-G pseudotyped lentiviruses were produced by transient cotransfection of the third generation packaging plasmids pMD2G, psPAX2, and transfer vector with X-tremeGENE HP DNA Transfection Reagent (Sigma-Aldrich) into HEK293T cells. The media was changed 6 h posttransfection. Supernatants were collected at 48 and 72 h after transfection, pooled, passed through a 0.45-µm filter, and frozen at −80 °C. For lentiviral transduction, 1 × 105 cells per well were seeded in six-well tissue culture plates and infected the following day with lentiviruses. Cells were trypsinized and processed for FACS analysis after 3 d of infection to determine the transduction efficiency.
Quantification of HEV RNA by RT-qPCR.
Total RNA was isolated from HepG2C3A cells using an RNeasy Mini Kit (Qiagen). cDNA was synthesized from 0.5 μg RNA using the SuperScript III First-Strand Synthesis System (Invitrogen) according to the manufacturer’s instructions. Quantitative PCR was performed using an Applied Biosystems SYBR Green PCR Master Mix and the following primer pairs:
| Gene | Forward primer | Reverse primer |
| HEVΔORF2/3[Gluc] | ATGGGAGTCAAAGTTCTGTTTGC | TCCATCTCTTTGAGCACCTC |
| GAPDH | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC |
For the data analysis, the threshold cycle (Ct) values for HEVΔORF2/3[Gluc] were normalized to those for GAPDH.
Confocal Microscopy.
HepG2C3A cells were washed with PBS 3 d after lentiviral transduction with HA-tagged wild-type or mutant ORF3 constructs, fixed with 4% (wt/vol) paraformaldehyde (PFA), and then permeabilized with 0.3% Triton X-100. The cells were blocked with 2% (wt/vol) BSA and immunolabeled with mouse anti-HA (Clone HA-7, Sigma-Aldrich) and rabbit anti-calnexin (Abcam) antibodies for 1 h at room temperature. Cells were washed with PBS and incubated with Alexa Fluor 488-conjugated goat anti-mouse antibody (Thermo Fisher Scientific) or Alexa Fluor 647-conjugated goat anti-rabbit antibody (Thermo Fisher Scientific) for 1 h. Nuclei were stained with Hoechst dye. To observe the localization of ORF3 in the Xenopus laevis oocytes, the oocytes were collected at 48 h after ORF3 mRNA injection for immunostaining assay as described above, and rabbit anti-ORF3 polyclonal antibody (a kind gift from Suzanne Emerson, NIH) and Alexa Fluor 555-conjugated goat anti-rabbit antibody (Thermo Fisher Scientific) were used. Images were taken using a Nikon A1 Spectral Confocal Microscope. ImageJ analysis was done using ImageJ software (National Institutes of Health).
Flow Cytometric Analysis.
Expression of lentivirally delivered transgenes was analyzed by flow cytometry. Kernow C1/p6 ORF2 or ORF3 (IAV M2 or its mutant) were transduced into target cells by bicistronic lentiviruses expressing zsGreen or mCherry. After 3 d transduction, cells were fixed in 4% (wt/vol) PFA in PBS for 15 min, then washed with PBS. The efficiencies of transduction of Kernow C1/p6 ORF2 and ORF3 (IAV M2 or its mutant) were determined by simultaneous expression zsGreen and mCherry. All samples were analyzed on a BD LSRII flow cytometer using FlowJo Software (FlowJo).
Quantification of Intracellular Virus Infectivity.
The Kernow C1/p6-ΔORF2/3[Gluc] RNA transfected HepG2C3A cells were cultured in the 12-well cell culture plate, and cells were trypsinized and washed with PBS and then lysed by adding 1 mL H2O per well and put on ice for 20 min, vortexing intermittently every 5 min. The samples were centrifuged at 13,000 × g for 10 min to remove cellular debris, and 0.9 mL of the supernatant was collected and 0.1 mL of 10× concentrated PBS was added to infect cells.
Statistical Analysis.
Student’s t test or one-way ANOVA with Tukey’s HSD test was used to test for statistical significance of the differences between the different group parameters. P values of less than 0.05 were considered statistically significant.
Acknowledgments
We thank Drs. Suzanne U. Emerson and Patricia Farci (National Institute of Allergy and Infectious Diseases) for providing the pKernow C1/p6 Gluc construct; Christina deCoste and John Grady (Molecular Biology Flowcytometry Core Facility, Princeton University) for outstanding technical help; and members of the A.P. laboratory, in particular Jenna Gaska, in addition to Shiqi Xie (University of Texas Southwestern Medical Center) and Andrew Harris (Rutgers University), for critical discussions and comments on the manuscript. The work was supported in part by grants from Princeton University and an Investigator in Pathogenesis Award by the Burroughs Wellcome Fund (to A.P.). Q.D. is supported by a postdoctoral fellowship from the New Jersey Commission on Cancer Research (DHFS16PPC007).
Footnotes
Conflict of interest statement: Q.D. and A.P. have filed a patent application on the use of the HEV transcomplementation system and ORF3’s ion channel function for anti-HEV drug screening.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614955114/-/DCSupplemental.
References
- 1.Dalton HR, Bendall R, Ijaz S, Banks M. Hepatitis E: An emerging infection in developed countries. Lancet Infect Dis. 2008;8(11):698–709. doi: 10.1016/S1473-3099(08)70255-X. [DOI] [PubMed] [Google Scholar]
- 2.Kamar N, et al. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med. 2008;358(8):811–817. doi: 10.1056/NEJMoa0706992. [DOI] [PubMed] [Google Scholar]
- 3.Schildgen O, Müller A, Simon A. Chronic hepatitis E and organ transplants. N Engl J Med. 2008;358(23):2521–2522, author reply 2522. doi: 10.1056/NEJMc080634. [DOI] [PubMed] [Google Scholar]
- 4.Dalton HR, Bendall RP, Keane FE, Tedder RS, Ijaz S. Persistent carriage of hepatitis E virus in patients with HIV infection. N Engl J Med. 2009;361(10):1025–1027. doi: 10.1056/NEJMc0903778. [DOI] [PubMed] [Google Scholar]
- 5.Colson P, Kaba M, Moreau J, Brouqui P. Hepatitis E in an HIV-infected patient. J Clin Virol. 2009;45(4):269–271. doi: 10.1016/j.jcv.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Colson P, Dhiver C, Gerolami R. Hepatitis E virus as a newly identified cause of acute viral hepatitis during human immunodeficiency virus infection. Clin Microbiol Infect. 2008;14(12):1176–1180. doi: 10.1111/j.1469-0691.2008.02102.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, et al. Long-term efficacy of a hepatitis E vaccine. N Engl J Med. 2015;372(10):914–922. doi: 10.1056/NEJMoa1406011. [DOI] [PubMed] [Google Scholar]
- 8.Mallet V, et al. Brief communication: Case reports of ribavirin treatment for chronic hepatitis E. Ann Intern Med. 2010;153(2):85–89. doi: 10.7326/0003-4819-153-2-201007200-00257. [DOI] [PubMed] [Google Scholar]
- 9.Kamar N, et al. Ribavirin therapy inhibits viral replication on patients with chronic hepatitis e virus infection. Gastroenterology. 2010;139(5):1612–1618. doi: 10.1053/j.gastro.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Debing Y, et al. Hepatitis E virus mutations associated with ribavirin treatment failure result in altered viral fitness and ribavirin sensitivity. J Hepatol. 2016;65(3):499–508. doi: 10.1016/j.jhep.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Cao D, Meng XJ. Molecular biology and replication of hepatitis E virus. Emerg Microbes Infect. 2012;1(8):e17. doi: 10.1038/emi.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad I, Holla RP, Jameel S. Molecular virology of hepatitis E virus. Virus Res. 2011;161(1):47–58. doi: 10.1016/j.virusres.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi M, et al. Monoclonal antibodies raised against the ORF3 protein of hepatitis E virus (HEV) can capture HEV particles in culture supernatant and serum but not those in feces. Arch Virol. 2008;153(9):1703–1713. doi: 10.1007/s00705-008-0179-6. [DOI] [PubMed] [Google Scholar]
- 14.Emerson SU, et al. Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J Virol. 2010;84(18):9059–9069. doi: 10.1128/JVI.00593-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagashima S, et al. A PSAP motif in the ORF3 protein of hepatitis E virus is necessary for virion release from infected cells. J Gen Virol. 2011;92(Pt 2):269–278. doi: 10.1099/vir.0.025791-0. [DOI] [PubMed] [Google Scholar]
- 16.Yamada K, et al. ORF3 protein of hepatitis E virus is essential for virion release from infected cells. J Gen Virol. 2009;90(Pt 8):1880–1891. doi: 10.1099/vir.0.010561-0. [DOI] [PubMed] [Google Scholar]
- 17.Shukla P, et al. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc Natl Acad Sci USA. 2011;108(6):2438–2443. doi: 10.1073/pnas.1018878108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenney SP, Wentworth JL, Heffron CL, Meng XJ. Replacement of the hepatitis E virus ORF3 protein PxxP motif with heterologous late domain motifs affects virus release via interaction with TSG101. Virology. 2015;486:198–208. doi: 10.1016/j.virol.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyagi S, Surjit M, Roy AK, Jameel S, Lal SK. The ORF3 protein of hepatitis E virus interacts with liver-specific alpha1-microglobulin and its precursor alpha1-microglobulin/bikunin precursor (AMBP) and expedites their export from the hepatocyte. J Biol Chem. 2004;279(28):29308–29319. doi: 10.1074/jbc.M402017200. [DOI] [PubMed] [Google Scholar]
- 21.Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69(3):517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 22.Cordes FS, Tustian AD, Sansom MS, Watts A, Fischer WB. Bundles consisting of extended transmembrane segments of Vpu from HIV-1: Computer simulations and conductance measurements. Biochemistry. 2002;41(23):7359–7365. doi: 10.1021/bi025518p. [DOI] [PubMed] [Google Scholar]
- 23.Li F, et al. Efficient genetic manipulation of the NOD-Rag1-/-IL2RgammaC-null mouse by combining in vitro fertilization and CRISPR/Cas9 technology. Sci Rep. 2014;4:5290. doi: 10.1038/srep05290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zafrullah M, Ozdener MH, Panda SK, Jameel S. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol. 1997;71(12):9045–9053. doi: 10.1128/jvi.71.12.9045-9053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emerson SU, Nguyen H, Torian U, Purcell RH. ORF3 protein of hepatitis E virus is not required for replication, virion assembly, or infection of hepatoma cells in vitro. J Virol. 2006;80(21):10457–10464. doi: 10.1128/JVI.00892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holsinger LJ, Nichani D, Pinto LH, Lamb RA. Influenza A virus M2 ion channel protein: A structure-function analysis. J Virol. 1994;68(3):1551–1563. doi: 10.1128/jvi.68.3.1551-1563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenney SP, et al. The PSAP motif within the ORF3 protein of an avian strain of the hepatitis E virus is not critical for viral infectivity in vivo but plays a role in virus release. J Virol. 2012;86(10):5637–5646. doi: 10.1128/JVI.06711-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surjit M, Oberoi R, Kumar R, Lal SK. Enhanced alpha1 microglobulin secretion from hepatitis E virus ORF3-expressing human hepatoma cells is mediated by the tumor susceptibility gene 101. J Biol Chem. 2006;281(12):8135–8142. doi: 10.1074/jbc.M509568200. [DOI] [PubMed] [Google Scholar]
- 29.Sze CW, Tan YJ. Viral membrane channels: Role and function in the virus life cycle. Viruses. 2015;7(6):3261–3284. doi: 10.3390/v7062771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nieva JL, Madan V, Carrasco L. Viroporins: Structure and biological functions. Nat Rev Microbiol. 2012;10(8):563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terwilliger EF, Cohen EA, Lu YC, Sodroski JG, Haseltine WA. Functional role of human immunodeficiency virus type 1 vpu. Proc Natl Acad Sci USA. 1989;86(13):5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwatsuki-Horimoto K, et al. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J Virol. 2006;80(11):5233–5240. doi: 10.1128/JVI.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu W, et al. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc Natl Acad Sci USA. 2006;103(33):12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol. 2007;81(16):8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinmann E, et al. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007;3(7):e103. doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki T, et al. The human polyoma JC virus agnoprotein acts as a viroporin. PLoS Pathog. 2010;6(3):e1000801. doi: 10.1371/journal.ppat.1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin X, Ambardekar C, Lu Y, Feng Z. Distinct entry mechanisms for nonenveloped and quasi-enveloped hepatitis E viruses. J Virol. 2016;90(8):4232–4242. doi: 10.1128/JVI.02804-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Z, et al. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496(7445):367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin X, Li X, Feng Z. Role of envelopment in the HEV life cycle. Viruses. 2016;8(8):E229. doi: 10.3390/v8080229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4(11):3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pavlović D, et al. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc Natl Acad Sci USA. 2003;100(10):6104–6108. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinmann E, et al. Antiviral effects of amantadine and iminosugar derivatives against hepatitis C virus. Hepatology. 2007;46(2):330–338. doi: 10.1002/hep.21686. [DOI] [PubMed] [Google Scholar]
- 43.Foster TL, et al. Resistance mutations define specific antiviral effects for inhibitors of the hepatitis C virus p7 ion channel. Hepatology. 2011;54(1):79–90. doi: 10.1002/hep.24371. [DOI] [PubMed] [Google Scholar]
- 44.Ewart GD, Mills K, Cox GB, Gage PW. Amiloride derivatives block ion channel activity and enhancement of virus-like particle budding caused by HIV-1 protein Vpu. Eur Biophys J. 2002;31(1):26–35. doi: 10.1007/s002490100177. [DOI] [PubMed] [Google Scholar]
- 45.Wilson L, Gage P, Ewart G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353(2):294–306. doi: 10.1016/j.virol.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]