Significance
The distribution of the hormone auxin controls most processes in plant development. Auxin distribution within the plant requires PIN transporters and efficient PIN-mediated transport requires PIN phosphorylation. Phosphorylation seemingly also controls auxin transport by targeting PINs to specific sides of the cell. Understanding how auxin is directed and activated through phosphorylation is essential to understand plant growth. Two different protein kinases targeting the same phosphosites in PIN1 can activate auxin efflux. Surprisingly, however, only one affects PIN1 polar distribution. Here, we show that the differential effects of the two kinases on PIN1 cannot be explained by phosphorylation at the established phosphosites, and suggest that a more complex model is needed to explain the effects of the kinase on PIN1 polarity.
Keywords: auxin transport, protein kinase, Arabidopsis, PIN1, polarity
Abstract
The directional distribution of the phytohormone auxin is essential for plant development. Directional auxin transport is mediated by the polarly distributed PIN-FORMED (PIN) auxin efflux carriers. We have previously shown that efficient PIN1-mediated auxin efflux requires activation through phosphorylation at the four serines S1–S4 in Arabidopsis thaliana. The Brefeldin A (BFA)-sensitive D6 PROTEIN KINASE (D6PK) and the BFA-insensitive PINOID (PID) phosphorylate and activate PIN1 through phosphorylation at all four phosphosites. PID, but not D6PK, can also induce PIN1 polarity shifts, seemingly through phosphorylation at S1–S3. The differential effects of D6PK and PID on PIN1 polarity had so far been attributed to their differential phosphosite preference for the four PIN1 phosphosites. We have mapped PIN1 phosphorylation at S1–S4 in situ using phosphosite-specific antibodies. We detected phosphorylation at PIN1 phosphosites at the basal (rootward) as well as the apical (shootward) plasma membrane in different root cell types, in embryos, and shoot apical meristems. Thereby, PIN1 phosphorylation at all phosphosites generally followed the predominant PIN1 distribution but was not restricted to specific polar sides of the cells. PIN1 phosphorylation at the basal and apical plasma membrane was differentially sensitive to BFA treatments, suggesting the involvement of different protein kinases or trafficking mechanisms in PIN1 phosphorylation control. We conclude that phosphosite preferences are not sufficient to explain the differential effects of D6PK and PID on PIN1 polarity, and suggest that a more complex model is needed to explain the effects of PID.
The phytohormone auxin is a central regulator of plant development and tropic growth (1, 2). Proper plant development strictly requires the directed cell-to-cell transport of auxin, which is achieved by a system of auxin influx and efflux transporters (1). AUXIN RESISTANT1 (AUX1)/LIKE-AUX1 (LAX) proteins are auxin influx transporters and PIN-FORMED (PIN) proteins are auxin efflux transporters that may act together with ABC transporters (3–8). Auxin transport gains its directionality through the often polar distribution of the plasma membrane-resident PIN auxin efflux carriers, PIN1–PIN4 and PIN7 in Arabidopsis thaliana (1, 9). Directional auxin transport results in the formation of cellular auxin maxima and minima that provide essential cues for plant growth and differentiation at the level of individual cells and tissues (1, 2). PIN1 localizes to the basal (rootward) plasma membrane in root stele cells and directs auxin transport toward the root tip (3). PIN2 localizes differentially to the basal (rootward) and apical (shootward) plasma membrane in cortex and epidermis cells, respectively, and the opposing auxin transport streams in cortex and epidermis are important for gravitropic root growth (10, 11).
Efficient PIN1-mediated auxin efflux requires activation by phosphorylation (12, 13). In the case of PIN1, the AGCVIII protein kinases D6 PROTEIN KINASE (D6PK) and PINOID (PID) activate auxin efflux through phosphorylation at the PIN1 serines S1 (S231), S2 (S252), S4 (S271), and S3 (S290), respectively (Fig. 1 A and B) (12). In in vitro kinase assays, PID preferentially phosphorylates PIN1 at S1–S3 (12, 14, 15). S1–S3 are embedded in motifs that share striking sequence similarity to each other and that are highly conserved between PIN1–PIN4 and PIN7 (Fig. 1 A and B). D6PK preferentially phosphorylates PIN1 at S4 but it also phosphorylates S1–S3, albeit less efficiently than PID (12). Functional analyses of protein kinase-dependent PIN1-mediated auxin efflux support the relevance of the respective phosphosite preferences detected in vitro for kinase-activated auxin transport (12). There, PIN1 S1–S3 mutations affect PIN1 activation by PID more strongly than PIN1 S4 mutations and, conversely, PIN1 S4 mutations affect PIN1 activation by D6PK more strongly than PIN1 S1–S3 mutations. However, mutations of all four phosphosites are required to fully impair PID- and D6PK-dependent PIN1 activation, indicating that both kinases are able to target all four PIN1 phosphosites (2, 12). S5, an additional phosphosite in PIN3, related to S4 and preferentially targeted by D6PK, is not conserved in PIN1 and will therefore not be discussed further (Fig. 1 A and B) (12, 13).
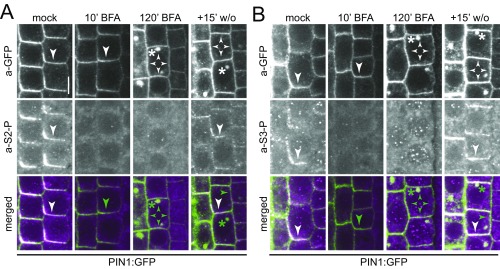
Fig. 1.
Phosphosite-specific PIN1 antibodies detect PIN1 phosphorylation in a BFA-sensitive manner. (A) Pretty box protein sequence alignment of a region of the intracellular hydrophilic loop of A. thaliana PIN1–PIN4 and PIN7. The S1–S5 phosphosites described for PINs are marked with asterisks. S5 is not conserved in PIN1, where it aligns with a D (aspartic acid; S5/D), and examining S5 phosphorylation was therefore not relevant for this study. (B) Predicted topology of PIN1 with 10 transmembrane domains and the intracellular hydrophilic loop with the relative positions of the phosphosites marked. (C) Dot blot immunoblot against unphosphorylated (S) and phosphorylated peptides S(-P) probed with anti-PIN1 S1-P through S4-P antibodies. (D and E) Representative confocal images of primary root stele cells after immunostaining of 4-d-old PIN1:GFP Arabidopsis seedlings with anti-GFP (a-GFP; PIN1:GFP) (green), a-PIN1 S1-P (D) (magenta) and a-PIN1 S4-P (E) (magenta) antibodies. Seedlings were mock-treated or BFA-treated (50 µM) for 10 and 120 min, respectively. A 15-min washout (w/o) followed the 120-min BFA treatment. Arrowheads mark strong and weak plasma membrane staining, asterisks mark intracellular BFA compartments. Overlap of green and magenta signals is indicated by white arrowheads in the merged images. Green arrowheads and asterisks indicate the absence of a corresponding magenta signal from a-PIN1 S1-P or a-PIN1 S4-P immunostaining. The corresponding experiments with a-PIN1 S2-P and a-PIN1 S3-P are shown in Fig. S1 A and B; images of entire roots immunostained with a-PIN1 S1-P through S4-P are shown in Fig. S4, supporting the representative nature of the images shown here. (Scale bar, 5 µm.) (F and G) Quantification of fluorescence intensity at the basal plasma membrane after immunostaining as shown in D and E. Data represent the average from two independent experiments (n = 50 cells). Student's t test datasets with no statistical difference fall in one group and were labeled accordingly. Upper and lowercase letters serve to distinguish the results obtained with a-GFP and a-PIN1 S1-P or a-PIN1 S4-P, respectively.
In vivo, inactivation of D6PK and the functionally related D6PKL1–3 (D6PK-LIKE1–3), achieved either by chemical inhibitor-mediated removal of D6PK and D6PKLs from the plasma membrane or by the gradual mutational inactivation of the D6PK genes, correlates strongly with decreases in PIN1 phosphorylation as well as with decreases in directional auxin transport in stems and hypocotyls (13, 16, 17). Thus, PIN1 phosphorylation is essential for auxin transport in planta and may allow predicting PIN1 activity in situ.
Although D6PK and PID activate PIN1 through phosphorylation of the same phosphosites in vitro, the two kinases have differential effects on PIN1 in vivo. Overexpression of PID but not of D6PK promotes the retargeting of PIN1 from the basal (rootward) to the apical (shootward) plasma membrane in root cells (14, 15). PID-dependent PIN1 phosphorylation has been proposed to mediate this effect because PIN1 becomes insensitive to PID overexpression in transgenic lines where PIN1 S1–S3 are replaced by the nonphosphorylatable A (alanine) (14, 15). Furthermore, PIN1 is targeted to the apical plasma membrane in a PID-independent manner when S1–S3 are replaced by the supposedly phosphorylation-mimicking E (glutamic acid) (14, 15). These observations suggested that PID-dependent PIN1 S1–S3 phosphorylation serves as a sorting signal for differential PIN1 intracellular transport and targeting. As yet, the differential effects of PID and D6PK on PINs could only be explained by the differential phosphosite preferences of the two kinases (2, 12). Here, we show that this distinction cannot be the main feature underlying their differential cell biological effects.
Besides their differential phosphosite preference, D6PK and PID also differ from each other in regard to a number of cell biological criteria. Whereas PID is localized to the plasma membrane in a nonpolar manner, D6PK localizes to the basal plasma membrane of most cells (13, 18). D6PK, as well as PIN1, are constitutively recycling to and from the plasma membrane and both proteins are internalized after treatment with Brefeldin A (BFA), a fungal inhibitor that blocks the GN (GNOM) ARF-GEF–dependent recycling of endosomal cargo from the trans-Golgi network to the plasma membrane (13, 19). However, whereas D6PK is rapidly and completely internalized within minutes after BFA treatment, PIN1 has a comparatively slow response and remains detectable at the plasma membrane, even after prolonged BFA treatments (13). In contrast, the plasma membrane targeting of PID cannot be efficiently blocked by BFA inhibitor treatments (18). Thus, D6PK, PID, and PINs differ from each other in regard to their cell biological behavior and their BFA sensitivity.
Here, we monitor PIN1 phosphorylation as a proxy for the spatial and temporal dynamics of PIN1 activation and localization at the subcellular level. To this end, we have generated PIN1 S1–S4 phosphosite-specific antibodies and examine PIN1 phosphorylation patterns in situ. We detected PIN1 phosphorylation at the basal (rootward) as well as the apical (shootward) plasma membranes in different root cell types and in embryos, as well as in shoot meristems. PIN1 phosphorylation generally followed the predominant PIN1 distribution but was not restricted to specific plasma membrane domains. PIN1 phosphorylation at the basal and apical plasma membrane was differentially sensitive to BFA treatments, suggesting the involvement of different protein kinases or trafficking mechanisms in PIN1 phosphorylation control. We concluded that phosphosite preferences are not sufficient to explain the differential effects of D6PK and PID on PIN1 polarity, and suggest that a more complex model is needed to explain PID-dependent PIN1 polarity control.
Results
Phosphosite-Specific Antibodies Detect PIN1 Phosphorylation in Situ.
PIN1 S1–S4 are phosphorylation targets for PID and D6PK (12, 14, 15). To monitor PIN1 phosphorylation at these specific sites individually in situ, we generated peptide antibodies directed against phosphorylated PIN1 S1–S4, designated a-PIN1 S1-P to a-PIN1 S4-P (Fig. 1 A and B). Dot blot immunoblotting showed that the antibodies are specific for the respective PIN1 phospho-peptides, with little or no cross-reactivity toward the respective other PIN1 phospho-peptides or sequence-related phospho-peptides from other PINs (Fig. 1C).
We tested the in planta specificity of these antibodies using transgenic lines expressing wild-type PIN1:GFP or PIN1:YFP and their variants carrying A (alanine) mutations at one of the four phosphosite serines (S1A, S2A, S3A, and S4A) in a pin1 loss-of-function mutant background (12). We detected PIN1:GFP or PIN1:YFP with all four phosphorylation-specific antibodies as well as with an a-GFP antibody at the basal (rootward) plasma membrane of root stele cells in the entire root (Fig. 1 D–G and Fig. S1). In turn, the a-PIN1 S1-P, a-PIN1 S2-P, and a-PIN1 S4-P antibodies did not recognize PIN1:GFP or PIN1:YFP with alanine substitutions at the respective S1, S2, or S4 phosphosites (Fig. S2 A–C). Only a-PIN1 S3-P still detected basally localized PIN1:YFP in PIN1:YFP S3A lines (Fig. S2D). We concluded that three phosphosite antibodies are highly specific to phosphorylated PIN1 serine residues and could thus be used to monitor PIN1 phosphorylation in situ.
Fig. S1.
Phosphosite-specific a-PIN1 S2-P and S3-P antibodies detect PIN1 phosphorylation in a BFA-sensitive manner. Representative confocal images of stele cells of the primary root after immunostaining of 4-d-old PIN1:GFP Arabidopsis seedlings with anti-GFP (a-GFP, PIN1:GFP; green), a-PIN1 S2-P (magenta; A) and a-PIN1 S3-P (magenta; B) antibodies. Seedlings were mock-treated or BFA-treated for 10 and 120 min, respectively. A 15-min washout (w/o) followed the 120-min BFA treatment. Arrowheads mark strong and weak plasma membrane staining, asterisks mark intracellular compartments. Overlap of green and magenta signals is indicated by white arrows in the merged images. Green arrowheads and asterisks indicate the absence of a corresponding magenta signal from a-PIN1 S1-P or a-PIN1 S4-P immunostaining. (Scale bar, 5 µm.)
Fig. S2.
In situ analysis of the PIN1 phosphosite-specificity. Representative confocal images of stele cells of the primary root after immunostaining of 4-d-old wild-type seedlings or pin1 mutant seedlings transformed with PIN1:GFP or PIN1:YFP transgenes with alanine replacement mutations of S1–S4. Seedlings were genotyped for homozygosity of the underlying pin1 mutant allele (pin1−/−) and stained with anti-GFP (a-GFP, PIN1:GFP; green) and a-PIN1 S1-P (magenta; A), a-PIN1 S2-P (magenta; B), a-PIN1 S4-P (magenta; C), and a-PIN1 S3-P (magenta; D) antibodies. Overlap of green and magenta signals is indicated by white arrowheads in the merged images. Green arrowheads indicate the absence of a corresponding magenta signal from phosphosite-specific antibody staining. (Scale bar, 5 µm.)
PIN1 S1–S4 Phosphorylation at the Basal Plasma Membrane Is BFA-Sensitive.
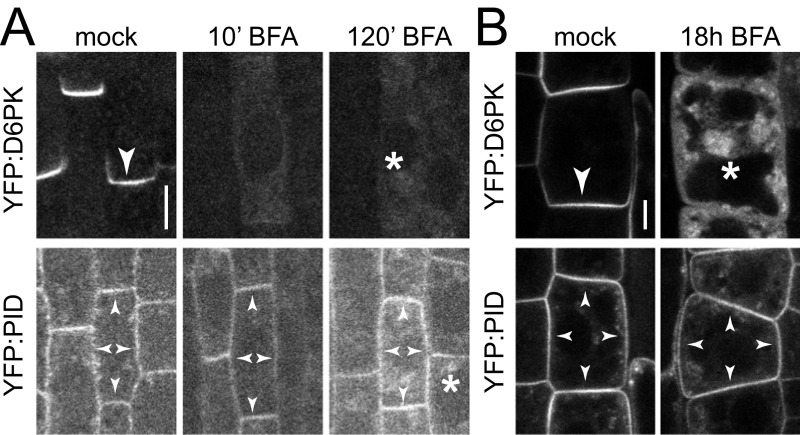
Short-term BFA treatments (10 min) result in the complete internalization of D6PK. In turn, D6PK rapidly relocalizes to the basal plasma membrane following short (15 min) washout treatments (Fig. S3) (13). Over the duration of a 10-min BFA treatment, PIN1 or PID abundance at the plasma membrane are not detectably affected. After a 120-min BFA treatment, however, PIN1 reportedly becomes apolarly distributed and internalized (Fig. 1 D–G and Figs. S1 and S3A) (13, 18). We exploited this differential BFA sensitivity of D6PK and PIN1 to examine PIN1 phosphorylation after the depletion of BFA-sensitive kinases from the plasma membrane. Interestingly, phosphorylation at all four PIN1 phosphosites was strongly reduced, if not lost, after a 10-min BFA treatment, when D6PK was completely internalized but PIN1 protein was still clearly detectable at the basal plasma membrane (Fig. 1 D–G and Figs. S1 and S3A). After a 120-min BFA treatment, PIN1 was apolarly distributed at the plasma membrane and also detected in intracellular BFA compartments (Fig. 1 D–G and Fig. S1). However, PIN1 phosphorylation at any of the four sites could not be detected, either at the plasma membrane or in the intracellular compartments (Fig. 1 D–G and Fig. S1). Although the phosphosite-specific antibodies recognized intracellular structures, these signals did not consistently overlap with a-GFP (PIN1) signals and also occurred outside of the PIN1 expression domain, indicating that they are unspecific background signals. When the 120-min BFA treatment was followed by a 15-min washout, resulting in the polar retargeting of D6PK to the basal plasma membrane, PIN1 phosphorylation was again detected preferentially at the basal plasma membrane at all four phosphosites (Fig. 1 D–G and Fig. S1) (12, 13). These phosphorylation patterns were observed throughout the entire root meristem (Fig. S4). The phosphosite-specific antibodies thereby mirrored the apparent phosphorylation pattern detected for PIN1 with an anti-GFP antibody in immunoblots of total root cell extracts exposed to the same BFA treatments (Fig. S5) (12, 13). This finding suggested that BFA-sensitive protein kinases may be required to maintain PIN1 phosphorylation at all four PIN1 phosphosites and that dephosphorylation by protein phosphatases rapidly antagonizes PIN1 phosphorylation. Even though we had observed a residual signal with the a-PIN1 S3-P antibody in PIN1:YFP S3A pin1 lines, a-PIN1 S3-P showed the same behavior as the other phosphosite-specific antibodies with regard to BFA sensitivity (Figs. S2D, S4, and S5). We therefore judged that a-PIN1 S3-P may interact with one of the highly related PIN phosphosites in one of the other functionally redundant PINs (Fig. 1A). Because all antibodies showed comparable behaviors in the experimental conditions tested to this point, PIN1 S1-P and PIN1 S4-P were chosen as representative phosphosites for further investigations.
Fig. S3.
D6PK and PID have differential BFA-sensitivity in root stele and epidermis cells. Representative confocal images of (A) stele cells and (B) epidermal cells of the primary root from 4-d-old YFP:D6PK and YFP:PID Arabidopsis seedlings after mock or BFA treatments as used for the experiments shown in Figs. 1 and 5 and Fig. S1, followed by immunostaining (A) or live imaging (B). Arrowheads mark strong and weak plasma membrane staining, asterisks mark intracellular compartments. (Scale bars, 5 µm.)
Fig. S4.
Phosphosite-specific PIN1 antibodies detect PIN1 phosphorylation in a BFA-sensitive manner. Representative confocal images of primary roots after immunostaining of 4-d-old PIN1:GFP Arabidopsis seedlings with anti-GFP (a-GFP, PIN1:GFP; green), a-PIN1 S1-P (magenta; A), a-PIN1 S2-P (magenta; B), a-PIN1 S4-P (magenta; C), and a-PIN1 S3-P (magenta; D) antibodies. Seedlings were mock-treated or BFA-treated for 10 and 120 min, respectively. A 15-min washout (w/o) followed the 120-min BFA treatment. White signal in merged images indicates overlap of green and magenta signals. (Scale bar, 50 µm.)
Fig. S5.
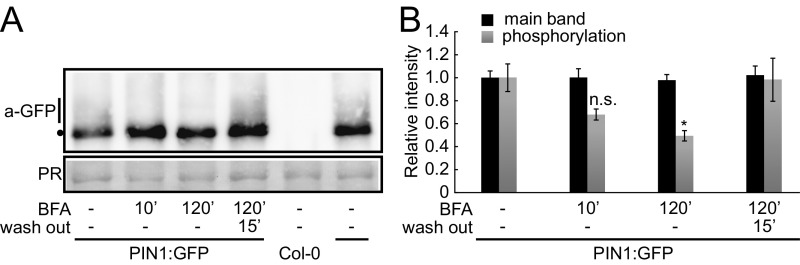
BFA-sensitive PIN1 phosphorylation can be detected in immunoblots. (A) Anti-GFP (a-GFP; PIN1:GFP) immunoblot against 10 µg total protein extract from 5-d-old PIN:GFP and wild-type (Col-0) seedling roots. PR (Ponceau Rouge) staining was used as a protein loading control. The dot marks the main band that correlates with the migration of the nonphosphorylated protein; the line marks the higher molecular weight smear that corresponds to phosphorylated forms of the protein. (B) Quantification of the density profiles of the main band and the phosphorylation-dependent smear from four immunoblots (two biological and two technical replicates) as shown in A. Student's t test: *P ≤ 0.05; n.s., not significant.
PIN1 Phosphorylation Is GNOM-Dependent.
The ARF-GEF GNOM is inhibited by BFA and functional GN is required for PIN1 and D6PK trafficking (13, 19). To test the GN-dependency of the observed BFA effects on PIN1 phosphorylation, we used transgenic lines expressing BFA-sensitive wild-type GNwt or the BFA-insensitive but functional GNM696L variant (19). Indeed, after 30-min BFA treatments, PIN1 S1 and S4 phosphorylation at the plasma membrane was maintained in GNM696L but impaired in GNwt. We concluded that PIN1 phosphorylation at the basal plasma membrane is mediated by BFA-sensitive GN trafficking-dependent kinases (Fig. S6).
Fig. S6.
The BFA-sensitivity of PIN1 phosphorylation is GNOM-dependent. Representative confocal images of root stele cells after immunostaining of 4-d-old BFA-sensitive GNwt and BFA-insensitive GNM696L transgenic mock- and BFA-treated Arabidopsis seedlings stained with a-PIN1 S1-P and a-PIN1 S4-P antibodies. Arrowheads mark the staining at the basal plasma membrane. (Scale bar, 5 µm.)
The Phosphatase Inhibitor Cantharidin Delays PIN1 Dephosphorylation.
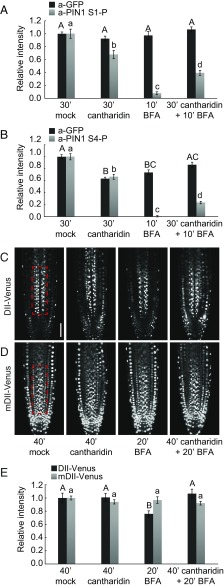
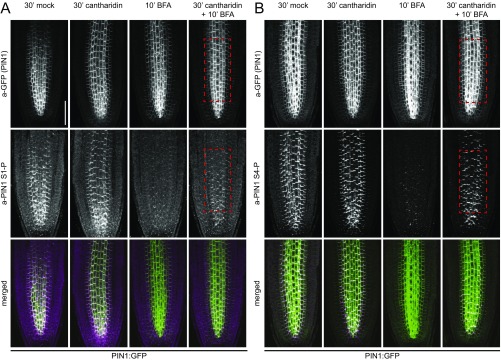
The rapid decrease in PIN1 phosphorylation following BFA treatment can be explained by the activity of phosphatases. We tested whether PIN1 phosphorylation was affected by cantharidin, a phosphatase inhibitor previously used to manipulate PIN-dependent auxin transport (20–22). Indeed, cantharidin pretreatments delayed PIN1 dephosphorylation following BFA treatments significantly, albeit not completely (Fig. 2 A and B and Fig. S7).
Fig. 2.
Cantharidin inhibits PIN1 dephosphorylation and alters auxin distribution patterns. (A and B) Quantification of fluorescence intensity in the root stele after immunostaining following BFA (50 µM, 10 min) and cantharidin or mock (30 min) treatments. The average from three independent experiments is shown (total n ≥ 44). Representative images of entire roots immunostained with a-PIN1 S1-P and a-PIN1 S4-P as used for quantifications are shown in Fig. S7. (C and D) Representative live fluorescence microscopy images of root tips of 4-d-old Arabidopsis seedlings expressing the auxin-sensitive DII-VENUS (C) or the auxin-insensitive mDIl-VENUS markers (D) subjected to BFA (50 µM, 20 min) and cantharidin or mock (40 min) treatments. (Scale bar, 50 µm.) (E) Quantification of fluorescence intensities in the framed areas of the root stele after live imaging as shown in C and D. The average from three independent experiments (n ≥ 21) is shown. The slightly longer treatments in C–E were chosen because the downstream responses of the primary effects shown in A and B were being examined. In A, B, and E, Student's t test datasets with no statistical difference fall in one group and were labeled accordingly. Upper and lowercase letters serve to distinguish the results obtained with the different series.
Fig. S7.
Cantharidin inhibits PIN1 dephosphorylation. Representative confocal fluorescence microscopy images of primary roots after immunostaining of 4-d-old PIN1:GFP Arabidopsis seedlings following BFA (50 µM, 10 min) and cantharidin or mock (30 min) treatments with anti-GFP (a-GFP, PIN1:GFP), a-PIN1 S1-P (A) or a-PIN1 S4-P (B) antibodies. (Scale bar, 50 µm.)
The effects of cantharidin on PIN1 phosphorylation should affect PIN1 auxin efflux activity and consequently auxin distribution patterns in the root. We monitored auxin distribution with the auxin-labile DII-VENUS reporter (23). Because, following the interference with auxin transport, changes in auxin responses are expected to occur with a delay, we conducted these experiments using slightly longer BFA (20 min vs. 10 min) and cantharidin (40 min vs. 30 min) treatments, respectively. As expected from an increase in cellular auxin levels as a consequence of reduced auxin efflux, DII-VENUS fluorescence intensities decreased rapidly following BFA treatments (Fig. 2 C–E). This decrease was attenuated when BFA was applied together with cantharidin, indicative for a comparatively more active auxin efflux (Fig. 2 C–E). At the same time, mDII-VENUS, the stabilized auxin-insensitive control variant of DII-VENUS, did not respond to any of the treatments (Fig. 2 C–E). Thus, the inhibition of PIN1 dephosphorylation may have direct effects on auxin efflux and result, as suggested by the changes in DII-VENUS abundance, in differential cellular accumulations of auxin.
PIN1 Phosphorylation Can Be Detected at Apical and Lateral Plasma Membranes.
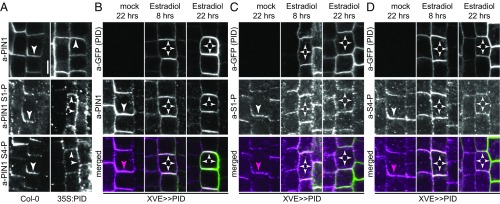
Ectopic PID expression causes PIN1 polarity shifts, which had been proposed to be promoted by PIN1 S1–S3 phosphorylation (14, 15, 24). We used PID overexpressing seedlings (35S:PID) to examine the consequences of altered PIN1 plasma membrane distribution on PIN1 phosphorylation. In 2-d-old PID overexpressing seedlings, PIN1 became detectable at the lateral and apical plasma membranes of many root cells. Interestingly, PIN1 S1 and S4 phosphorylation, after PID overexpression, was not found exclusively at either the apical or basal end of the cell, as had been hypothesized (18, 25), but followed the general distribution pattern of PIN1 (Fig. 3A). Similarly, PIN1 phosphorylation patterns followed the distribution of PIN1 in lines expressing PID under control of an estradiol-inducible system (XVE>>PID:GFP) (15). Whereas PIN1 often lost its initial basal polarity and became apolar after 8 h of PID induction, PIN1 became detectable at the apical plasma membrane only after prolonged inductions (22 h). The distribution of phosphorylated PIN1, as detected with a-PIN1 S1-P or a-PIN1 S4-P, followed again the distribution of PIN1 but was not associated with specific plasma membrane subdomains (Fig. 3 B–D). Thus, PIN1 polar distribution is not associated with specific PIN1 phosphorylation patterns.
Fig. 3.
PIN1 phosphorylation follows PIN1 polarity changes after PID overexpression. (A) Representative confocal images of primary root stele cells after immunostaining of 4-d-old Col-0 wild-type and 35S:PID seedlings with anti-PIN1 (a-PIN1), a-PIN1 S1-P, and a-PIN1 S4-P antibodies. (B–D) Representative confocal images of primary root stele cells after immunostaining of 4-d-old estradiol-inducible XVE>>PID seedlings with anti-GFP (PID) (green) and (B) anti-PIN1 (a-PIN1; magenta), (C) a-PIN1 S1-P (a-S1-P; magenta), and (D) a-PIN1 S4-P (a-S4-P; magenta). Seedlings were mock- or estradiol-treated for 8 and 22 h, respectively. Arrowheads mark strong and weak plasma membrane staining. Overlap of green and magenta signals is indicated by white arrowheads in merged images. Magenta arrowheads indicate the absence of a corresponding green signal from a-GFP (PID) immunostaining. (Scale bar, 5 µm.)
PIN1 Phosphorylation in Cortex and Epidermal Cells Is Independent of PIN1 Polar Localization.
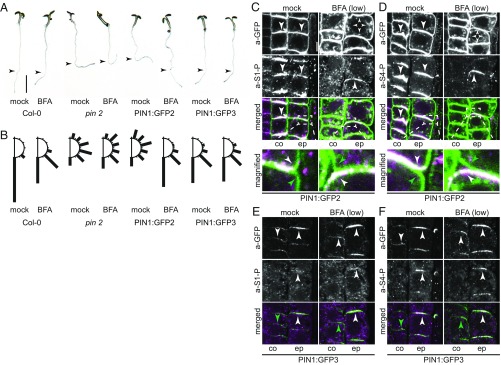
Root epidermis and cortex cells differentially localize PIN2 at the apical and basal plasma membranes, respectively (10). The same differential polar distribution in cortex and epidermis cells is obtained when PIN1, harboring a GFP-tag in the cytoplasmic loop, is expressed from the PIN2 promoter fragment in a pin2 mutant background (PIN2p::PIN1:GFP3) (26). Differential positioning of the GFP-tag in PIN2p::PIN1:GFP2, however, results in the divergent polar targeting, where PIN1:GFP2 is preferentially localized at the basal plasma membrane in both cortex and epidermis cells (26). We examined PIN1:GFP2 and PIN1:GFP3 phosphorylation and detected, with both types of transgenic lines, PIN1:GFP at the respective apical or basal domains with phosphorylations at S1 and S4 (Fig. 4). Thus, PIN1 phosphorylation followed PIN1 distribution, but did not correlate with the presence of PIN1 at either the apical or basal end of the cell, as could be hypothesized (18, 25).
Fig. 4.
PIN1 phosphorylation at the basal and apical plasma membranes is differentially BFA-sensitive. Representative confocal images of root cortex (c) and epidermal (e) cells after immunostaining of 4-d-old PIN1:GFP2 and PIN1:GFP3 seedlings following mock or BFA treatments (50 µM; 10 and 30 min) with anti-GFP (PIN1:GFP2 or PIN1:GFP3; green) and (A and B) a-PIN1 S1-P (a-S1-P; magenta) or (C and D) a-PIN1 S4-P (a-S4-P; magenta). Arrowheads mark plasma membrane staining, asterisks mark intracellular compartments. Overlap of green and magenta signals is indicated by white arrowheads in merged images. Green arrowheads and asterisks indicate the absence of a corresponding magenta signal from a-PIN1 S1-P or a-PIN1 S4-P immunostaining. (Scale bar, 5 µm.)
Whereas PID is apolarly distributed at the plasma membranes of root epidermis and cortex cells, D6PK localizes to the basal plasma membrane in all cell types of the root meristem (Fig. S3) (13, 18). We reasoned that the differential BFA sensitivity of the PIN1 regulatory kinases should have differential effects on PIN1 phosphorylation after BFA treatment. Indeed, PIN1 phosphorylation of the basally localized PIN1:GFP2 in cortex and epidermis cells, as well as PIN1 phosphorylation of basally localized PIN1:GFP3 in the cortex, were strongly BFA-sensitive. Importantly, however, phosphorylation of apical PIN1:GFP3 in the epidermis was strongly insensitive to BFA, suggesting that a BFA-insensitive protein kinase or a BFA-insensitive trafficking machinery promotes PIN1 phosphorylation at the apical side of the cell (Fig. 4 C and D). With regard to the strong BFA sensitivity of PIN1 phosphorylation at the basal plasma membrane, these results correlate well with the known behavior and basal distribution of D6PK. With regard to the BFA-insensitive PIN1 phosphorylation at the apical plasma membrane, PIN1 phosphorylation may be maintained by the BFA-insensitive PID. This would, however, also lead to the question why the apolarly distributed BFA-insensitive PID, after BFA treatment, cannot maintain PIN1 phosphorylation at the basal plasma membrane and may invite the hypothesis that additional BFA-sensitive cofactors function together with PID at this side of the cell.
Differential Effects of BFA on Root Gravitropism Correlate with PIN1 Phosphorylation Patterns and Hypothetical Auxin Transport in the PIN2 Expression Domain.
PIN1:GFP3 displays the same plasma membrane distribution as PIN2 in root cortex and epidermis cells and PIN1:GFP3 can suppress the pin2 gravitropism defect in PIN2p::PIN1:GFP3 (26). PIN1:GFP3 can thus functionally replace PIN2 in these contexts. Because the root gravitropism response is dependent on shoot-directed auxin transport in the root epidermis (10, 11, 27), the rescue of pin2 by PIN1:GFP3 should only occur when the apically localized epidermal PIN1:GFP3 carried auxin transport-activating phosphorylation. Because PIN1:GFP3 phosphorylation at the epidermal apical plasma membrane was not impaired after BFA treatment, we hypothesized that the rescue of the pin2 gravitropism defect by PIN1:GFP3 should be maintained following BFA treatments. We could indeed confirm this hypothesis and show that root gravitropism was maintained in PIN1:GFP3 when seedlings were grown on BFA-containing medium (Fig. 5 A and B).
Fig. 5.
Differential BFA sensitivity of apical and basal PIN1 phosphorylation correlate with BFA-sensitive rescue of the gravitropism defect in pin2 seedlings. (A) Representative photographs of 5-d-old light-grown Arabidopsis seedlings transferred for 18 h to medium with or without BFA (10 µM). The arrowheads point to the position of the root tip at the point of transfer. (Scale bar, 4 mm.) (B) Quantification of root gravitropism in 30° windows from seedlings as shown in A. The sum of seedlings with root angles corresponding to 30° windows left and right from the vertical axis were combined. Cumulative data from three independent experiments are shown (n ≥ 128 seedlings). (C–F) Representative confocal images of root cortex (co) and epidermis (ep) cells after immunostaining of 5-d-old PIN1:GFP2 (C and D) and PIN1:GFP3 (E and F) seedlings following the same treatment as in A with anti-GFP (a-GFP; PIN1:GFP; green), a-PIN1 S1-P (magenta), and a-PIN1 S4-P (magenta). Arrowheads mark strong and weak plasma membrane staining. Overlap of green and magenta signals is indicated by white arrowheads in merged images. Green arrowheads indicate the absence of a corresponding magenta signal from a-PIN1 S1-P or a-PIN1 S4-P immunostaining. In C, immunostained epidermal cells (51 of 79) showed a basal plasma membrane signal following the mock treatment and an apical signal after BFA treatment (92 of 240). In D, immunostained cells showed a basal plasma membrane signal (128 of 140) following the mock treatment and an apical signal after BFA treatment (81 of 177). In all cases, the remaining cells showed either no signal for a-PIN1 S1-P or a-PIN1 S4-P, or the polarity of that signal could not be unequivocally determined, based on the criteria as shown in the magnifications. (Scale bar, 5 µm.)
In pin2 PIN2p::PIN1:GFP2, BFA treatments lead to a dephosphorylation of basally localized PIN1:GFP2 in epidermis and cortex cells. BFA treatments should thus impair root-directed PIN1:GFP2-dependent auxin transport in this line. We reasoned that the inability of PIN1:GFP2 to rescue the gravitropism defect of pin2 should, at least in part, be suppressed by BFA treatments in pin2 PIN2p::PIN1:GFP2. Interestingly, we detected, specifically after BFA treatment, a surprisingly strong rescue of the gravitropism defect in BFA-treated pin2 PIN2p::PIN1:GFP2 seedlings (Fig. 5 A and B). To understand the molecular mechanism underlying this rescue, we analyzed the localization and phosphorylation of PIN1:GFP2 by immunostaining in seedlings grown on BFA-containing medium or a corresponding control medium. We found that this resulted in a depolarization and sometimes apicalization of PIN1:GFP2 from the basal plasma membrane and, in all cells with an immunostaining signal (n = 92, a-PIN1 S1-P; n = 81; a-PIN1 S4-P) also to PIN1:GFP2 phosphorylation at the apical membrane (Fig. 5 C and D). This finding suggested that the BFA-induced rescue of pin2 by PIN1:GFP2 may, at least in part, be because of a reversal of PIN1:GFP2-mediated and BFA-sensitive root-directed auxin transport to BFA-insensitive shoot-directed auxin transport in the epidermis.
PIN1 Is Not Activated by Phosphomimicking Mutations of PIN1 S1–S4.
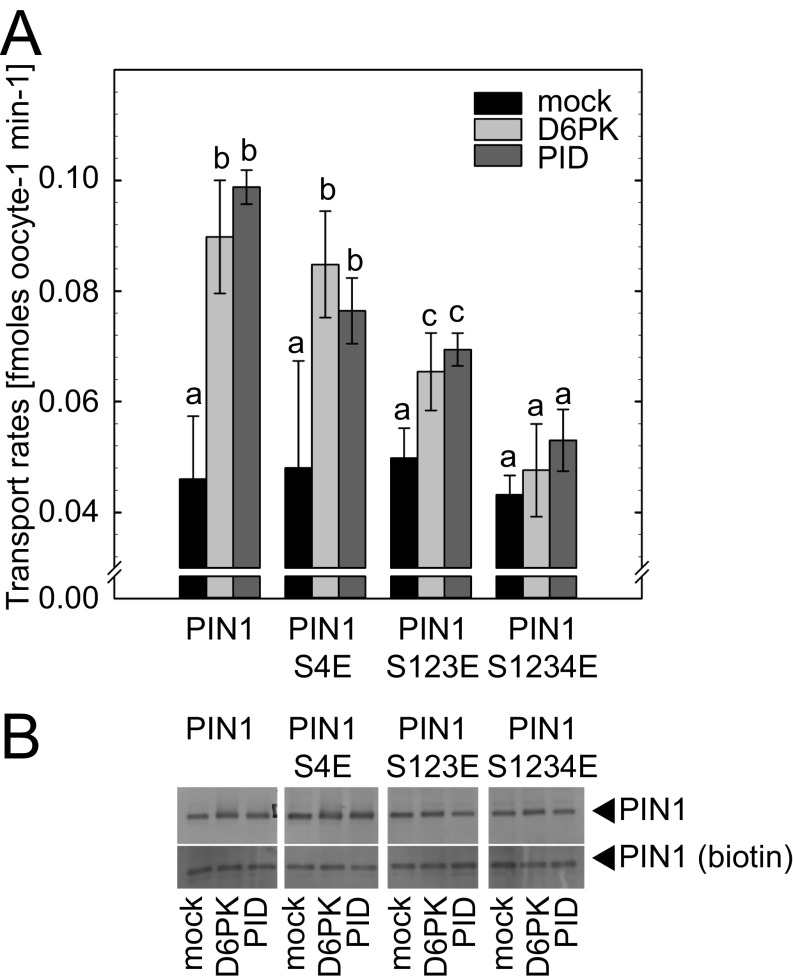
Phosphorylation events at phosphosites may be mimicked by replacing the phosphorylated residues by negatively charged D (aspartic acid) or E (glutamic acid). Phosphorylation-mimicking mutations of PIN1 S1–S3 (PIN1 S123E) were reported to result in PIN1 targeting to the apical plasma membrane (14, 15). Because PIN1 phosphorylation is also required for PIN1 auxin efflux activation, we examined the activity of PIN1 S123E in Xenopus laevis oocyte-based auxin transport assays. Neither PIN1 S123E nor the additionally generated PIN1 S4E and PIN1 S1234E variants showed any kinase-independent auxin efflux activity in this assay (Fig. S8). What is more, the PIN1 S123E and PIN1 S1234E could not be efficiently activated by D6PK or PID, and therefore behaved like loss-of-function rather than gain-of-function variants with regard to auxin transport activity (Fig. S8). Thus, phosphorylation-mimicking mutations that were reported to be sufficient for PIN1 polarity changes in planta are not sufficient for the activation of PIN1-mediated auxin efflux in a heterologous auxin transport assay (14).
Fig. S8.
Glutamic acid exchange mutations of PIN1 phosphosites do not result in protein kinase-independent auxin transport activation. (A) Auxin (indole-3-acetic acid, IAA) transport rates of Xenopus oocytes injected with PIN1 or PIN1 variants where S (serine) phosphosites had been replaced by possibly phosphorylation-mimicking E (glutamic acid) residues, together with protein kinases or a mock control injection as specified in the legend. (B) Immunoblot analysis of Xenopus oocytes injected with PIN1 variants and protein kinases (or mock) as specified. (Upper) Membrane fraction; (Lower) immunoblots of proteins following surface biotinylation demonstrating the presence of equal amounts of PIN1 in the plasma membrane of all oocytes.
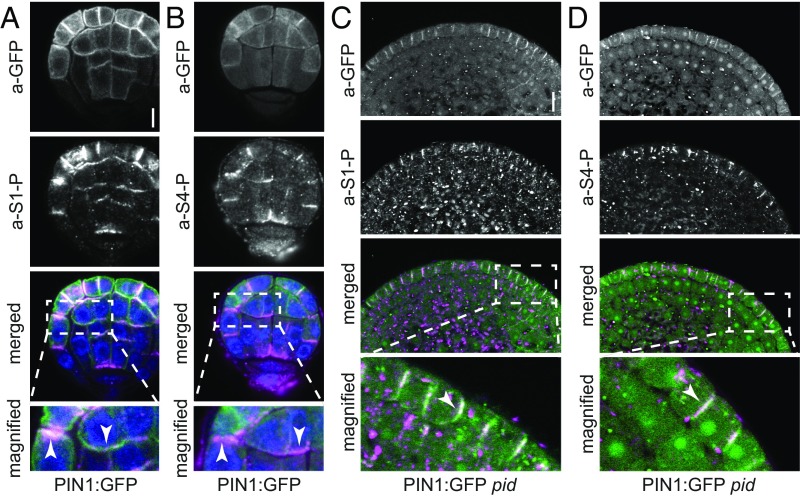
PIN1 Phosphorylation Can Be Detected at the Basal Domains in Arabidopsis Embryos and in Shoot Meristems.
Differential auxin transport and differential PIN polarity are crucial during early plant development (28). Cotyledon-directed PIN1 in the epidermis directs auxin transport toward incipient cotyledon initiation sites, whereas root-directed PIN1 in the inner cells helps to establish a critical auxin maximum that forms above the hypophysis (29). Because the Arabidopsis embryo is amenable to immunostaining and differential PIN1 polarities can be examined in the embryo in a wild-type context, we examined PIN1 phosphorylation in different embryo developmental stages. There, we observed PIN1 S1 and S4 phosphorylation at the basal as well as apical plasma membranes in PIN1:GFP embryos (Fig. 6 A and B). Similar to our observations made in stele cells of the primary root, PIN1 was phosphorylated at S1 and S4 at the basal membrane of embryonic inner cells (Fig. 6 A and B). In each case, PIN1 phosphorylation correlated with PIN1 distribution but not with specific PIN1 localization at the basal or apical plasma membrane. Furthermore, we examined PIN1 phosphorylation in the undifferentiated shoot apical meristems of pid mutants and showed that S1- and S4-phosphorylated PIN1, contrary to the expectation based on published literature (24, 25), accumulated at the basal membrane of epidermal cells (Fig. 6 C and D).
Fig. 6.
PIN1 phosphorylation follows PIN1 distribution in Arabidopsis embryos and pid shoot apical meristems. (A and B) Representative confocal images of Arabidopsis PIN1:GFP embryos stained with a-GFP (PIN1; green) and a-PIN1 S1-P (A; magenta) or a-PIN1 S4-P antibodies (B; magenta). Arrowheads mark the immunostaining at the plasma membrane. Merged images also include DAPI staining of nuclei (blue). (Scale bar, 5 µm.) (C and D) Representative confocal images of nondifferentiated pid PIN1:GFP mutant shoot apical meristems stained with a-GFP (PIN1; green) and a-PIN1 S1-P (C; magenta) or a-PIN1 S4-P antibodies (D; magenta). White arrowheads mark the immunostaining at the plasma membrane in the merged images. (Scale bar, 10 µm.)
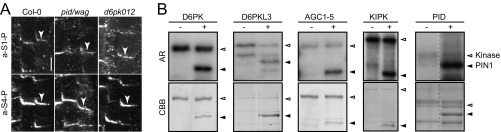
Unknown Kinases Act Redundantly with D6PK and PID in Phosphorylating PIN1 in the Root.
D6PK and PID belong to the family of AGCVIII protein kinases, which is comprised of 23 sequence-related kinases in Arabidopsis (30). D6PK and PID supposedly act redundantly with their closest homologs, D6PK-LIKE1 through D6PK-LIKE3, and PID2, WAG1, and WAG2, respectively (15, 31). When we examined PIN1 phosphorylation in loss-of-function mutants of d6pk d6pkl1 d6pkl2 (d6pk012) and pid pid2 wag1 wag2 (pid/wag), we found that PIN1 phosphorylation at S1 and S4 was maintained, suggesting that other kinases may act redundantly with D6PKs and PID/WAGs in PIN1 phosphorylation, at least in the root stele cells examined here (Fig. S9A). Because many other kinases of the AGCVIII family had not been tested with regard to their phosphorylation activity toward PIN proteins, we performed phosphorylation experiments with previously uncharacterized family members, namely D6PKL3 (AT3G27580) but also AGC1–5 (AT3G12690) and KIPK (AT3G52890), using the PIN1 cytoplasmic loop as a substrate. Each of these protein kinases phosphorylated the PIN1 substrate and therefore qualifies as additional kinase that can phosphorylate PIN1 in the d6pk012 or pid/wag mutants (Fig. S9B).
Fig. S9.
PIN1 phosphorylation is maintained in pid/wag quadruple and d6pk triple mutants. (A) Representative confocal images of primary root stele cells after immunostaining of 4-d-old wild-type seedlings (Col-0) as well as d6kp012 (d6pk d6pkl1 d6pkl2) and pid/wag (pid pid-like2 wag1 wag2) mutant seedlings with a-PIN1 S1-P and a-PIN1 S4-P antibodies. Arrowheads mark the staining at the basal plasma membrane. (Scale bar, 5 µm.) (B) Results from a phosphorylation experiment using recombinant AGCVIII kinases (kinase) as specified and a PIN1 cytoplasmic loop substrate (PIN1). AR, autoradiograph; CBB, Coomassie Brilliant Blue-stained gel as a loading control.
Discussion
PIN1 phosphorylation was previously linked to the control of PIN1 activity and polarity (2, 12, 13, 24, 25, 32, 33). In the present study, we established phosphosite-specific antibodies against PIN1 S1–S4. At least three of these antibodies, directed against PIN1 S1, S2, and S4, enabled us to examine specific PIN1 phosphorylation events in situ in the context of PIN1 activity and polarity control.
PIN1 phosphorylation at PIN1 S1–S3 by the BFA-insensitive PID kinase has been proposed to control PIN1 polarity (15, 33). Because phosphorylated PIN1 should be targeted to the apical plasma membrane in root cells, it would be expected that PIN1 S1–S3 phosphorylation is detected at the apical but not at the basal plasma membrane. In contrast, we detected PIN1 phosphorylation at S1–S3 at the basal and, after PID overexpression, also at the apical plasma membrane of root stele cells. Thereby, PIN1 phosphorylation followed the general distribution pattern of PIN1 and was not associated with the plasma membrane at a specific side of the cell. This finding was also true in lines expressing differentially GFP-tagged PIN1 in the PIN2 expression domain, where PIN1 phosphorylation could be detected at the basal and the apical plasma membrane in root epidermis or cortex cells (26). In addition, in the complex tissue of a differentiating Arabidopsis embryo, we observed PIN1 phosphorylation at the apical and basal plasma membrane of embryonic cells and no correlation between PIN1 phosphorylation and specific sides of the cell. In epidermal cells of pid mutant shoot apical meristems where, according to previous models (24, 25), PIN1 phosphorylation should lead to PIN1 apicalization, we detected PIN1 phosphorylation also at the basal plasma membrane. In addition, we also obtained no evidence that PIN1 might be phosphorylated during intracellular transport, for example when it accumulated in BFA compartments after BFA treatment. There is thus no obvious correlation between PIN1 polar distribution and PIN1 phosphorylation at the phosphosites examined.
Previous studies had supported the model explaining PIN1 polarity changes by PIN1 S1–S3 phosphorylation by examining PIN1 variants, in which S1–S3 phosphorylation was prevented through mutations to alanines (S123A) or mimicked through mutations to glutamic acid (S123E). There, PIN1 S123A and PIN1 S123E were detected at the basal and apical plasma membranes of root cells, respectively (33). In line with the expectation that dynamic PIN1 trafficking to different polar sides of the cell, and consequently differential and dynamic auxin distribution, was required for the implementation of complex developmental processes, neither of these mutant variants could rescue pin1 mutant phenotypes (14). We had previously shown that PIN1 S123A variants were, as expected, impaired in kinase-dependent PIN1 activation and auxin efflux when tested in Xenopus oocytes (12). Interestingly, we now found that PIN1 S123E variants were functionally impaired in their ability to promote auxin export from oocytes when coexpressed with D6PK or PID. In contrast to the expectation that the glutamic acid replacement mutations would mimic PIN1 phosphorylation and—as we would assume—activation, neither PIN1 S123E nor the more complex PIN1 S1234E variant exported auxin from oocytes in a protein kinase-independent manner. Thus, at least with regard to auxin transport activation and the responsiveness to protein kinases, the phosphomimicking variants behaved like variants with reduced PIN1 function. Of course, this observation cannot be easily related to the mechanisms controlling PIN1 polarity because it cannot be excluded that the PIN1 trafficking machinery may recognize the phosphomimicking variants as a result of distinct affinity requirements for phosphorylated PIN1.
We had previously argued that the phosphosite preferences of the two kinases may be the relevant distinguishing feature that could explain the differential effects of PID and D6PK on PIN1 polarity control (2, 12, 13, 15). Previous work had established that PID and D6PK phosphorylate PIN1 S1–S4 but that PID preferentially phosphorylated S1–S3, whereas D6PK preferentially phosphorylated S4 (2, 12, 13, 15). However, we obtained no evidence for a differential phosphorylation at the PIN1 phosphosites in any experimental setting. Rather, all four PIN1 phosphorylations, or at least PIN1 S1 and S4 phosphorylation, coincided in all cases tested. Taken together, these findings suggested that the phosphorylation of all four serines may be mediated by the same protein kinases. We thus propose that the differential phosphorylation at these sites, as detected in vitro, may not be biologically relevant in vivo. Hence, differential phosphosite preference may not be suitable to explain the differences between D6PK and PID in PIN1 polarity control. The fact that differential antibody affinities preclude stringent quantitative comparisons of phosphorylation levels between the PIN1 phosphosites in immunostaining experiments is a limiting technical factor in this analysis.
Our observations thus show that the prevalent model explaining PIN1 polarity by PID-dependent phosphorylation can no longer hold true. As previously reported, and confirmed here with transgenic lines generated independently for our study, PIN1 polarity is clearly altered after PID overexpression. However, phosphorylation of the preferential PID phosphorylation site PIN1 S1 was not specifically associated with the respective polarity change. Therefore, as yet unknown factors must control PIN1 polarity together with PID. These could be PID-specific protein interactors that may not be able to bind or act in concert with D6PK. Although PID and D6PK belong to the same class of AGCVIII protein kinases, D6PK and PID have substantial differences that would allow for such regulatory protein interactions to take place (e.g., in their N and C termini, as well as in an insertion domain that resides within the kinase domains of both proteins) (30). PID has already been reported to engage in interactions with proteins other than PINs and these may play an additional role in controlling PID activity (34). Based on our experiments with an inducible PID expression line, we also argue that the respective mechanism does not have the fast dynamics that could be expected for a phosphorylation-controlled PIN1 repolarization process. Instead, and in line with previous reports, we detected in our experiments a clear change in PIN1 polarity only after an extended induction period of 22 h (24).
PIN1 phosphorylation controls PIN1 auxin efflux activity (12). Here, we report on several settings where predictions on auxin transport activity and consequently auxin distribution could be made based on PIN1 phosphorylation. We observed in root stele cells that the interference with PIN1 phosphorylation resulted in the apparent accumulation of auxin within cells, as judged with the auxin-sensitive DII-VENUS marker. We could further explain the suppression of the gravitropism defect of a pin2 mutant expressing PIN1:GFP2 after BFA treatment, not only by the effects of BFA on BFA-sensitive kinases at the basal plasma membrane, but also by the effects of activated phosphorylated PIN1 at the apical plasma membrane, where PIN1 phosphorylation is maintained after BFA treatment by apparently BFA-insensitive protein kinases or trafficking machineries. Therefore, the availability of phosphosite-specific PIN1 antibodies will, in the future, allow understanding PIN1-mediated auxin efflux in a given cell or tissue context based on PIN1 phosphorylation rather than based solely on PIN1 abundance.
Genetic studies indicate that D6PK acts in a functionally redundant manner with at least three other sequence-related D6PK-LIKE genes (12, 16, 17). Based on genetic and cell biological studies, functional redundancy was also proposed between PID, WAG1, and WAG2, and possibly with PID2, which are all members of a distinct subclade within the Arabidopsis AGCVIII protein kinase family (30, 31). We found that PIN1 phosphorylation was not detectably impaired in complex mutants of the D6PK and PID-related kinases, suggesting that other protein kinases (e.g., other protein kinases of the AGCVIII family) also regulate PIN phosphorylation in addition to D6PKs and PID/WAGs. This hypothesis finds indirect support in the observation that root growth and gravitropism are not as severely impaired in the complex d6pk or pid/wag mutants as may be expected from mutants with severely impaired PIN and auxin transport function (10, 17, 20, 31, 35). Several of the phosphorylation events observed in our experiments support the notion that these, as yet, unknown PIN1-regulatory protein kinases are related—in their cell biological behavior—to D6PK and PID. D6PK is a strongly BFA-sensitive protein kinase and PIN1 phosphorylation at the basal plasma membrane of root cells was very sensitive to BFA treatments. In contrast, PID is a BFA-insensitive kinase and may maintain PIN1 phosphorylation at the apical plasma membrane where PIN1 phosphorylation was not responsive to BFA treatments. Solely based on structural and sequence criteria, several hitherto uncharacterized members of the AGCVIII protein kinase family could qualify as candidate PIN regulators (30). We showed that uncharacterized AGCVIII kinases can phosphorylate PIN1 in vitro and these may control PINs by phosphorylation in vivo (e.g., in the d6pk012 or pid/wag backgrounds examined here). In other recently published work, we provide evidence that these kinases also localize to the plasma membrane (36), providing further support for the hypothesis that also these kinases may regulate activities at the plasma membrane. We also noted with interest that the BFA-insensitive PID was seemingly not sufficient to maintain PIN1 phosphorylation at the basal plasma membrane after BFA treatment, even though it is apolarly distributed in these cells and expected to function at the basal plasma domain. Thus, phosphorylation control of PIN1 activity and polarity by PID must be more complex than previously anticipated. Understanding the transcriptional and posttranslational mechanisms controlling the expression and abundance of the different functionally related protein kinases will therefore be an important topic for future research aiming at mapping the complexities of PIN regulation. Phosphosite-sepcific antibodies, as established here for PIN1, will then be an important tool to monitor the activity and effects of these phosphorylation events.
Materials and Methods
Biological Material.
All A. thaliana lines used in this study are in the Columbia ecotype. The following lines have been described previously: pin1 (SALK_047613) PIN1p::PIN1:GFP (PIN1:GFP) and pin1 (SALK_047613) PIN1p::PIN1:YFP (PIN1:YFP) (37); pin2 PIN2p::PIN1:GFP2 and pin2 PIN2p::PIN1:GFP3 (26); pin1 PIN1p::PIN1:GFP S1A (14); estradiol-inducible G10-90::XVE>>PID (15); BFA-sensitive GNOMwt and BFA-insensitive GNOMM696L (19); DII-VENUS and mDII-VENUS (38); d6pk d6pkl1 d6pkl2 (d6pk012) and 35S::YFP:D6PK (17); and pid/PID pid2 wag1 wag2 (31). pid pid2 wag1 wag2 quadruple mutants were isolated from the progreny of pid/PID pid2 wag1 wag2 based on the cotyledon formation-deficient phenotype specific for the quadruple mutant (31). The pin1 (SALK_047613) mutant backgrounds were genotyped for homozygosity using primers as listed in Table S1.
Table S1.
Primers used in this study
| Name | Purpose | Primer sequence (5′–3′) |
| PIN1 S2A | Mutagenesis of S252 (S2) to A252 (S2A) in PIN1:YFP | phos-CCACGTGGCGCTAGTTTTAATCATAC |
| PIN1 S3A | Mutagenesis of S290 (S3) to A290 (S3A) in PIN1:YFP | phos-ACTCCGAGACCTGCCAACTACG |
| PIN1 S1E | Mutagenesis of S231 (S1) to E231 (S1E) in p002:PIN1 | phos-ACACCTAGACCTGAAAATCTAAC |
| PIN1 S2E | Mutagenesis of S252 (S2) to E252 (S2E) in p002:PIN1 | phos-GCCACGTGGCGAAAGTTTTAATCATAC |
| PIN1 S3E | Mutagenesis of S290 (S3) to E290 (S3E) in p002:PIN1 | phos-ACTCCGAGACCTGAAAACTACG |
| PIN1 S4E | Mutagenesis of S271 (S4) to E271 (S4E) in p002:PIN1 | phos-TGGTCGGAACGAAAACTTTGGTC |
| GW-PID-FW | Amplification and cloning of PID CDS in pGW-35S-MYC and pExtag-YFP-GW | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGTTACGAGAATCAGACGGT |
| GW-PID-RV1 | Amplification and cloning of PID CDS in pGW-35S-MYC and pExtag-YFP-GW | GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAAAAGTAATCGAACGCCGCTG |
| pin1 LP | pin1 (SALK_047613) mutant genotyping: pin1 LP | CAAAAACACCCCCAAAATTTCTC |
| pin1 RP | pin1 (SALK_047613) mutant genotyping: pin1 RP | AATCATCACAGCCACTGATCC |
| LBb1.3 | pin1 (SALK_047613) mutant genotyping: LBb1.3 | ATTTTGCCGATTTCGGAAC |
| AGC1-5 FW | Amplification and cloning of AGC1-5 | GGGGACAAGTTTGTACAAAAAAAGCAGGTTAATGGACTTAGCTTCTAAGAAGAACAC |
| AGC1-5 RV | Amplification and cloning of AGC1-5 | GGGGACCACTTTGTACAAGAAAGCTGGGTACTAAAAGTACTCGAAATCTATATACGCC |
| KIPK FW | Amplification and cloning of KIPK | GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGGGGTCGTTTGCAGG |
| KIPK RV | Amplification and cloning of KIPK | GGGGACCACTTTGTACAAGAAAGCTGGGTATTAAAACAACTCGAACTCAAGATGATC |
| D6PKL3 | Amplification and cloning of D6PKL3 | GGGGACCACTTTGTACAAGAAAGCTGGGTCCTGATAGTGAAAGAAATCACAAATCAG |
| D6PKL3 | Amplification and cloning of D6PKL3 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGGATTCTTCTTCATCAGT |
phos, phosphorylation.
Molecular Cloning.
PIN1p::PIN1:YFP (PIN1:YFP) is a previously described plant transformation construct for the expression of a functional YFP-tagged PIN1 under control of the PIN1 promoter fragment (12). Mutagenesis of the PIN1 S2 and S3 phosphosites to alanine yielded PIN1:YFP S2A and S3A, which were introduced in PIN1:YFP as previously described for PIN1:YFP S4A (12). The constructs were transformed into heterozygous PIN1/pin1 (SALK_047613) plants by Agrobacterium-mediated transformation and pin1 homozygous lines carrying the PIN1:YFP transgenes were isolated from the progeny and used for immunostaining (39). PIN1:GFP S1A was previously described (33).
For auxin transport assays in X. laevis oocytes, PIN1 S (serine) phosphosites were mutagenized in p002:PIN1 to potentially phosphorylation-mimicking E (glutamic acid) through several rounds of PCR mutagenesis with primers PIN1 S1E, PIN1 S2E, PIN1 S3E, and PIN1 S4E (12).
The PID overexpression construct 35S:PID was obtained by PCR amplification of the PID coding sequence with primers PID-GW-FW and PID-GW-RV. The resulting fragment was inserted into pDONR201 (LifeTechnologies) and from there into pGW-35S-MYC (a gift from Jane Parker, Max Planck Institute for Plant Breeding Research, Cologne, Germany). A stop codon in PID-GW-RV prevents the in-frame fusion with the C-terminal MYC-tag of this vector. 35S:PID was introduced in the Col-0 wild-type background using the floral dip method (39).
A construct for the expression of YFP-tagged PID (YFP:PID) in Arabidopsis was obtained by PCR amplification of the PID coding sequence, insertion of the PCR fragment into pDONR201 followed by the transfer into pExtag-YFP-GW (a gift from Jane Parker, Max Planck Institute for Plant Breeding Research, Cologne, Germany). The transgenic construct was introduced into the Col-0 wild-type using the floral-dip method (39).
cDNAs of protein kinases were amplified using forward and reverse primers for the respective gene and amplification products were transferred, via pDONR201, into pDEST15 using the Gateway cloning system (Invitrogen). Expression constructs for D6PK and PID were previously described (12).
All primer sequences are listed in Table S1.
Phosphosite-Specific Antibodies.
Phosphosite-specific antibodies were generated by Eurogentec using the following peptides for immunization and affinity purification of rabbit sera: PIN1 S1 N′-LSATPRP-S(P)-NLTNA-C′, PIN1 S2 N′-RNPTPRG-S(P)-SFNHT-C′, PIN1 S3 N′-GPTPRP-S(P)-NYEEDG-C′, and PIN1 S4 N′-SGGGRN-S(P)-NFGPGE-C′.
SI Materials and Methods
Dot Blot Analysis.
For dot blot analysis, 20 µg of each peptide used for immunization or its nonphosphorylated counterpart were spotted onto PVDF membranes. Membranes were then blocked with PBS/2% (wt/vol) BSA and incubated with a-PIN1 S1-P through a-PIN1 S4-P (1:500). Anti-rabbit HRP (1:100,000; Sigma-Aldrich) was used as a secondary antibody.
Immunohistochemistry.
For whole-mount immunostaining of primary root meristems, seedlings were grown in continuous light on standard growth medium (4.2 g/L Murashige and Skoog salts, 1% sucrose, 0.5 g/L 2-[N-morpholino-]ethanesulfonic acid, 5.5 g/L agar, pH 5.8) for 4–5 d. Where applicable, seedlings were treated in liquid growth medium supplemented with 50 µM BFA (ThermoFisher), 25 µM cantharidin (Sigma-Aldrich), 10 µM β-estradiol (Sigma-Aldrich), or a DMSO mock solvent control (40). Seedlings were fixed for 1 h at room temperature under vacuum in 4% (vol/vol) paraformaldehyde in PBS at pH 7.4. Cell walls were partially digested for 30 min at 37 °C with 2% (wt/vol) Driselase (Sigma). Plasma membranes were permeabilized for 1 h at room temperature with 3% (vol/vol) Nonidet P-40 (AppliChem) in 10% (vol/vol) DMSO/PBS. The samples were blocked for 1 h at room temperature with 4% (wt/vol) BSA in PBS. Primary and secondary antibodies were diluted in blocking solution and incubated for 4 h at 37 °C. All washes were performed with 0.1% Triton X-100 (AppliChem) in PBS. For pid1 immunostainings, plants were cultivated under sterile conditions on 1/2 MS medium without sugar for 3 wk and undifferentiated shoot apical meristems were dissected and immunostained as described above with minor modifications: fixation was performed for 3 h, cell wall digestion was extended to 60 min, antibody incubation times were extended to 6 h for the primary antibodies and 6 h for the secondary antibodies. The following primary and secondary antibodies were used: mouse anti-GFP (1:400; Roche), rabbit anti-PIN1 (1:1,000) (41), goat anti-PIN1 (1:400; Nottingham Arabidopsis Stock Centre), rabbit anti-PIN1 S1-P (1:100), rabbit anti-PIN1 S2-P (1:100), rabbit anti-PIN1 S3-P (1:100), rabbit anti-PIN1 S4-P (1:300), ALEXA488-conjugated anti-mouse (1:500; ThermoFisher), Cy3-conjugated goat anti-rabbit (1:500; Dianova), and FITC-conjugated rabbit anti-goat (1:100; Dianova). Samples were mounted in mounting solution [90% (vol/vol) glycerol, 10% (vol/vol) PBS, 25 mg/mL 1,4-diazabicyclo[2.2.2]octane, pH 9.0], and images were acquired with an Olympus BX61 microscope equipped with a FV1000 confocal laser scanning unit (Olympus). Images were processed only for brightness and contrast adjustments using Adobe Photoshop software. PIN1 distribution and PIN1 polarity were determined using lateral signal distribution as a secondary parameter or obvious cell morphologies.
For whole-mount immunostaining of embryos, ovules were fixed for 1 h at room temperature under vacuum in 4% (wt/vol) paraformaldehyde in microtubule-stabilizing buffer (50 mM Pipes, 5 mM EGTA, 5 mM MgSO4, pH 6.9–7.0). Embryos and endosperm were dissected from washed ovules and squashed onto gelatin-coated microscopic slides. Coverslips were removed after dipping the slides into liquid nitrogen. After drying for at least 15 min, specimens were rehydrated in 1× PBS for 10 min and cell walls were partially digested for 40 min with 2% (wt/vol) Driselase (Sigma). The plasma membrane was permeabilized for 1 h with 3% (vol/vol) Nonidet P-40 in 10% (vol/vol) DMSO/PBS. Unspecific interactions were blocked with PBS/3% (wt/vol) BSA at room temperature. Antibodies were diluted in PBS/3% (wt/vol) BSA and incubated for 3 h at 37 °C or overnight at 4 °C. Nuclear DNA was stained with 1 μg/mL DAPI (Sigma). Mounting was done in Citifluor.
All quantitative fluorescence intensity measurements were performed in Fiji (ImageJ, NIH). Plasma membrane signal intensities were measured by drawing a line across the plasma membrane of individual cells. An additional line measurement for every root was performed outside the PIN1 expression domain and used for background correction. The total root stele signal intensities were measured from areas as indicated by the framed area shown in Fig. S7. Signal intensities from a corresponding area outside the PIN1 expression domain were also measured for each root and used for background correction. For statistical evaluation, one-way ANOVA or Student's t tests were performed in Microsoft Excel and post hoc Tukey HSD analysis using an online calculator (astatsa.com/).
Live Imaging.
For live imaging of DII-VENUS, seedlings were grown on 1/2 MS growth medium without sucrose in continuous light at 18 °C for 4 d. Treatments were performed in liquid medium at 18 °C as described for immunohistochemistry. Image acquisition and quantification of fluorescence signal intensities were performed as previously described (13). Quantitative fluorescence intensity measurements were performed as described for immunohistochemistry from areas specified in the figure.
Protein Extraction and Immunoblotting.
For anti-GFP immunoblots, 5-d-old seedlings were treated as described for immunohistochemistry. Total protein extracts were prepared from roots in extraction buffer [50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 0.1 mM MG132, 1 mM PMSF, protease inhibitor mixture (Sigma)] and PhosSTOP phosphatase inhibitor mixture (Roche). Total protein content was measured with Bradford reagent and 10 µg protein were resuspended in 5× Laemmli buffer and heated to 45 °C for 15 min before loading onto gels for immunoblotting with rabbit anti-GFP (1:3,000; Life Technologies) and anti-rabbit HRP (1:100,000; Sigma). Membranes were stained with Ponceau Rouge to control for equal loading and transfer. Signal quantification was performed using Fiji (ImageJ, NIH) by drawing a box around the main band as well as the phosphorylation smear.
Protein extraction from Xenopus laevis oocytes was performed as previously described (12).
Physiological Experiments.
For gravitropic root growth assays, 4-d-old seedlings grown vertically on standard growth medium without sucrose were transferred to plates containing 10 µM BFA or the equivalent amount of solvent (DMSO) and the position of the root tip was marked. Plates were then transferred to the dark and root angles were determined from scanned images after 18 h growth using Fiji (ImageJ, NIH). Root angles were assessed by measuring the angle between the point of transfer and final position of the root tip.
Auxin Transport Experiment.
Auxin transport experiments in X. laevis oocytes and the corresponding protein extractions, membrane fractionations, and immunoblotting were conducted as previously described (12). Protein extraction and surface biotinylation, followed by immunoblotting, were carried out as described previously (42).
Protein Kinase Experiments.
Protein phosphorylation experiments using recombinant purified proteins were performed as previously described (12).
Acknowledgments
The authors thank Jutta Elgner for excellent technical assistance; Jiri Friml (Institute of Science and Technology Austria, Klosterneuburg, Austria), Remko Offringa (University of Leiden, Leiden, The Netherlands), Teva Vernoux (Ecole normale supérieur de Lyon, Lyon, France), and Tatsuya Sakai (Niigata University, Niigata, Japan) for mutant or transgenic seed lines and constructs; and Hannelore Daniel (Technical University of Munich, Freising, Germany) for the generous and uncomplicated supply of Xenopus oocytes. Financial support for this work came from Deutsche Forschungsgemeinschaft Grants SCHW751/12-1 (to C.S.), SFB1101 (to G.J.), and SFB924 and HA3468/6-1 (to U.Z.H.); as well as a Fundação para a Ciência e a Tecnologia Fellowship SFRH/BD/73187/2010 (to I.C.R.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614380114/-/DCSupplemental.
References
- 1.Teale WD, Paponov IA, Palme K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol. 2006;7(11):847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- 2.Barbosa IC, Schwechheimer C. Dynamic control of auxin transport-dependent growth by AGCVIII protein kinases. Curr Opin Plant Biol. 2014;22:108–115. doi: 10.1016/j.pbi.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Gälweiler L, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282(5397):2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- 4.Friml J, et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell. 2002;108(5):661–673. doi: 10.1016/s0092-8674(02)00656-6. [DOI] [PubMed] [Google Scholar]
- 5.Noh B, Bandyopadhyay A, Peer WA, Spalding EP, Murphy AS. Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature. 2003;423(6943):999–1002. doi: 10.1038/nature01716. [DOI] [PubMed] [Google Scholar]
- 6.Geisler M, et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005;44(2):179–194. doi: 10.1111/j.1365-313X.2005.02519.x. [DOI] [PubMed] [Google Scholar]
- 7.Bainbridge K, et al. Auxin influx carriers stabilize phyllotactic patterning. Genes Dev. 2008;22(6):810–823. doi: 10.1101/gad.462608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Péret B, et al. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell. 2012;24(7):2874–2885. doi: 10.1105/tpc.112.097766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krecek P, et al. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009;10(12):249. doi: 10.1186/gb-2009-10-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller A, et al. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 1998;17(23):6903–6911. doi: 10.1093/emboj/17.23.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahman A, et al. Gravitropism of Arabidopsis thaliana roots requires the polarization of PIN2 toward the root tip in meristematic cortical cells. Plant Cell. 2010;22(6):1762–1776. doi: 10.1105/tpc.110.075317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zourelidou M, et al. Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife. 2014 doi: 10.7554/eLife.02860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbosa IC, Zourelidou M, Willige BC, Weller B, Schwechheimer C. D6 PROTEIN KINASE activates auxin transport-dependent growth and PIN-FORMED phosphorylation at the plasma membrane. Dev Cell. 2014;29(6):674–685. doi: 10.1016/j.devcel.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Huang F, et al. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell. 2010;22(4):1129–1142. doi: 10.1105/tpc.109.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhonukshe P, et al. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development. 2010;137(19):3245–3255. doi: 10.1242/dev.052456. [DOI] [PubMed] [Google Scholar]
- 16.Willige BC, et al. D6PK AGCVIII kinases are required for auxin transport and phototropic hypocotyl bending in Arabidopsis. Plant Cell. 2013;25(5):1674–1688. doi: 10.1105/tpc.113.111484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zourelidou M, et al. The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development. 2009;136(4):627–636. doi: 10.1242/dev.028365. [DOI] [PubMed] [Google Scholar]
- 18.Kleine-Vehn J, et al. PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell. 2009;21(12):3839–3849. doi: 10.1105/tpc.109.071639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geldner N, et al. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112(2):219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 20.Sukumar P, Edwards KS, Rahman A, Delong A, Muday GK. PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiol. 2009;150(2):722–735. doi: 10.1104/pp.108.131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin H, et al. Complex regulation of Arabidopsis AGR1/PIN2-mediated root gravitropic response and basipetal auxin transport by cantharidin-sensitive protein phosphatases. Plant J. 2005;42(2):188–200. doi: 10.1111/j.1365-313X.2005.02369.x. [DOI] [PubMed] [Google Scholar]
- 22.Rashotte AM, DeLong A, Muday GK. Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell. 2001;13(7):1683–1697. doi: 10.1105/TPC.010158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vernoux T, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friml J, et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science. 2004;306(5697):862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- 25.Michniewicz M, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130(6):1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 26.Wisniewska J, et al. Polar PIN localization directs auxin flow in plants. Science. 2006;312(5775):883. doi: 10.1126/science.1121356. [DOI] [PubMed] [Google Scholar]
- 27.Abas L, et al. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol. 2006;8(3):249–256. doi: 10.1038/ncb1369. [DOI] [PubMed] [Google Scholar]
- 28.Lau S, Slane D, Herud O, Kong J, Jürgens G. Early embryogenesis in flowering plants: Setting up the basic body pattern. Annu Rev Plant Biol. 2012;63:483–506. doi: 10.1146/annurev-arplant-042811-105507. [DOI] [PubMed] [Google Scholar]
- 29.Steinmann T, et al. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;286(5438):316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- 30.Galván-Ampudia CS, Offringa R. Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci. 2007;12(12):541–547. doi: 10.1016/j.tplants.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Haga K, Hayashi K, Sakai T. PINOID AGC kinases are necessary for phytochrome-mediated enhancement of hypocotyl phototropism in Arabidopsis. Plant Physiol. 2014;166(3):1535–1545. doi: 10.1104/pp.114.244434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Nodzynski T, Pencík A, Rolcík J, Friml J. PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc Natl Acad Sci USA. 2010;107(2):918–922. doi: 10.1073/pnas.0909460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Offringa R, Huang F. Phosphorylation-dependent trafficking of plasma membrane proteins in animal and plant cells. J Integr Plant Biol. 2013;55(9):789–808. doi: 10.1111/jipb.12096. [DOI] [PubMed] [Google Scholar]
- 34.Benjamins R, Ampudia CS, Hooykaas PJ, Offringa R. PINOID-mediated signaling involves calcium-binding proteins. Plant Physiol. 2003;132(3):1623–1630. doi: 10.1104/pp.103.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blilou I, et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433(7021):39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 36.Barbosa IC, et al. Phospholipid composition and a polybasic motif determine D6 PROTEIN KINASE polar association with the plasma membrane and tropic responses. Development. 2016;143(24):4687–4700. doi: 10.1242/dev.137117. [DOI] [PubMed] [Google Scholar]
- 37.Benková E, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115(5):591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 38.Brunoud G, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482(7383):103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 39.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 40.Sauer M, Friml J. Immunolocalization of proteins in plants. Methods Mol Biol. 2010;655:253–263. doi: 10.1007/978-1-60761-765-5_17. [DOI] [PubMed] [Google Scholar]
- 41.Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413(6854):425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- 42.Bröer S. Xenopus laevis oocytes. Methods Mol Biol. 2010;637:295–310. doi: 10.1007/978-1-60761-700-6_16. [DOI] [PubMed] [Google Scholar]