Significance
The importance of arthropod microbiota in the capacity of pathogens (including malaria and flaviviruses, among others) to persist in vectors and cause infection is just beginning to be appreciated. The influence of pathogens, either directly or indirectly, to manipulate vector microbiota for their own benefit, has not been described. In this study, we demonstrate that a pathogen can use an arthropod molecule to alter vector microbiota and enhance infection. We believe that this work will help others consider that pathogens are not passive microbes when they enter the arthropod vector but actively influence vector gene expression that can manipulate the local environment (in this case the microbiota) and facilitate pathogen infection of the vector.
Keywords: Anaplasma, Ixodes scapularis, antifreeze protein, biofilm, microbiome
Abstract
Arthropods transmit diverse infectious agents; however, the ways microbes influence their vector to enhance colonization are poorly understood. Ixodes scapularis ticks harbor numerous human pathogens, including Anaplasma phagocytophilum, the agent of human granulocytic anaplasmosis. We now demonstrate that A. phagocytophilum modifies the I. scapularis microbiota to more efficiently infect the tick. A. phagocytophilum induces ticks to express Ixodes scapularis antifreeze glycoprotein (iafgp), which encodes a protein with several properties, including the ability to alter bacterial biofilm formation. IAFGP thereby perturbs the tick gut microbiota, which influences the integrity of the peritrophic matrix and gut barrier—critical obstacles for Anaplasma colonization. Mechanistically, IAFGP binds the terminal d-alanine residue of the pentapeptide chain of bacterial peptidoglycan, resulting in altered permeability and the capacity of bacteria to form biofilms. These data elucidate the molecular mechanisms by which a human pathogen appropriates an arthropod antibacterial protein to alter the gut microbiota and more effectively colonize the vector.
Arthropods serve as vectors of numerous pathogens that cause important human and animal diseases. Many of these arthropods are blood feeders and transmit or acquire microorganisms during engorgement (1, 2). Arthropods harbor a diverse group of native microbes ranging from bacteria to fungi, and the microbiota play critical functions in vector physiology, nutrition, and digestion (3–8). Microbial communities within vectors have important roles in the ability of specific pathogens to colonize and persist within the arthropod, and also to be efficiently transmitted to the mammalian host (9–11). In arthropods, the microbiota is associated with a biofilm, generated from diverse bacterial species. These microbial biofilms are necessary for establishing successful symbiotic relationships with their arthropod hosts (12–14).
Ticks are obligate blood-sucking ectoparasites that host a variety of disease-causing pathogens, including Borrelia, Rickettsia, Anaplasma, Ehrlichia, Francisella, and Babesia (15). The diversity of microbial communities has been characterized in different ticks (16–21). The effect of tick microbiota on the transmission of human infectious diseases, and the molecular mechanisms underlying these processes, has only recently received attention. We have demonstrated the importance of the normal gut microbiota of the blacklegged tick, Ixodes scapularis, a major vector of the Lyme disease agent, Borrelia burgdorferi, in the effective colonization of the tick by the spirochete (10). Understanding the interactions between arthropod gut microbiota and the pathogens that ticks transmit, in both a positive and negative manner, may provide new strategies for controlling and preventing vector-borne diseases.
Anaplasma phagocytophilum is an obligate intracellular pathogen transmitted by I. scapularis. This pathogen causes human granulocytic anaplasmosis, an illness that can result in fever, headache, muscle pains, and pancytopenia. In the northeastern United States, A. phagocytophilum is primarily maintained within I. scapularis and Peromyscus leucopus, the white-footed mouse (22). Ixodes ticks acquire Anaplasma during the blood meal. Upon entering the tick, the bacterium rapidly migrates from the gut and establishes itself in the salivary glands, where it is transstadially maintained through different stages of the arthropod life cycle (23, 24). The molecular mechanisms that Anaplasma uses to colonize and penetrate the tick gut barrier are not known. We recently demonstrated that the I. scapularis antifreeze glycoprotein (IAFGP) inhibits bacterial biofilms (25). In this study, we delineate how A. phagocytophilum appropriates IAFGP to alter the tick gut microbiota by inhibiting biofilm development, thereby enabling this pathogen to efficiently colonize the tick vector.
Results
Tick Microbiota Are Involved in A. phagocytophilum Colonization of the Gut.
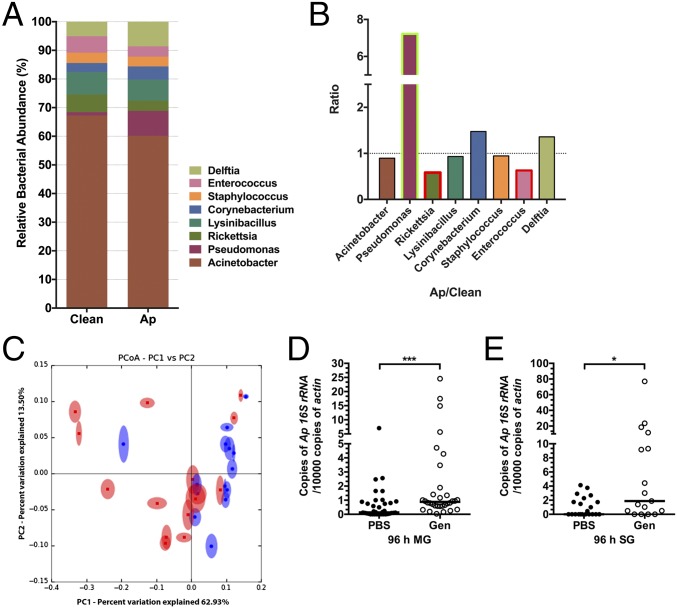
We first examined whether A. phagocytophilum modulates the tick microbiota. Illumina sequencing of amplified 16S rDNA assessed the diversity of bacterial species in A. phagocytophilum-infected and uninfected nymphs. At the taxonomic rank of order, distinct differences were noted in the relative abundance of bacteria between uninfected and A. phagocytophilum-infected nymphal guts (SI Appendix, Fig. S1 A and B). Post analysis and screening (see SI Appendix, SI Materials and Methods and Tables S1 and S2) we observed eight bacterial genera including Acinetobacter, Pseudomonas, Rickettsia, Lysinibacillus, Cornybacterium, Staphylococcus, Enterococcus, and Delftia that were identified in both experimental groups, but at different relative levels of abundance (Fig. 1A). Enterococcus and Rickettsia were decreased in A. phagocytophilum-infected ticks, whereas Pseudomonas was increased in A. phagocytophilum-infected ticks, in comparison with uninfected ticks (Fig. 1B). Principal coordinate analysis of the distance of microbial communities showed a clear separation between uninfected and A. phagocytophilum-infected nymphs (Fig. 1C). These results show that A. phagocytophilum alters the relative composition of the tick microbiota when it infects the tick gut. To examine the role of tick microbiota in A. phagocytophilum colonization, clean nymphal ticks were fed on gentamicin-treated mice that had been infected with A. phagocytophilum. Gentamicin kills many bacteria but not A. phagocytophilum. Quantitative reverse transcription (qRT)-PCR analysis of the bacterial burden in ticks engorging on gentamicin-treated mice, that was demonstrated to possess an altered microbiome (SI Appendix, Figs. S1 C−E and S2), significantly increased A. phagocytophilum colonization, compared with control mice (Fig. 1 D and E). These results demonstrate that the gut microbiota plays an important role in A. phagocytophilum colonization of ticks.
Fig. 1.
A. phagocytophilum changes the tick microbiota and dysbiosis enhances A. phagocytophilum colonization. (A and B) Comparison of the gut microbial composition of uninfected and A. phagocytophilum-infected fed nymphs. (A) Total bacterial abundance and (B) ratio (Ap/Clean), at the taxonomic rank of genus, of uninfected (Clean) and A. phagocytophilum-infected (Ap) fed nymphs. Green and red box outlines indicate the genera with increased and decreased bacterial abundance in A. phagocytophilum-infected nymphs. (C) Principal coordinate analysis (PCoA) of the microbial communities from uninfected (blue) and A. phagocytophilum-infected (red) fed nymphs based on weighted UniFrac. Statistical significance was calculated using ANOSIM method, P = 0.028. (D and E) The qRT-PCR assessment of the A. phagocytophilum burden in normal (PBS) and dysbiosed gentamicin-treated (Gen) nymphal tick (D) guts (MG) and (E) salivary glands (SG). Horizontal bars represent the median. Results were pooled from three independent experiments. Statistical significance was calculated using a two-tailed nonparametric Mann−Whitney test (***P < 0.001; *P < 0.05).
The Peritrophic Matrix Is Essential for Colonization of the Tick by A. phagocytophilum.
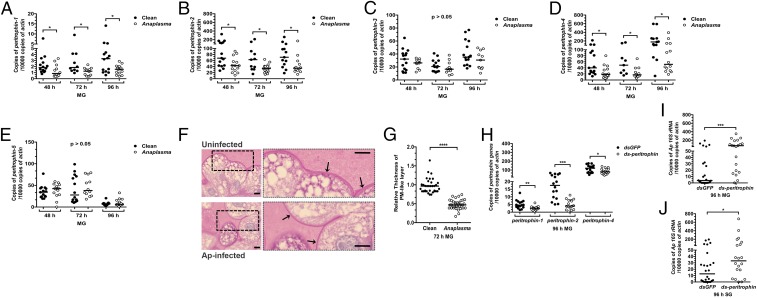
The peritrophic matrix (PM) is a glycoprotein-rich layer that separates the epithelial cells from the tick gut lumen, and peritrophin is the major component of the PM. We determined whether the PM plays a role in A. phagocytophilum colonization of ticks. The presence of A. phagocytophilum influenced the expression of several peritrophin genes, as assessed by qRT-PCR. The expression levels of peritrophin-1, peritrophin-2, and peritrophin-4 were significantly decreased in A. phagocytophilum-infected nymphs compared with uninfected nymphs (Fig. 2 A, B, and D). In contrast, expression of peritrophin-3 and peritrophin-5 was not altered in A. phagocytophilum-infected ticks (Fig. 2 C and E). Nymphs that fed on A. phagocytophilum-infected mice showed a decrease in thickness of the PM compared with nymphs that were fed on control mice (Fig. 2 F and G). To further examine the role of the PM in A. phagocytophilum colonization, RNAi was used to silence peritrophin expression. A decrease in peritrophin expression, using a dsRNA mixture of peritrophin-1, peritrophin-2, and peritrophin-4 injected into nymphal ticks (Fig. 2H), significantly enhanced the colonization of A. phagocytophilum in the tick gut and salivary glands, compared with control nymphs (Fig. 2 I and J). These results demonstrate that A. phagocytophilum infection induces, via the host, changes in the gut barrier.
Fig. 2.
The PM influences colonization of the tick by A. phagocytophilum. (A−E) A. phagocytophilum influences the expression of (A) peritrophin-1, (B) peritrophin-2, (C) peritrophin-3, (D) peritrophin-4, and (E) peritrophin-5 in the tick gut (MG) as assessed by qRT-PCR. Results were pooled from three independent experiments, and statistical significance was calculated using a two-tailed nonparametric Mann−Whitney test (*P < 0.05). (F) PAS staining of Carnoy’s fixed and sectioned fed guts from uninfected and A. phagocytophilum-infected nymphs. Boxed outlines within the images on the left have been magnified 2.5×. Arrows indicate the PAS-positive PM-like layer. (Scale bar, 10 μm.) (G) Quantification of the relative thickness of the PM-like layer from uninfected and A. phagocytophilum-infected nymphal guts (MG). (H−J) The qRT-PCR examination of the expression of (H) peritrophin-1, peritrophin-2, and peritrophin-4 upon peritrophin dsRNA mixture injection of nymphal ticks, and qRT-PCR expression of the A. phagocytophilum burden in (I) guts (MG) and (J) salivary glands of dsgfp- and ds-peritrophin-injected nymphs fed on A. phagocytophilum-infected mice. Each dot represents one nymph. Horizontal bars represent the median. Statistical significance was calculated from three independent experiments using a two-tailed nonparametric Mann−Whitney test (****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05).
IAFGP Modulates the Gut PM.
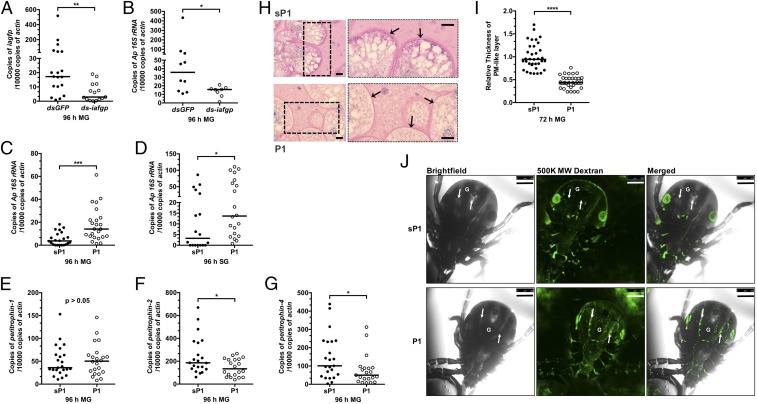
IAFGP, a secreted I. scapularis antifreeze glycoprotein, was recently determined to inhibit bacterial biofilm formation (25). The expression of iafgp is induced by cold exposure and A. phagocytophilum infection (26). The qRT PCR demonstrated that iafgp is highly induced in the tick gut by the presence of A. phagocytophilum (26), and silencing iafgp expression by RNAi diminishes the A. phagocytophilum burden in tick guts (Fig. 3 A and B). These data suggest that IAFGP plays an important role in A. phagocytophilum colonization of ticks.
Fig. 3.
IAFGP and its peptide derivative, P1, influence the PM and A. phagocytophilum colonization of tick gut. (A and B) The qRT-PCR examination of the expression of (A) iafgp and (B) the A. phagocytophilum burden in dsgfp- and ds-iafgp-injected nymphs [gut (MG)] fed on A. phagocytophilum-infected mice. Each dot represents one nymph. (C and D) The qRT-PCR assessment of the A. phagocytophilum burden in the (C) guts (MG) and (D) salivary glands with P1- or control [scrambled P1(sP1)]-injected nymphs fed on A. phagocytophilum-infected mice. Each dot represents one nymph. (E−G) The qRT-PCR analysis of expression levels of (E) peritrophin-1, (F) peritrophin-2, and (G) peritophin-4 in P1- and sP1-injected nymphs [gut )MG)]. Each dot represents one nymph. (H) PAS staining of Carnoy’s fixed and sectioned fed guts from P1- and sP1-injected nymphs. Arrows indicate the PAS-positive PM-like layer. Boxed outlines within the images on the left have been magnified 2×. (Scale bar, 10 μm.) (I) Quantification of relative thickness of the PM-like layer from P1- and sP1-injected nymphal guts (MG). Statistical significance was calculated using a two-tailed nonparametric Mann−Whitney test from three pooled experiments (****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05). (J) P1 injection of ticks improves the permeability of the PM. Confocal microscopy of 24-h P1- or sP1-injected nymphs that were capillary-fed Fluorescein-conjugated 500,000 MW dextran (500K MW Dextran). Magnification is 10×. G marks the gut diverticula within the tick. The arrows point to the hemocoel outside the gut. (Scale bar, 250 μm.)
To further investigate the role of IAFGP, a peptide (P1) (PARKARAATAATAATAATAATAAT) derived from IAFGP, which retains the antibiofilm properties (25), was injected into tick guts, and those nymphs were then fed on A. phagocytophilum-infected mice. A scrambled P1 (sP1) peptide (AATAATATAAARRAAAAPTTAKTT) served as control (25). The A. phagocytophilum burden in P1-injected tick guts was significantly increased compared with sP1-injected ticks, and more A. phagocytophilum migrated to the salivary glands as a consequence (Fig. 3 C and D). Because the PM was identified to play a role in A. phagocytophilum infection of tick guts (Fig. 2 F−I), we examined whether P1 affected the tick gut PM. The qRT-PCR showed that expression of peritrophin-2 and peritophin-4, but not peritrophin-1, was decreased in P1-injected ticks compared with sP1-injected control ticks (Fig. 3 E−G). Periodic acid−Schiff staining of 72-h-fed, P1-injected nymphs showed decreased thickness of the PM compared with sP1-injected control nymphs (Fig. 3 H and I). Having previously demonstrated that alterations in the PM thickness compromises the barrier integrity of the PM (10), we similarly evaluated the improved permeability of the PM by introducing Fluorescein-conjugated 500,000 molecular weight (MW) dextran into the guts of P1- or sP1-injected 24-h-fed nymphs and examined the unfixed guts by confocal microscopy. In contrast to sP1-injected ticks, the 500,000 MW dextran was predominantly localized outside gut lumen, suggesting that P1 injection of ticks reduces the thickness of the PM, permitting the large-sized dextran beads to pass from the gut lumen through the PM and out of the gut (Fig. 3J). We propose that this improvement in A. phagocytophilum infection is attributed to the thinning of the PM, which potentially allows for improved access of the pathogen through the PM to the midgut epithelium and subsequently it’s egress to the salivary glands. Cumulatively, these results indicate that IAFGP, or its peptide derivative P1, facilitates A. phagocytophilum colonization of the tick gut by indirectly modulating the PM.
IAFGP Alters the Tick Microbiota and Influences the Gut Barrier.
In Ixodes ticks, the integrity of the PM is indirectly modulated by the gut microbiota, in part, by the normal microbiota’s influence on the STAT pathway (10). Our previous study showed that IAFGP and P1 have antivirulence properties against diverse bacteria, and specific antibiofilm activity was observed using Staphylococcus aureus as our model organism (25). Based on these two lines of evidence, we hypothesized that IAFGP influences the tick gut microbiota, which results in alterations in the thickness and permeability of the PM.
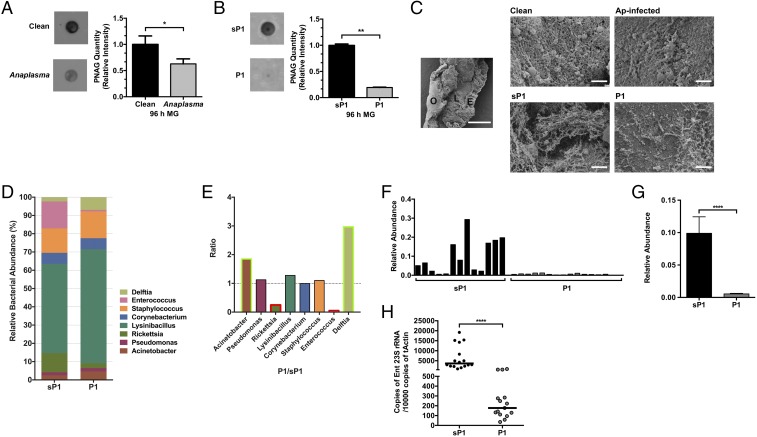
Exopolysaccharides such as poly-N-acetylglucosamine (PNAG) are a major component of bacterial biofilms. PNAG is broadly distributed in diverse microbes and can be detected by a specific antibody (27). Having observed native biofilm formation in tick guts using a PNAG antibody (SI Appendix, Fig. S2A), we examined the presence of biofilms using the PNAG antibody in A. phagocytophilum-infected nymphal guts. The guts from A. phagocytophilum-infected and uninfected fed nymphs were collected, and PNAG was extracted and detected by immunoblot. The amount of detectable PNAG in A. phagocytophilum-infected nymphs was significantly decreased in comparison with uninfected nymphs (Fig. 4A). Moreover, endogenous IAFGP induced within A. phagocytophilum-infected nymphs was found to colocalize with these biofilms within tick guts, as detected by confocal microscopy (SI Appendix, Fig. S2B). These results suggest that A. phagocytophilum infection and induction of IAFGP within ticks influences native biofilm formation. To examine whether P1 can similarly influence biofilm formation in ticks, P1-injected guts from 96-h-fed nymphal ticks were collected. The amount of PNAG was significantly decreased in P1-injected nymphs compared with sP1-injected control ticks, which confirms that P1 can also modify biofilms in ticks (Fig. 4B). To visualize biofilms in the tick guts, we performed scanning electron microscopy (SEM). In normal tick guts, the commensal bacteria could form intact biofilm structures; however, A. phagocytophilum-infected tick guts showed disrupted biofilms (Fig. 4C, Top). Similarly, P1-injected tick guts showed a significant depreciation in biofilms compared with sP1-injected ticks (Fig. 4C, Bottom).
Fig. 4.
IAFGP and P1 alter biofilm formation and microbiota in ticks. (A and B) The relative amount of PNAG extracted from (A) uninfected and. A. phagocytophilum-infected nymphs and (B) P1- vs. sP1-injected nymphs and their respective quantification. Relative intensity of PNAG blots were analyzed by ImageJ (graphs). Graphical data were pooled from two independent experiments, and statistical significance was calculated using a two-tailed nonparametric Mann−Whitney test (**P < 0.01; *P < 0.05). (C) SEM of tick gut biofilm from uninfected (Clean) and A. phagocytophilum (Ap) infected nymphs (Top) and sP1- vs. P1-injected nymphs (Bottom). Representative images are shown for all samples. A representative uninfected (Clean) sample is provided (Left, Middle) at low magnification (500×) to visualize a dissected gut revealing the outside of the gut (O), epithelial cells lining the gut (E), and luminal content (L) (scale bar, 50 μm). Representative images displayed were taken at high magnification (∼8,500×; scale bar, 2 μm) focusing on the luminal content. Ap-infected and P1-injected guts show looser biofilms without dense connecting fibers or matrix-like material associated with reduced biofilms. Clean and sP1 guts, contrastingly, have thicker and well-connected matrix like material with embedded bacteria (cocci) representing robust biofilms. (D and E) Genus level (D) total bacterial composition of sP1- and P1-injected nymphs and (E) fold changes (P1/sP1) of major genera. Green and red box outlines represent genera with increased or decreased abundance in P1-injected nymphs. Statistical significance was calculated using a two-tailed nonparametric Mann−Whitney test (****P < 0.0001). (F–H) Comparison of the levels of Enterococcus species within the tick gut microbiota between sP1- and P1-injected nymphal ticks. (F and G) Relative abundance of Enterococci (F) among individual samples and (G) across all samples (pooled). (H) P1-injected nymphs show reduced levels of Enterococci as determined by qRT-PCR using Enterococci specific primers compared with sP1-injected ticks.
The microbiota of arthropods and mammals has long been associated with robust biofilms (12, 28). To further explore whether altering the biofilm has an effect on the whole tick gut microbial community, we compared the microbiota profiles of P1-injected and sP1-injected ticks. Bacteria of the genera Acinetobacter and Delftia were more abundant in P1-injected nymphs than sP1-injected nymphs, whereas Enterococcus and Rickettsia genera were significantly decreased in P1-injected nymphs (Fig. 4 D and E). Enterococci, as an example, are robust biofilm formers (29, 30). Microbiota and Enterococcus-specific qPCR analysis from individual and pooled samples further confirmed that the relative abundance of Enterococci was dramatically decreased in P1-injected tick guts compared with sP1-injected ticks (Fig. 4 F−H). The reduction of Enterococcus in P1-injected ticks could therefore contribute to the change in biofilm formation and, thus, the overall architecture of the gut microbiome. Because we previously observed that A. phagocytophilum-infected ticks were also decreased in Enterococci sp. (Fig. 1B and SI Appendix, Fig. S2 C−E), this would further suggest a role for IAFGP in manipulation of the tick microbiota and their associated biofilms.
IAFGP Binds to the d-alanine Residue of Bacterial Peptidoglycan.
As alterations in biofilm formation and the microbiota impacted Anaplasma colonization, we examined the mechanism by which IAFGP interferes with biofilm development. Enterococci, a genus known to produce tenacious biofilms, were consistently observed as being negatively impacted within ticks through A. phagocytophilum colonization or P1 injection (see SI Appendix, Fig. S2 C−E and Fig. 4 F−H). Because we had previously documented that binding of IAFGP’s binding to S. aureus adversely impacted the bacterium’s capacity to form biofilms (25), we similarly tested a panel of Enterococci species including Enterococci faecalis, Enterococci faecium, and vancomycin-resistant E. faecalis to bind either IAFGP or P1.
Each of these species has been represented in different arthropod populations including Rhipicephalus ticks, some of which are commonly known as cattle ticks (3, 31, 32). Recombinant GST-tagged IAFGP, but not GST alone, bound to each of the three species with varying affinities: Binding to E. faecalis was the strongest, followed by E. faecium and then vancomycin-resistant E. faecalis (Fig. 5A). Similar binding affinities were observed with P1 relative to sP1 (SI Appendix, Fig. S3A). Previous work showed that IAFGP binds to the bacterial surface and, more importantly, that both IAFGP and P1 bind S. aureus peptidoglycan in vitro (25) (SI Appendix, Fig. S3C). Binding of IAFGP or P1 to peptidoglycan extracted from each Enterococcus species was assessed by using a wheat germ agglutinin (WGA) antibody that specifically detects bacterial peptidoglycan. The strongest binding to biotinylated GST-tagged IAFGP or peptide P1 (immobilized to streptavidin beads) was detected with peptidoglycan muropeptides (associated fraction) extracted from E. faecalis, followed by E. faecium (Fig. 5B and SI Appendix, Fig. S3B). Minimal binding was noted with vancomycin-resistant E. faecalis (Fig. 5B and SI Appendix, Fig. S3B). Comparing the previously obtained results with S. aureus (25) and the current data from each of the three Enterococci species, we recognized that S. aureus and E. faecalis are very similar in their peptidoglycan structure (except for their cross-bridge) (33). We therefore postulated that IAFGP binds the pentapeptide chain of bacterial peptidoglycan at its terminal amino acid residues (Fig. 5C). E. faecium and vancomycin-resistant E. faecalis show greater variability in their stem peptides, with only 3% of E. faecium having the same terminal d-alanyl-d-alanine amino acids as S. aureus or E. faecalis (34); vancomycin-resistant E. faecalis replaces its terminal d-alanine residue with lactate, contributing to its vancomycin resistance.
Fig. 5.
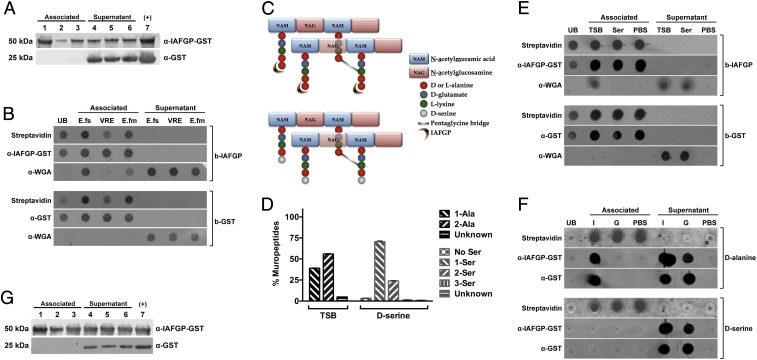
IAFGP binds to the terminal d-alanine residue of Enterococcus sp. and S. aureus peptidoglycan. (A) E. faecalis (lanes 1 and 4), E. faecium (lanes 2 and 5), and vancomycin-resistant E. faecalis (lanes 3 and 6) bacteria were incubated with either recombinant GST-IAFGP or GST alone. Bound (Associated) and unbound (Supernatant) protein was detected by immunoblot. Recombinant GST-IAFGP or GST alone (lane 7) was used as a positive control. (B) IAFGP displays varied binding to Enterococcal peptidoglycan. Streptavidin-coated magnetic Dynabeads bound to biotinylated IAFGP-GST (b-IAFGP) or GST alone (b-GST) were incubated with peptidoglycan muropeptides extracted from E. faecalis (E.fs), vancomycin-resistant E. faecalis (VRE), and E. faecium (E.fm) cells grown in BHIG. Biotinylated proteins bound to beads were detected using an infrared-labeled streptavidin probe. IAFGP-GST was detected using a polyclonal murine primary antibody, and GST was detected using a monoclonal murine primary antibody. Muropeptides from cells were detected using a polyclonal WGA antibody. Unbound protein (UB) was also collected and spotted. Associated or Supernatant fractions represent the fractions that were either pulled down with the magnetic bead or remained unbound. (C) Schematic describing S. aureus peptidoglycan and binding site of IAFGP. The S. aureus peptidoglycan backbone is a polymer consisting of glycan strands of alternating sugar subunits: NAG and NAM. The NAM subunits have short peptides (muropeptides) comprising l-alanine, d-glutamate, l-lysine, and two d-alanines. The cross-link between neighboring muropeptides is mediated by a transpeptidase enzyme and allows for the formation of a short peptide interbridge consisting of five glycines (pentaglycine bridge). IAFGP is hypothesized to bind to the terminal d-alanine residue of the stem pentapeptide (Top). Growth of S. aureus in an alternative amino acid like d-serine (Bottom) replaces the terminal d-alanine residue with the newly supplied d-serine. (D) Muropeptide composition of S. aureus peptidoglycan. S. aureus grown in either TSB or medium supplemented with 125 mM d-serine were analyzed for their muropeptide residue composition. Data represent the percentage of total bacterial muropeptides that incorporate a given residue(s) at its carboxyl terminus. “No Ser” represents d-alanine terminating muropeptides. Data were pooled from three independent experiments ± SEM. (E and F) IAFGP binds to the terminal d-alanine residue of the S. aureus muropeptide chain. (E) Streptavidin-coated magnetic Dynabeads bound to b-IAFGP or b-GST were incubated with muropeptides obtained from cells grown in either TSB or medium supplemented with 125 mM d-serine. (F) Experiment was similarly performed as in E using synthetic biotinylated pentapeptides containing either alanine (d-alanine) or serine (d-serine) residues bound to beads and incubated with IAFGP-GST (I) or GST alone (G). PBS was used as a negative binding control. Associated or Supernatant fractions represent the fractions that were either pulled down with the magnetic bead or remained unbound. (G) IAFGP does not bind WTA. Wildtype S. aureus strain SA113 (lanes 1 and 3) and its isogenic WTA-deficient mutant SA113 ΔtagO (lanes 2 and 5) and complemented strain SA113 pRBtagO (lanes 3 and 7) were tested for binding to recombinant GST-IAFGP or control GST alone. Bound (Associated) and unbound (Supernatant) protein was detected by immunoblot analysis. Recombinant GST-IAFGP or GST alone (lane 7) without any bacteria was used as a positive control.
Observing similar bacterial and peptidoglycan binding between S. aureus and E. faecalis, we determined the specific residues required for IAFGP or peptide binding using S. aureus as our model bacteria. S. aureus peptidoglycan grown in tryptic soy broth (TSB), typically shares a 3:2 ratio of d-alanyl-d-alanine as its terminal pentapeptide residues versus a single terminal d-alanine, resulting from the hydrolytic transpeptidation or carboxypeptidation reaction (Fig. 5D and SI Appendix, Fig. S4 and Table S4). Supplementing medium with an alternate d-amino acid such as d-serine causes S. aureus to incorporate the newly supplemented d-amino acid into the carboxyl terminus of the stem peptides (35). This results in the d-alanyl-d-serine terminating peptidoglycan stem residue in ∼75% of all muropeptides compared with only 6% of cells retaining the native d-alanyl-d-alanine (Fig. 5 C and D and SI Appendix, Fig. S4 B and C and Table S4). Ultra performance liquid chromatography (UPLC) analysis also revealed a significantly lower percentage of crosslinking between the glycan strands for d-serine and IAFGP cultured bacteria (SI Appendix, Fig. S4E).
Observing an overwhelming shift toward replacement of the terminal alanine residue with serine at the terminal muropeptide site with supplementation of 125 mM d-serine, we determined IAFGP binding to muropeptides grown from either medium, similar to what was previously done with the Enterococci (Fig. 5B and SI Appendix, Fig. S3B). Biotinylated GST-tagged IAFGP, but not biotinylated GST alone, successfully bound muropeptides (associated fraction) obtained from TSB medium, as detected by the WGA antibody (Fig. 5E). There was no signal detected by WGA antibody with muropeptides obtained from d-serine cultured cells. The WGA antibody, which recognizes the N-acetylglucosamine (NAG) sugar, was used because all of the muropeptides obtained from either TSB or d-serine cultures were associated with both the N-acetylmuramic acid (NAM) and NAG sugar residues (SI Appendix, Fig. S4C). To confirm this binding interaction between IAFGP and muropeptides with a terminal (fifth) d-alanine residue, we generated five amino acid synthetic pentapeptides with either d-alanine or d-serine as their terminal (fifth) residue. Using these biotinylated pentapeptides, bound to streptavidin beads, GST-tagged IAFGP, but not GST alone or the PBS buffer control, could be detected in the associated fraction of pentapeptides with d-alanine residue (Fig. 5F). This confirmed IAFGP binding to the muropeptides generated with the d-alanine terminal residue, but not to those with the d-serine residue. Histidine (His)-tagged P1, and not scramble peptide sP1 or PBS, was also found to similarly bind synthetic muropeptides with a terminal d-alanine residue (SI Appendix, Fig. S3D). Similarly, neither IAFGP nor P1 could bind E. faecalis or E. faecium whole bacteria cultured in media supplemented with125 mM d-serine (SI Appendix, Fig. S3 E and F).
Beyond the peptidoglycan layer, other cell wall glycopolymers including teichoic acids (TAs) decorate the surface of Gram-positive bacteria. TAs including wall teichoic acids (WTAs) and lipoteichoic acids (LTAs) are a constant motif of Gram-positive cell envelopes, and they form part of the fabric of the cell wall attaching either to the peptidoglycan or to membrane lipids, respectively (36). Both WTAs and LTAs are usually modified with d-alanine residues (37). To determine whether binding of IAFGP is specific for terminal d-alanine residues of peptidoglycan and not those of WTAs, we tested IAFGP and P1 binding to S. aureus mutants deficient in producing WTA (Fig. 5G and SI Appendix, Fig. S3G). IAFGP and P1 demonstrated consistent binding across wild-type and mutant S. aureus strains without any significant depreciation. In addition, there was no significant binding of IAFGP to LTA (SI Appendix, Fig. S3H). These data demonstrate a direct interaction between IAFGP (or P1) and the terminal d-alanine residue of the peptidoglycan chain of Gram-positive bacteria like S. aureus or E. faecalis. Gram-negative bacteria have been previously demonstrated to possess this same terminal d-alanine amino acid moiety (38, 39). However, multiple reasons including (i) access to peptidoglycan being limited, due to the presence of the outer membrane, and (ii) the efficient action of the DD-carboxypeptidase enzymes, that converts almost all of their pentapeptide (NAG-NAM-l-ala-d-glu-mDap-d-ala-d-ala) to tetrapeptides (NAG-NAM-l-ala-d-glu-mDap-d-ala), skews the binding specificity of either IAFGP or its peptide derivative P1 more toward Gram positives. These two major criteria notwithstanding, muropeptides from Gram-negative bacteria, as demonstrated by the Escherichia coli DV9000 mutant, would also be able to bind either IAFGP or P1 (SI Appendix, Fig. S5 A and B).
Alterations in Peptidoglycan Reduce S. aureus and E. faecalis Biofilm Formation.
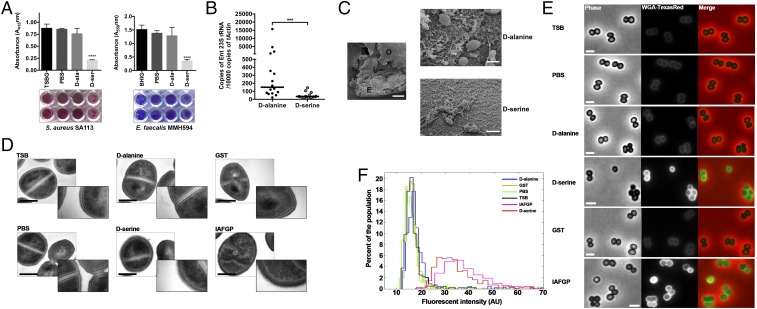
Because IAFGP binds the terminal d-alanine of the peptidoglycan peptide stem, we investigated the relationship between this observation and the antibiofilm activity of IAFGP (25). Our studies in serine-supplemented medium provided a model system in which to study how S. aureus or E. faecalis behaves with an altered peptidoglycan. We established a connection between biofilm formation and peptidoglycan by performing in vitro biofilm assays with bacterial cultures grown in traditional rich medium (native peptidoglycan) versus medium supplemented with 125 mM d-serine (errant peptidoglycan) (Fig. 5C). S. aureus and E. faecalis cultures passaged in 125 mM d-serine showed a 75% decrease in biofilm formation (Fig. 6A). Supplementation with 125 mM d-alanine, the native residue at the carboxyl terminus, did not have a significant decrease in biofilms compared with medium alone or supplementation with d-serine (Fig. 6A). This result suggested that changing the terminal residue from alanine to serine resulted in the drastic decrease in biofilm formation. Growth and viability for each bacterium were unaffected under any of these treatment conditions (SI Appendix, Fig. S6).
Fig. 6.
d-serine incorporation causes alterations to bacterial biofilms and peptidoglycan. (A) Biofilm formation of S. aureus SA113 (Left) and E. faecalis MMH594 (Right) cultures was determined in respective media. Dissolved stains were pooled from three independent experiments ± SEM; representative images of the biofilm plates with two representative wells per condition are shown below the graphs. Statistical significance was calculated using one-way ANOVA with Dunnett’s multiple comparison posttest (***P < 0.0001). (B) d-serine-injected nymphal ticks show reduced levels of Enterococci compared with control d-alanine-injected ticks, as determined by qRT-PCR using Enterococci specific primers. Statistical significance was calculated using a two-tailed nonparametric Mann−Whitney test (****P < 0.0001; ***P = 0.0003). (C) SEM of tick gut biofilms from d-alanine- versus d-serine-injected nymphal ticks. d-alanine-treated representative sample taken at low magnification (500×) is provided to visualize a dissected gut revealing the outside of the gut (O), epithelial cells lining the gut (E), and luminal content (L). (Scale bar, 50 μm.) Representative images are shown for both samples taken at high magnification (∼8,500×; scale bar, 2 μm) focusing on the luminal content. Compared with dense matrix biofilms with d-alanine treatment, there is a severe scarcity of the biofilm-associated polymeric matrix and bacteria with the d-serine treatment. (D) Overnight cultures of S. aureus SA113 in TSB medium alone or medium supplemented with 125 mM d-alanine, 125 mM d-serine, 0.1 mg/mL GST-IAFGP, or 0.1 mg/mL GST alone were processed for transmission electron microscopy. Representative images are shown. Cultures were also grown in an equivalent volume of PBS (buffer control). (Left) Representative images were taken at 60,000 magnification. (Scale bar, 500 nm.) Inset images were taken at 150,000 magnification (box length = scale bar = 800 nm). (E) Bacterial cells cultured in TSB or medium supplemented with PBS, d-alanine, d-serine, GST, or IAFGP were stained with the NAG binding analog Wheat Germ Agglutinin–Texas Red and imaged by phase-contrast and epifluorescence microscopy. (Scale bar, 2 μm.) (F) Population analysis, at the single-cell level, demonstrates that IAFGP and d-serine treatments result in an increase in fluorescent signal intensity. Total fluorescent signal intensities were normalized by cell area, and histograms were depicted as a function of population frequency; n > 500 cells for each treatment was pooled from two independent experiments.
Because IAFGP, P1, and now d-serine severely alter bacterial biofilms, we next investigated whether d-serine’s antibiofilm phenotype could be observed within ticks. Microbiota and Enterococcus-specific qRT-PCR analysis for individual and pooled samples using d-serine-treated nymphal ticks further demonstrated that the relative abundance of Enterococci was dramatically decreased with d-serine compared with injection with control d-alanine (Fig. 6B and SI Appendix, Fig. S7 A–D). Visualization of tick gut biofilms by SEM showed a severe depreciation in biofilm-like structures, associated fibers, and matrix material within d-serine-treated ticks compared with d-alanine-treated ticks (Fig. 6C). To confirm that the changes within the microbiota and reductions in biofilms were attributed to the administered treatment—d-serine, P1 (Fig. 4), or gentamicin (Fig. 1)—IAFGP expression levels within these ticks were determined (SI Appendix, Fig. S8A). Our data indicate that alterations to the tick microbiota and deleterious effects on biofilms, previously observed with IAFGP through A. phagocytophilum acquisition, in these instances was not due to increased iafgp expression (SI Appendix, Fig. S8A). Moreover, in contrast to recent work (40), we have additionally ruled out the possibility of host-related factors like IFN-γ to induce expression of IAFGP or improved infection of A. phagocytophilum (SI Appendix, Fig. S8 B and C).
Because IAFGP or d-serine is able to bind or incorporate into peptidoglycan, thereby significantly affecting biofilms, we hypothesized that these treated cells would show a change in cellular morphology. Therefore, we examined physical differences in bacterial cell morphology using transmission electron microscopy. For this, S. aureus cells were grown in rich versus serine-supplemented media. S. aureus cells passaged in d-serine exhibited a more diffuse peptidoglycan layer with distinct ruffled features (Fig. 6D). This may be associated with the decrease in cross-linked peptidoglycan, as shown by UPLC (see SI Appendix, Fig. S4E). Interestingly, a similar morphological phenotype was exhibited by cells grown in 0.1 mg/mL of IAFGP but not by those grown in GST alone (Fig. 6D). Although neither IAFGP- nor GST-treated cells showed any difference in the peptidoglycan composition (SI Appendix, Fig. S4D), IAGFP-treated cells, similar to d-serine, were significantly reduced in their cross-linking (SI Appendix, Fig. S4E). The peptidoglycan-occupied area for cells grown with either d-serine or IAFGP was significantly higher compared with controls (SI Appendix, Fig. S9A). It is likely that this phenomenon extends beyond S. aureus, as we also observed biofilm inhibition by P1 in other Gram-positive bacteria. Biofilm biomass was dramatically reduced, in vitro, by two other Staphylococcus sp., Streptococcus mutans, and Corynebacterium pseudodiptheriticum in the presence of P1 (SI Appendix, Fig. S7E).
Biofilm inhibition by IAFGP or P1, mediated by peptidoglycan binding, is likely to be important in reducing the barrier for clearance of bacteria by the immune system, antimicrobial peptides, or antibiotics. We tested this hypothesis by determining the permeability of the peptidoglycan layer using a fluorescent WGA protein. S. aureus cells grown in d-serine or IAFGP showed significantly higher fluorescent intensities compared with TSB or other control growth conditions (Fig. 6E). The diffused peptidoglycan layer with IAFGP or d-serine (Fig. 6D and SI Appendix, Fig. S9A) would suggest better penetration by the fluorescent WGA. WGA intercalates through the layers of peptidoglycan to gain better access to their native binding site, the NAG sugar residues. Alternatively, however, the increase in fluorescence with IAFGP or d-serine could implicate more peptidoglycan within these respective treatment conditions. To differentiate between these two plausible hypotheses, we determined the permeability of the cell using the cell membrane as a binding target. Permeability past the peptidoglycan, with subsequent access to the cell membrane, was tested using the membrane-binding fluorescent dye, FM 4-64. Treatment with FM 4-64 revealed increased fluorescent signal intensities for cells cultured with IAFGP or d-serine compared with controls (SI Appendix, Fig. S9B), indicating an increase in permeability. These data demonstrate that alterations in peptidoglycan, mediated by d-serine incorporation or via IAFGP binding, inhibit biofilm formation and alter the peptidoglycan in a nonlethal manner, allowing for enhanced permeability.
Discussion
A. phagocytophilum, an obligate intracellular pathogen, enters its tick host following a blood meal and rapidly penetrates the gut barrier and migrates to salivary glands, where it persists (41, 42). Gut commensal microbes play critical roles in enabling hosts to resist invading bacteria, and alterations of the microbial gut community of vectors can influence infection loads with pathogens (43). In our A. phagocytophilum−I. scapularis model, distinct differences were noted in the relative abundance and composition of the microbiota of ticks feeding on uninfected, or A. phagocytophilum-infected, mice, indicating that the presence of A. phagocytophilum alters the tick gut microbial community. Numerous factors including exposure to environmental bacteria, geographic location, tick species, gender, developmental status, and time since molting and feeding, among others, are expected to play an important role in contributing to the microbiome composition in ticks (10, 44, 45). Therefore, although the tick gut-associated microbes might not be conserved across different regions, with several published reports suggesting a large diversity (46–49), and others suggesting that inflated diversities are due to environmental contaminants (50), we used multiple criteria (see SI Appendix) to accurately assess the microbial composition of our laboratory-reared and murine host-fed I. scapularis nymphs. Based on our stringencies and exclusion criteria, we observed eight genera including Acinetobacter, Pseudomonas, Rickettsia, Lysinibacillus, Corynebacterium, Staphylococcus, Enterococcus, and Delftia that were consistently observed, independent of treatment condition. Comparing the unfed nymphal microbiome in our earlier work (10) to the fed nymphal microbiome in this work, genera such as Streptococcus [observed in some tick cohorts, although excluded from this main analysis (SI Appendix, Table S3)], Rickettsia, Pseudomonas, Acinetobacter, Staphylococcus, and Delftia are observed in both, albeit at different proportions. Genera such as Acidovorax, Brevundimonas, Commamonas, Stenotrophomonas, and Sphingobium were observed in the unfed nymphs but not in the fed nymphs, and genera such as Lysinibacillus, Corynebacteria, and Enterococcus were observed only in the fed nymphs. Some of these differences could be attributed to the fed versus unfed status of the nymphs or differences in the sequencing platforms used: 454 sequencing versus Illumina MiSeq platform for the 16S rDNA sequencing used in this study. Within the rigors of our experimental framework, we demonstrate that there are significant variations in the relative abundance of gut microbial genera during A. phagocytophilum colonization of ticks. Our paper unravels the potential mechanism by which these changes occur and invokes the tick protein, IAFGP. Additionally, we highlight the mechanism by which A. phagocytophilum coopts this tick protein to best potentiate its infection in ticks.
IAFGP, the I. scapularis antifreeze glycoprotein, is induced by ticks in the presence of A. phagocytophilum (26), and silencing iafgp impairs A. phagocytophilum colonization of the gut. Independently, we demonstrated IAFGP to inhibit biofilm formation among Gram-positive pathogens such as S. aureus (25). Because A. phagocytophilum colonization reduced the abundance of Gram-positive biofilm-forming species like the Enterococci, we hypothesized that IAFGP alters the tick microbiota by inhibiting bacterial biofilms in the gut. Biofilms have been demonstrated to serve important roles in facilitating pathogen colonization in arthropod intestines (12–14). To elucidate the importance of IAFGP in shaping the tick gut microbiota, we injected the antibiofilm peptide, P1, derived from IAFGP (25), and showed that this improved A. phagocytophilum infection, in ticks. Exopolysaccharides such as PNAG, a major component of bacterial biofilms, are broadly distributed among diverse microbes (27). Using antibodies directed against PNAG as a proxy to gauge biofilms within the tick, P1-injected I. scapularis guts showed a marked reduction of biofilms, similar those with A. phagocytophilum infection. SEM confirmed this phenotype with normal tick guts (clean or sP1-injected) having thicker and well-connected matrix-like material with embedded bacteria such as cocci representing robust biofilms, whereas A. phagocytophilum-infected or P1-injected tick guts showed looser biofilms without connecting fibers or matrix-like representative material indicating reduced biofilms. In an effort to delineate the molecular mechanism of IAFGP expression and P1 injection adversely affecting biofilms, we sought to use tick gut commensal Enterococci, a species that was markedly reduced within our microbiome analysis during A. phagocytophilum infection or P1-treatment, and compare results with S. aureus, which also forms robust biofilms. Using three different Enterococci species (E. faecalis, vancomycin-resistant E. faecalis, and E. faecium), we identified a difference in the capacity of IAFGP and its peptide homolog to bind the individual species. Because IAFGP binds S. aureus peptidoglycan (25), we hypothesized that the variability in binding to the Enterococci could be attributed to the difference in the peptidoglycan composition. We now demonstrate that IAFGP binds to the terminal d-alanine residue of the bacterial peptidoglycan peptide moiety—common to E. faecalis and S. aureus.
d-amino acids are readily incorporated into bacterial peptidoglycan by replacing the terminal residue with the newly provided d-amino acid (35), and these types of alterations inhibit biofilm formation (51) including those by tick gut commensals like E. faecalis. Because IAFGP and P1 can bind the terminal d-alanine residue of bacterial peptidoglycan, use of compounds like d-serine that prevent biofilms and biochemically alter peptidoglycan provide an independent assessment of changes within the tick gut in an Anaplasma (IAFGP) free system. Similarities in the relative abundance of Enterococci within the tick gut, found by microbiome analysis or SEM imaging of tick gut biofilms when treated with d-serine, to those during A. phagocytophilum infection or P1 injection of tick guts support the mechanistic action of IAFGP’s peptidoglycan binding activity on modulating the tick microbiota. d-amino acids interfere with the linking of biofilm adhesion proteins, and with cell wall remodeling (51–54). Therefore, we hypothesized that IAFGP binding to peptidoglycan could cause alterations in bacterial physiology and morphology that would reduce the ability of Gram-positive bacteria to form tenacious biofilms—adversely affecting the relative composition of the gut microbiota. IAFGP binding to peptidoglycan resulted in a more permeable peptidoglycan with significantly reduced cross-linking between the glycan strands; this is of particular importance because the increased permeability increases the sensitivity of bacteria to host immune defenses such as antimicrobial peptides and other toxins (55, 56), which could eventually cause a change in the composition of the tick guts microbial community.
Effects of IAFGP against the microbiome and specific genera are critical for optimal A. phagocytophilum infection of ticks. In an effort to tease out the specificity of IAFGP’s or P1’s binding, multiple factors were evaluated. During tick feeding, depending on the host and other biological factors, blood ingested can contain varying levels of free amino acids, including free d-alanine (nanomolar range) (57). Concentrations only greater than 100 mM of free d-alanine were able to outcompete IAFGP’s binding to peptidoglycan and inhibit the antibiofilm properties of IAFGP or P1, highlighting IAFGP’s or P1’s strong binding efficiency to the terminal d-alanine residues of the bacterial peptidoglycan (SI Appendix, Fig. S10). Moreover, using synthetic variations of the IAFGP or P1 sequence (SI Appendix, Table S5), we could confirm the binding specificity of either protein or peptide to the terminal d-alanine residue of peptidoglycan and its inextricable link to inhibiting biofilms among Gram-positive bacteria (SI Appendix, Figs. S11 and S12). These data, taken cumulatively, not only confirm a level of specificity for IAFGP or P1, for Gram-positive bacterial peptidoglycan, that terminate in a d-alanine residue, but also in restricting biofilm formation. Although Gram-positive bacteria are adversely affected, Gram-negative bacteria like Pseudomonas sp. or E. coli neither bind IAFGP or P1 nor are inhibited in their biofilms on account of their internalized peptidoglycan and absence of the terminal (fifth) d-alanine residue on their peptidoglycan (SI Appendix, Fig. S5) (58–61). This is particularly pertinent because A. phagocytophilum is technically categorized as a Gram-negative organism. However, unique physical characteristics and previously determined molecular evidence suggest the absence of peptidoglycan from A. phagocytophilum (62–64), which would nullify any effect IAFGP (or P1) would have on A. phagocytophilum. Our earlier work described the role for elevated levels of IAFGP, upon A. phagocytophilum infection of Ixodes ticks, to protect ticks from the cold environment (26). Therefore, IAFGP aids Anaplasma in two manners: by increasing the ability of ticks to survive in the winter and therefore indirectly increasing the chance of Anaplasma transmission to mice, and by altering the tick microbiomes and their associated biofilms, thereby enhancing its colonization within the tick vector.
Understanding the functional consequence of tick–microbiome interactions is fundamental to developing new paradigms to control tick-transmitted pathogens. We previously demonstrated that dysbiosis causes the tick gut microbiota to interact differently with epithelial cells and diminishes the activity of a global transcriptional regulator, STAT (10). STAT controls expression of different downstream target genes involved in immunity and cell repair, including peritrophin, the major component of the PM (10). Interestingly, we previously showed that A. phagocytophilum infection of Ixodes ticks reduces expression of stat, and that colonization is enhanced when stat is silenced (65); this suggests a role for the PM as a contributing factor during A. phagocytophilum infection of the tick. The glycan-rich PM layer separates the gut lumen from the epithelial cells and provides a protective barrier in arthropods against microbes and components of the incoming blood meal (66). Periodic acid-Schiff (PAS) staining of A. phagocytophilum-infected and P1-injected nymphal guts showed a significant decrease of the PM-like layer, which leads to increased PM permeability. We propose that the improvement in A. phagocytophilum infection is attributed to the thinning of the PM, which potentially allows for improved access of the pathogen through the PM to the midgut epithelium and, subsequently, it’s egress to the salivary glands. One could hypothesize that, during A. phagocytophilum infection, a series of events that include (i) alterations in the microbiome and reduction in biofilms through IAFGP and (ii) the thinning of the PM through stat, barriers that would otherwise prevent effective contact with the midgut epithelium, are accommodated so as to allow for the strongest colonization of the tick. Although thinning of the PM and the consequent increased PM permeability is conducive to increased A. phagocytophilum colonization, B. burgdorferi colonization is jeopardized by a compromised PM barrier (10). B. burgdorferi migrates through the PM to colonize the epithelial cells, and persists in this location, where an intact PM protects the spirochete from the toxic components of the gut lumen. The contrasting influence of PM integrity on two tick-borne pathogens could potentially result in competition during a natural cocolonization setting. An earlier study by Levin and Fish (67) suggests interference between these two agents during transfer from coinfected mice to ticks. These observations might also explain, in part, the generally lower prevalence of A. phagocytophilum and B. burgdorferi-coinfected ticks (<1 to 6%) versus ticks coinfected with B. burgdorferi and Babesia microti (2 to 19%) (68). Nevertheless, further studies, using both laboratory and field coinfected ticks, will be required to delineate the role of microbiota on infections with multiple pathogens.
Defining the interactions between the tick (gut) and A. phagocytophilum colonization furthers our understanding of the key events that determine prevalence of this common tick-borne infectious disease. Although we have yet to elucidate how acquisition of this pathogen induces expression of IAFGP, our current findings highlight a functional link between IAFGP, the tick microbiota and biofilm formation, and the PM and reveal their functional consequence on A. phagocytophilum colonization. Similar to the gut microbiota of mammals, the microbiota associated with arthropods plays an important role in host development, pathogen resistance, nutrition, and physiology (69). Microbial symbionts can reduce vector competence, and specific gut microbiota play a protective role in Anopheles mosquitoes against Plasmodium colonization (70–73). Although knowledge about the function of microbiota in diverse arthropods is still limited, cross-talk between the arthropod, its gut microbiota, and incoming pathogens is likely to be important (9–11, 69). This study demonstrates how a pathogen induces the arthropod to express a protein that directly inhibits bacterial biofilms, and that the effects of this protein on the tick microbiota enable the microbe to effectively colonize the vector. This paradigm—delineated with A. phagocytophilum and I. scapularis—is likely to be applicable to other vector-borne diseases.
Materials and Methods
All studies with mice were carried out following the animal protocol number 2014-07941 approved by Yale University's Institutional Animal Care and Use Committee (IACUC). The IACUC is governed by applicable Federal and State regulations, including those of the Animal Welfare Act (AWA), Public Health Service (PHS), NIH Office of Laboratory Animal Welfare Assurance Number A3230-01, and the United States Department of Agriculture (USDA), License and Registration Number 16-R-0001, and is guided by the US Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research and Training. Experimental details on Anaplasma infection of mice, microbiome sample preparation, Illumina pyrosequencing, sequence analysis and selection criteria, sectioning and staining of nymphs, bacterial strains and growth conditions, immunoblot analysis, tick RNA isolation, quantitative RT-PCR and microinjection, extraction and UPLC and MS/MS analysis of bacterial peptidoglycan, binding assays, biofilms assays, and microscopy can be found in SI Appendix. Details of primers used in this study can be found in SI Appendix, Table S6.
Supplementary Material
Acknowledgments
We thank Dr. Can Bruce for microbiota sequence analysis, Michael Schadt for excellent technical assistance with histology, Huihui Dong for technical assistance with assorted in vitro experiments, and Morven Graham (Yale School of Medicine, Center for Cellular and Molecular Imaging) for help with the microscopic imaging. We thank William Gray and Bradley Parry for critical input on the analysis strategy for the fluorescence microscopy. The S. aureus WTA mutants were provided by Dr. Andreas Peschel (University of Tubingen, Tubingen, Germany). This work was supported, in part, by a gift from the John Monsky and Jennifer Weis Monsky Lyme Disease Research Fund. The F.C. laboratory received funding support from Laboratory for Molecular Infection Medicine Sweden, Knut and Alice Wallenberg Foundation, Kempe and the Swedish Research Council (VR). A.K.Y. was supported by a Umeå Center for Microbial Research/VR postdoctoral position. This work was supported by National Institutes of Health Grant 41440. E.F. and C.J.-W. are investigators of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.P. is a Guest Editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the NCBI Sequence Read Archive (SRA; accession no. PRJNA353730).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613422114/-/DCSupplemental.
References
- 1.Goddard J. Infectious Diseases and Arthropods. Humana; Totowa, NJ: 2000. [Google Scholar]
- 2.Hill CA, Kafatos FC, Stansfield SK, Collins FH. Arthropod-borne diseases: Vector control in the genomics era. Nat Rev Microbiol. 2005;3(3):262–268. doi: 10.1038/nrmicro1101. [DOI] [PubMed] [Google Scholar]
- 3.Andreotti R, et al. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol. 2011;11(1):6. doi: 10.1186/1471-2180-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boissière A, et al. Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 2012;8(5):e1002742. doi: 10.1371/journal.ppat.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broderick NA, Lemaitre B. Gut-associated microbes of Drosophila melanogaster. Gut Microbes. 2012;3(4):307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dillon RJ, Dillon VM. The gut bacteria of insects: Nonpathogenic interactions. Annu Rev Entomol. 2004;49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 7.Engel P, Moran NA. Functional and evolutionary insights into the simple yet specific gut microbiota of the honey bee from metagenomic analysis. Gut Microbes. 2013;4(1):60–65. doi: 10.4161/gmic.22517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Gilbreath TM, 3rd, Kukutla P, Yan G, Xu J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS One. 2011;6(9):e24767. doi: 10.1371/journal.pone.0024767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirimotich CM, Ramirez JL, Dimopoulos G. Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe. 2011;10(4):307–310. doi: 10.1016/j.chom.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narasimhan S, et al. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe. 2014;15(1):58–71. doi: 10.1016/j.chom.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss B, Aksoy S. Microbiome influences on insect host vector competence. Trends Parasitol. 2011;27(11):514–522. doi: 10.1016/j.pt.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maltz MA, Weiss BL, O’Neill M, Wu Y, Aksoy S. OmpA-mediated biofilm formation is essential for the commensal bacterium Sodalis glossinidius to colonize the tsetse fly gut. Appl Environ Microbiol. 2012;78(21):7760–7768. doi: 10.1128/AEM.01858-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JK, et al. Purine biosynthesis, biofilm formation, and persistence of an insect-microbe gut symbiosis. Appl Environ Microbiol. 2014;80(14):4374–4382. doi: 10.1128/AEM.00739-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purdy AE, Watnick PI. Spatially selective colonization of the arthropod intestine through activation of Vibrio cholerae biofilm formation. Proc Natl Acad Sci USA. 2011;108(49):19737–19742. doi: 10.1073/pnas.1111530108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Fuente J, Estrada-Pena A, Venzal JM, Kocan KM, Sonenshine DE. Overview: Ticks as vectors of pathogens that cause disease in humans and animals. Fronts Biosci. 2008;13:6938–6946. doi: 10.2741/3200. [DOI] [PubMed] [Google Scholar]
- 16.Almeida AP, et al. Coxiella symbiont in the tick Ornithodoros rostratus (Acari: Argasidae) Ticks Tick Borne Dis. 2012;3(4):203–206. doi: 10.1016/j.ttbdis.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Carpi G, et al. Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS One. 2011;6(10):e25604. doi: 10.1371/journal.pone.0025604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heise SR, Elshahed MS, Little SE. Bacterial diversity in Amblyomma americanum (Acari: Ixodidae) with a focus on members of the genus Rickettsia. J Med Entomol. 2010;47(2):258–268. doi: 10.1603/me09197. [DOI] [PubMed] [Google Scholar]
- 19.Lalzar I, Harrus S, Mumcuoglu KY, Gottlieb Y. Composition and seasonal variation of Rhipicephalus turanicus and Rhipicephalus sanguineus bacterial communities. Appl Environ Microbiol. 2012;78(12):4110–4116. doi: 10.1128/AEM.00323-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menchaca AC, et al. Preliminary assessment of microbiome changes following blood-feeding and survivorship in the Amblyomma americanum nymph-to-adult transition using semiconductor sequencing. PLoS One. 2013;8(6):e67129. doi: 10.1371/journal.pone.0067129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponnusamy L, et al. Diversity of Rickettsiales in the microbiome of the lone star tick, Amblyomma americanum. Appl Environ Microbiol. 2014;80(1):354–359. doi: 10.1128/AEM.02987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woldehiwet Z. The natural history of Anaplasma phagocytophilum. Vet Parasitol. 2010;167(2-4):108–122. doi: 10.1016/j.vetpar.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 23.Noda H, Munderloh UG, Kurtti TJ. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl Environ Microbiol. 1997;63(10):3926–3932. doi: 10.1128/aem.63.10.3926-3932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuval B, Spielman A. Duration and regulation of the developmental cycle of Ixodes dammini (Acari: Ixodidae) J Med Entomol. 1990;27(2):196–201. doi: 10.1093/jmedent/27.2.196. [DOI] [PubMed] [Google Scholar]
- 25.Heisig M, et al. Antivirulence properties of an antifreeze protein. Cell Reports. 2014;9(2):417–424. doi: 10.1016/j.celrep.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neelakanta G, Sultana H, Fish D, Anderson JF, Fikrig E. Anaplasma phagocytophilum induces Ixodes scapularis ticks to express an antifreeze glycoprotein gene that enhances their survival in the cold. J Clin Invest. 2010;120(9):3179–3190. doi: 10.1172/JCI42868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cywes-Bentley C, et al. Antibody to a conserved antigenic target is protective against diverse prokaryotic and eukaryotic pathogens. Proc Natl Acad Sci USA. 2013;110(24):E2209–E2218. doi: 10.1073/pnas.1303573110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macfarlane S. Microbial biofilm communities in the gastrointestinal tract. J Clin Gastroenterol. 2008;42(Suppl 3 Pt 1):S142–S143. doi: 10.1097/MCG.0b013e31816207df. [DOI] [PubMed] [Google Scholar]
- 29.Mohamed JA, Huang DB. Biofilm formation by enterococci. J Med Microbiol. 2007;56(Pt 12):1581–1588. doi: 10.1099/jmm.0.47331-0. [DOI] [PubMed] [Google Scholar]
- 30.Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155(Pt 6):1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 31.Channaiah LH, Subramanyam B, McKinney LJ, Zurek L. Stored-product insects carry antibiotic-resistant and potentially virulent enterococci. FEMS Microbiol Ecol. 2010;74(2):464–471. doi: 10.1111/j.1574-6941.2010.00949.x. [DOI] [PubMed] [Google Scholar]
- 32.Ahmad A, Ghosh A, Schal C, Zurek L. Insects in confined swine operations carry a large antibiotic resistant and potentially virulent enterococcal community. BMC Microbiol. 2011;11(1):23. doi: 10.1186/1471-2180-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouhss A, et al. Identification of the UDP-MurNAc-pentapeptide:L-alanine ligase for synthesis of branched peptidoglycan precursors in Enterococcus faecalis. J Bacteriol. 2001;183(17):5122–5127. doi: 10.1128/JB.183.17.5122-5127.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patti GJ, Chen J, Schaefer J, Gross ML. Characterization of structural variations in the peptidoglycan of vancomycin-susceptible Enterococcus faecium: Understanding glycopeptide-antibiotic binding sites using mass spectrometry. J Am Soc Mass Spectrom. 2008;19(10):1467–1475. doi: 10.1016/j.jasms.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Jonge BL, Gage D, Xu N. The carboxyl terminus of peptidoglycan stem peptides is a determinant for methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46(10):3151–3155. doi: 10.1128/AAC.46.10.3151-3155.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weidenmaier C, Peschel A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 2008;6(4):276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 37.Neuhaus FC, Baddiley J. A continuum of anionic charge: Structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67(4):686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32(2):149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 39.Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev. 2008;32(2):287–306. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- 40.Smith AA, et al. Cross-species interferon signaling boosts microbicidal activity within the tick vector. Cell Host Microbe. 2016;20(1):91–98. doi: 10.1016/j.chom.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hodzic E, et al. Granulocytic ehrlichiosis in the laboratory mouse. J Infect Dis. 1998;177(3):737–745. doi: 10.1086/514236. [DOI] [PubMed] [Google Scholar]
- 42.Liu L, et al. Ixodes scapularis salivary gland protein P11 facilitates migration of Anaplasma phagocytophilum from the tick gut to salivary glands. EMBO Rep. 2011;12(11):1196–1203. doi: 10.1038/embor.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koch H, Schmid-Hempel P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci USA. 2011;108(48):19288–19292. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halos L, et al. Ecological factors characterizing the prevalence of bacterial tick-borne pathogens in Ixodes ricinus ticks in pastures and woodlands. Appl Environ Microbiol. 2010;76(13):4413–4420. doi: 10.1128/AEM.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zolnik CP, Prill RJ, Falco RC, Daniels TJ, Kolokotronis SO. Microbiome changes through ontogeny of a tick pathogen vector. Mol Ecol. 2016;25(19):4963–4977. doi: 10.1111/mec.13832. [DOI] [PubMed] [Google Scholar]
- 46.Budachetri K, Gaillard D, Williams J, Mukherjee N, Karim S. A snapshot of the microbiome of Amblyomma tuberculatum ticks infesting the gopher tortoise, an endangered species. Ticks Tick Borne Dis. 2016;7(6):1225–1229. doi: 10.1016/j.ttbdis.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dittmer J, et al. Disentangling a holobiont—Recent advances and perspectives in Nasonia wasps. Front Microbiol. 2016;7:1478. doi: 10.3389/fmicb.2016.01478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang XC, et al. The composition and transmission of microbiome in hard tick, Ixodes persulcatus, during blood meal. Ticks Tick Borne Dis. 2014;5(6):864–870. doi: 10.1016/j.ttbdis.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 49.Narasimhan S, Fikrig E. Tick microbiome: The force within. Trends Parasitol. 2015;31(7):315–323. doi: 10.1016/j.pt.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salter SJ, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hochbaum AI, et al. Inhibitory effects of D-amino acids on Staphylococcus aureus biofilm development. J Bacteriol. 2011;193(20):5616–5622. doi: 10.1128/JB.05534-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conlon BP, et al. Role for the A domain of unprocessed accumulation-associated protein (Aap) in the attachment phase of the Staphylococcus epidermidis biofilm phenotype. J Bacteriol. 2014;196(24):4268–4275. doi: 10.1128/JB.01946-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. Giving structure to the biofilm matrix: An overview of individual strategies and emerging common themes. FEMS Microbiol Rev. 2015;39(5):649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dubrac S, Boneca IG, Poupel O, Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol. 2007;189(22):8257–8269. doi: 10.1128/JB.00645-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bulet P, Hetru C, Dimarcq JL, Hoffmann D. Antimicrobial peptides in insects; structure and function. Dev Comp Immunol. 1999;23(4-5):329–344. doi: 10.1016/s0145-305x(99)00015-4. [DOI] [PubMed] [Google Scholar]
- 56.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 57.Serra F, Johnston J, Carnie J, Palou A. Altered blood amino acid distribution in genetically obese mice. Biochim Biophys Acta. 1991;1097(4):289–292. doi: 10.1016/0925-4439(91)90083-l. [DOI] [PubMed] [Google Scholar]
- 58.Glauner B, Höltje JV, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263(21):10088–10095. [PubMed] [Google Scholar]
- 59.Amanuma H, Strominger JL. Purification and properties of penicillin-binding proteins 5 and 6 from Escherichia coli membranes. J Biol Chem. 1980;255(23):11173–11180. [PubMed] [Google Scholar]
- 60.Höltje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62(1):181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 2011;30(16):3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin M, Rikihisa Y. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect Immun. 2003;71(9):5324–5331. doi: 10.1128/IAI.71.9.5324-5331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rikihisa Y, Perry BD, Cordes DO. Ultrastructural study of ehrlichial organisms in the large colons of ponies infected with Potomac horse fever. Infect Immun. 1985;49(3):505–512. doi: 10.1128/iai.49.3.505-512.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rikihisa Y, et al. Ultrastructural and antigenic characterization of a granulocytic ehrlichiosis agent directly isolated and stably cultivated from a patient in New York state. J Infect Dis. 1997;175(1):210–213. doi: 10.1093/infdis/175.1.210. [DOI] [PubMed] [Google Scholar]
- 65.Liu L, et al. Ixodes scapularis JAK-STAT pathway regulates tick antimicrobial peptides, thereby controlling the agent of human granulocytic anaplasmosis. J Infect Dis. 2012;206(8):1233–1241. doi: 10.1093/infdis/jis484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuraishi T, Binggeli O, Opota O, Buchon N, Lemaitre B. Genetic evidence for a protective role of the peritrophic matrix against intestinal bacterial infection in Drosophila melanogaster. Proc Natl Acad Sci USA. 2011;108(38):15966–15971. doi: 10.1073/pnas.1105994108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levin ML, Fish D. Interference between the agents of Lyme disease and human granulocytic ehrlichiosis in a natural reservoir host. Vector Borne Zoonotic Dis. 2001;1(2):139–148. doi: 10.1089/153036601316977741. [DOI] [PubMed] [Google Scholar]
- 68.Belongia EA. Epidemiology and impact of coinfections acquired from Ixodes ticks. Vector Borne Zoonotic Dis. 2002;2(4):265–273. doi: 10.1089/153036602321653851. [DOI] [PubMed] [Google Scholar]
- 69.Engel P, Moran NA. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol Rev. 2013;37(5):699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 70.Gonzalez-Ceron L, Santillan F, Rodriguez MH, Mendez D, Hernandez-Avila JE. Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J Med Entomol. 2003;40(3):371–374. doi: 10.1603/0022-2585-40.3.371. [DOI] [PubMed] [Google Scholar]
- 71.Cirimotich CM, et al. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332(6031):855–858. doi: 10.1126/science.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bahia AC, et al. Exploring Anopheles gut bacteria for Plasmodium blocking activity. Environ Microbiol. 2014;16(9):2980–2994. doi: 10.1111/1462-2920.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bian G, et al. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340(6133):748–751. doi: 10.1126/science.1236192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.