Abstract
BACKGROUND
Fluorescence in situ hybridisation (FISH) associated with Tyramide Signal Amplification (TSA) using oligonucleotides labeled with non-radioactive fluorophores is a promising technique for detection and differentiation of fungal species in environmental or clinical samples, being suitable for microorganisms which are difficult or even impossible to culture.
OBJECTIVE
In this study, we aimed to standardise an in situ hybridisation technique for the differentiation between the pathogenic species Paracoccidioides brasiliensis and Paracoccidioides lutzii, by using species-specific DNA probes targeting the internal transcribed spacer-1 (ITS-1) of the rRNA gene.
METHODS
Yeast and mycelial phase of each Paracoccidioides species, were tested by two different detection/differentiation techniques: TSA-FISH for P. brasiliensis with HRP (Horseradish Peroxidase) linked to the probe 5’ end; and FISH for P. lutzii with the fluorophore TEXAS RED-X® also linked to the probe 5’ end. After testing different protocols, the optimised procedure for both techniques was accomplished without cross-positivity with other pathogenic fungi.
FINDINGS
The in silico and in vitro tests show no reaction with controls, like Candida and Cryptococcus (in silico) and Histoplasma capsulatum and Aspergillus spp. (in vitro). For both phases (mycelial and yeast) the in situ hybridisation showed dots of hybridisation, with no cross-reaction between them, with a lower signal for Texas Red probe than HRP-TSA probe. The dots of hybridisation was confirmed with genetic material marked with 4’,6-diamidino-2-phenylindole (DAPI), visualised in a different filter (WU) on fluorescent microscopic.
MAIN CONCLUSION
Our results indicated that TSA-FISH and/or FISH are suitable for in situ detection and differentiation of Paracoccidioides species. This approach has the potential for future application in clinical samples for the improvement of paracoccidioidomycosis patients prognosis.
Keywords: paracoccidioidomycosis, fluorescence in situ hybridisation, tyramide signal amplification, Paracoccidioides spp
Paracoccidioides species present a relatively slow growth in culture, which difficult their clinical and environmental isolation by direct culture. Therefore, many efforts have been concentrated on molecular detection techniques (Theodoro et al. 2005, Terçarioli et al. 2007, Arantes et al. 2013) such as polymerase chain reaction (PCR) and its variations, which have been largely used for Paracoccidioides spp. detection in several environmental samples (soil, plants and animal tissues) (Bagagli et al. 1998, Corredor et al. 2005, Theodoro et al. 2005, Richini-Pereira et al. 2008, Taylor 2011, Arantes et al. 2013).
Clinically, the reference laboratorial method for diagnosis of Paracoccidioides spp. is the direct visualisation of the pathogen in tissue or its isolation in culture (Mendes & Shikanai-Yasuda 2003, Shikanai-Yasuda et al. 2006), but in cases of few infective fungal cells or uncultivable material, molecular techniques such as in situ detection of specific target DNA could be a promising choice (Perlin & Zhao 2009 , Ampel 2010 , Koepsell et al. 2012 , Litvintseva et al. 2015) for disease diagnosis because it acts directly in the biological sample (Nakagome et al. 1991 , Moter & Gobel 2000 , Liehr 2009). This method has opened a new perspective not only for environmental research of Paracoccidioides species (de Brito 1999 , Arantes et al. 2016), but also for their detection in patient’s tissues. This might also be an interesting strategy mainly for uncultivable fungi, such as Lacazia loboi another systemic pathogen from Ajellomycetacea family (Lacaz et al. 2002).
The fluorescence in situ hybridisation - Fluorescence in situ hybridisation (FISH) technique is usually associated with tyramide signal amplification (TSA), which is based on the ability of peroxidase (HRP), in the presence of low concentrations of H2O2, to convert the labeled tyramide in a substrate containing highly reactive oxidised free radicals that can covalently bind to tyrosine residues at or near to the HRP (Adams 1992). This technique is an alternative for research with environmental samples, because it exponentially increases the fluorescence signal. For instance, this technique was effective in the detection of an entire new phylum within the fungal kingdom, named Cryptomycota (Jones et al. 2011).
In situ hybridisation (ISH) has already been carried out for Paracoccidioides detection in patient’s oral biopsies, targeting the 28S rRNA coding gene with a 14 nucleotide probe (de Brito 1999). However, despite of the interesting results, when compared to the large current available database for fungal rRNA sequences, this probe cannot distinguish between P. brasiliensis and P. lutzii and also shows complementarity to 28S gene from many other fungal species. This can be explained by the high conservative nature of the 28S rRNA coding gene (White et al. 1990). So, despite of the high sensitivity of this probe, whose target is a multi-copy gene, its specificity is compromised.
In this work we purposed the use of probes whose target, the internal transcribed spacer 1 (ITS1), from the rRNA coding sequence, is multi-copy and also variable in the fungal genomes, being conserved within P. brasiliensis and also within P. lutzii. The specificity of this probes was tested in silico and in vitro, mainly in the yeast phase of these fungi. We also described the detailed standardisation of the in situ hybridisation protocol for Paracoccidioides differentiation in mycelial (saprophyte) and also in yeast (parasitic) phase, which we have already successfully applied for ecological studies on the geographical distribution of the Paracoccidioides species (Arantes et al. 2016), and now the propose of the use of in situ hybridisation for yeast phase to differentiate both pathogens directly on clinical samples.
MATERIALS AND METHODS
In silico analysis for probes design - The rRNA coding gene, more specifically the ITS-1 sequence (White et al. 1990), was used for the differential probe design as previously described for environmental aerosol samples (Arantes et al. 2016). In addition, the partial rRNA sequences from different Paracoccidioides genotypes (Teixeira et al. 2009, Arantes et al. 2013) were aligned in order to select conserved regions within species, but polymorphic between them, so that one probe hybridises to the species complex P. brasiliensis and the other to P. lutzii. GenBank sequences used were EU870314; EU870315; AY631235; EU118561; EU118560; EU118548; EU118554; EU118553; EU118549; EU118546; EU118547; EU118545; EU118543; EU118542 (P. brasiliensis) and EU870298; EU870303; EU870306; EU870309; EU870310; EU870311; AF092903; EU870299 (P. lutzii). In order to check the specificity of the probes for Paracoccidioides genus, ITS1-5.8S-ITS2 sequences of phylogenetically nearby fungi were also used in the in silico analysis, such as Histoplasma capsulatum and Emmonsia sp., access number AF129547 and AF038322 respectively, from Ajellomycetaceae family, as well as sequences of common fungal pathogens in clinical specimens, as Candida albicans (EF192231), Candida parapsilosis (EF68035), Candida tropicalis (EF190225), Candida glabrata (KX130866) and Candida metapsilosis (EF190228), Cryptococcus neoformans (KT585710; KT958228; KT958227) and Cryptococcus gattii (FJ534878; FJ534877; KC171355). The sequences were aligned in MEGA 6.0 software by using the ClustalW algorithm (Thompson et al. 1994, Tamura et al. 2013). Thirty-eight ITS sequences from P. brasiliensis, 19 from P. lutzii, two from Histoplasma capsulatum and two from Emmonsia sp. were used. The designed probes were submitted to the similarity analysis on NCBI site database by using the Blastn tool (Altschul et al. 1990) to check and confirm their specificity. The in silico analysis was used to exclude any possible cross hybridisation to other fungal species.
Fungal samples for standardisation of FISH and TSA-FISH methods - Three P. brasiliensis isolates (T16B1, Pb192 and T15LN1) and three P. lutzii (Pb01, Pb66 and PbEE) were used as positive controls for standardising hybridisation and for specificity test (cross-hybridisation between then). Isolates were obtained from the mycology collections of the Fungal Biology Lab (Department of Microbiology and Immunology, Biosciences Institute, UNESP, Botucatu, SP) and Fungal Molecular Biology Lab (University of Brasilia - UnB, DF). These isolates are from clinical (Pb192, Pb01, Pb66 and PbEE) and armadillo samples (T16B1 and T15LN1), and have been previously identified as P. brasiliensis or P. lutzii (Arantes et al. 2013).
For specificity test, the fungi Histoplasma capsulatum, Aspergillus flavus, Aspergillus fumigatus and Trichophyton mentagrophytes (Lacaz et al. 2002) are tested with Paracoccidioides spp. probes. They were used because have a higher positivity rate in clinical samples, in addition to being possible interferences as hybridisation probes in mycological laboratories and as previously described (Arantes et al. 2016) in environmental samples. Fungi were maintained on agar Sabouraud petri plates until their use in experiments, with incubation time and temperature variables for each genus and/or species.
FISH and TSA-FISH methods - Reagent preparation for TSA-FISH method was adapted from the TSA PerkinElmer® commercial kit and from the Sampling-Protocol Analysis: Parasitic-Host Dynamics Study, Nautset Marsh (Salt Pond and Mill Pond) and protocol described in Biegala et al. (2002). To colonies of mycelial phase, a fragment of 4mm2 was removed and added to the fixation solution, for the yeast phase, two calibrated handles (50 µL) were added to the fixative solution. The volume of fixing solution used in both cases was 3.0 mL.
For FISH and TSA-FISH fungal cultures (yeast and mycelia) were inactivated in fixation solution (4% paraformaldehyde plus 0.1 M phosphate buffer) for maintenance of their structure and genetic material. A sequence of 50%, 80% and 100% ethanol solutions were used to remove the fixation solution and dehydrate cells for posterior probe absorption in the hybridisation step. After dehydration, 10 mL of pre-hybridisation buffer [2.0 mL of ultra pure H2O; 4.0 mL of 40% Formamide; 1.8 mL of 5M NaCl; 200 μL of 1M Tris (pH 7.5); 100 μL of 1% SDS; 2 mL of 10% Buffer Blocking Agent] were added to the samples for stabilising and improving their permeability by differences in osmotic pressure. After this first step, the cells were hybridised with probes prepared in hybridisation buffer [2.85 mL MilliQ H20; 4.0 mL deionised formamide (40%); 1.8 mL NaCl 5M; 200 µL Tris 1M (pH 7.5); 100 µL 1% SDS; 2.0 mL Blocking buffer 10%] at a final concentration of 50 ng/μL. After 16-17 hours of incubation at 42ºC, the slides with fungal controls were washed with 50 mL of Washing Buffer [47.54 mL of ultra-pure H2O; 460 μL of 5M NaCl, 500 μL of 0.5M EDTA, 500 μL of 1% SDS and 1 mL of 1M Tris (pH 7.5)] for removal of non-specific binding probes. After washing, the slides were stabilised with 250 mL of TNT buffer [217.315 mL of ultra-pure H2O; 25 mL of 1M Tris (pH 7.5); 7.5 mL of 5M NaCl and 0.185 mL of Tween 20].
After equilibrating and washing the slides with TNT buffer, for TSA-FISH protocol, 30 μL of TSA solution from the commercial kit (TSA Plus PerkinElmer®) were added onto each slide, which were then incubated for 30 min in a dark humid chamber at room temperature. Then, the slides were washed again and dried at room temperature, following the addition of DAPI (4’,6-diamidino-2-phenylindole), a fluorescent dye that binds to DNA, mainly in A-T regions. We used DAPI in fluorescence microscopy tests to observe the DNA regions with integrity of cellular structures and to mark the genetic material and indicate the real dots of probe hybridisation, for distinguishing between the points of intracellular accumulation of the probes and actual hybridisation of the same. The slides were covered with a cover slip and observed under a fluorescence microscope. For FISH method, after addition of TNT buffer, the steps were the same applied for TSA-FISH slides but without addition of TSA solution.
RESULTS
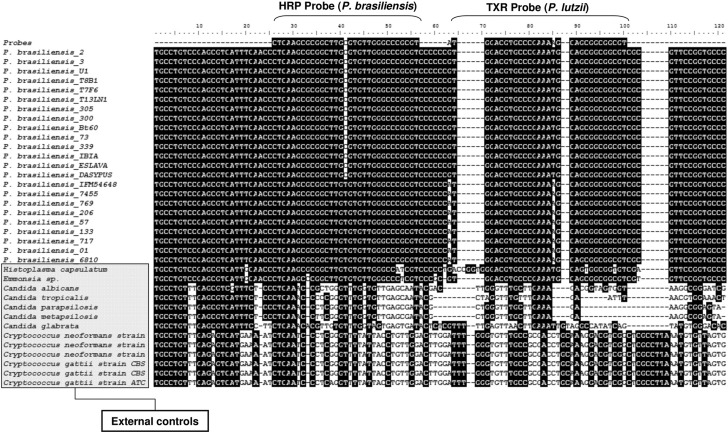
In silico analysis - Probes were designed to target the ITS1 region of rRNA in P. brasiliensis and P. lutzii. The ITS probe/HRP for P. brasiliensis distinguishes it from P. lutzii due to the C/T SNP (Single Nucleotide Polymorphism) and the ITS probe/Texas Red for P. lutzii distinguishes it from P. brasiliensis due the G/A and T/A SNPs. Both probes are specific for Paracoccidioides, and therefore, not complementary to the ITS1 from Histoplasma, Emmonsia, Candida or Cryptococcus as demonstrated by the sequence alignment (Fig. 1). The in silico analysis of probes performed on the NCBI database using the Blastn tool showed no cross-reactivity with other fungi.
Fig. 1. : in silico inferred probe specificity for in situ hybridisation of Paracoccidioides brasiliensis and P. lutzii. Aligned sequences of the internal transcribed spacer 1 (ITS1) rRNA region of Paracoccidioides spp., Histoplasma capsulatum and Emmonsia sp. used for the in silico tests of probe designing for in situ hybridisation of P. brasiliensis and P. lutzii, with fluorophores HRP + Fluorescein and Texas-Red, respectively. The design of specific probes was based on the divergences between the Paracoccidioides species and between the Paracoccidioides genus and other Ajellomycetacea species (H. capsulatum and Emmonsia spp), and other pathogenic yeasts like Cryptococcus sp. (Tremellaceae) and Candida spp. (Saccharomycetaceae).
FISH for P. lutzii and TSA-FISH for P. brasiliensis mycelia and yeast cells - The Texas-Red probe, specific for P. lutzii, successfully hybridised with mycelial cells of the Pb01, Pb66 and PbEE isolates (Supplementary data, Fig. 1A, C, E, G (1.6MB, pdf) ), while the HRP probe specific for P. brasiliensis hybridised mycelial cells of T16B1, T15LN9 and Pb192 (Supplementary data, Fig. 1I, K, M, O (1.6MB, pdf) ). The genetic material was labeled with DAPI as positive control for the nuclear hybridisation (Supplementary data, Fig. 1B, D, F, H, J, L, N, P (1.6MB, pdf) ).
Similarly, the species-specific probes Texas-Red and HRP-TSA hybridise to the yeast phase of P. lutzii (Fig. 2A-D), and P. brasiliensis (Fig. 2E-H), respectively. Some of these yeast cells hybridisation images for P. lutzii (Fig. 3A) and P. brasiliensis (Fig. 3B) were merged to the corresponding DAPI images obtained under the WU filter, using Image J software (Schineider et al. 2012) for a better visualisation of the hybridisation dots in yeast cells.
Fig. 2. : Paracoccidioides spp. yeast cells hybridisation by FISH and TSA-FISH techniques. (A, C) Yeasts of P. lutzii isolate Pb01 hybridised with Texas-Red probe by FISH technique; (B, D) yeasts of P. lutzii stained with DAPI for genetic material visualisation in cellular structures; (E, G) yeasts of P. brasiliensis isolate T16B1 hybridised with HRP-Fluorescein probe by TSA-FISH technique; (F, H) yeasts of P. brasiliensis stained with DAPI for genetic material visualisation in cellular structures. (Magnification 400x and 1000x).
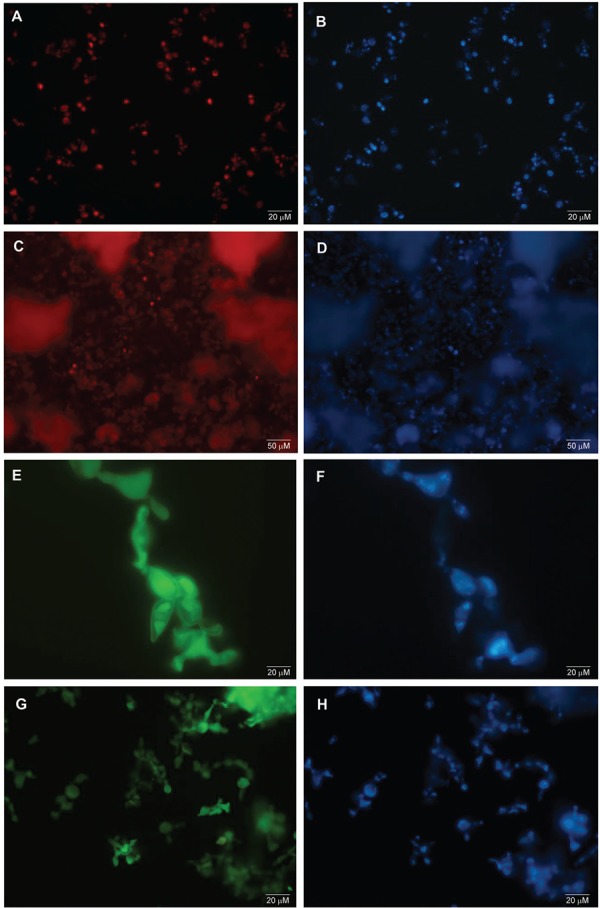
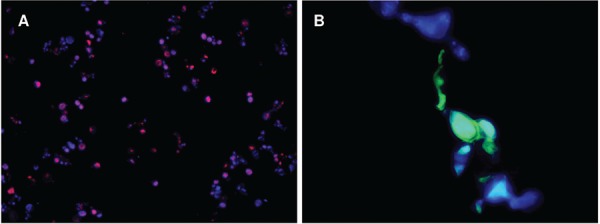
Fig. 3. : merged images of the yeast hybridisation with Paracoccidioides lutzii and P. brasiliensis specific probes. (A) Yeasts of P. lutzii hybridised with TEXAS-Red probe indicated by intracellular red dots (400x); (B) yeasts of P. brasiliensis hybridised with HRP-TSA probe indicated by intracellular green dots (1000x); (A, B) cellular walls can be visualised by structures marked in blue colour.
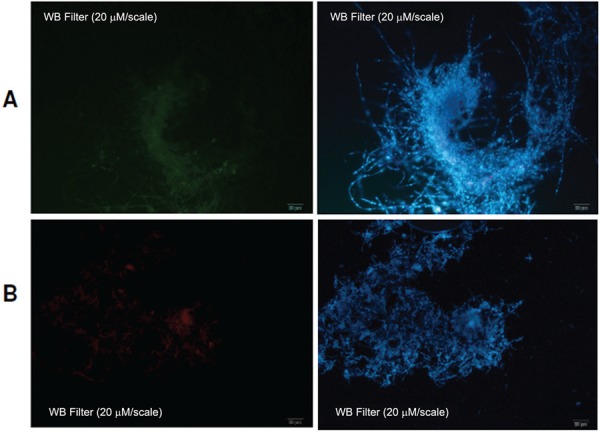
Specificity and sensitivity in vitro tests - There was no cross-hybridisation of Texas Red-probe (used in FISH for P. lutzii detection) or the HRP-probe (used in TSA-FISH for P. brasiliensis) to P. brasiliensis and P. lutzii isolates, respectively (Fig. 4). In Fig. 4A-B, we can observe the natural fluorescence of the fungal cells in a few points, due to accumulation of probes and/or fluorophores without signal emission points for hybridised probes. Also, no cross-reaction was observed for each probe against H. capsulatum (Arantes et al. 2016) or the other fungi tested, such as Aspergillus flavus, A. fumigatus and the dermatophyte T. mentagrophytes (Supplementary data, Fig. 2 (1.6MB, pdf) ). Twenty four slides were prepared for this test (six for each isolate), and after observation under fluorescence microscope, no hybridisation was visualised for each probe, proving their specificity for Paracoccidioides spp. detection. However, retention of Texas Red probe within hyphae cells was detected on a single slide of A. flavus (Supplementary data, Fig. 2 (1.6MB, pdf) ).
Fig. 4. : test for cross-hybridisation of ITS HRP and ITS Texas-Red probes against Paracoccidioides lutzii and. P. brasiliensis respectively. (A) P. lutzii isolate Pb01 (400x), subjected to ITS HRP probe by TSA-FISH technique. No hybridisation was observed; (B) P. brasiliensis isolate T16B1 (400x), subjected to ITS Texas Red probe by FISH technique. No hybridisation was observed. For both isolates genetic material was visualised with DAPI (blue) under WU filter in a fluorescence microscope.
DISCUSSION
In this study we modified existing protocols described in literature and commercial kits in order to use HRP and Texas-Red labeled oligonucleotides probes for identification of Paracoccidioides spp. cells in mycelial phase, as previously described (Arantes et al. 2016) and mainly in yeast phase, as a new approach.
During the optimisation of FISH (with Texas-Red probe) and TSA-FISH (with HRP-Fluorescein probe) techniques, a final protocol were adapted for satisfactory differentiation between Paracoccidioides species. Both probes were hybridised to their targets and showed no cross-reactivity within the genus and between Paracoccidioides spp. and other fungal species according to the in silico and in vitro analysis performed. For the probes’ specificity test against other fungi (Supplementary data, Fig. 2 (1.6MB, pdf) ), we visualised some dots of probes signal, which were considered points of probes accumulation, mainly due to non-removal of not hybridised probes during the washing steps of the fungal cells, which were confirmed due to non-genetic material stained with DAPI inside these cell, when viewed with WU filter. So, we attributed this result to the retention of crystallised probe, even after the washing steps that follow the hybridisation. Therefore, both tested probes were considered specific and suitable for fungal detection.
The signal emitted by HRP-labeled (P. brasiliensis) was higher than the Texas Red-labeled (P. lutzii) probe signal because the TSA-FISH technique significantly increases the signal emission of hybridised probes after addition of Fluorescein in the final step of TSA (Speel et al. 1999, Kubota et al. 2006). Despite this signal difference, only the application of the FISH technique using a probe labeled with Texas Red showed a satisfactory signal emission of the hybridised fungal structures. However, the fluorescence incidence tends to be difficult to measure and interpret; it depends on the observer’s experience, which requires time and dedication to identify the hybridised target cells. In our experiment, we did not visualise the slides for a long time after hybridisation because no stabilisation solutions were used. Therefore, we indicate the use of any anti-fade reagent for longer microscopic analysis in such cases (Longin et al. 1993, Wu & Luo 2006, Fouquet et al. 2015).
The in situ hybridisation technique showed to be an alternative to the molecular detection of Paracoccidioides species by PCR and its variants, such as Nested PCR. Besides, this technique also distinguishes between P. brasiliensis and P. lutzii. In fact, the FISH or TSA-fish technique implemented here has been successfully used for detection of P. brasiliensis and P. lutzii in armadillo burrows, as demonstrated in our recent study (Arantes et al. 2016). Furthermore, the detection and differentiation of P. brasiliensis and P. lutzii, directly in the analysed sample make the in situ hybridisation technique a potential tool for clinical diagnosis as well. In addition to the great specificity, this approach maintains the classic diagnostic standard reference for any mycosis, which consists in displaying the fungal agent in the biological sample. Moreover, it also has the potential to monitoring the clinical evolution of paracoccidioidomycosis, as well as to assist researches on clinical and antifungal response differences between P. brasiliensis and P. lutzii during infection.
ACKNOWLEDGEMENTS
To all research groups who directly or indirectly participated in this study.
Footnotes
Financial support: FAPESP (grant nº 2012/03233-3 and 2012/14047-6), CAPES.
REFERENCES
- Adams JC. Biotin amplification of biotin and horseradish peroxidase signals in histochemical stains. J Histochem Cytochem. 1992;40(10):1457–1463. doi: 10.1177/40.10.1527370. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ampel NM. The diagnosis of coccidioidomycosis. 2F1000 Med Rep. 2010;2 doi: 10.3410/M2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes TD, Theodoro RC, Macoris SAG, Bagagli E. Detection of Paracoccidioides sp. in environmental aerosol samples. Med Mycol. 2013;51(1):83–92. doi: 10.3109/13693786.2012.698444. [DOI] [PubMed] [Google Scholar]
- Arantes TD, Theodoro RC, Teixeira MM, Bosco SMG, Bagagli E. Environmental mapping of Paracoccidioides spp. in Brazil reveals new clues into genetic diversity, biogeography and wild host association. PLoS Negl Trop Dis. 2016;10(4):e0004606. doi: 10.1371/journal.pntd.0004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagagli E, Sano A, Coelho KL, Alquati S, Miyaji M, de Camargo ZP, et al. Isolation of Paracoccidioides brasiliensis from armadillos (Dasypus novemcinctus) captured in an endemic area of paracoccidioidomycosis. Am J Trop Med Hyg. 1998;58(4):505–512. doi: 10.4269/ajtmh.1998.58.505. [DOI] [PubMed] [Google Scholar]
- Biegala IC, Kennaway G, Alverca E, Lennon JF, Vaulot D, Simon N. Identification of bacteria associated with dinoflagellates (dinophyceae) Alexandrium spp. using tyramide signal amplification–fluorescent in situ hybridization and confocal microscopy. J Phycol. 2002;38(2):404–411. [Google Scholar]
- Corredor GG, Peralta LA, Castaño JH, Zuluaga JS, Henao B, Arango M, et al. The naked-tailed armadillo Cabassous centralis (Miller 1899): a new host to Paracoccidioides brasiliensis. Molecular identification of the isolate. Med Mycol. 2005;43(3):275–280. doi: 10.1080/13693780412331271090. [DOI] [PubMed] [Google Scholar]
- de Brito T. In situ hybridization in paracoccidioidomycosis. Med Mycol. 1999;37(3):207–211. [PubMed] [Google Scholar]
- Fouquet C, Gilles JF, Heck N, dos Santos M, Schwartzmann R, Cannaya V, et al. Improving axial resolution in confocal microscopy with new high refractive index mounting media. PLoS ONE. 2015;10(3):e0121096. doi: 10.1371/journal.pone.0121096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MD, Forn I, Gadelha C, Egan MJ, Bass D, Massana R, et al. Discovery of novel intermediate forms redefines the fungal tree of life. Nature. 2011;474(7350):200–203. doi: 10.1038/nature09984. [DOI] [PubMed] [Google Scholar]
- Koepsell SA, Hinrichs SH, Iwen PC. Applying a real-time PCR assay for Histoplasma capsulatum to clinically relevant formalin-fixed paraffin-embedded human tissue. J Clin Microbiol. 2012;50(10):3395–3397. doi: 10.1128/JCM.01705-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K, Ohashi A, Imachi H, Harada H. Visualization of mcr mRNA in a methanogen by fluorescence in situ hybridization with an oligonucleotide probe and two-pass tyramide signal amplification (two-pass TSA FISH) J Microbiol Methods. 2006;66(3):521–528. doi: 10.1016/j.mimet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Lacaz CS, Porto E, Martins JEC, Heins-Vaccari EM, de Melo NT. Tratado de micologia médica Lacaz. São Paulo: Sarvier; 2002. [Google Scholar]
- Liehr T. Fluorescence in situ Hybridization (FISH) - Application guide. Berlin Heidelberg: Springer-Verlag; 2009. [Google Scholar]
- Litvintseva AP, Marsden-Haug N, Hurst S, Hill H, Gade L, Driebe EM, et al. Valley fever: finding new places for an old disease: Coccidioides immitis found in Washington state soil associated with recent human infection. Clin Infect Dis. 2015;60(1):e1–e3. doi: 10.1093/cid/ciu681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longin A, Souchier C, French M, Bryon PA. Comparison of anti-fading agents used in fluorescence microscopy: image analysis and laser confocal microscopy study. J Histochem Cytochem. 1993;41(12):1833–1840. doi: 10.1177/41.12.8245431. [DOI] [PubMed] [Google Scholar]
- Mendes RP, Shikanai-Yasuda MA. Cimerman S, Cimerman B. Medicina Tropical. São Paulo: Atheneu; 2003. Paracoccidioidomicose; pp. 505–545. [Google Scholar]
- Moter A, Gobel UB. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J Microbiol Methods. 2000;41(2):85–112. doi: 10.1016/s0167-7012(00)00152-4. [DOI] [PubMed] [Google Scholar]
- Nakagome Y, Nagafuchi S, Seki S, Nakahori Y, Tamura T, Yamada M, et al. A repeating unit of the DYZ1 family on the human Y chromosome consists of segments with partial male-specificity. Cytogenet Cell Genet. 1991;56(2):74–77. doi: 10.1159/000133053. [DOI] [PubMed] [Google Scholar]
- Perlin DS, Zhao Y. Molecular diagnostic platforms for detecting Aspergillus. Med Mycol. 2009;47(Suppl. 1):S223–S232. doi: 10.1080/13693780802126583. [DOI] [PubMed] [Google Scholar]
- Richini-Pereira VB, Bosco SM, Griese J, Theodoro RC, Macoris SA, da Silva RJ, et al. Molecular detection of Paracoccidioides brasiliensis in road-killed wild animals. Med Mycol. 2008;46(1):35–40. doi: 10.1080/13693780701553002. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH image to imageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai-Yasuda MA, Telles FQ, Filho, Mendes RP, Colombo AL, Moretti ML. Guidelines in paracoccidioidomycosis. Rev Soc Bras Med Trop. 2006;39:297–310. doi: 10.1590/s0037-86822006000300017. [DOI] [PubMed] [Google Scholar]
- Speel EJM, Hopman AHN, Komminoth P. Amplification methods to increase the sensitivity of in situ hybridization: play CARD(S) J Histochem Cytochem. 1999;47(3):281–288. doi: 10.1177/002215549904700302. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW. One fungus = One name: DNA and fungal nomenclature twenty years after PCR. IMA fungus. 2011;2(2):113–120. doi: 10.5598/imafungus.2011.02.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MM, Theodoro RC, de Carvalho MJ, Fernandes L, Paes HC, Hahn RC, et al. Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol Phylogenetic Evol. 2009;52(2):273–283. doi: 10.1016/j.ympev.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Terçarioli GR, Bagagli E, Reis GM, Theodoro RC, Bosco SM, Macoris SA, et al. Ecological study of Paracoccidioides brasiliensis in soil: growth ability, conidia production and molecular detection. 92BMC Microbiol. 2007;7 doi: 10.1186/1471-2180-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoro RC, Candeias JM, Araújo JP, Jr, Bosco SM, Macoris SA, Padula LO, et al. Molecular detection of Paracoccidioides brasiliensis in soil. Med Mycol. 2005;43(8):725–729. doi: 10.1080/13693780500129418. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribossomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors, editors. PCR protocols: a guide to methods and applications. San Diego: Academic; 1990. pp. 315–322. [Google Scholar]
- Wu JS, Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat Protoc. 2006;1(4):2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.