Abstract
Background:
Nonsyndromic cleft lip with or without cleft palate (NSCL/P) is a common congenital malformation among live births, and depends on race and ethnic background. The CDH1 gene plays a vital role in orofacial development. Our research was conducted to examine the association between 3 single-nucleotide polymorphisms in the CDH1 gene and NSCL/P.
Methods:
Three single-nucleotide polymorphisms (rs16260, rs9929218, and rs1801552) of the CDH1 gene were genotyped using the Snapshot mini-sequencing technique in 331 patients with NSCL/P and 271 controls from the northern Chinese Han population.
Results:
The investigation indicated that presence of the CDH1 rs1801552 TT genotype under the assumption of a recessive model is related to the decreased risk for NSCL/P (odds ratio 0.53, 95% confidence interval 0.34–0.81, P = 0.003). The results were still significant after the Bonferroni correction for multiple comparisons. However, nonsignificant differences in rs16260 and rs9929218 were found between cases and controls.
Conclusion:
Our study demonstrates that the CDH1 polymorphisms were significantly associated with the risk of NSCL/P in the northern Chinese Han population. We provide further evidence regarding the role of CDH1 variations in the development of NSCL/P in a northern Chinese Han population.
Keywords: association, CDH1, NSCL/P, polymorphism
1. Introduction
Nonsyndromic cleft lip with or without cleft palate (NSCL/P) is one of the most common congenital malformations in human beings. The average incidence globally is 1.7 per 1000 live births, with regard to differences in terms of geographical position and ethnicity,[1] with higher proportions in Asian and Native American populations than in African populations. NSCL/P is regarded as a complex genetic disease attributed to multiple genes that may interact with environmental agents.[2] Because environmental factors such as maternal smoking, maternal alcohol use, poor nutrition, viral infection, medical drugs, and teratogens in the workplace and at home have been identified as risk factors for NSCL/P, much more attention has been paid to unraveling the genetic contribution of these factors.[3]
Recently, several studies have offered proof that the CDH1 gene may serve as a new candidate gene for NSCL/P.[4–6] Located on chromosome 16q22.1, the CDH1 gene consists of 16 exons separated by 15 introns. It encodes E-cadherin, which belongs to the family of cell–cell adhesion molecules.[7,8] E-cadherin plays an important role in cell adhesion, which is vital to establishing intercellular junction complexes and is required for the adhesive properties of epithelial cells.[9] The protein is highly expressed in human embryos in the front nasal prominence in the fourth and fifth week, and in the lateral and medial nasal prominences in the sixth week of human embryogenesis, indicating that it is expressed during the key phases of lip and palate development.[10] These results conformed to those obtained in mouse embryos by immunohistochemistry.[11] Therefore, the expression profile of the protein is closely related to the epithelial–mesenchymal transformation (EMT), a phenomenon that removes epithelial cells from the palatal medial edge epithelia throughout the process of palatogenesis and allows for mesenchymal continuity and palatal fusion. Due to the close relationships between EMT and E-cadherin, some researchers have regarded the CDH1gene as the “master gene” with the function of switching EMT on and off.[12–14] As a result, defects in CDH1 involved in cell–cell adhesion system dysfunction could lead to the complex events that bring about human orofacial clefts.[12]
We performed this case-control study to examine whether 3 single-nucleotide polymorphisms (SNPs) of the CDH1 gene were associated with NSCL/P in a northern Chinese Han population.
2. Material and methods
2.1. Sample
In all, 331 NSCL/P patients and 271 healthy controls, who were all Han Chinese and unrelated, were recruited for this study. They were patients at the Affiliated Stomatology Hospital of Harbin Medical University, the Second Affiliated Hospital of Harbin Medical University, Harbin Children's Hospital, and Heilongjiang Provincial Hospital. Based on the selection standard for the controls, there was no evidence of any other serious illness, particularly hereditary diseases or a family history of NSCL/P. The control group matched the case group in terms of age and sex distribution, socioeconomic status, and ethnic background.
The subjects suffering from NSCL/P were divided into 2 categories: cleft lip with or without cleft palate (CL/P) and cleft palate only (CPO). The patient group consisted of 216 individuals with CL/P and 115 individuals with CPO. The Ethics Committee of the First Affiliated Hospital of Harbin Medical University approved this hospital-based case-control study. Each participant or their guardians signed the written informed consent forms.
2.2. SNP selection, DNA extraction, target DNA amplified and genotyping
Three SNPs in CDH1 were selected in this research based on the following standard: prior evidence of association with CL/P and minor allele frequency of the SNP of at least 20% in the Chinese population in accordance with the HapMap data.
DNA was extracted from peripheral blood using an AxyPrep-96 DNA Isolation kit (Axygen Scientific, Santa Clara, CA) following the manufacturer's protocol for genetic analysis. The DNA pellet was dissolved in Tris-EDTA buffer, and its purity and concentration were identified by the spectrophotometric measurement of the absorbance at 260 and 280 nm.
Through multiplex polymerase chain reactions (PCRs), the target DNAs were amplified. PCRs were performed in 15-μL solutions containing 1 μL DNA, 1.5 μL of 10× buffer, 1.5 μL MgCl2, 0.3 μL dNTP, 0.3 μL of Taq polymerase (Fermentas, Canada), and 0.15 μL of each primer. PCR conditions were as follows: an initial 94°C for 3 minutes, then 35 cycles of 94°C for 15 seconds, 55°C for 15 seconds, 72°C for 30 seconds, and a final at 72°C for 3 minutes.
Genotyping of the 3 SNPs was performed using the Snapshot mini-sequencing technique. Three microliter of the PCR product was purified with 0.8 μL of FastAP (Fermentas, Canada) and 0.2 μL of ExoI (Fermentas, Canada), and incubated at 37°C for 15 minutes in turn at 80°C for 15 minutes. The extension reactions were conducted in a total volume of 6 μL containing 1 μL of Snapshot Mix (Applied Biosystems), 2 μL of purified PCR product, and 0.2 μL of each extension primers. The cycling conditions were 96°C for 1 minute, then 30 cycles of 96°C for 10 seconds, 52°C for 5 seconds, and 60°C for 30 seconds. One microliter of the extension products were mixed with 9 μL of HIDI (Applied Biosystems), and then, the compound was denatured at 95°C for 3 minutes. Then, the 3 SNPs were analyzed using an ABI PRISM 3730 DNA Sequencer (Applied Biosystems). For the sake of quality control, reactions were conducted repeatedly in 10% of the samples at random for each SNP, and the results were the same.
2.3. Statistical analysis
Between the case and control groups, the differences in the genotype and allele frequencies of the 3 polymorphisms were evaluated through a chi-square test. Conditional logistic regression models produced genotypic odds ratios (ORs) and 95% confidence intervals (CIs) for genotypes. All allele and genotype frequencies, Hardy–Weinberg equilibrium, pair-wise linkage disequilibrium, and haplotype analyses were conducted online using a web-based association study program.[15] A P value of <0.05 (2-sided) served as the criterion of statistical significance. We applied Bonferroni correction to adjust for multiple testing.[16]
3. Results
All of the SNPs were in Hardy–Weinberg equilibrium (Table 1). The primary information of the 3 SNPs and the minor allele frequencies are listed in Table 1. Between the NSCL/P group and the control groups, there were obvious differences in the allele frequencies of rs1801552 (P = 0.003). The allele frequencies of rs16260 and rs9929218 in the NSCL/P group were not substantially different from those among the controls. There were also differences in the allele frequencies of rs1801552 in the CPO subgroup (P = 0.001).
Table 1.
Primary information for single-nucleotide polymorphisms of CDH1 and allelic distribution.
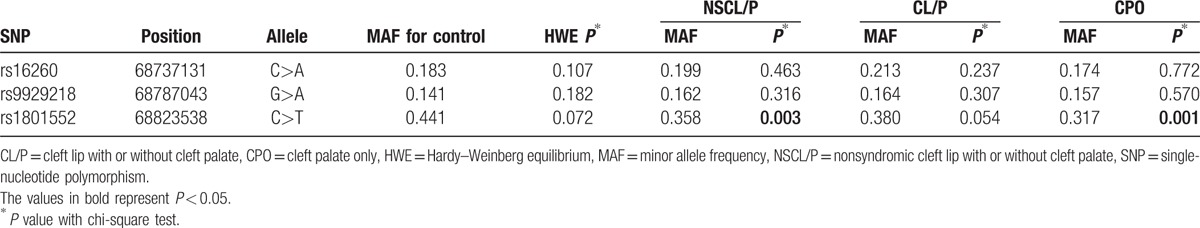
From data in Tables 2 and 3, statistical analyses showed that the rs1801552 TT genotype was related to decreased NSCL/P susceptibility (OR 0.48, 95% CI 0.30–0.77, P = 0.002) in comparison with the CC genotype. Further analyses in the recessive genetic models revealed that CDH1rs1801552 was differentially distributed between cases and controls. Combining the CT and CC genotypes of CDH1 rs1801552 under a recessive genetic model, it was found that the homozygous genotype (TT) was associated with a decreased risk of NSCL/P (OR 0.53, 95% CI 0.34–0.81). Moreover, the distribution of this genotype was significantly different between NSCL/P cases and controls (P = 0.003), and the above results were still significant after the Bonferroni correction for multiple comparisons. However, for rs16260 and rs9929218, no significant discrepancies of genotype were observed.
Table 2.
Distribution of CDH1 polymorphisms genotypes in cases and controls.

Table 3.
Association of CDH1 polymorphisms with the risk of nonsyndromic cleft lip with or without cleft palate.
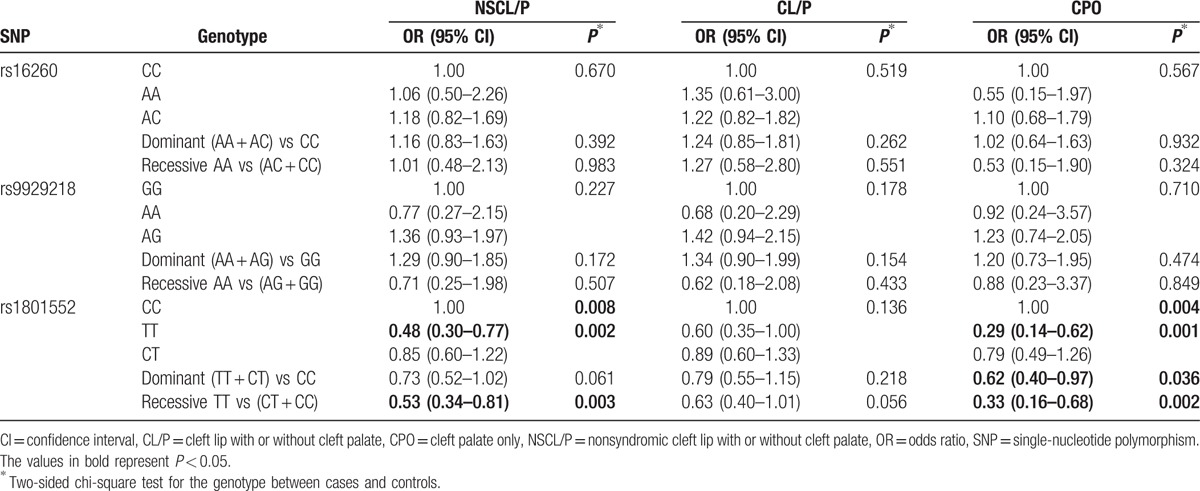
For the patients who were divided into the subgroups, there was a similar obvious trend between the patients and controls for rs1801552 in the CPO subgroup (Table 3). In this subgroup, there were significant differences in the genotypes of rs1801552. In contrast to NSCL/P, under the dominant genetic models, the results showed that CDH1 rs1801552 was differentially distributed between cases and controls (OR 0.62, 95% CI 0.40–0.97, P = 0.036) in the CPO subgroup, although these associations did not remain significant after correction for multiple testing. In the CL/P and CPO subgroups, there were no significant discrepancies in the genotypes of rs16260 and rs9929218.
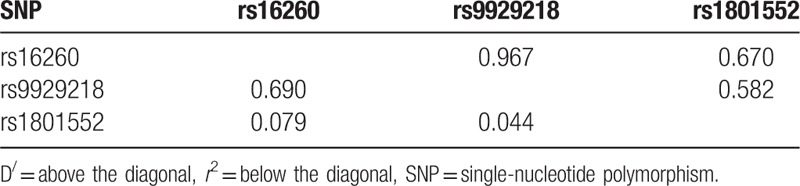
There was strong pair-wise linkage disequilibrium between rs16260 and rs9929218 (D = 0.967; Table 4), revealed by the observation of linkage disequilibrium between 3 SNPs. Therefore, we performed a haplotype analysis and did not observe any effect for any of the tested haplotypes.
Table 4.
Linkage disequilibrium analyses between CDH1 markers.
4. Discussion
Nonsyndromic oral clefts are among the most common congenital anomalies among live births[1] and are the result of complex interactions between genetic and environmental risk factors, which may exert different influences on distinct populations.[2] Therefore, identification of the causative genetic alterations will present further information on the craniofacial development.[1] This study aimed to investigate whether 3 polymorphisms in the CDH1 gene were associated with NSCL/P in a northern Chinese Han population.
CDH1 is particularly attractive as a candidate gene for NSCL/P (4–6). Vogelaar et al[4] showed that germ line CDH1 mutations added to the susceptibility of nonsyndromic oral clefts in a cohort of European descent. It was found that there were associations between nonsynonymous variants of CDH1 and oral clefts in a Thai population.[6] In addition, research on the entire exome sequencing in multiple cleft families showed novel and damaging single-nucleotide variants in the CDH1 gene in an Indian family. These results further demonstrated that oral clefts may occur as a result of mutations in CDH1.[17]
In our study, the impact of 3 polymorphisms of the CDH1 gene on the susceptibility to NSCL/P in a sample of a northern Chinese Han population was investigated. A significant difference was found between cases with NSCL/P and control subjects regarding CDH1 rs1801552. This result conforms to the findings of Hozyasz et al,[5] who revealed that the CDH1 rs1801552 variant was in association with the decreased risk of NSCL/P in a Polish population. In this study, the SNP marker rs1801552 (between alleles C and T [C > T]) presented statistical significance for TT genotype frequency among all 331 patients and 271 healthy control subjects (OR 0.53, 95% CI 0.34–0.81, in comparison with CC + CT), which suggested that rs1801552 may be associated with NSCL/P. The results indicated that the presence of the CDH1 rs1801552 TT genotype under the assumption of a recessive model is related to the decreased risk for NSCL/P in the northern Chinese Han population. However, nonsignificant differences in rs16260 and rs9929218 were found between cases and controls in the northern Chinese Han population. As for the southern Chinese population, Song and Zhang[18] revealed that rs16260 overall genotype frequencies in CPO groups were significantly different, whereas no significant associations between rs16260and NSCL/P were identified. Research by Rafighdoost et al[19] indicated that the rs16260 AC and AA genotypes were risk factors of NSCL/P in Iranians, but not rs9929218. Krasone et al[20] did not find any association between rs9929218 SNP and NSCL/P in the studied Latvian cohort.
In addition, Letra et al[21] discovered that in a Brazilian population, 2 genetic variants in CDH1 were associated with susceptibility to NSCL/P, a result found in a sample cohort containing 500 NSCL/P individuals and 500 unrelated controls. Different consequences may result from substantially multiplex genetic backgrounds and environmental exposure among various populations. When we further divided the patients into 2 subgroups (CL/P and CPO), it was found that only rs1801552 was in association with CPO and that the minor allele exhibited a protective effect. Although strong pair-wise LD existed between the rs16260 and rs9929218, we did not observe any effects for any of the tested haplotypes.
As far as we know, this report is the first on the association between rs1801552 SNP in CDH1 and NSCL/P in a northern Chinese Han population. To some extent, it revealed an association between variations of CDH1 and NSCL/P risk. Unfortunately, the functional influence of the 1801552 SNP on CDH1 activity in craniofacial tissues is still uncertain.
Recent studies show that the causative genes for NSCL/P are also involved in carcinogenesis, implying a hypothesis that the same genes may be involved in embryonic development and later in cancer development.[4,21,22] The association with the CDH1 mutation has strong biological plausibility, as CDH1 encodes E-cadherin, a cell–cell adhesion protein that is overexpressed during the critical stages of lip and palate development.[10,23] The loss of the expression of this protein is the reason for the increased capacity of cells to invade surrounding tissues.[7,24] In families presenting CL/P and hereditary diffuse gastric cancer (HDGC), mutations in CDH1 gene have been found. Frebourg et al[10] was the first to report 2 different splicing site mutations in CDH1 in 2 families where some relatives had CL/CP and diffuse gastric cancer. Frebourg et al also noted the increased risk of CL/CP in HDGC patients with a CDH1 mutation. Kluijt et al,[25] who observed high incidence of CL/P from the Dutch HDGC-families, also agreed with these results. A French team had reported similar results in 5 carriers from 2 families. In that case, there was a young patient with a history of CL, CP, and HDGC, who was demonstrated to carry a germ line mutation in CDH1.[26] Exploration of the presented correlations with cancer history would be relevant.
In the future, a personal or family history of craniofacial clefts may be incorporated into the updated HDGC-defining standard.[26] Unfortunately, there was not enough data concerning the personal and familial cancer history of participants. In our opinion, from this point on, we should rigorously and systematically monitor a personal or family history of NSCL/P in any individual with a suspicion of HDGC, as it would positively increase the probability of identifying a mutation in CDH1.
In summary, although the SNP coverage of CDH1 and the number of study participants were limited, our study demonstrates that the CDH1 polymorphisms were significantly associated with the risk of NSCL/P in the northern Chinese Han population, which is consistent with previous findings in a Polish population. This work is a supplement to the CDH1 gene studies. In the future, our studies will confirm the current data using a larger sample base and investigate their functional significance in NSCL/P development.
Acknowledgments
Our deepest gratitude should go first and foremost to all of the participants who donated samples for this study. Second, we express our appreciation to the Affiliated Stomatology Hospital of Harbin Medical University, the Second Affiliated Hospital of Harbin Medical University, Harbin Children's Hospital, and Heilongjiang Provincial Hospital.
Footnotes
Abbreviations: CI = confidence interval, CL/P = cleft lip with or without cleft palate, CPO = cleft palate only, EMT = epithelial–mesenchymal transformation, HDGC = hereditary diffuse gastric cancer, HWE = Hardy–Weinberg equilibrium, MAF = minor allele frequency, NSCL/P = nonsyndromic cleft lip with or without cleft palate, OR = odds ratio, PCR = polymerase chain reaction, SNP = single-nucleotide polymorphism.
The authors report no conflicts of interest.
References
- [1].Mossey PA, Little J, Munger RG, et al. Cleft lip and palate. Lancet 2009;374:1773–85. [DOI] [PubMed] [Google Scholar]
- [2].Dixon MJ, Marazita ML, Beaty TH, et al. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet 2011;12:167–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Stanier P, Moore GE. Genetics of cleft lip and palate: syndromic genes contribute to the incidence of non-syndromic clefts. Hum Mol Genet 2004;13(Spec No 1):R73–81. [DOI] [PubMed] [Google Scholar]
- [4].Vogelaar IP, Figueiredo J, van Rooij IA, et al. Identification of germline mutations in the cancer predisposing gene CDH1 in patients with orofacial clefts. Hum Mol Genet 2013;22:919–26. [DOI] [PubMed] [Google Scholar]
- [5].Hozyasz KK, Mostowska A, Wójcicki P, et al. Nucleotide variants of the cancer predisposing gene CDH1 and the risk of non-syndromic cleft lip with or without cleft palate. Fam Cancer 2014;13:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ittiwut R, Ittiwut C, Siriwan P, et al. Variants of the CDH1 (E-cadherin) gene associated with oral clefts in the Thai population. Genet Test Mol Biomarkers 2016;20:406–9. [DOI] [PubMed] [Google Scholar]
- [7].Stemmler MP, Bedzhov I. A Cdh1HA knock-in allele rescues the Cdh1-/- phenotype but shows essential Cdh1 function during placentation. Dev Dyn 2010;239:2330–44. [DOI] [PubMed] [Google Scholar]
- [8].Chen B, Zhou Y, Yang P, et al. CDH1-160C>A polymorphism is an ethnicity-dependent risk factor for gastric cancer. Cytokine 2011;55:266–73. [DOI] [PubMed] [Google Scholar]
- [9].Kerrigan JJ, Mansell JP, Sengupta A, et al. Palatogenesis and potential mechanisms for clefting. J R Coll Surg Edinb 2000;45:351–8. [PubMed] [Google Scholar]
- [10].Frebourg T, Oliveira C, Hochain P, et al. Cleft lip/palate and CDH1/E-cadherin mutations in families with hereditary diffuse gastric cancer. J Med Genet 2006;43:138–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Montenegro MA, Rojas M, Dominguez S, et al. Cytokeratin, vimentin and E-cadherin immunodetection in the embryonic palate in two strains of mice with different susceptibility to glucocorticoid-induced clefting. J Craniofac Genet Dev Biol 2000;20:137–43. [PubMed] [Google Scholar]
- [12].Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat 1995;154:8–20. [DOI] [PubMed] [Google Scholar]
- [13].Katoh M. Epithelial-mesenchymal transition in gastric cancer. Int J Oncol 2005;27:1677–83. [PubMed] [Google Scholar]
- [14].Tan EJ, Kahata K, Idås O, et al. The high mobility group A2 protein epigenetically silences the Cdh1 gene during epithelial-to-mesenchymal transition. Nucl Acids Res 2015;43:162–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 2005;15:97–8. [DOI] [PubMed] [Google Scholar]
- [16].Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. Br Med J 1995;310:1073–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bureau A, Parker MM, Ruczinski I, et al. Whole exome sequencing of distant relatives in multiplex families implicates rare variants in candidate genes for oral clefts. Genetics 2014;197:1039–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Song Y, Zhang S. Association of CDH1 promoter polymorphism and the risk of non-syndromic orofacial clefts in a Chinese Han population. Arch Oral Biol 2011;56:68–72. [DOI] [PubMed] [Google Scholar]
- [19].Rafighdoost H, Hashemi M, Narouei A, et al. Association between CDH1 and MSX1 gene polymorphisms and the risk of nonsyndromic cleft lip and/or cleft palate in a southeast Iranian population. Cleft Palate Craniofac J 2013;50:e98–104. [DOI] [PubMed] [Google Scholar]
- [20].Krasone K, Lāce B, Akota I, et al. IRF6 AP-2a binding site promoter polymorphism is associated with oral clefts in Latvia. Stomatologija 2014;16:132–6. [PubMed] [Google Scholar]
- [21].Letra A, Menezes R, Granjeiro JM, et al. AXIN2 and CDH1 polymorphisms, tooth agenesis, and oral clefts. Birth Defects Res A Clin Mol Teratol 2009;85:169–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gurzu S, Sugimura H, Orlowska J, et al. New insights in histogenetic pathways of gastric cancer. Medicine (Baltimore) 2015;94:e1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cavallaro U, Dejana E. Adhesion molecule signalling: not always a sticky business. Nat Rev Mol Cell Biol 2011;12:189–97. [DOI] [PubMed] [Google Scholar]
- [24].Cattaneo F, Venesio T, Molatore S, et al. Functional analysis and case-control study of -160C/A polymorphism in the E-cadherin gene promoter: association with cancer risk. Anticancer Res 2006;26:4627–32. [PubMed] [Google Scholar]
- [25].Kluijt I, Siemerink EJ, Ausems MG, et al. CDH1-related hereditary diffuse gastric cancer syndrome: clinical variations and implications for counseling. Int J Cancer 2012;131:367–76. [DOI] [PubMed] [Google Scholar]
- [26].Benusiglio PR, Caron O, Consolino E, et al. Cleft lip, cleft palate, hereditary diffuse gastric cancer and germline mutations in CDH1. Int J Cancer 2013;132:2470. [DOI] [PubMed] [Google Scholar]


