Abstract
Suicide is among the leading causes of death worldwide. The polyamine system has been increasingly implicated in the neurobiology of suicide. Previous research has indicated that epigenetic mechanisms play a role in explaining dysregulation of polyamine genes in suicide completers. Nevertheless, regulatory mechanisms explaining polyamine biosynthetic genes displaying dysregulated expression in suicide completers, including ornithine decarboxylase antizymes 1 and 2 (OAZ1 and OAZ2), S-adenosylmethionine decarboxylase (AMD1), and arginase 2 (ARG2), have yet to be elucidated. In this study, we investigated methylation patterns in the promoter region of OAZ1, OAZ2, AMD1, and ARG2 in Brodmann area 44 from a group of 33 suicide completers and 31 non-suicide controls. We found significant site-specific differences in methylation in the promoter of ARG2 and AMD1 that were also significantly negatively correlated with gene expression. These findings provide further support for a role for the involvement of epigenetic modifications in the regulation of genes associated with polyamine biosynthesis, and which may contribute to the complexity of suicidal behaviors.
Keywords: Suicide, polyamines, epigenetics, DNA methylation
Introduction
Suicide is an important cause of premature death around the world (Nock et al., 2008) and it is clear that biological factors play a role in underlying the suicide process. However, the exact molecular mechanisms at play remain largely unclear. Recently, the polyamine system has become an interesting target of research aimed towards understanding the neurobiological alterations associated with suicide (Fiori & Turecki, 2008).
Polyamines are ubiquitous aliphatic molecules, which include putrescine, spermine, spermidine, and agmatine, each of which is incorporated into a highly regulated metabolic pathway. This pathway includes 3 rate-limiting enzymes, one of which is S-adenosylmethionine decarboxylase (AMD1). The initial support for the involvement of the polyamine system in suicide came from observations that spermidine/spermine N1-acetyltransferase (SAT1) gene expression was downregulated in numerous brain regions in suicide completers as compared to control subjects (Guipponi et al., 2009; Sequeira et al., 2006). Moreover, functionally characterized genetic variants in the promoter region of SAT1 were associated with suicide (Fiori, Mechawar, & Turecki, 2009; Sequeira et al., 2006). More recently, gene expression studies have identified altered expression of additional polyamine-related genes, including ornithine decarboxylase antizymes 1 and 2 (OAZ1 and OAZ2), arginase 2 (ARG2), and AMD1 in Brodmann area 44 (BA44) of suicide completers with a history of mood disorders (Fiori et al., 2011a).
Gene expression is controlled by a variety of factors including epigenetic modifications, which are of great interest due to their regulation by environmental factors (Nestler, 2009). Post-translational histone modifications and DNA methylation are examples of epigenetic mechanisms that modify gene expression without altering the DNA sequence (Nestler, 2009). To date, epigenetic modifications have been implicated in several psychiatric phenotypes, including schizophrenia (Akbarian et al., 2005), bipolar disorder (Hobara et al., 2010), and suicide (McGowan et al., 2008). Recently, our group found a significant increase in trimethylated histone 3-lysine 4 (H3K4me3), a marker of open chromatin, in suicide completers for OAZ1, and the levels of this modification were significantly correlated with expression of this gene (Fiori, Gross, & Turecki, 2011b). However, we found no evidence for elevated levels of H3K4me3 in the promoters of OAZ2, ARG2, or AMD1. To further investigate the effects of epigenetic factors on the altered expression levels of these four genes, we investigated the potential role of DNA methylation.
Materials and Methods
Subjects
Post-mortem brain tissue from BA44, which has been previously shown to demonstrate altered levels of polyamine genes in suicides (Fiori et al., 2011a), was obtained from the Quebec Suicide Brain Bank. Tissue was dissected at 4°C, snap-frozen in liquid nitrogen, and stored at −80°C following standard procedures. The Quebec Coroner’s office assessed the cause of death for each subject, cases and controls alike, and psychiatric diagnoses were obtained using psychological autopsy following the Structured Clinical Interviews for DSM-IV Axis 1 (Dumais et al., 2005). Control subjects deceased from either natural (n=18) or accidental (n=16) causes. Methods of suicide included: hanging (n=25), overdose (n=4), jumping (n=2), cutting (n=1), drowning (n=1), and shooting (n=1). Of the 34 subjects in the suicide group, 28 were known to have an axis I disorder, which included schizophrenia (n=3), major depressive disorder (n=12), depressive disorder not otherwise specified (n=3), bipolar disorder (n=7), and substance abuse (n=3). Of those, 9 subjects had prescriptions for psychiatric medications. Eight control subjects had prescriptions for non-psychiatric medications. All subjects, including controls, were French-Canadian males and were matched for age, post-mortem interval (PMI), and brain pH. Detailed information on subjects is found in Table 1. Written informed consent was obtained from next-of-kin for all subjects, and our local institutional review board approved this study.
Table 1.
Subject information
| AGE | BRAIN pH | PMI | RIN | DNA QUALITY | SEX | ETHNICITY | |
|---|---|---|---|---|---|---|---|
| CONTROLS | 43.6 ± 2.89 | 6.53 ± 0.0487 | 45.2 ± 4.72 | 6.4 ± 0.11 | 2.05 ± 0.0105 | 100% Males (34/34) | 100% Caucasian (34/34) |
| SUICIDES | 38.6 ± 2.25 | 6.61 ± 0.0464 | 33.9 ± 3.31 | 6.7 ± 0.11 | 2.06 ± 0.00616 | 100% Males (34/34) | 100% Caucasian (34/34) |
Deatiled information on subjects used in both the gene expression and DNA methylation analyses.
Groups were controlled for age, brain pH, and PMI, as values did not differ significantly between groups (p > 0.05).
Values are expressed as either mean ± SEM or percentage.
RIN: RNA Integrity number; DNA Quality is expressed as the 260/280 ratio.
Gene Expression
RNA extracted from BA44 of 34 non-suicide controls and 34 suicide completers using QIAGEN’s RNeasy Mini kit was used to synthesize cDNA. All subjects had RNA Integrity Number (RIN) values greater than 5.0, as determined using the Agilent 2100 Bioanalyzer and Agilent RNA 6000 Nano Assay protocol (Table 1). Gene expression of all four genes was measured using quantitative real-time polymerase chain reaction (qRT-PCR) on Applied Biosystems’ 7900HT Fast Real-Time PCR System. SYBR green primers for each gene are listed in Table 2, where glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. The 9600 emulation thermal cycle protocol was: 50°C for 2 minutes, 95°C for 10 minutes, and 40 repetitions of 95°C for 15 seconds and 60°C for 1 minute. The data was extracted by relative quantification using Applied Biosystems’ SDS 2.4 and RQ Manager 1.2.1 software.
Table 2.
Gene expression primers
| GENE | FORWARD PRIMER | REVERSE PRIMER |
|---|---|---|
| GAPDH | TTGTCAAGCTCATTTCCTGG | TGTGAGGAGGGGAGATTCAG |
| AMD1 | GATGGAACTTATTGGACTATTCACATCAC | CTGTGCGACATTTAGAACTCTGATTAAC |
| ARG2 | TTGCTGAGGAAATACACAATACAGG | GGTTAGCTGTAGTCTTCGCCTC |
| OAZ1 | GACAGCTTTGCAGTTCTCCTGG | TTCGGAGCAAGGCGGCTC |
| OAZ2 | GCTGATGGGAGCAAAGAAGG | AGCTGAAGGTCTTCAGGAGTG |
Primers used to assess gene expression by qRT-PCR. Glyceraldehyde-3-phosphate (GAPDH) was used as the endogenous control. All primers are listed from 5′ → 3′
Methylation Analysis
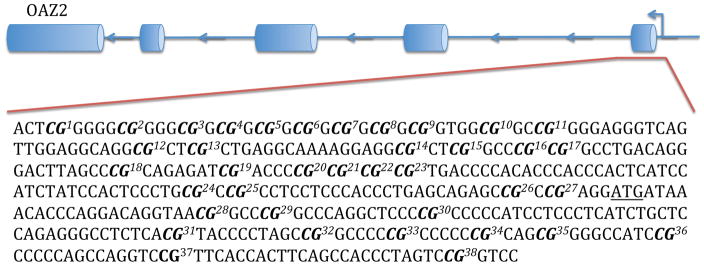
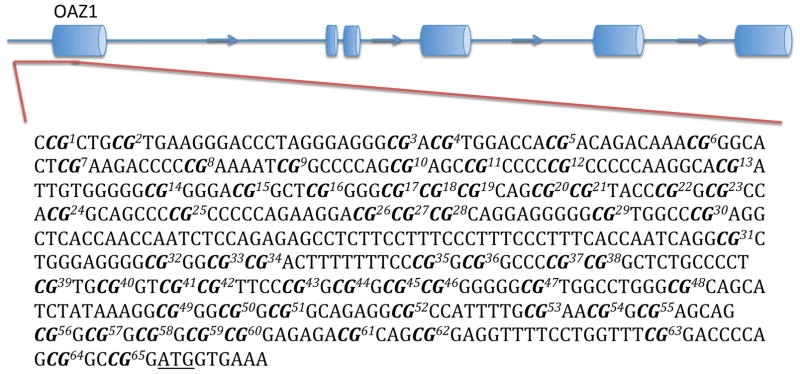
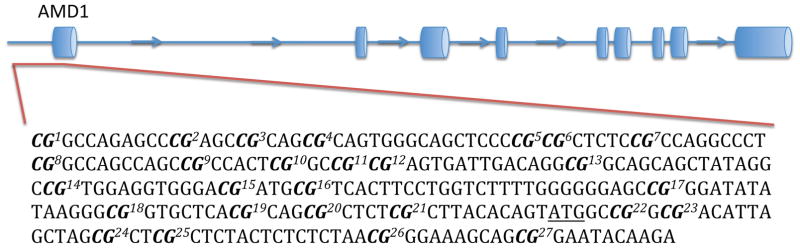
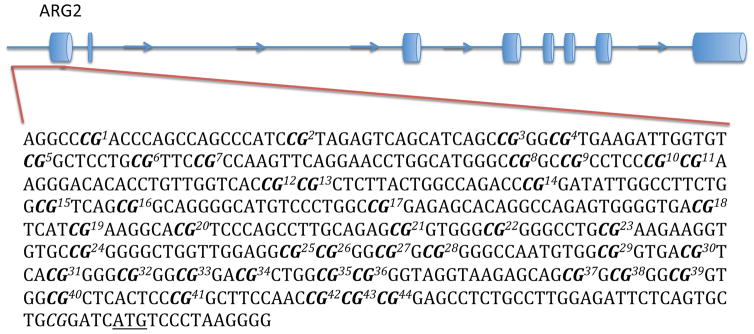
Genomic DNA was extracted from BA44 of 31 non-suicide controls and 33 suicide completers using QIAGEN’s QIAamp DNA Mini Kit. Concentrations of genomic DNA were assessed using the Thermo Scientific Nanodrop 1000 spectrophotometer and each sample had a 260/280 ratio greater than 1.8 (Table 1). Three non-suicide controls and one suicide completer were excluded from this analysis due to poor DNA quality. Genomic DNA from each sample was bisulfite-treated using QIAGEN’s Epitect Bisulfite Kit following the manufacturer’s guidelines. At the Génome Québec Innovation Centre, Sequenom’s EpiTYPER was used to characterize differentially methylated CpGs in the promoter region of each gene. Promoter regions were defined as approximately 500 base pairs (bp) upstream of the transcription start site (TSS) (Fig. 1). In order to meet the mass threshold necessary for accurate measurement by Sequenom’s EpiTYPER, CpGs within the promoter region were clustered together by the EpiTYPER software. As such, certain analyzed fragments contained more than one CpG. Levels of methylation were assessed for each cluster of CpGs.
Figure 1. DNA regions for methylation analysis.
The figure shows the predicted structure of AMD1, ARG2, OAZ1, and OAZ2, as well as the DNA sequences that were analyzed. Each sequence represents the promoter, which was defined as a region of approximately 500 base pairs upstream of the TSS. Bolded CpGs are numbered sequentially to correspond with figures 3 and 4. The underlined ATG represents the predicted TSS.
Statistics
Statistical analyses were performed on SPSS version 18 and GraphPad Prism 5. Differences in methylation between groups and across all CpGs were assessed using a 2-way, mixed model ANOVA followed by LSD post-hoc analyses. In all cases, p-values were considered statistically significant for p ≤ 0.05, and all p-values were corrected for age, PMI, and psychiatric medication.
Results
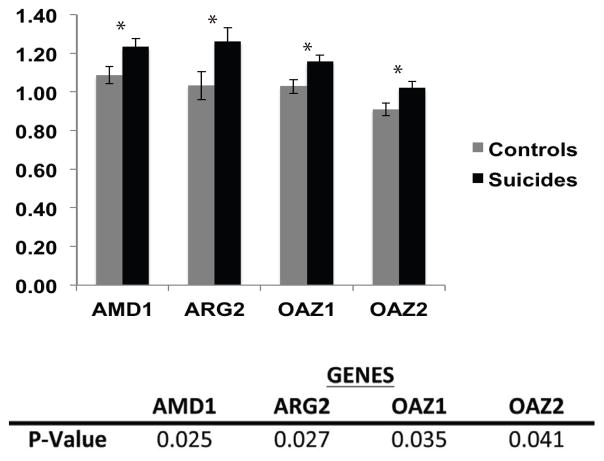
In this study, we first examined the level of gene expression of OAZ1, OAZ2, AMD1, and ARG2 (Fig. 2). For all of these genes, we found increased expression levels in suicides as compared to the non-suicide controls (p=0.035, 0.041, 0.025, and 0.027, respectively). We then examined the methylation levels in a region of the promoter, upstream of the TSS, for each gene. Due to the repressive function of methylation in gene promoters (Maunakea et al., 2010), we expected generally low levels in these functionally active promoters. Indeed, mean methylation levels across the promoter of the 4 genes were between 3% and 8%, with OAZ1 being the only gene to show overall methylation group differences (p=0.029). OAZ2 also showed an overall group difference, although it was a trend towards significance (p=0.057) Each of the four genes, however, showed CpG site-specific group differences, some of which were significantly correlated with gene expression.
Figure 2. OAZ1, OAZ2, AMD1, and ARG2 gene expression is increased in suicide completers as compared to non-suicide controls.
Gene expression levels ± SEM in the promoter region of OAZ1, OAZ2, AMD1, and ARG2 between non-suicide controls (grey) and suicide completers (black). One-tailed t-test p-values for expression of each gene are listed in the table.
* = significant differences of p ≤ 0.05 in a one-tailed t-test.
OAZ1 and OAZ2
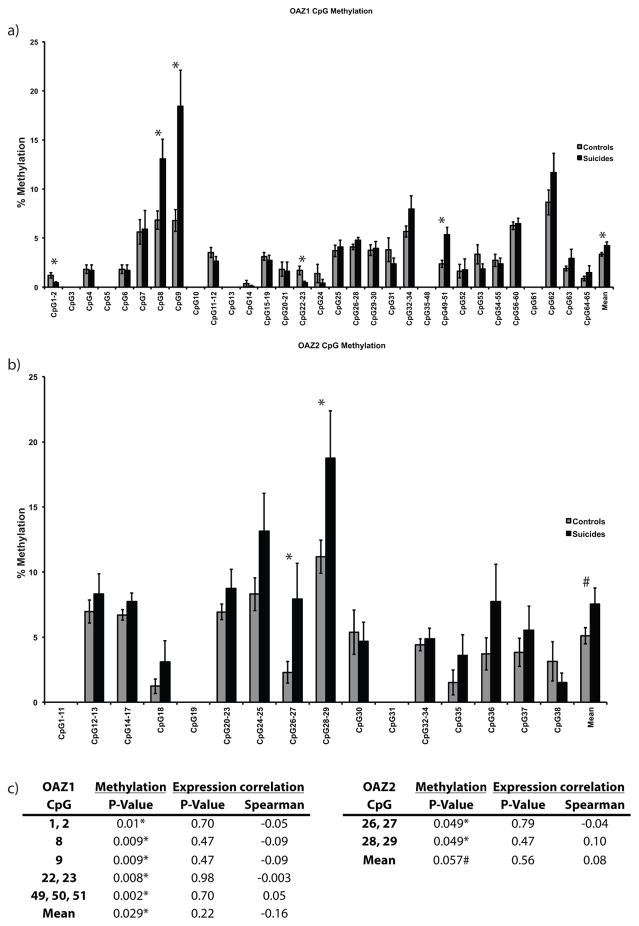
The investigated regions for OAZ1 and OAZ2 were 481 and 480 base pairs (bp) in length, respectively. Within these regions, there were 65 and 38 CpGs, respectively. Methylation levels in OAZ1 ranged between 0.1% to 18.4% while those in OAZ2 were between 1.2% and 18.7%. Methylation data for OAZ1 showed a significant main effect of CpG sites (F(26, 113) = 31.8, p<0.001) and a significant interaction between phenotype and CpG sites (F(53, 113) = 2.53, p=0.001). Post-hoc tests showed significant hypomethylation in suicide completers for CpGs1-2 (p=0.01) and CpGs22-23 (p=0.008). In addition, there was significant hypermethylation in suicides completers compared to controls for CpG8 (p=0.009), CpG9 (p=0.008), CpGs49-51 (p=0.002), and mean levels across all CpGs (p=0.029) (Fig. 3a). However, none of these differences correlated with OAZ1 gene expression (Fig. 3c). Data for OAZ2 revealed a significant main effect of CpG sites (F(13,61) = 9.06, p<0.001) and of phenotype (F(1, 61) = 13.2, p<0.001). Post-hoc tests highlighted significant hypermethylation in suicide completers for CpGs26-27 (p=0.049) and CpGs28-29 (p=0.049), while there was a trend toward significance of the mean levels across all CpGs (p=0.057) (Fig. 3b). Similar to OAZ1, none of these differences correlated significantly with OAZ2 gene expression (Fig. 3c).
Figure 3. Methylation differences across individual CpGs and across all CpGs are not correlated with gene expression for OAZ1 and OAZ2.
a, b) Mean percent levels of methylation ± SEM in the promoter region of OAZ1 and OAZ2, respectively, for 31 non-suicide controls (grey) and 33 suicides (black).
c) P-value for % methylation and Spearman correlations between significantly different CGs and gene expression.
* = significant differences of p ≤ 0.05.
# = trend towards significance, with p = 0.057
AMD1
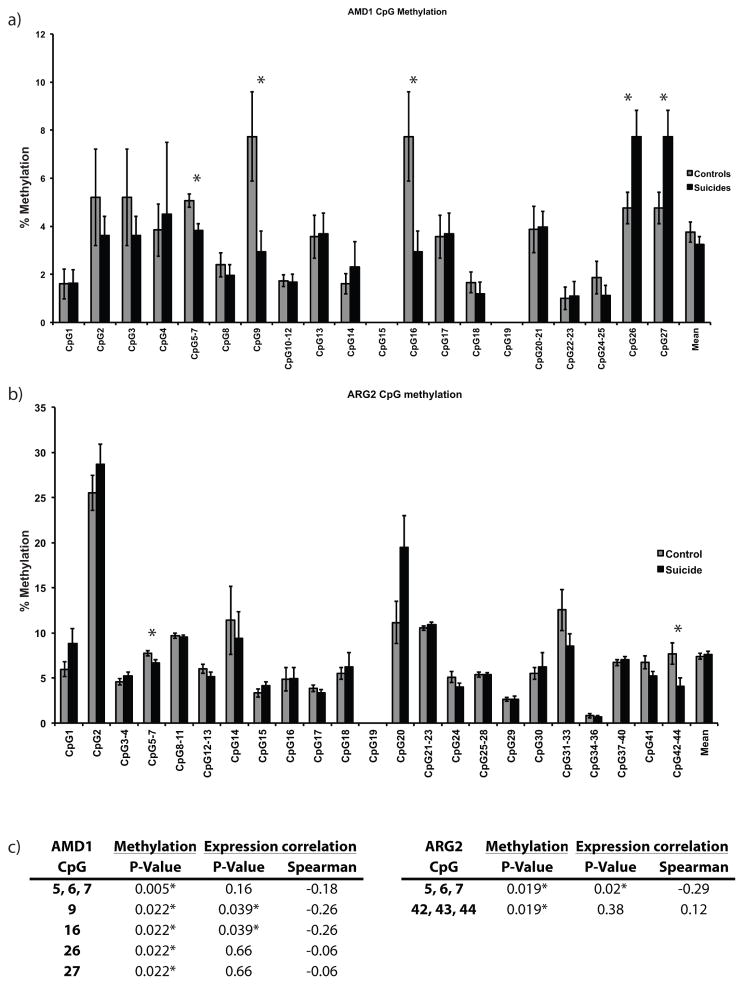
The investigated region for AMD1 was 479 bp in length, and contained 44 CpGs. Methylation levels across this region ranged from 1.0% to 7.7%. Statistical analyses highlighted a significant main effect of CpG sites (F(17,77) = 14.5, p<0.001) and a significant interaction between phenotype and CpG sites (F(35, 77) = 1.86, p=0.028). Although we found no significant differences in overall methylation between groups, post-hoc tests did show significant hypomethylation in suicide completers for CpGs5-7 (p=0.005), CpG9 (p=0.022), and CpG 16 (p=0.022). In addition, CpG26 and CpG27 were hypermethylated (p=0.022) in this group (Fig. 4a). Of particular interest, hypomethylation of CpG9 and CpG16 showed significant negative correlations with AMD1 gene expression (Fig. 4c).
Figure 4. Methylation differences across individual CpGs for AMD1 and ARG2 are significantly correlated with gene expression.
a, b) Mean percent levels of methylation ± SEM in the promoter region of AMD1 and ARG2, respectively, for 31 non-suicide controls (grey) and 33 suicides (black).
c) P-value for % methylation and Spearman correlations between significantly different CGs and gene expression.
* = significant differences of p ≤ 0.05.
ARG2
The investigated region for ARG2 was 495 bp in length, and contained 27 CpGs. Mean methylation levels across CpGs in this region were between 0.7% and 28.7%. Data for ARG2 showed a significant main effect of CpG sites (F(21, 93) = 160.1, p<0.001) and a trend toward significant interaction between phenotype and CpG sites (F(43, 93) = 1.60, p=0.062). Similar to the results of AMD1, we found no significant differences in overall methylation. However, we did find significant hypomethylation in CpGs5-7 (p=0.019) and CpGs42-44 (p=0.019) in suicide completers (Fig. 4b), the former being significantly negatively correlation with ARG2 gene expression (Fig. 4c).
Although we found both site-specific and overall differences in methylation between groups for all genes, only DNA methylation in the promoter region of ARG2 and AMD1 shows a relationship to their respective gene’s levels of expression. These effects were all independent of age, PMI, and prescription of psychiatric medication. Brain pH was not assessed as a covariate in the methylation analyses as it has been previously shown that brain pH does not significantly impact the DNA methylation landscape (Ernst et al., 2008).
Discussion
Overall, our results suggest that the increases in gene expression of AMD1 and ARG2 in suicide completers are correlated with decreases in methylation of specific CpGs in the promoter of these genes. Phenotypic changes in gene expression that are elicited by mechanisms other than DNA sequence variation, including epigenetic factors, have been increasingly investigated. To date, DNA methylation, histone modifications, and non-coding RNAs are among the most well-studied mechanisms of epigenetic regulation (Dudley, Li, Kobor, Kippin, & Bredy, 2011).
Of particular interest is the silencing effect of DNA methylation at gene promoters, which is mediated primarily by preventing the binding of transcription factors (Elliott, Ezra-Nevo, Regev, Neufeld-Cohen, & Chen, 2010). In this study, we show significant site-specific decreases in DNA methylation in the promoter region of ARG2 and AMD1 that are correlated with the increases in levels of expression of these genes in suicide completers. Although we have not investigated the exact mechanism by which DNA methylation may act on the expression of ARG2 and AMD1, a recent study by Kubota and colleagues showed that decreases in E2F transcription factor 1 (E2F-1) are associated with decreased expression of genes related to the pathogenesis of major depression (Kubota et al., 2009). Interestingly, our results show decreases in DNA methylation in the promoter region of ARG2 and AMD1 in an area where E2F-1 is predicted to bind. This modification may, in turn, lead to increased binding of E2F-1 and, therefore, increased gene expression. Certainly, further experimentation is necessary, however these results provide insight into possible mechanisms explaining the relationship between ARG2 and AMD1 expression and suicide.
Although DNA methylation is the best characterized epigenetic mechanism (Dudley et al., 2011), post-translational histone modifications represent an equally important avenue to investigate, as these modifications may also affect gene expression by altering binding of transcription factors (Elliott et al., 2010). Previously, our group showed significant increases in H3K4me3 in OAZ1 of suicide completers. Levels of this modification, representative of open and actively transcribed DNA, were positively correlated with the increases in OAZ1 gene expression in suicide completers. Since there were no significant differences in this modification in OAZ2, AMD1, or ARG2, it was hypothesized that the increases in gene expression may be associated with other mechanisms, such as DNA methylation. The results from this study suggest that at least part of the differential expression in AMD1 and ARG2 may be explained by differential DNA methylation. However, this does not seem to be the case for OAZ2, at least not for the region investigated.
The combined effect of epigenetic modifications is not limited to histone modifications. Undoubtedly, there is a relationship between DNA methylation and histone modifications in determining gene expression. Interestingly, this relationship can work in both directions, where DNA methylation can either be directed by or be a template for histone modifications (Cedar & Bergman, 2009). These complex interactions are likely to be mediated by histone and DNA methyltransferases, as well as other methylation-related enzymes. Previous work has shown a significant correlation between OAZ1 promoter levels of H3K4me3 and OAZ1 gene expression in BA 44 of suicide completers (Fiori et al., 2011b). In this same brain region and also in suicide completers, here we also show an overall increase in mean methylation levels in the promoter of OAZ1. Taken together, it may be proposed that one epigenetic modification may be dependent on another, and that the combined effects may lead to the observed phenotypic differences in gene expression.
The results of this study provide insight into the molecular mechanisms associated with altered polyamine gene expression in suicide completers. However, the results of this study should be interpreted taking into account their limitations. For one, we investigated DNA methylation in a 500 bp region of the promoters of the genes investigated. As such, it is possible that for OAZ1 and OAZ2, DNA methylation changes in other regulatory regions of these genes are associated with their observed upregulation in suicide completers. Similarly, although we have observed in this study results suggesting a potential mechanism for the overexpression of ARG2 and AMD1 in suicide completers, further research is necessary to find the exact action of DNA methylation on these genes. It is also possible that the expression of these genes is regulated by other epigenetic mechanisms such as non-coding RNAs or other post-translational histone modifications (Bannister & Kouzarides, 2011). Finally, due to the use of post-mortem brain tissue, it is possible that there were confounding post-mortem or clinical variables that were not accounted for in this study. It is important to note that we controlled for the effects of the most important confounding post-mortem variables.
Future research should not only investigate larger regions of the promoter, but should also take into consideration specific cell types. In this study, we investigated DNA extracted from brain tissue homogenates; however, DNA methylation patterns are tissue and cell-type specific (Iwamoto et al., 2011). Therefore, differentiating the epigenetic landscapes of different brain cell types may lead to further insight into the relationship between these genomic marks and suicidal behaviors.
In its entirety, this study provides insight into the effects of promoter methylation on increased expression of polyamine biosynthetic genes in suicide completers. We showed that DNA methylation at specific CpGs plays an important role in the upregulation of ARG2 and AMD1. These findings lend further evidence toward the importance of the polyamine system in the neurobiology of suicidal behavior.
References
- Akbarian S, Ruehl MG, Bliven E, Luiz LA, Peranelli AC, Baker SP, Roberts RC, Bunney WE, Jr, Conley RC, Jones EG, Tamminga CA, Guo Y. Chromatin alterations associated with down-regulated metabolic gene expression in the prefrontal cortex of subjects with schizophrenia. Archives of general psychiatry. 2005;62:829–840. doi: 10.1001/archpsyc.62.8.829. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell research. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature reviews Genetics. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Dudley KJ, Li X, Kobor MS, Kippin TE, Bredy TW. Epigenetic mechanisms mediating vulnerability and resilience to psychiatric disorders. Neuroscience and biobehavioral reviews. 2011 doi: 10.1016/j.neubiorev.2010.12.016. [DOI] [PubMed] [Google Scholar]
- Dumais A, Lesage AD, Lalovic A, Seguin M, Tousignant M, Chawky N, Turecki G. Is violent method of suicide a behavioral marker of lifetime aggression? The American journal of psychiatry. 2005;162:1375–1378. doi: 10.1176/appi.ajp.162.7.1375. [DOI] [PubMed] [Google Scholar]
- Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nature neuroscience. 2010;13:1351–1353. doi: 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- Ernst C, McGowan PO, Deleva V, Meaney MJ, Szyf M, Turecki G. The effects of pH on DNA methylation state: In vitro and post-mortem brain studies. Journal of neuroscience methods. 2008;174:123–125. doi: 10.1016/j.jneumeth.2008.06.027. [DOI] [PubMed] [Google Scholar]
- Fiori LM, Bureau A, Labbe A, Croteau J, Noel S, Merette C, Turecki G. Global gene expression profiling of the polyamine system in suicide completers. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2011a:1–11. doi: 10.1017/S1461145710001574. [DOI] [PubMed] [Google Scholar]
- Fiori LM, Gross JA, Turecki G. Effects of histone modifications on increased expression of polyamine biosynthetic genes in suicide. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2011b:1–6. doi: 10.1017/S1461145711001520. [DOI] [PubMed] [Google Scholar]
- Fiori LM, Mechawar N, Turecki G. Identification and characterization of spermidine/spermine N1-acetyltransferase promoter variants in suicide completers. Biological psychiatry. 2009;66:460–467. doi: 10.1016/j.biopsych.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Fiori LM, Turecki G. Implication of the polyamine system in mental disorders. Journal of psychiatry & neuroscience: JPN. 2008;33:102–110. [PMC free article] [PubMed] [Google Scholar]
- Guipponi M, Deutsch S, Kohler K, Perroud N, Le Gal F, Vessaz M, Laforge T, Petit B, Jollant F, Guillaume S, Baud P, Courtet P, La Harpe R, Malafosse A. Genetic and epigenetic analysis of SSAT gene dysregulation in suicidal behavior. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2009;150B:799–807. doi: 10.1002/ajmg.b.30901. [DOI] [PubMed] [Google Scholar]
- Hobara T, Uchida S, Otsuki K, Matsubara T, Funato H, Matsuo K, Suetsugi M, Watanabe Y. Altered gene expression of histone deacetylases in mood disorder patients. Journal of psychiatric research. 2010;44:263–270. doi: 10.1016/j.jpsychires.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Ueda J, Oldham MC, Ukai W, Hashimoto E, Saito T, Geschwind DH, Kato T. Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome research. 2011;21:688–696. doi: 10.1101/gr.112755.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K, Inoue K, Hashimoto R, Kumamoto N, Kosuga A, Tatsumi M, Kamijima K, Kunugi H, Iwata N, Ozaki N, Takeda M, Tohyama M. Tumor necrosis factor receptor-associated protein 1 regulates cell adhesion and synaptic morphology via modulation of N-cadherin expression. Journal of neurochemistry. 2009;110:496–508. doi: 10.1111/j.1471-4159.2009.06099.x. [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, Huang TC, Unterberger A, Suderman M, Ernst C, Meaney MJ, Turecki G, Szyf M. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PloS one. 2008;3:e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ. Epigenetic mechanisms in psychiatry. Biological psychiatry. 2009;65:189–190. doi: 10.1016/j.biopsych.2008.10.030. [DOI] [PubMed] [Google Scholar]
- Nock MK, Borges G, Bromet EJ, Cha CB, Kessler RC, Lee S. Suicide and suicidal behavior. Epidemiologic reviews. 2008;30:133–154. doi: 10.1093/epirev/mxn002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira A, Gwadry FG, Ffrench-Mullen JM, Canetti L, Gingras Y, Casero RA, Jr, Rouleau G, Benkelfat C, Turecki G. Implication of SSAT by gene expression and genetic variation in suicide and major depression. Archives of general psychiatry. 2006;63:35–48. doi: 10.1001/archpsyc.63.1.35. [DOI] [PubMed] [Google Scholar]