Our data indicate that in the context of HCC, miR-214 acts as a putative tumour suppressor by targeting UCP2 and defines a novel mechanism of regulation of UCP2.
Keywords: cell, hepatocellular carcinoma (HCC), HepG2, uncoupling proteins, uncoupling protein 2 (UCP2)
Abstract
Gemcitabine (GEM), a commonly used chemotherapeutic agent in hepatocellular carcinoma (HCC) patients, uses oxidative stress induction as a common effector pathway. However, GEM alone or in combination with oxaliplatin hardly renders any survival benefits to HCC patients. We have recently shown that this is part due to the overexpression of the mitochondrial uncoupling protein 2 (UCP2) that in turn mediates resistance to GEM in HCC patients. However, not much is known about regulatory mechanisms underlying UCP2 overexpression in HCC. Differential protein expression in HCC cell lines did not show a concomitant change in UCP2 transcript level, indicating post-transcriptional or post-translational regulatory mechanism. In situ analysis revealed that UCP2 is a putative target of miR-214. miR-214 expression is significantly down-regulated in HCC patient samples as compared with normal adjacent tissues and in cell line, human hepatoblastoma cells (HuH6), with high UCP2 protein expression. We demonstrated using miR-214 mimic and antagomir that the miRNA targeted UCP2 expression by directly targeting the wild-type, but not a miR-214 seed mutant, 3’ UTR of UCP2. Overexpression of miR-214 significantly attenuated cell proliferation. Finally, analysis in 20 HCC patients revealed an inverse correlation in expression of UCP2 and miR-214 (Pearson's correlation coefficient, r=−0.9792). Cumulatively, our data indicate that in the context of HCC, miR-214 acts as a putative tumour suppressor by targeting UCP2 and defines a novel mechanism of regulation of UCP2.
INTRODUCTION
A widely expressed subcategory of mitochondrial anion-carrier in animals and plants is the uncoupling protein (UCP) family, with the mammalian genome encoding uncoupling protein 1 (UCP1) to uncoupling protein 5 (UCP5) homologues [1–3]. The most ubiquitous among these five homologues is uncoupling protein 2 (UCP2), with detectable expression in skeletal muscle, brain, pancreas, liver and immune cells [4]. UCP2 is located in chromosome 11q13.4 and encodes for a protein of 309 amino acids and predicted molecular mass of 33.299 kDa. UCP2 is largely expressed in the inner mitochondrial membrane, but expression is also noted in the nucleus, peroxisome, cytosol and plasma membrane [4].
UCP2 in conjunction with uncoupling protein 3 (UCP3) function in suppressing electron transport chain mediated generation of reactive oxygen species (ROS) [5,6]. Physiological levels of ROS are involved in a multitude of cellular functions, inclusive of inflammation, apoptosis, phagocytosis and proliferation [7]. However, overproduction of ROS leads to oxidative damage [8].
Given this intricate role of UCPs in maintaining ROS homoeostasis and cell cycle progression, it is hardly surprising that their aberrant expression have pro-tumorigenic effects on the cell [9]. UCP2 is found to be overexpressed in hepatocellular carcinoma (HCC) [10] and colon cancer [11]. In colon cancer cells, UCP5 is also overexpressed [12]. Current evidence suggests that UCP2 targets p53 and reverses pro-apoptotic signals initiated by p53 in response to oxidative stress [13]. We have recently shown that UCP2 expression mediates resistance to Gemcitabine (2’,2’-difluoro-2’-deoxycytidine; GEM), which is used in combination with oxaliplatin as chemotherapeutic agents in HCC and that inhibition of UCP2 makes HCC cell lines susceptible to treatment with GEM [14].
Given the important role of UCP2 in HCC, it is imperative to understand the regulatory mechanisms that dictate expression of UCP2 in HCC. Our experiments have cumulatively shown that UCP2 transcript is post-transcriptionally regulated by miR-214 in normal hepatic cells and that down-regulation of miR-214 in HCC induces UCP2 expression in these HCC cells.
MATERIALS AND METHODS
Clinical samples, tissue processing and ethical considerations
Fresh-frozen and paraffin-embedded HCC tissues and corresponding adjacent non-tumorous HCC tissue samples were obtained from 25 Chinese patients at Qilu Hospital of Shandong University between 2010 and 2014. All cases were included post review by pathologist and histological confirmation as HCC and only where complete clinical pathology and follow-up data were available. None of the 25 included patients underwent preoperative local or systemic treatment. The study protocol was approved by the Institutional Review Board of the Qilu Hospital of Shandong University. Freshly harvested samples were immersed in RNAlater (Life Technologies) before snap freezing within 30 min post-surgery. All tissue samples were stored in liquid nitrogen until further use.
Cell culture
HCC cell lines human hepatoblastoma cells (HuH6) and human lens epithelial cells (HLE) were obtained from the A.T.C.C. and maintained at 37°C in a CO2 incubator in Dulbecco's modified Eagle's media (DMEM) supplemented with 10% FBS (Gibco) and 100 I.U./ml penicillin and 100 μg/ml streptomycin (Gibco).
Isolation of mitochondria
Isolation of mitochondria from different cell lines was as recently and previously described [14,15].
RNA and miRNA extraction and quantitative real-time PCR
Total RNA was isolated from cultured cells and tumour tissues using Trizol reagent. First strand cDNA was synthesized using the RevertAid™ First Strand cDNA synthesis Kit (Life Technologies), which was then used for real-time PCR using TaqMan Gene Expression probes (Life Technologies). 18s rRNA (TaqMan Assay ID: Hs03003631_g1) was used as an internal control for assessing UCP2 (TaqMan Assay ID: Hs01075227_m1) transcript level. Data were normalized to 18s rRNA expression and analysed by the −ΔΔCt method. According to the manufacturer's instructions, miRNA from tissues and cells was extracted using the mirVana miRNA isolation kit (Life Technologies) and the expression levels of hsa-miR-214 and U6 small nuclear RNA (RNU6B) were detected by TaqMan miRNA assays (Life Technologies) (TaqMan Assay IDs: 002306 and 001093 respectively). Data were normalized to RNU6B expression and analysed by the −ΔΔCt method.
Determination of mRNA stability
HuH6 and HLE cells were treated with 10 μM Actinomycin-D (Sigma–Aldrich) for 0.5, 2, 4, 6, 8, 10 or 12 h before RNA isolation. Amount of UCP2 levels in the isolated mRNA samples were determined by quantitative real-time PCR as described above and compared with levels in untreated samples from the same cells. Relative expression was normalized to TBP (TaqMan Assay ID: Hs00427620_m1) in the same samples and data were converted into percent mRNA left at the indicated time points.
Western blot analysis
Western blot analysis was performed as described previously using rabbit anti-UCP2 antibody (Santa Cruz Biotechnology) [16,17]. All membranes were probed with anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibody (Santa Cruz Biotechnology) to confirm equal protein loading.
Cell proliferation assays
Cell proliferation was quantified using a mitochondrial colorimetric assay (MTT assay, Sigma–Aldrich) as per the manufacturer's recommendations and as described recently [14]. Results from three independent triplicates were expressed as mean ± S.D.
Plasmids
The UCP2 3' UTR clone in pMirTarget was obtained from Origene. The UCP2 3' UTR reporter was constructed by amplifying the endogenous UCP2 3' UTR from the Origene clone. XhoI and ApaI sites were added to the 5'- and 3'- ends of the fragment during the preceding PCR reaction and cloned into the XhoI and ApaI site on the Rr-luc-6XCXCR4 (Addgene plasmid 11308) Renilla luciferase vector. To make the UCP2 3’ UTR mutant construct, site-directed mutagenesis was used to delete 6–16 region, corresponding to the hsa-miR-214-binding site. A firefly luciferase vector was used as transfection and normalization control in all luciferase assays. Constructs were sequence verified to University of California Santa Cruz (UCSC) human genome reference version human genome (hg19).
Transfection and luciferase assays
Cells (4×104) were transiently transfected with the luciferase reporter constructs using Lipofectamine LTX (Life Technologies) as per the manufacturer's instructions. Where indicated, cells were transfected with the miR-214 mimic or antagomir (Life Technologies) along with the UCP2 3’ UTR constructs. Forty-eight hours after transfection, the renilla and firefly luciferase activities were measured consecutively using Dual-luciferase reporter assay system (Promega) as per manufacturer's protocol. Each reporter plasmid was transfected at least twice (on different days) in triplicate. Post-normalization, the data were represented as relative fluorescence units (RFU) ± S.D.
Statistical analyses
SPSS version 20.0 (IBM) was used for all statistical analysis. Two-sided P-values <0.05 were considered statistically significant.
RESULTS
UCP2 transcript is targeted by miR-214
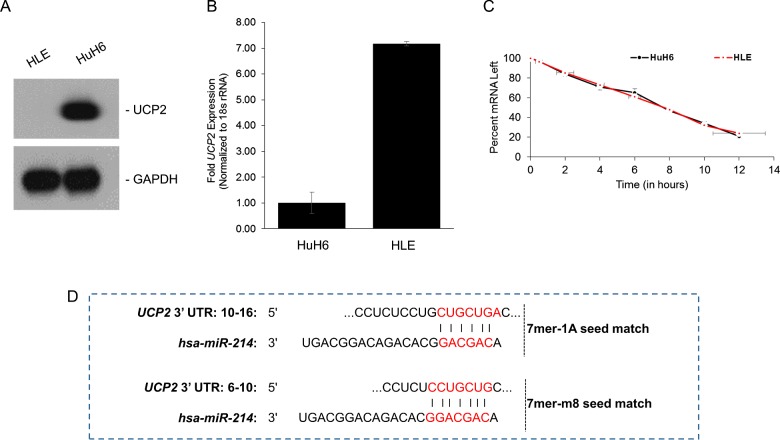
We have recently observed differential UCP2 protein expression among different HCC cell lines [14]. Whereas robust steady state UCP2 protein expression was detected in HuH6 cells, it was suppressed in the HLE cells (Figure 1A). Assessment of UCP2 transcript levels indicated that the difference in protein expression was not due to differential transcription rates. In fact, UCP2 transcript was significantly overexpressed (7±0.3-fold, P<0.05) in HLE cells as compared with the HuH6 cells (Figure 1B). This indicated a post-transcriptional or post-translational regulatory mechanism underlying differential UCP2 protein expression in these cells.
Figure 1. UCP2 expression is post-transcriptionally regulated in liver cancer cells.
(A) Basal expression levels of UCP2 in mitochondrial extracts obtained from indicated HCC cell lines. The blot was stripped and probed with GAPDH to serve as a loading control. (B) Steady state expression of UCP2 mRNA was determined in indicated cell lines. Data were normalized to 18s rRNA expression. (C) HuH6 and HLE cells were treated with Actinomycin-D for indicated times to determine relative stability of UCP2 transcript in the two cell lines. The slope of the two cell lines showed that degradation of the UCP2 mRNA in either cell lines followed similar kinetics. (D) Complementary 7mer-1A and 7mer-m8 seed match between miR-214 and the 3’ UTR of UCP2 as predicted by TargetScan software.
Evaluation of mRNA stability following Actinomycin-D treatment did not reveal any significant difference in UCP2 half-life in the two cell lines (Figure 1C). We next wanted to determine if UCP2 is being targeted by miRNAs. In situ prediction using TargetScan platform [18] showed that miR-214 have two putative and adjacent binding sites in the 3’ UTR of UCP2 (Figure 1D).
miR-214 is down-regulated in HCC samples
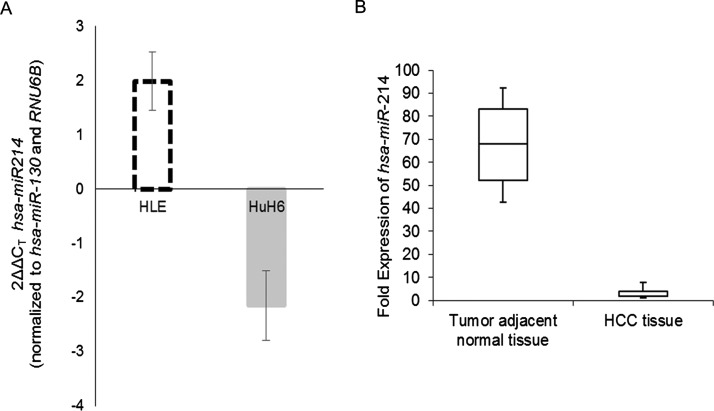
Quantitative real-time PCR showed that miR-214 expression was up-regulated in HLE cells and suppressed in HuH6 cells (Figure 2A). Evaluation of miR-214 expression in 25 paired HCC and adjacent normal tissue specimens showed that miR-214 expression was significantly down-regulated in HCC tissue (median, 6.39; range, 1.25–9.01) compared with normal counterparts (median, 68.87; range, 38.17–91.42) (P<0.001) (Figure 2B).
Figure 2. miR-214 expression is down-regulated in HCC.
Steady state expression of miR-214 in indicated cell lines (A) or paired tumour and adjacent non-tumour tissue (B) were determined. Data were normalized to RNU6B expression.
Modulating miR-214 levels impacted proliferation in the HCC cells
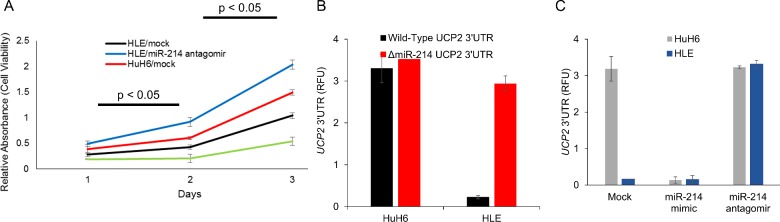
Since UCP2 can inhibit ROS-induced apoptosis [19], we rationalized that altering UCP2 transcript levels by modulating miR-214 expression might affect proliferation rates. This led us to examine whether overexpression via transfection of miR-214 mimic in HuH6 cells and suppression via transfection of miR-214 antagomir in HLE cells would impact proliferation rates. miR-214 mimic significantly decreased cell viability of HuH6 cells at 24, 48 and 72 h post-transfection respectively, compared with the mock control (P<0.05 in each case). Vice versa, miR-214 antagomir induced significantly more cell proliferation in HLE cells at the indicated time points (P<0.05 in each case) (Figure 3A).
Figure 3. UCP2 is a bona fide target of miR-214, expression level of which control cell viability.
(A) Cell viability was measured in HLE and HuH6 cells at 24, 48 and 72 h after transfection with miR-214 antagomir or mimic respectively, by the MTT assay. (B) Relative luciferase activity of transiently transfected luciferase reporter constructs containing either full-length or mutated (miR-214-binding sites deleted) UCP2 3’ UTR in indicated cells. (C) Relative luciferase activity of transiently transfected luciferase reporter constructs containing full-length UCP2 3’ UTR in indicated cells, alone or in combination with miR-214 mimic and antagomir.
UCP2 is a direct target of miR-214 in HCC cells
We next determined if UCP2 is a bona fide target of miR-214 in HCC cell lines. To test this putative interaction, luciferase reporter constructs containing the wild-type UCP2 3’ UTR were transfected in HuH6 and HLE cells (Figure 3B). UCP2 3’ UTR containing reporter were inhibited 9.8±0.34-fold (P=0.0037) in HLE cells compared with the HuH6 cell line. To confirm that the effects observed was due to miR-214 targeting the UCP2 3’ UTR, we generated and tested a miR-214 binding mutant of the UCP2 3’ UTR reporter, in which both the putative binding sites between 6–16 nucleotides were deleted. The miR-214 binding mutant UCP2 3’ UTR reporter did not show any difference in relative luciferase activity between HLE and Huh cells (Figure 3B), confirming that UCP2 mRNA was being targeted by the miR-214 in these cells. This was further corroborated by reporter assays performed in HLE cells transfected with miR-214 mimic and HuH6 cells transfected with miR-214 antagomir. Whereas miR-214 mimic inhibited UCP2 3’ UTR reporter (P<0.05) in HuH6 cells, miR-214 antagomir rescued reporter activity in the HLE cells (P<0.05) (Figure 3C).
miR-214 expression is inversely correlated with UCP2 levels and HCC disease
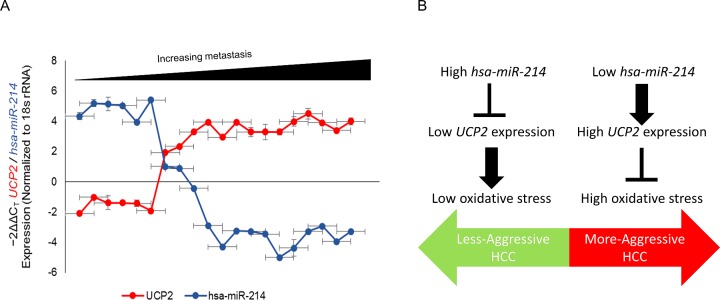
Given that our experiments indicated that UCP2 is a bona fide target of miR-214, we hypothesized that suppression of miR-214 expression might be an underlying feature of human prostate cancer. We determined miR-214 and UCP2 expression in 20 HCC patients, ten with high miR-214 and ten with low miR-214 expression. The ones with high miR-214 expression corresponded to N0, N1 cases (non-metastatic) whereas those with low miR-214 expression corroborated to highly metastatic disease. Our results indicated a dynamic and inverse correlation between down-regulation in the levels of miR-214 and the observed increase in the expression of UCP2 in HCC tissue specimens (Figure 4A) (P<0.005, Pearson correlation, r=−0.9792).
Figure 4. UCP2 mRNA and miR-214 are inversely correlated in patients with HCC.
(A) Pearson correlation demonstrating the inverse relation between UCP2 and miR-214 in paired samples (P<0.005, Pearson correlation, r=−0.9792). (B) Model illustrating the relationship between expression level of miR-214 and UCP2, mitochondrial superoxide generation and HCC.
DISCUSSION
miRNAs are evolutionarily conserved 21–23 nucleotides RNAs that regulate post-transcriptional gene expression either by blocking translation or degrading target mRNAs and have been increasingly shown to function as tumour suppressors or oncogenes [20,21]. miRNAs can function in both normal and transformed cells and have even been shown to play a role in metastasis [22–25].
Regulation of factors participating in ROS homoeostasis by miRNA is not without prior precedence. It has been shown that during progression from adaptive hypertrophy to heart failure, miR-214 and miR-30* together regulate cardiac vascular endothelial growth factor (VEGF) expression and angiogenesis by targeting X-box-binding protein-1 (XBP1), a key transcription factor of the unfolded protein response in mammalian cells [26].
Our results suggest that in the context of HCC, miR-214 functions as a tumour suppressor (Figure 4B). However, along with miR-126, miR-214 has been shown to be overexpressed in malignant endothelial proliferative disease [27]. This presents a unique case where the same miRNA can function as a tumour suppressor or oncomir in a context-dependent fashion. Elucidating the underlying mechanism regulating miR-214 expression will help explain the differential functional readouts of miR-214 in malignant proliferative disease and HCC.
Our prediction of miR-214 targeting UCP2 mRNA was based on the TargetScan algorithm. However, according to the miRTarBase, miRNAs that target UCP2 are hsa-miR-15a-5p and hsa-miR-484. Similarly, according to the miRanda algorithm miR-497, miR-15a, miR-424, miR-195, miR-16 and miR-15b can target UCP2, accessed on February 25, 2016. Experiments have shown that five programmes, namely TargetScan, TargetScanS, PicTar, DIANA-microT and miRNA target genes database (EIMMO) had a specificity of approximately 50% and sensitivity ranging from 6–12% [28]. Our prediction of miR-214 was through one of these five algorithms. However, it is important for future studies to validate if other miRNAs target UCP2 mRNA in the context of HCC or otherwise.
UCP2 is known to suppress ROS level which is overexpressed by various types of cancer cells including HCC cell lines. Inhibition of UCP2 in cancer cells have been shown to increase susceptibility of drug-resistant cancer cells to cytotoxic agents [19,29], indicating UCP2 is overexpressed in these cancers. It will be important to confirm if miR-214 levels are also down-regulated in these cells or UCP2 expression is controlled by additional mechanism.
Abbreviations
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GEM
Gemcitabine
- HCC
hepatocellular carcinoma
- HLE
human lens epithelial cells
- HuH6
human hepatoblastoma cells
- MTT
mitochondrial colorimetric assay
- RNU6B
U6 small nuclear RNA
- ROS
reactive oxygen species
- UCP2
uncoupling protein 2
- UCP5
uncoupling protein 5
AUTHOR CONTRIBUTION
Guangsheng Yu and Jianlu Wang designed, acquired, analysed, interpreted the data and wrote the manuscript; Kesen Xu and Jiahong Dong oversaw the project and worked to finalize the manuscript. All authors approved the final version of the manuscript.
FUNDING
This work was supported by the National Natural Science Foundation of China [grant numbers 81100206, 81373172 and 81302124]; the Shandong Provincial Natural Science Foundation [grant numbers ZR2014HQ019 and ZR2014HP065]; and the Science and Technology Development Plan Project of Medicine and Health Care in Shandong Province [grant number 2014WS0078].
References
- 1.Krauss S., Zhang C.Y., Lowell B.B. The mitochondrial uncoupling-protein homologues. Nature Rev. 2005;6:248–261. doi: 10.1038/nrm1592. [DOI] [PubMed] [Google Scholar]
- 2.Fleury C., Neverova M., Collins S., Raimbault S., Champigny O., Levi-Meyrueis C., Bouillaud F., Seldin M.F., Surwit R.S., Ricquier D., Warden C.H. Uncoupling protein-2: a novel gene linked to obesity and hyperinsulinemia. Nat. Genetics. 1997;15:269–272. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- 3.Boss O., Samec S., Paoloni-Giacobino A., Rossier C., Dulloo A., Seydoux J., Muzzin P., Giacobino J.P. Uncoupling protein-3: a new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997;408:39–42. doi: 10.1016/S0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- 4.Donadelli M., Dando I., Fiorini C., Palmieri M. UCP2, a mitochondrial protein regulated at multiple levels. Cell. Mol. Life Sci. 2014;71:1171–1190. doi: 10.1007/s00018-013-1407-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richard D., Picard F. Brown fat biology and thermogenesis. Front. Biosci. (Landmark Ed.) 2011;16:1233–1260. doi: 10.2741/3786. [DOI] [PubMed] [Google Scholar]
- 6.Garlid K.D., Jaburek M., Jezek P., Varecha M. How do uncoupling proteins uncouple? Biochim. Biophys. Acta. 2000;1459:383–389. doi: 10.1016/s0005-2728(00)00175-4. [DOI] [PubMed] [Google Scholar]
- 7.Salganik R.I. The benefits and hazards of antioxidants: controlling apoptosis and other protective mechanisms in cancer patients and the human population. J. Am. Coll. Nutr. 2001;20(5 Suppl):464S–472S. doi: 10.1080/07315724.2001.10719185. discussion 73S-75S. [DOI] [PubMed] [Google Scholar]
- 8.Lobo V., Patil A., Phatak A., Chandra N. Free radicals, antioxidants and functional foods: impact on human health. Pharmacogn. Rev. 2010;4:118–126. doi: 10.4103/0973-7847.70902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valle A., Oliver J., Roca P. Role of uncoupling proteins in cancer. Cancers. 2010;2:567–591. doi: 10.3390/cancers2020567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carretero M.V., Torres L., Latasa U., Garcia-Trevijano E.R., Prieto J., Mato J.M., Avila M.A. Transformed but not normal hepatocytes express UCP2. FEBS Lett. 1998;439:55–58. doi: 10.1016/S0014-5793(98)01335-0. [DOI] [PubMed] [Google Scholar]
- 11.Horimoto M., Resnick M.B., Konkin T.A., Routhier J., Wands J.R., Baffy G. Expression of uncoupling protein-2 in human colon cancer. Clin. Cancer Res. 2004;10(18 Pt 1):6203–6207. doi: 10.1158/1078-0432.CCR-04-0419. [DOI] [PubMed] [Google Scholar]
- 12.Santandreu F.M., Valle A., Fernandez de Mattos S., Roca P., Oliver J. Hydrogen peroxide regulates the mitochondrial content of uncoupling protein 5 in colon cancer cells. Cell. Physiol. Biochem. 2009;24:5, 379–6. doi: 10.1159/000257430. [DOI] [PubMed] [Google Scholar]
- 13.Baffy G., Derdak Z., Robson S.C. Mitochondrial recoupling: a novel therapeutic strategy for cancer? Br. J. Cancer. 2011;105:469–474. doi: 10.1038/bjc.2011.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu G., Liu J., Xu K., Dong J. Uncoupling protein 2 mediates resistance to gemcitabine-induced apoptosis in hepatocellular carcinoma cell lines. Biosci. Rep. 2015;35:e00231. doi: 10.1042/BSR20150116. doi: 10.1042/BSR20150116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bookelman H., Trijbels J.M., Sengers R.C., Janssen A.J., Veerkamp J.H., Stadhouders A.M. Pyruvate oxidation in rat and human skeletal muscle mitochondria. Biochem. Med. 1978;20:395–403. doi: 10.1016/0006-2944(78)90089-3. [DOI] [PubMed] [Google Scholar]
- 16.Patel D., Nan Y., Shen M., Ritthipichai K., Zhu X., Zhang Y.J. Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J. Virol. 2010;84:11045–11055. doi: 10.1128/JVI.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nan Y., Wang R., Shen M., Faaberg K.S., Samal S.K., Zhang Y.J. Induction of type I interferons by a novel porcine reproductive and respiratory syndrome virus isolate. Virology. 2012;432:261–270. doi: 10.1016/j.virol.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Pfefferle A., Mailloux R.J., Adjeitey C.N., Harper M.E. Glutathionylation of UCP2 sensitizes drug resistant leukemia cells to chemotherapeutics. Biochim. Biophys. Acta. 2013;1833:80–89. doi: 10.1016/j.bbamcr.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esquela-Kerscher A., Slack F.J. Oncomirs–microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 22.Aleckovic M., Kang Y. Regulation of cancer metastasis by cell-free miRNAs. Biochim. Biophys. Acta. 2015;1855:24–42. doi: 10.1016/j.bbcan.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaur A., Jewell D.A., Liang Y., Ridzon D., Moore J.H., Chen C., Ambros V.R., Israel M.A. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 24.Kumar M.S., Lu J., Mercer K.L., Golub T.R., Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 25.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 26.Duan Q., Chen C., Yang L., Li N., Gong W., Li S., Wang D.W. MicroRNA regulation of unfolded protein response transcription factor XBP1 in the progression of cardiac hypertrophy and heart failure in vivo. J. Transl. Med. 2015;13:363. doi: 10.1186/s12967-015-0725-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heishima K., Mori T., Ichikawa Y., Sakai H., Kuranaga Y., Nakagawa T., Tanaka Y., Okamura Y., Masuzawa M., Sugito N., et al. MicroRNA-214 and microRNA-126 are potential biomarkers for malignant endothelial proliferative diseases. Int. J. Mol. Sci. 2015;16:25377–25391. doi: 10.3390/ijms161025377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witkos T.M., Koscianska E., Krzyzosiak W.J. Practical aspects of microRNA target prediction. Curr. Mol. Med. 2011;11:93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mailloux R.J., Adjeitey C.N., Harper M.E. Genipin-induced inhibition of uncoupling protein-2 sensitizes drug-resistant cancer cells to cytotoxic agents. PLoS One. 2010;5:e13289. doi: 10.1371/journal.pone.0013289. [DOI] [PMC free article] [PubMed] [Google Scholar]