Abstract
Infection with Zika virus has been associated with serious neurological complications and fetal abnormalities. However, the dynamics of viral infection, replication and shedding are poorly understood. Here we show that both rhesus and cynomolgus macaques are highly susceptible to infection by lineages of Zika virus that are closely related to, or are currently circulating in, the Americas. After subcutaneous viral inoculation, viral RNA was detected in blood plasma as early as 1 d after infection. Viral RNA was also detected in saliva, urine, cerebrospinal fluid (CSF) and semen, but transiently in vaginal secretions. Although viral RNA during primary infection was cleared from blood plasma and urine within 10 d, viral RNA was detectable in saliva and seminal fluids until the end of the study, 3 weeks after the resolution of viremia in the blood. The control of primary Zika virus infection in the blood was correlated with rapid innate and adaptive immune responses. We also identified Zika RNA in tissues, including the brain and male and female reproductive tissues, during early and late stages of infection. Re-infection of six animals 45 d after primary infection with a heterologous strain resulted in complete protection, which suggests that primary Zika virus infection elicits protective immunity. Early invasion of Zika virus into the nervous system of healthy animals and the extent and duration of shedding in saliva and semen underscore possible concern for additional neurologic complications and nonarthropod-mediated transmission in humans.
Zika virus (ZIKV) is a member of the flavivirus family first isolated from a febrile sentinel rhesus monkey in Uganda in 1947 (ref. 1). Primary ZIKV infection is often mild in adults, with only a small percentage of those infected presenting as clinically symptomatic2–4. Diagnosis is complicated by clinical similarity and geographic overlap with dengue and chikungunya infection4–7. However, ZIKV is associated with serious neurological complications, such as Guillain–Barré syndrome8–11, as well as with pediatric microcephaly, that have been reported to overlap with ZIKV endemicity in South America12–15.
The primary mode of ZIKV transmission is via an arthropod vector16. Limited reports have indicated that sexual transmission occurs, but the frequency, transmissibility and relative risk of male-to-female versus female-to-male acquisition is poorly described beyond single case reports17,18. Notably, ZIKV has been detected in the blood of human patients for only short periods of time after the onset of symptoms19–21. The detection of viral shedding in bodily fluids such as the saliva21, urine22 and semen17,18, or invasion into the central nervous system (CNS), have been documented11, but the viral dynamics and pathogenicity are poorly understood.
ZIKV has been shown to infect human fibroblasts, keratinocytes and dendritic cells in vitro and might use one of several adhesion molecules (DC-SIGN, AXL, TYRO3 or TIM-1) as possible entry receptors23. In mother-to-fetus transmission, Tabata and colleagues24 have demonstrated that ZIKV isolates can infect primary human placenta cells (primary human cytotrophoblasts, umbilical endothelial cells, placental fibroblasts, Hofbauer cells, amniotic epithelial cells and trophoblast progenitors) and that entry into these cells is at least partially determined by the expression of T cell immunoglobulin mucin (TIM)-1, which is expressed on select cell populations. However, the specific host cell(s) that support ZIKV replication during early infection in vivo are presently unknown. Similarly, ZIKV replication dynamics, distribution in anatomic tissues and the duration of infectious shedding are unclear. Moreover, the immune response to primary ZIKV in humans is largely uncharacterized.
Recently, Dudley et al.25 conducted an elegant demonstration showing that rhesus macaques are permissive to infection by Asian lineages of ZIKV. They showed that viral replication in the blood is rapid, and that infected rhesus macaques display signs of immune activation and proliferation that result in the development of virus-specific T cells. The authors demonstrate that the virus was detectable in both the urine and saliva, with kinetics that are delayed and blunted when compared to those in blood plasma. They also detected ZIKV, albeit infrequently, in the CSF or vaginal secretions.
Here we experimentally infected a total of 28 rhesus and eight cynomolgus macaques using two different ZIKV strains, and we evaluated the dynamics of ZIKV in the blood, tissues, CSF and mucosal secretions of these macaque species. We also assessed the temporal distribution of ZIKV RNA in anatomic tissues and parallel immune responses during early acute infection and up until 4 weeks after virus challenge, and then anamnestic immune responses after secondary ZIKV infection.
RESULTS
ZIKV dynamics in the blood
Our experimental infection studies used sequence-defined ZIKV isolates of Thai and Puerto Rican (PR) origin (GeneBank accession numbers KF993678 and KU501215) that were expanded on Vero cell lines. To establish the ZIKV dynamics in blood and other body fluids, we challenged ten Indian rhesus monkeys (five females and five males) with 1 × 106 plaque-forming units (PFU) of a Thai isolate via a subcutaneous (SC) route. The ZIKV dose used for SC infection was estimated on the basis of the viral burden present in arthropod vectors for West Nile virus or ZIKV, as described26–28. Blood, saliva and urine samples were collected for days 1–5, 7, 10 and then weekly until day 28 after infection. CSF, semen and vaginal fluids were collected weekly for 4 weeks. In the blood, high-level viremia was detected in nine of ten animals by day 1 after infection, and peak viremia was achieved in most animals by 2 d after infection (Fig. 1a). Peak ZIKV RNA levels ranged from 8.8 × 105 to 4.9 × 107 RNA copies/ml, with the exception of one female monkey that exhibited delayed viremia, which reached peak levels at 3–4 d after infection (5.0 × 104 RNA copies/ml). Blood plasma viral RNA levels fell rapidly after day 2 and reached undetectable levels in nine animals by day 5. In the animal (5690) with the highest peak viremia (4.9 × 107 RNA copies/ml), ZIKV RNA remained detectable at day 7 but was undetectable by day 10. Although the viral load in males showed a trend toward higher peak viremia, there was no significant difference between ZIKV RNA levels in the blood plasma of males and females at peak, or the total ZIKV RNA burden, calculated as the area under the curve (AUC) (P = 0.056) (Fig. 1b). We also assessed the dynamics of a PR ZIKV isolate in the blood and secretions in an additional five Indian-origin rhesus macaques (two females and three males) that were challenged with 1 × 105 TCID50. Blood plasma ZIKV RNA reached peak levels by 4 d after infection (range 1.2 × 106 to 1.2 × 107 RNA copies/ml) and resolved by day 8 after infection in all monkeys. Overall, the viral kinetics of the PR isolate are similar to those of the Thai variant, although slight delays can be observed in time to peak viremia, consistent with the lower infecting dose used (Supplementary Fig. 1a).
Figure 1.
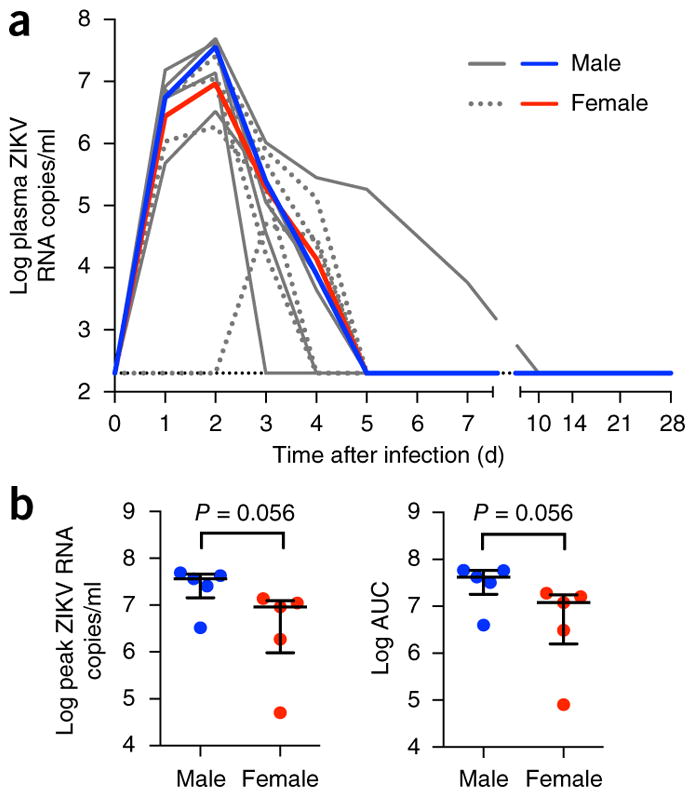
Blood plasma ZIKV RNA kinetics during primary infection. (a) Viral replication kinetics after subcutaneous infection of ten Indian-origin rhesus macaques with 1 × 106 PFU of a Thai isolate. ZIKV replication was monitored between days 1 and 28 after infection. Log10 virus RNA copies per ml in blood plasma samples from individual animals are shown in gray. Median log10 ZIKV RNA copies for male (n = 5) and female (n = 5) are shown in red and blue, respectively. The limit of detection (<200 RNA copies/ml) for blood plasma is indicated (black hatched line). (b) The blood plasma ZIKV RNA levels at peak viremia or combined burden as AUC were compared between male and female monkeys. The results are expressed as the median with interquartile range. Each dot represents one animal. Comparison between groups was determined using a Mann–Whitney test. Error bars indicate interquartile range with median.
Clinical metrics and immune responses to ZIKV infection
Blood chemistries were monitored throughout primary ZIKV infection. Similarly to observations made by other investigators25, aspartate aminotransferase, alanine aminotransferase, alkaline phosphatase and creatinine phosphatase increased and peaked by day 3 (Supplementary Fig. 2). Modulation in peripheral body temperature during infection was recorded by subcutaneously implanted transponders (every 30 min) for the first 14 d after ZIKV infection (Supplementary Fig. 3). In all animals, body temperature was significantly elevated by day 1 after infection and remained significantly elevated until day 8, although no animal’s temperature recording exceeded 39.5 °C, owing to the peripheral location of the implant (day 1, P = 0.004; day 2, P = 0.005; days 3–7, P < 0.001; day 8, P = 0.01).
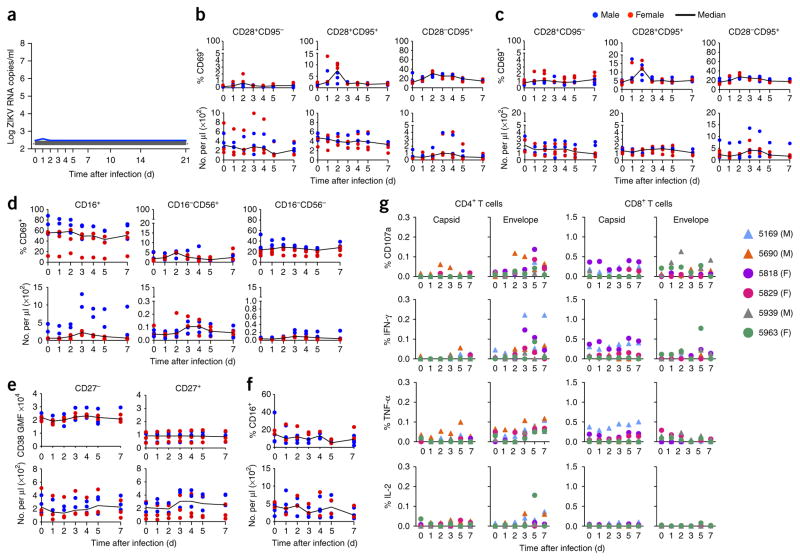
Complete blood counts were monitored, and within 24 h of infection, increases in white blood cell count, largely attributed to neutrophil mobilization, were noted (Supplementary Fig. 4). These counts returned to baseline in most animals by day 2 after infection. We also observed significant increases in the percentage of activated monocytes (P = 0.002 on day 2) as compared to that at baseline (measured as percentage of CD16+ cells); these peaked by day 2 after infection, with activation levels in males significantly higher than those in females (P = 0.008) (Supplementary Fig. 5a,b). Total monocyte numbers peaked between days 1 and 4 of infection.
We also evaluated changes in the abundance of lymphocytes as a result of ZIKV infection and observed transient declines in this population (in the peripheral blood) during the first day after infection, which is consistent with lymphocyte infiltration into infected tissues (Supplementary Fig. 4). Most major lymphocyte subsets (B, T and natural killer (NK) cells) exhibited declines during this period (naive and central memory CD4+ T cells, all CD8+ T cell subsets, naive B cells and all NK cell subsets; Supplementary Fig. 5c–f).
Increased levels of CD69+ T cell activation, as compared to those at baseline, were observed and occurred predominantly in the CD28−CD95+ subset, which includes effector and effector memory T cells, and absolute numbers peaked between days 2 and 5 after infection (Supplementary Fig. 5c,d). B cell activation, as measured by the expression of CD38, was observed in both naive (CD27−) and memory (CD27+) B cell subsets, and this peaked on approximately day 3. NK cell activation, as measured by the percentage of CD69+ expression, was detected predominantly in CD16+ and CD16−CD56− NK cell subsets, and peaked on day 2. The absolute numbers of CD16+ NK cells peaked on day 4, whereas the numbers of CD16−CD56− cells peaked on day 10, after infection (Supplementary Fig. 5e,f).
We next evaluated the development of virus-specific T cells from peripheral blood mononuclear cells (PBMCs) and lymph nodes on days 7, 14, 21 and 28 after infection by intracellular cytokine staining, after stimulation with overlapping peptide pools covering the ZIKV capsid and envelope proteins. By day 7 after ZIKV infection, peptide-stimulated PBMCs from eight of ten monkeys elicited both capsid- and envelope-specific CD4+ T cell responses (Supplementary Fig. 6a). ZIKV capsid and envelope responses were of similar magnitude, with peak responses delayed until day 28.
CD8+ T cells responses specific to capsid or envelope peptides were variable between animals, with most responses being low in magnitude (<0.3% of CD8+ T cells producing cytokine). However, a few monkeys developed high-magnitude responses (>1%). Monkeys 5169 and 5818 developed high-magnitude responses to capsid, and animals 5829 and 5939 developed high-magnitude, envelope-specific responses. By contrast, both CD4+ and CD8+ T cell responses to ZIKV peptides in the lymph node were weak (Supplementary Fig. 6b). Prior to day 28, only four animals (of ten) had developed detectable capsid- or envelope-specific CD8+ T cell responses. Antibody responses were generated rapidly, with detectable changes in IgG and IgM as early as days 3–7 after infection (Supplementary Table 1).
We also used a multiplex bead immunoassay to assess the blood plasma cytokine levels of rhesus macaques on days 1, 3 and 7 after ZIKV infection. Blood plasma samples were analyzed for multiple cytokines using multiplex bead immunoassays (Online Methods). Heat-map clusters (from levels of each cytokine on days 1, 3 and 7 after infection) were calculated as the relative ratio to baseline (day 0) and are shown from high to low values (Supplementary Fig. 7a). We observed significantly increased concentrations of C-C motif chemokine ligand 2 (CCL2, also known as MCP-1) (day 1, P < 0.001; day 3, P = 0.017), interleukin (IL)-15 (day 1, P = 0.011; day 3, P = 0.005), vascular endothelial growth factor (VEGF; P = 0.007), IL-10 (P = 0.05) and IL-1α (P < 0.0001) during days 1–3 after infection as compared to the levels at baseline, whereas soluble CD40 ligand (sCD40L; P = 0.04) and IL-8 (day 1, P = 0.04; day 3, P = 0.03) levels were significantly decreased (Supplementary Fig. 7b). Our data demonstrate substantial early CD16+ monocyte and NK cell activation that is in agreement with the notable rise in MCP-1 and IL-15 levels. Other cytokine modulations, such as changes in IL-10 levels, have also been reported during dengue infection29–31.
Modeling of ZIKV infection
To further understand the dynamics of ZIKV infection as reflected in the blood, we fit our data to a mathematical acute-viral-infection model. This model incorporates an intracellular eclipse phase (after infection) during which an infected cell does not produce virus. After the eclipse period, infected cells produce new virus. Notably, despite a homogeneous challenge inoculum, the estimated values of the initial blood plasma ZIKV RNA levels, V(0), in the best-fit parameter sets varied greatly across animals. However, we found that the quality of the fits and the best-fit values of other model parameters were insensitive to the value of V(0) (Supplementary Fig. 8a). According to the parameter estimates, our model predicts an average eclipse phase for ZIKV of 4 h or less, and that productively ZIKV-infected cells have an average half-life of approximately 3.0 h in vivo; the macaque (5690) with the longest-persisting viremia was an outlier (Supplementary Fig. 8c). We assessed all activated cell populations, but noted an association only between reductions in primary ZIKV RNA levels in the blood and levels of NK cell subset activation. Our modeling also confirmed a significant association between activated (CD69+) CD16−CD56+ NK cells and the overall rate of post-peak decline of ZIKV RNA levels in blood plasma (P = 0.025) (Supplementary Figs. 5e and 8b).
ZIKV invasion of the CNS and infectious shedding into mucosal fluids
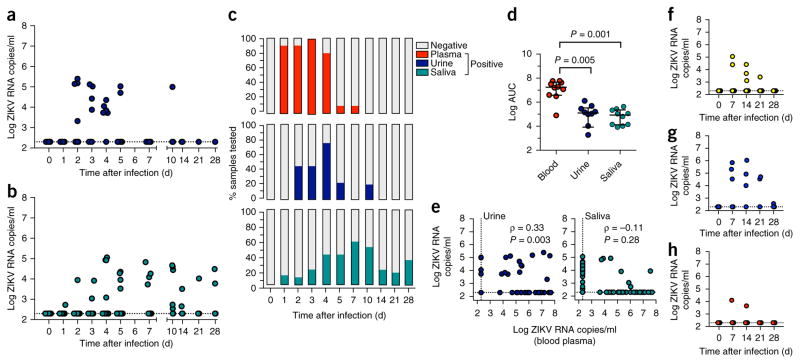
We next assessed the magnitude and temporality of ZIKV shedding into mucosal fluids (urine, saliva, semen and vaginal secretions) and CSF. Urine and saliva samples were collected at the same frequency as blood samples (at days 1–5, 7, 10, 14, 21 and 28). Clean urine samples were obtained by cystocentesis, and saliva was acquired by cotton roll salivette32. In the urine of monkeys infected with ZIKV, viral RNA was detectable as early as day 2 after infection in 44% of animals, and ranged from 5.3 × 103 copies/ml to 2.5 × 105 RNA copies/ml. ZIKV shedding in urine was detected in 71% of animals but became rapidly undetectable as primary viremia in the blood resolved (Fig. 2). However, sporadic shedding in urine was detected in one animal as late as 10 d after infection.
Figure 2.
Timing and magnitude of ZIKV shedding in CSF and mucosal fluids during primary infection. (a,b) ZIKV RNA was extracted from each specimen and quantitated by RT–qPCR in urine (a) or saliva (b). (c) The percentage of ZIKV RNA positive blood plasma, urine and saliva samples are shown over time after infection. (d) The total ZIKV RNA burden found in blood, urine and saliva, as expressed as AUC. (e) The association between ZIKV RNA levels in blood plasma and each of urine or saliva is shown. The strength and direction of any association was assessed. Spearman correlation coefficient (ρ) and P value are indicated. (f–h) Longitudinal (weekly) ZIKV RNA measurements in CSF (f), semen (g) and vaginal fluids (h) are shown. Error bars indicate interquartile range with median. The comparison of the values from the groups of animals was determined using a nonparametric Kruskal–Wallis test with multiple-comparison test.
In saliva, ZIKV RNA was detectable in 10–20% of animals by day 3 after infection, ranging from 3.5 × 102 to 1.1 × 105 RNA copies/ml. In comparison to the blood, the duration of ZIKV RNA in saliva was extended, and viral shedding was variable, but nevertheless detectable, in 33–50% of animals until the end of the study at 28 d after infection (Fig. 2b,c). During primary infection, the average total viral burden (AUC) in urine and saliva were similar but approximately 2 logs lower than RNA levels measured in the blood plasma (Fig. 2d). ZIKV RNA levels in the blood were significantly correlated with ZIKV shed into the urine (P = 0.003), consistent with the finding that ZIKV RNA was generally detected in the urine when ZIKV blood levels exceeded 1 × 104 RNA copies/ml. ZIKV RNA levels in urine were correlated with ZIKV blood plasma levels most significantly on days 2 (P = 0.01) and 3 (P = 0.04) after infection. Conversely, no direct relationship was observed between ZIKV RNA levels in saliva and levels detected in the blood (P = 0.28; Fig. 2e). ZIKV shedding in the saliva was detectable for approximately 3 weeks after the resolution of primary blood plasma viremia. We also examined this relationship between ZIKV RNA levels in the blood with those in the urine and saliva for each individual animal. In urine, significant correlations (positive) were found in five animals (P = 0.002), whereas the other five animals did not show any direct relationship. Again, for individual animals, we found no correlation between ZIKV RNA levels in blood plasma and saliva shedding in nine of ten animals with the infection.
Lumbar punctures were performed weekly to quantify possible ZIKV RNA invasion into the CNS. ZIKV burden in the CSF ranged between 1.1 × 103 and 1.1 × 105 RNA copies/ml, and was detectable in 50% of animals between days 7 and 21 after infection. ZIKV RNA in CSF was detected only in animals with peak viremia greater than median peak blood plasma viral RNA levels (Fig. 2f).
We observed similar patterns of ZIKV shedding into mucosal fluids (urine and saliva) or CSF in monkeys that were infected using a PR isolate. We detected brief ZIKV shedding in urine and prolonged shedding in saliva samples even after blood plasma ZIKV RNA became undetectable. ZIKV RNA in the CSF was detected in 100% of animals infected with the PR isolate on day 14 after infection, and in 40–60% of infected animals on days 7 through 21 after infection (Supplementary Fig. 1b–d).
We next evaluated ZIKV RNA levels present in male and female genital secretions, again from weekly samples. Plasma samples from the semen were evaluated and found to harbor ZIKV RNA at high levels. Seminal plasma ZIKV values ranged between 3.4 × 104 and 1.1 × 106 RNA copies/ml on days 7–21 after infection. Notably, ZIKV RNA levels were detected in semen samples until day 28 after infection (3.5 × 104 to 5.1 × 104 RNA copies/ml), 3 weeks after the resolution of ZIKV viremia in the blood (Fig. 2g). Conversely, in females, only sporadic, low-level ZIKV RNA shedding was detected in vaginal swab samples at a single time point (in two of five female animals) at any point during post-infection monitoring (Fig. 2h). We also examined inter-animal variability by comparing the frequency of ZIKV RNA detected in various mucosal fluids and CSF from individual monkeys. ZIKV RNA was detected in all samples, including semen, urine, saliva and CSF, from three males, with the highest peak viremia. Conversely, no ZIKV RNA was detected in urine, cervicovaginal lavage (CVL) or CSF from the one female that exhibited delayed viremia and lower peak ZIKV RNA levels in blood plasma. Among six animals (two males and four females) with a median range of peak viremia in blood plasma, ZIKV RNA was detected in CSF from two animals, in semen from one male, in CVL from two females and in urine from five animals. Notably, ZIKV RNA was detected in saliva from all ten animals, regardless of their ZIKV RNA levels in blood plasma.
We then evaluated the infectivity in vitro of ZIKV that was shed into saliva, semen and CSF samples collected from rhesus monkeys with ZIKV infection on days 7 and 14 after infection. Dilutions of these mucosal fluids and CSF were used to inoculate Vero cell monolayers (described in Online Methods) and compared against infected blood plasma samples from day 3 after infection. Increases in ZIKV RNA levels in culture supernatants were observed in all fluids (saliva, semen, CSF and blood plasma) over 72 h of monitoring, which confirmed the infectivity and replicative ability of ZIKV in these fluids (Supplementary Fig. 9).
ZIKV dynamics and tropism in tissues
At 28 d after infection, we humanely euthanized four (two males and two females) of the ten animals infected with Thai ZIKV isolates to evaluate any potential ZIKV RNA in anatomic tissues by RNAscope in situ hybridization (Online Methods). We identified regions in the male genital tract that harbored persistent foci of ZIKV-infected cells localized in the testes, prostate and seminal vesicles (data not shown).
To determine the routes of viral spread and tissue sequestration of ZIKV during early infection required for productive infection, we used a PR ZIKV isolate that has been titrated in vivo by subcutaneously dosing 13 Indian-origin rhesus monkeys (two or three per group) with five (1-log) dilutions of the original challenge stock (Fig. 3a). Although infection was delayed by challenge with the lower infectious doses, ZIKV acquisition was still observed in all animals, even after administering doses as low as 1 × 102 TCID50. Moreover, despite these differences in inoculum size, all monkeys generated comparable peak ZIKV RNA levels once blood plasma viremia was detected.
Figure 3.
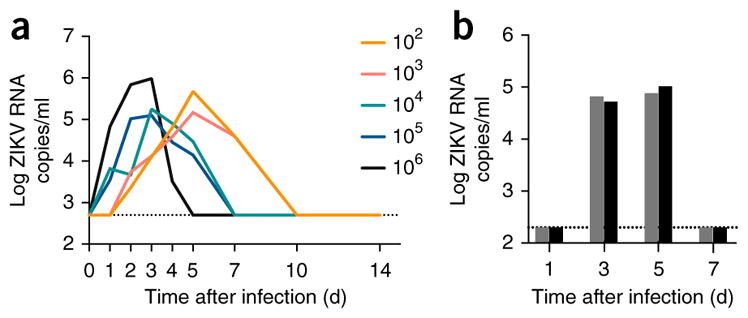
Infection of rhesus macaques with a Puerto Rican ZIKV isolate. (a) 13 Indian-origin rhesus monkeys were used to titrate a PR ZIKV isolate. Two animals per group were infected with 1 × 102 or 1 × 106 TCID50 units, and three animals per group were infected with 1 × 103, 1 × 104 or 1 × 105 TCID50 units, delivered subcutaneously. Viral load was monitored, and the median blood plasma ZIKV RNA levels are shown between days 1 and 14 after infection. Eight cynomolgus monkeys were then subcutaneously infected with 1 × 105 TCID50 units of the same PR ZIKV isolate, and then two monkeys were euthanized on each of days 1, 3, 5 and 7 for the detection of ZIKV RNA in various tissues by RNAscope in situ hybridization. (b) The blood plasma ZIKV RNA levels at the time of euthanasia are shown. The limit of detection (<200 RNA copies/ml) for blood plasma is indicated (black hatched line).
We also sought to determine the ability of cynomolgus monkeys to support ZIKV infection. Eight animals were infected subcutaneously using the previously mentioned PR isolate at a dose (1 × 105 TCID50) within the range described for other flaviviruses when transmitted by a mosquito bite26–28. Animals were then humanely euthanized, two (n = 2) per time point on days 1, 3, 5 and 7 after infection. Blood plasma viremia was detected readily in animals on days 3 and 5 after infection (Fig. 3b). Multiple anatomic tissues from each animal were prepared and analyzed by RNAscope, including lymph nodes (LNs) (inguinal, axillary, mesenteric), male genital tract (seminal vesicles, testes, prostate), female genital tract (uterus, ovaries), gastrointestinal tract (jejunum, colon), liver, kidney, bladder, lung, bone marrow and CNS (parietal lobe, basal nuclei, hippocampus, cerebellum).
Consistent with the subcutaneous route of infection and presumptive lymphatic draining pattern of ZIKV, we first observed ZIKV RNA positive cells in the inguinal LNs at 1 d after infection (data not shown). By 3 d after infection, there was an expansion of ZIKV RNA in the cells of the inguinal LNs and evidence of systemic spread to the distal axillary LNs (Fig. 4a). Viral replication in the LNs peaked by 5 d after infection within the inguinal LNs, and evidence of viral clearance was present by 7 d after infection (Fig. 4a). Confocal microscopy analysis of lymph nodes demonstrated that productively infected ZIKV RNA positive cells were primarily of the CD163+CD68+ myeloid lineage and the MPO+ neutrophil lineage (Fig. 4), and with rare, but infected, CD163−CD68− DC-SIGN+ dendritic cells (DCs). By contrast, we did not detect ZIKV RNA in either CD20+ B cells or CD3+ T cells (Supplementary Fig. 10).
Figure 4.
Detection of ZIKV RNA in lymphoid, neurologic and reproductive tissues. (a–c) Detection of ZIKV RNA by RNAscope in lymph nodes on days 3 (top), 5 (middle) and 7 (bottom) (a), lymphoid and reproductive tissues on day 7 (b) and cerebellum on days 5 (left) and 7 (right) (c). Red spots indicate ZIKV RNA. (d,e) Subsequent immunofluorescence staining was as follows: lymph nodes on day 7 were stained for CD163 and CD68 (myeloid cells, green), ZIKV RNA (red), myeloperoxidase (MPO, neutrophils, blue) and DAPI (DNA, gray) (d); cerebellum was stained on day 5 for NeuN (neurons, green), ZIKV RNA (red), Iba1 (microglia, blue) and DAPI (e). White arrows indicate ZIKV-infected cells. Images are representative of (n = 2) per time point. All images are shown at 60× magnification. All scale bars, 200 μm.
Although ZIKV infection was detected in other anatomic sites besides lymphoid tissue, the level of viral replication was limited in nature (data not shown). Notably, this was not the case in the CNS; the cerebellum contained the highest levels of ZIKV RNA among all non-lymphoid tissues evaluated, localized specifically within cerebellar granule cells (Fig. 4c,e). Although ZIKV RNA was cleared from the blood plasma by day 7 after infection, ZIKV RNA positive cells persisted in multiple anatomic tissues, including the LNs and the male and female genital tract (Fig. 4b), to the end of the monitoring period of our study (day 28; data not shown). Thus, we have identified tissue CD163+CD68+ myeloid cells, neutrophils and cerebellar neurons as important ZIKV target cell populations in vivo.
Protection from heterologous ZIKV infection
Six monkeys (three females and three males), infected initially with a Thai isolate, were then re-challenged with a heterologous PR isolate (1 × 105 TCID50) on day 45 after the primary infection. We monitored blood plasma viremia on days 1–21 after infection. Notably, blood plasma viral RNA levels remained negative (<200 copies/ml) in five of six animals. One exception was a single animal that showed low levels of RNA (275 ZIKV RNA copies/ml) at a single time point (day 1) after infection (Fig. 5a).
Figure 5.
Protection against heterologous challenge with a Puerto Rican ZIKV isolate. (a) Six animals (three females and three males) were re-challenged subcutaneously with 1 × 105 TCID50 of a PR ZIKV isolate 45 d after primary infection. Viral RNA levels in blood plasma were monitored between days 1 and 21 after the secondary challenge. ZIKV RNA copies in blood plasma are indicated. A single positive sample, detected in one male monkey 1 d after infection, is indicated (blue solid line). Lymphocyte activation and frequencies were measured during re-challenge. Activation and numbers of T cells, B cells, NK cells and monocytes were measured by flow cytometry on days 0, 1, 2, 3, 4, 5 and 7 after ZIKV infection of three male and three female monkeys. (b,c) Percentage of activated (CD69+) and total numbers of naive (CD28+CD95−) (left), central memory (CD28+CD95+) (middle), and effector and effector memory (CD28−CD95+) (right) CD4+ (b) and CD8+ (c) T cell subsets in whole blood. (d) Percentage of activated (CD69+) and total numbers of CD16+ (left), CD16−CD56+ (middle) and CD16−CD56− (right) NK cells in whole blood. (e) CD38 expression (geometric mean fluorescence of CD38) and total numbers of naive (CD27−) (left) and memory (CD27+) (right) B cells in whole blood. (f) Percentage of activated and total numbers of monocytes in whole blood. Black line indicates median. Individual values are shown for males (blue) and females (red). (g) ZIKV-specific T cell responses were measured during ZIKV re-challenge. T cell degranulation and cytokine production (in PBMCs) was measured by flow cytometry after the stimulation of cells on days 0, 1, 2, 3, 5 and 7 after re-challenge with overlapping peptide pools spanning the entire ZIKV capsid and envelope proteins. Percentage of cells expressing CD107a, interferon (IFN)-γ, tumor necrosis factor (TNF)-α and IL-2 after a 6-h peptide stimulation are shown. The threshold for determining positive values, after background subtraction of unstimulated corresponding samples, was applied as previously described43. Values below this threshold were set to a value of 0. Values greater than 1.5-fold above that of the day of re-challenge were considered to be anamnestic. Each monkey is indicated by a different color symbol. Triangles indicate males, and circles indicate females.
After heterologous ZIKV challenge, hemoglobin and hematocrit decreased in a similar manner to that observed in primary infection. However, white blood cell counts did not increase to the levels observed during primary infection, nor did neutrophil number increase. Total lymphocytes changed, but to a lesser extent than that seen during primary infection (Supplementary Fig. 4).
Central memory T cells showed a sharp peak in activation on day 2 after re-challenge (Fig. 5b,c). Effector memory cells also showed increased activation, which was slightly more prolonged than that seen for central memory cells. The numbers of naive and central memory T cells in the blood did not change after re-challenge, but there was an expansion of the effector memory populations that peaked between days 3 and 4 after challenge. Both CD16−CD56+ and CD16−CD56− NK cells displayed small increases in the percentage of activation level, peaking around day 2, whereas the percentage of activated CD16+ NK cells continually decreased (in the blood) for the first 5 d after re-challenge (Fig. 5d). B cells did not display increased activation, and there were no substantial changes in B cell numbers (Fig. 5e). Monocyte activation was not detected (Fig. 5f). The development of virus-specific T cells from PBMCs on days 1, 2, 3, 5 and 7 following re-challenge was analyzed. All six monkeys developed weak anamnestic CD4+ T cell responses and similarly weak CD8+ T cell recall responses after re-challenge (Fig. 5g). We did note significant increases in neutralizing-antibody titers measured on day 28 post infection from primary infection (P = 0.006) that persisted until the time of secondary challenge, as assessed by a PRNT90 assay (Online Methods). Titers were also increased in four animals on day 7 after secondary challenge (Supplementary Table 2).
DISCUSSION
By using our tractable model, we demonstrate—at least in the nonhuman primates studied here—rapid ZIKV replication and viral turnover rates that engender a limited productive half-life for infected cells. The major host cells sustaining ZIKV replication in vivo are of myeloid lineages found in the blood and tissues. It is formally possible that some portion of the ZIKV RNA positive neutrophils represent the phagocytosis of infected material or cells, although evidence from other flaviviruses seems to contradict this notion33. The demonstration of rapid and high-level ZIKV infection of cerebellar neurons is concerning, and might help to explain the neurologic outcomes associated with ZIKV infection observed in adults and early gestational neonates34–36. These findings might also warrant enhanced clinical vigilance in Zika-endemic areas for adults who present with neurologic symptoms37,38. Notably, ZIKV RNA invasion of the CNS was detected in animals with the highest peak blood plasma viremia, which perhaps implies that at least the initial means of ZIKV seeding of the CNS might be passive spillover of virus from the periphery.
Similar to the findings of Dudley et al.25, we demonstrate that rapid innate and adaptive immune responses limit viral replication in the blood and that these responses offer protection from future reinfection. Neither study has accounted for limiting numbers of target cells as a means of RNA reduction during acute infection. However, both studies clearly show that despite ZIKV-specific immunity, viral shedding continues unabated in anatomic tissues and fluids. Our data suggest that anatomic sites such as the male genital tract (MGT) and oral mucosa might harbor persistent reservoirs of ZIKV that, under certain conditions, could facilitate viral transmission over extended periods to uninfected hosts39. Particularly, the sustained high-level production of infectious virus in the MGT, highlights the possibility of sexual transmission long after the resolution of symptomatic infection40,41. Notably, we observed very high levels of ZIKV RNA in the uterus of infected animals but unexpectedly low levels in vaginal fluids. If these observations translate to ZIKV infection of humans, then it might indicate a reduced potential for female-to-male sexual transmission, despite posing substantial risk to pregnancy42. However, in both male and female macaques, extended ZIKV shedding in the saliva implies that the oral mucosa can sustain viral replication over extended periods. The observation of persistent infectious shedding in oral fluids underscores a possible concern for transmission in household settings. Collectively, these findings further highlight the need for the rapid development of vaccines and therapeutics against ZIKV.
ONLINE METHODS
Study design and ZIKV challenge
28 outbred, adult (male and female) Indian-origin rhesus monkeys (Macaca mulatta) and eight (male and female) cynomolgus monkeys (Macaca fascicularis) were used in these studies. All animals were housed at Bioqual. Animals used in these studies were infected via a subcutaneous route using Thai (KF993678) or Puerto Rican (KU501215) ZIKV challenge strains. The described studies were approved by the Bioqual Institutional Animal Care and Use Committee (IACUC).
Sample collection and processing
Viral levels were monitored in the blood, urine, saliva, CSF, semen and vaginal secretions for 4 weeks after infection. Blood, saliva and urine were collected daily for the first week and then weekly. Semen, CSF and vaginal secretions were collected weekly. Blood was collected into EDTA anticoagulant. Clean urine was obtained by direct bladder cystocentesis. Saliva was collected by cotton roll Salivette (Sarstedt, Nümbrecht, Germany). CSF was obtained by lumbar puncture. Semen was collected by electro-ejaculation via rectal probe electrostimulation, as described44. Briefly, stimulation is provided by rhythmic pulsation from the stimulator. Seminal emission is collected into a sterile container. Seminal plasma was centrifuged immediately after collection at 4 °C and then frozen and stored at −80 °C. Vaginal secretions were collected by Wek-Cel sponges according to the manufacturer’s instructions. Samples were immediately frozen at −80 °C.
ZIKV detection in blood, CSF and mucosal fluids
Viral RNA levels in peripheral blood, urine, saliva, semen, CSF and cervicovaginal lavage (CVL) during primary infection (28 d after infection) were monitored by quantitative real-time PCR using primers and probe sets specific for ZIKV capsid. Viral RNA was isolated from cell-free blood plasma using QIAamp Viral RNA mini kit (Qiagen), as previously described32. Viral RNA extraction from other samples were performed using the NucliSENS miniMAG System (BioMérieux) by following the manufacturer’s suggested instructions. Briefly, 500 μl of each sample was directly added to 2 ml of lysis buffer; a cervical vaginal swab was mixed in lysis buffer and eluted. Extracted RNA was used for amplification on a 7300 ABI Real-Time PCR system (Applied Biosystems).
ZIKV infectivity in saliva, semen and CSF
The African green monkey kidney (Vero) cell line (ATCC-CCL81) was grown at 37 °C, under an atmosphere containing 5% CO2, in Eagle’s Minimum Essential Medium supplemented with 5% FBS (FBS), penicillin and streptomycin. The Vero cells were then seeded at a density of 40,000 cells/cm2 in 12-well plates 24 h before virus inoculation. Semen, saliva and CSF samples isolated from infected monkeys on day 7 and 14 after infection were thawed and then centrifuged at 3,000 rpm for 5 min to separate any remaining cells. Clarified samples were then diluted to 1:2 and 1:4 in serum-free Eagle’s Minimum Essential medium. Each dilution was used to infect monolayers (200 μl/well). After 2 h of incubation at 37 °C, the inoculum was removed from cells, and the cells were washed thoroughly with 37 °C PBS, twice. Then, 2 ml of culture medium were added in the presence of penicillin and streptomycin. As controls, Vero cells were incubated with either the heat-inactivated blood plasma from uninfected monkey (mock infected) or blood plasma samples of infected monkeys isolated on day 3 after infection (positive). Culture supernatants were taken at 24, 48 and 72 h after infection, and ZIKV RNA levels were quantified by RT–qPCR.
ZIKA virus RNA in situ hybridization
RNAscope probes were designed on the basis of a consensus sequence among four genomes: one consensus derived from sequencing the viral stock; two strains isolated from the blood of patients in Thailand in 2014 (KU681081.3) and the Philippines in 2012 (KU681082.3); and the 1947 MR 766 Ugandan sentinel monkey sequence (AY632535.2). The consensus was determined by majority rule among aligned nucleotide sites. Because KU681081.3 was not annotated, probes were designed on the basis of the annotations of Zika virus from the NCBI reference sequence NC_012532.1 (accession number, NC_012532), which consisted of 87 double-Z pairs (ZZ) targeting coding regions from 101–9773 bp in capsid (Ca), envelope (Env) and nonstructural proteins (NS) NS1, NS3 and NS5 genes. RNAscope was performed essentially as previously described45.
Phenotypic characterization of Zika-virus-RNA+ cells
Immunofluorescence phenotypic analysis for cells positive for Zika virus RNA was performed after RNAscope in situ hybridization, essentially as previously described, using TSA detection. We identified the different potential target cells by using the following antibodies: myeloid cells (CD163, Novo Castra-NCL-CD163 and CD68, Biocare-CM33C), neutrophils (myeloperoxidase, Dako-A0398), cerebellar granule cells (NeuN, Abcam ab104225), microglia cells (Iba1, Biocare CP290 A, B), T cells (CD3 clone SP7; Labvision/Thermo Scientific), B cells (CD20, Dako-MO755) and DC-SIGN-positive cells (DC-SIGN, Abcam ab59192). Slides were cover slipped with #1.5 GOLD SEAL cover glass (EMS) using Prolong Gold reagent (Invitrogen) and imaged on an Olympus FV10i confocal microscope using a 60× phase-contrast oil-immersion objective (NA 1.35) in sequential mode to separately capture the fluorescence from the different fluorochromes at an image resolution of 1024 × 1024 pixels.
ZIKV NS1-specific enzyme-linked immunosorbent assay (ELISA)
IgM and IgG Zika-specific antibody responses were assessed using the Euroimmun diagnostic kit assay. Briefly, a 1:100 dilution of nonhuman primate serum was performed in duplicate and added to the precoated plates. The assay was performed using the manufacturer’s instructions, with photometric measurements taken at 450 nm.
Plaque-reduction neutralization test (PRNT90)
NHP blood plasma samples were collected, heat inactivated and stored at −80 °C. NHP plasma and positive control ZIKV-neutralizing sera (of known titer) were serially diluted. Undiluted and diluted samples were added to an equal volume of 2,000 PFU/ml of ZIKV (Puerto Rico PRVABC59; NCBI accession number, KU501215). The virus–antibody mixture was incubated at 37 °C for 1 h before 100 μl (100 PFU ZIKV) was added to each well of a confluent 6-well tissue-culture dish seeded with Vero cells. After a 1-h virus adsorption at 37 °C, cells were overlaid with 1% noble agar in supplemented minimal essential medium (MEM). At 4 d after infection, an additional overlay with 0.02% (wt/vol) neutral red in MEM with 1% agar was added, and plaques were counted and recorded the following day. The assay was performed in triplicate and control wells were also incorporated. The neutralizing-antibody titer was expressed as the maximum dilution of blood plasma that yielded a 90% plaque reduction.
Immunophenotyping
PBMCs and blood plasma were collected following Ficoll density gradient centrifugation. 0.5 × 106 PBMCs were stained with three different antibody cocktails to assess T cells, B cells and NK cell subpopulations. For T cells, CD3 (clone SP34-2), CD4 (L200), CD8 (SK1), CD14 (M5E2), CD95 (DX2), CD28 (CD28.2, eBioscience), CD69 (TP1.55.3, Beckman Coulter) and CD20 (L27). For NK cells, CD3, CD8, NKG2A (Z199, Beckman Coulter), CD16 (3G8), CD56 (N901, Beckman Coulter) and CD69. For B cells, CD3, CD20, CD38 (OKT10, Nonhuman Primate Reagents Resource Program), CD40 (5C3), CD69, CD95, CD27 (O323, eBioscience) and CD86 (2331 (FUN-1)). All antibodies are from BD Biosciences unless indicated otherwise. After staining, cells were washed with 2% FBS in PBS (wash buffer), re-suspended in 2% formaldehyde and acquired on a BD LSRII within 24 h. All events were gated first on FSC singlets. For T, B and NK cell analysis, lymphocytes were first selected according to FSC-A and SSC-A profiles. Monocytes were selected on the basis of their relatively higher FSC-A and SSC-A profile, as compared to lymphocytes. B cells were identified as CD3−CD20+, and CD27 was used to define naive (CD27−) and memory (CD27+) subsets46. NK cells were identified as CD3−CD8+NKG2A+ and CD16 and CD56 were used to define subsets47,48. CD3+CD4+ and CD3+CD8+ T cell memory subsets were identified by CD28 and CD95 (ref. 49). Percentage of CD69 measured activation NK cell subsets and on naïve (CD28+CD95−), central memory (CD28+CD95+) and effector/effector memory (CD28−CD95+) T cells. CD38 geometric mean fluorescence (GMF) measured activation on naïve and memory B cells. Percentage of CD16 measured activation on monocytes. Absolute counts were derived by multiplying the percentage of the subset of interest per total lymphocytes by the absolute number of total lymphocytes per μl of blood provided by the complete blood count.
Intracellular cytokine staining
Measurements of CD107a, IL-2, IFN-γ and TNF-α on CD4+ and CD8+ T cells from cryopreserved PBMCs and lymph node mononuclear cells were performed as previously described50. After thawing, cells rested for 4 h before stimulation. Approximately 3 × 106 cells were stimulated with DMSO alone (unstimulated) or 2 μg/ml overlapping peptide (15 aa long, 11 aa overlap) pools (Sigma-Aldrich) covering the entire ZIKV capsid and envelope proteins and 1 μg/ml of anti-CD49d antibodies (BD Biosciences) in the presence of anti-CD107a (H4A3) antibody. The following conjugated antibodies and staining reagents were used for surface staining: CD4 (L200), CD95 (DX2), CD28 (L293), CD8 (SK1) and Yellow LIVE/DEAD Fixable Dead Cell Stain (Invitrogen). The following conjugated antibodies were used during intracellular staining: CD3 (SP34-2), IL-2 (MQ1-17H12), IFN-γ (B27), TNF-α (Mab11) and CD69 (TP1.55.3, Beckman Coulter). All antibodies are from BD Biosciences unless indicated otherwise. Value of percentage of cytokine-positive cells in unstimulated samples was subtracted from the corresponding value of stimulated samples. Responses were considered positive if 1.5-fold or greater above baseline. The threshold for determining positive responses was applied as described43.
ZIKV dynamic modeling
To understand the dynamics of viral infection as reflected in the blood plasma VL, we used a standard acute viral infection model that includes an eclipse phase after viral infection of a cell, during which time the infected cell does not produce virus51–53. After the eclipse phase, infected cells produces new virus. In the model, target cells, T, are infected at rate βVT by free virus V. On infection, a cell becomes nonproductively infected (I1) and then transitions to a productively infected state I2 at rate kI1. Productively infected cells are cleared at rate δI2 and produce new virus at rate pI2. Free virus is cleared from the system at rate cV. Under these assumptions, the model has the form: dT/dt = −βVT, dI1/dt = βVT–kI1, dI2/dt = kI1–δI2, dV/dt = pI2−cV. We set initial conditions at T(0) = 107 cells per mL, I1(0) = I2(0) = 0, and V(0) is allowed to take one of a set of values, V(0) = 101, 102, 103, 104 or 105 RNA copies per ml. There is uncertainty in the value of V(0) because the virus was injected subcutaneously, and the amount that initiates the infection in blood is unknown. We used a value of T(0) within the range of the estimated number of target cells available in West Nile virus infection52. The parameter describing the clearance of free virus was fixed at c = 25/day, similarly to the values determined for HIV and HCV54,55. Finally, the parameter describing the transition rate from nonproductively infected to productively infected was allowed to take one of a set of values, k = 4, 6, 12 or 18 per d, equivalent to a mean eclipse-phase time of 6, 4, 2 or 1.33 h. For each animal, the model was fitted to the data by a nonlinear least-squares approach using a differential evolution algorithm in R for each combination of k and V(0). In our analysis, we excluded the single outlier monkey that had delayed viremia, because there were only two viral load measurements above the limit of detection.
Statistical analyses
The analyses of virologic or immunologic data were conducted using a two-tailed Mann–Whitney test. All statistics between grouped variables or transformed log10 values were calculated using GraphPad Prism (version 6.0) by use of the exact Mann–Whitney U test and the Kruskal–Wallis test with corrections for multiple comparisons. R was used to generate heat maps. All P values were two-tailed, unless indicated, and considered to be significant when <0.05.
Supplementary Material
Acknowledgments
We thank B. Finneyfrock, Z. Pippin, A. Dodson and A. Cook for their expert animal husbandry and care, and J. Guedj for suggestions about the model simulations. CD38 antibodies were obtained from the NIH Nonhuman Primate Reagent Resource supported by HHSN272200900037C and OD010976. The data presented in this study are tabulated in the main paper and in the supplementary materials. This work was supported in part by federal funds from the National Cancer Institute (NIH Contract HHSN261200800001E). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products or organizations imply endorsement by the US Government. A.S.P. acknowledges support from NIH grants AI028433 and OD0110995. D.S. acknowledges support from the Public Health Agency of Canada.
Footnotes
AUTHOR CONTRIBUTIONS
J.B.W. designed the studies. C.E.O., H.A.-E. and S.-Y.L. led the virologic assays. S.-Y.L. and C.E.O. led the immunologic assays. C.D. and J.D.E. led all tissue analysis. L.T.S., R.W.O. and M.L. conducted the cytokine assays. P.T.H, M.L., C.E.O. and S.-Y.L. led the cytokine and chemokine analysis. S.-Y.L. and P.T.H. led the peptide design analysis. B.D.G., D. Stein and D. Safronetz led the antibody assays. D.L.V., S. Huang, S. Higgs and D. Safronetz produced the ZIKV stocks K.B., E.F.C.O. and A.S.P. led the mathematical modeling. H.A.-E. and M.G.L. led the clinical care of the macaques. J.B.W. led the studies and wrote the paper with all co-authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Dick GW, Kitchen SF, Haddow AJ. Zika virus. I Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 2.Brasil P, et al. Zika virus outbreak in Rio de Janeiro, Brazil: clinical characterization, epidemiological and virological Aspects. PLoS Negl Trop Dis. 2016;10:e0004636. doi: 10.1371/journal.pntd.0004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerbino-Neto J, et al. Clinical manifestations of Zika virus infection, Rio de Janeiro, Brazil, 2015. Emerg Infect Dis. 2016;22:1318–1320. doi: 10.3201/eid2207.160375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duffy MR, et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009;360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 5.Campos GS, Bandeira AC, Sardi SI. Zika virus outbreak, Bahia, Brazil. Emerg Infect Dis. 2015;21:1885–1886. doi: 10.3201/eid2110.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao-Lormeau VM, Musso D. Emerging arboviruses in the Pacific. Lancet. 2014;384:1571–1572. doi: 10.1016/S0140-6736(14)61977-2. [DOI] [PubMed] [Google Scholar]
- 7.Cardoso CW, et al. Outbreak of exanthematous illness associated with Zika, chikungunya, and dengue viruses, Salvador, Brazil. Emerg Infect Dis. 2015;21:2274–2276. doi: 10.3201/eid2112.151167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oehler E, et al. Zika virus infection complicated by Guillain-Barre syndrome—case report, French Polynesia, December 2013. Euro Surveill. 2014;19:20720. doi: 10.2807/1560-7917.es2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- 9.Millon P. In: Epidémiologie des syndromes de Guillain-Barré en Nouvelle-Calédonie entre 2011 et 2014: influence des arboviroses. Fourier UJ, editor. 2015. [Google Scholar]
- 10.Cao-Lormeau VM, et al. Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brasil P, et al. Guillain-Barré syndrome associated with Zika virus infection. Lancet. 2016;387:1482. doi: 10.1016/S0140-6736(16)30058-7. [DOI] [PubMed] [Google Scholar]
- 12.Schuler-Faccini L, et al. Possible association between Zika virus infection and microcephaly—Brazil, 2015. MMWR Morb Mortal Wkly Rep. 2016;65:59–62. doi: 10.15585/mmwr.mm6503e2. [DOI] [PubMed] [Google Scholar]
- 13.European Centre for Disease Prevention and Control. Microcephaly in Brazil potentially linked to the Zika virus epidemic. Stockholm: 2015. [Google Scholar]
- 14.Mlakar J, et al. Zika virus associated with microcephaly. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 15.Calvet G, et al. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis. 2016;16:653–660. doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- 16.Peterson AT, Osorio J, Qiao H, Escobar LE. Zika virus, elevation, and transmission risk. PLoS Curr. 2016 doi: 10.1371/currents.outbreaks.a832cf06c4bf89fb2e15cb29d374f9de. http://currents.plos.org/outbreaks/article/zika-virus-elevation-and-transmission-risk/ [DOI] [PMC free article] [PubMed]
- 17.Foy BD, et al. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis. 2011;17:880–882. doi: 10.3201/eid1705.101939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musso D, et al. Potential sexual transmission of Zika virus. Emerg Infect Dis. 2015;21:359–361. doi: 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bearcroft WG. Zika virus infection experimentally induced in a human volunteer. Trans R Soc Trop Med Hyg. 1956;50:442–448. [PubMed] [Google Scholar]
- 20.Bingham AM, et al. Comparison of test results for Zika virus RNA in urine, serum, and saliva specimens from persons with travel-associated Zika virus disease—Florida, 2016. MMWR Morb Mortal Wkly Rep. 2016;65:475–478. doi: 10.15585/mmwr.mm6518e2. [DOI] [PubMed] [Google Scholar]
- 21.Musso D, et al. Detection of Zika virus in saliva. J Clin Virol. 2015;68:53–55. doi: 10.1016/j.jcv.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 22.Gourinat AC, O’Connor O, Calvez E, Goarant C, Dupont-Rouzeyrol M. Detection of Zika virus in urine. Emerg Infect Dis. 2015;21:84–86. doi: 10.3201/eid2101.140894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamel R, et al. Biology of Zika virus infection in human skin cells. J Virol. 2015;89:8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabata T, et al. Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell Host Microbe. 2016;20:155–166. doi: 10.1016/j.chom.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dudley DM, et al. A rhesus macaque model of Asian-lineage Zika virus infection. Nat Commun. 2016;7:12204. doi: 10.1038/ncomms12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Styer LM, et al. Mosquitoes inoculate high doses of West Nile virus as they probe and feed on live hosts. PLoS Pathog. 2007;3:1262–1270. doi: 10.1371/journal.ppat.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gubler DJ, Rosen L. A simple technique for demonstrating transmission of dengue virus by mosquitoes without the use of vertebrate hosts. Am J Trop Med Hyg. 1976;25:146–150. doi: 10.4269/ajtmh.1976.25.146. [DOI] [PubMed] [Google Scholar]
- 28.Dutra HL, et al. Wolbachia blocks currently circulating Zika virus isolates in Brazilian Aedes aegypti mosquitoes. Cell Host Microbe. 2016;19:771–774. doi: 10.1016/j.chom.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rathakrishnan A, et al. Cytokine expression profile of dengue patients at different phases of illness. PLoS One. 2012;7:e52215. doi: 10.1371/journal.pone.0052215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandey N, et al. Serum levels of IL-8, IFNγ, IL-10, and TGF β and their gene expression levels in severe and non-severe cases of dengue virus infection. Arch Virol. 2015;160:1463–1475. doi: 10.1007/s00705-015-2410-6. [DOI] [PubMed] [Google Scholar]
- 31.Kumar Y, et al. Serum proteome and cytokine analysis in a longitudinal cohort of adults with primary dengue infection reveals predictive markers of DHF. PLoS Negl Trop Dis. 2012;6:e1887. doi: 10.1371/journal.pntd.0001887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitney JB, et al. Monitoring HIV vaccine trial participants for primary infection: studies in the SIV/macaque model. AIDS. 2009;23:1453–1460. doi: 10.1097/QAD.0b013e32832b43d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai F, et al. A paradoxical role for neutrophils in the pathogenesis of West Nile virus. J Infect Dis. 2010;202:1804–1812. doi: 10.1086/657416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soares CN, et al. Fatal encephalitis associated with Zika virus infection in an adult. J Clin Virol. 2016;83:63–65. doi: 10.1016/j.jcv.2016.08.297. [DOI] [PubMed] [Google Scholar]
- 35.Ventura CV, Maia M, Bravo-Filho V, Góis AL, Belfort R., Jr Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet. 2016;387:228. doi: 10.1016/S0140-6736(16)00006-4. [DOI] [PubMed] [Google Scholar]
- 36.Mlakar J, et al. Zika virus associated with microcephaly. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 37.Barcellos C, et al. Increased hospitalizations for neuropathies as indicators of Zika virus infection, according to Health Information System Data, Brazil. Emerg Infect Dis. 2016 doi: 10.3201/eid2211.160901. http://dx.doi.org/10.3201/eid2211.160901. [DOI] [PMC free article] [PubMed]
- 38.Mécharles S, et al. Acute myelitis due to Zika virus infection. Lancet. 2016;387:1481. doi: 10.1016/S0140-6736(16)00644-9. [DOI] [PubMed] [Google Scholar]
- 39.Turmel JM, et al. Late sexual transmission of Zika virus related to persistence in the semen. Lancet. 2016;387:2501. doi: 10.1016/S0140-6736(16)30775-9. [DOI] [PubMed] [Google Scholar]
- 40.D’Ortenzio E, et al. Evidence of sexual transmission of Zika virus. N Engl J Med. 2016;374:2195–2198. doi: 10.1056/NEJMc1604449. [DOI] [PubMed] [Google Scholar]
- 41.Deckard DT, et al. Male-to-male sexual transmission of Zika virus—Texas, January 2016. MMWR Morb Mortal Wkly Rep. 2016;65:372–374. doi: 10.15585/mmwr.mm6514a3. [DOI] [PubMed] [Google Scholar]
- 42.Prisant N, et al. Zika virus in the female genital tract. Lancet Infect Dis. 2016 doi: 10.1016/S1473-3099(16)30193-1. http://dx.doi.org/10.1016/S1473-3099(16)30193-1. [DOI] [PubMed]
- 43.Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrison RM. Semen parameters in Macaca mulatta: ejaculates from random and selected monkeys. J Med Primatol. 1980;9:265–273. doi: 10.1159/000460152. [DOI] [PubMed] [Google Scholar]
- 45.Deleage C, et al. Defining HIV and SIV reservoirs in lymphoid tissues. Pathog Immun. 2016;1:68–106. doi: 10.20411/pai.v1i1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuhrt D, et al. Naïve and memory B cells in the rhesus macaque can be differentiated by surface expression of CD27 and have differential responses to CD40 ligation. J Immunol Methods. 2011;363:166–176. doi: 10.1016/j.jim.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webster RL, Johnson RP. Delineation of multiple subpopulations of natural killer cells in rhesus macaques. Immunology. 2005;115:206–214. doi: 10.1111/j.1365-2567.2005.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves RK, et al. CD16− natural killer cells: enrichment in mucosal and secondary lymphoid tissues and altered function during chronic SIV infection. Blood. 2010;115:4439–4446. doi: 10.1182/blood-2010-01-265595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pitcher CJ, et al. Development and homeostasis of T cell memory in rhesus macaque. J Immunol. 2002;168:29–43. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- 50.Donaldson MM, et al. Optimization and qualification of an 8-color intracellular cytokine staining assay for quantifying T cell responses in rhesus macaques for pre-clinical vaccine studies. J Immunol Methods. 2012;386:10–21. doi: 10.1016/j.jim.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baccam P, Beauchemin C, Macken CA, Hayden FG, Perelson AS. Kinetics of influenza A virus infection in humans. J Virol. 2006;80:7590–7599. doi: 10.1128/JVI.01623-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Banerjee S, Guedj J, Ribeiro RM, Moses M, Perelson AS. Estimating biologically relevant parameters under uncertainty for experimental within-host murine West Nile virus infection. J R Soc Interface. 2016;13:20160130. doi: 10.1098/rsif.2016.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fukuhara M, et al. Quantification of the dynamics of enterovirus 71 infection by experimental-mathematical investigation. J Virol. 2013;87:701–705. doi: 10.1128/JVI.01453-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guedj J, et al. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc Natl Acad Sci USA. 2013;110:3991–3996. doi: 10.1073/pnas.1203110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramratnam B, et al. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet. 1999;354:1782–1785. doi: 10.1016/S0140-6736(99)02035-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.