Abstract
Biosimilar drugs must closely resemble the pharmacological attributes of innovator products to ensure safety and efficacy to obtain regulatory approval. Glycosylation is one critical quality attribute that must be matched, but it is inherently difficult to control due to the complexity of its biogenesis. This usually implies that costly and time-consuming experimentation is required for clone identification and optimization of biosimilar glycosylation. Here, we describe a computational method that utilizes a Markov model of glycosylation to predict optimal glycoengineering strategies to obtain a specific glycosylation profile with desired properties. The approach uses a genetic algorithm to find the required quantities to perturb glycosylation reaction rates that lead to the best possible match with a given glycosylation profile. Furthermore, the approach can be used to identify cell lines and clones that will require minimal intervention while achieving a glycoprofile that is most similar to the desired profile. Thus, this approach can facilitate biosimilar design by providing computational glycoengineering guidelines that can be generated with a minimal time and cost.
Keywords: biosimilars, CHO cells, erythropoietin, glycoengineering, Markov model
Graphical Abstract
1 Introduction
During biosimilar development, process parameters are adjusted to reproduce the pharmacological and biochemical properties of the original approved innovator protein drug. Protein glycosylation is a critical post-translational modification on most secreted mammalian proteins, and variations in glycan structure can significantly impact the bioactivity of a protein (Dalziel et al., 2014; Griebenow and Sola, 2009; Jefferis, 2009; Li and d’Anjou, 2009; Raju, 2008; Solá and Griebenow, 2011). Thus, regulatory agencies require that biosimilar drug glycoforms match the approved drug. Consequently, engineering the glycoprofile, i.e. the relative frequencies of glycans present on the protein, is a critical part of biosimilar production (Chiang et al., 2016; Niwa and Satoh, 2015; Tsuruta et al., 2015; Zhang et al., 2016).
Reproducing a glycoprofile can be difficult since, in theory, a cell could synthesize thousands of different glycans. Protein glycosylation is a non-template driven process whose outcome follows a statistical distribution. It is often difficult to predict this process with pure intuitive reasoning since perturbation of a glycosyltransferase will affect the abundances of glycans and the rates of other reactions connected to it, often leading to non-obvious glycoforms. As a consequence, expansive profiling of diverse clones and elaborate titration experiments are usually necessary. Therefore, computer simulations could provide valuable guidance by predicting the required quantities of reaction rate perturbation or aid in clone selection to facilitate glycoengineering efforts.
We previously developed a model of glycosylation that captured the stochastic nature of its biogenesis by conceptualizing it as a Markov chain process (Spahn et al., 2016). The advantage of this probabilistic approach is that the many factors influencing the kinetics of glycosylation reactions are subsumed in probabilities, thus avoiding the necessity to estimate kinetic parameters. Instead, the model uses an initial known glycoprofile from a starting production cell line. The observed glycan frequencies in this profile are used to empirically reconstruct the probabilities for each glycosylation reaction in the network. After this fitting process, the model can simulate how the glycoprofile will change after perturbing one or more enzyme-dependent reaction sets (see Methods). These perturbations can include various experimental techniques that can either involve cell engineering (knock-downs or overexpression of glycosylation genes) or bioprocess control (media supplementation with nutrients or inhibitors). Although this and other glycosylation models (Spahn and Lewis, 2014; Villiger et al., 2016) allow one to make a priori predictions of glycoprofiles, there is still a great need for easy-to-use computational approaches that would directly address glycoengineering efforts and help to quantitatively predict optimal perturbation strategies to recapitulate a desired glycoprofile.
Here we present a novel optimization-based implementation of the glycosylation Markov model that predicts the quantitative amount by which glycosylation reaction rates must be perturbed to achieve a desired glycoprofile. We demonstrate how this approach can guide glycoengineering in the context of biosimilar production, where the desired glycoprofile is already known, but the quantitative perturbations necessary to achieve it are not. To accomplish this, we reverse the simulation workflow such that, rather than predicting an unknown glycoprofile, the simulation predicts the optimal level of reaction rate perturbation that achieves a glycoprofile. We demonstrate the capabilities of this approach to predict experimental interventions needed to mimic the glycoprofile of Rituximab and Erythropoietin (EPO). These model-derived predictions can then serve as guidance to efficiently adjust the experimental means (e.g. the concentration range of an inhibitor). In addition, these simulations can help to assess which glycoprofiles will be harder or easier to engineer towards desired properties. This can help identify production cell lines that produce proteins with glycosylation profiles that will require minimal experimental modification to match the innovator drug.
2 Materials and methods
2.1 Glycoprofiling
Glycoprofiling was performed as previously published (Grav et al., 2015). Exponentially growing cells were seeded at 1×106 cells/mL and supernatant was harvested after 4 days by centrifugation. Supernatant for all samples was filtered before further processing. Rituximab was purified using HiTrap MabSelect columns (GE Healthcare) according to the manufacturer’s instructions. EPO containing supernatants (175 mL) were buffer exchanged with 400 mL 20 mM TRIS-HCl pH 8.0 using a VivaFlow 50 30 kDa MWCO cross flow cassette (Sartorius). Samples were loaded onto a Mono Q 5/50 GL column (GE Healthcare) and EPO was eluted using a gradient increase to 20 mM TRIS-HCl pH 8.0, 300 mM NaCl over 30 column volumes. EPO containing fractions were pooled and analyzed by SDS PAGE. Concentrations of Rituximab and EPO were measured by Nanodrop (Thermo Scientific). N-glycans from purified proteins were released and fluorescently labeled with GlykoPrep Rapid N-Glycan kit (ProZyme Inc., Hayward, CA). Labeled N-glycans were analyzed by LC-MS on a Thermo Ultimate 3000 HPLC with fluorescence detector coupled on-line to a Thermo Velos Pro Iontrap MS. Glycan abundance was measured by integrating the areas under normalized fluorescence spectrum peaks with Xcalibur software (Thermo Fisher Scientific) giving the relative amount of the glycans.
2.2 Simulation
The Markov model of glycosylation, including the rationale to computationally predict perturbations, is implemented as previously published (Spahn et al., 2016). Based on the data given in the initial glycoprofile, reaction probabilities are reconstructed to generate the model for the glycosylation reaction network present in the cell line from which the initial glycoprofile was obtained. An objective function is defined that takes this initial model, a list of perturbations of it, and the provided target profile as input variables. A perturbation is defined as the name of a glycosylation enzyme and a quantity q by which its reaction set is to be scaled up (if q > 1) or down (if 0 < q < 1). The term “reaction set” refers to the entirety of reactions in the glycosylation reaction network that depend upon a specific glycosylation enzyme. The objective function returns the mismatch between the computed perturbed profile and the target profile. To find a minimum of this mismatch, a genetic algorithm, as implemented in MATLAB (function ga), is set up that infers the optimal quantity of perturbation over the space of all possible quantities of perturbation within a given range between 0 and a maximum upper bound (qmax = 10 used in this study). Specifying this upper bound is also helpful in situations where it is clear that certain reaction sets need to be scaled down rather than up in which case the upper bound can be set to 1. The stopping criterion is set to a maximum of 20+(Number of reaction sets)*20 generations while premature stopping occurs if the best objective function value stalls for more than 10 generations. Population size is set to 20 individuals with default settings for selection, crossover and mutation. Refer to standard literature (e.g. (Mitchell, 1998) as well as the MATLAB website (www.mathworks.com) for an explanation of genetic algorithm rationale and terminology. Mismatch between two profiles is calculated as the absolute distance between average glycan frequencies, summed over all glycans in the profiles. Individual (non-negative) weights can be defined for individual glycans to adjust their impact on the overall mismatch between two profiles. If all weights are set to 1 (as used throughout this paper), the metric equals the Manhattan metric. A least 10 independent runs of the genetic algorithm were evaluated for each glycoprofile that all yielded the same minimal possible profile mismatch. In order to assess robustness, the predicted optimal quantities of perturbation were allowed to randomly vary by 10% and the resulting glycoprofiles were recalculated.
3 Results
3.1 An application of the glycosylation Markov model predicts how to achieve a target glycoprofile
The Markov model provides a platform to predict changes in an initial glycoprofile (i.e. the profile obtained from the production cell line as it stands) following reaction rate perturbations (Spahn et al., 2016). To predict the optimal engineering strategy to achieve a desired glycoprofile, here we reverse the workflow such that, in addition to the initial glycoprofile, the target glycoprofile (i.e. the desired profile of the innovator product) needs to be provided. In addition, it can be specified which enzyme-dependent glycosylation reaction sets are supposed to be perturbed. This choice is optional, but since often not all glycosylation reactions require perturbation, or only a subset is amenable to perturbation with the experimental means at hand, specification will speed up computation. For example, if a chemical inhibitor of a fucose transporter is to be used in combination with a supplementation of UDP-galactose, FucT and GalT would be specified as glycosylation reaction sets since fucosylation and galactosylation reactions are being affected through these treatments.
The simulation starts by first fitting the Markov model to the initial glycoprofile to infer the reaction probabilities in the production cell line (Spahn et al., 2016). To find an optimal quantitative strategy to engineer this profile towards the target profile, the simulation runs a genetic algorithm that randomly modifies the levels of perturbation in the specified reaction set, computes the resulting changes in the glycoprofile and retains the best solutions. By mixing and randomly changing these solutions, the algorithm iteratively converges on the target profile and reaches an optimal level of perturbation (see Methods). The final quantitative levels of perturbations leading to this optimum are returned. The similarity between glycoprofiles is measured by using a profile mismatch metric. This metric can be flexibly defined for the requirements of the specific glycoengineering problem. For example, if certain glycans are more important than others in the profile (due to regulatory or functional reasons), weights can be assigned to them so these priorities are reflected by the simulation (see Methods).
3.2 Engineering strategies can be designed to match the glycoprofile of IgGs
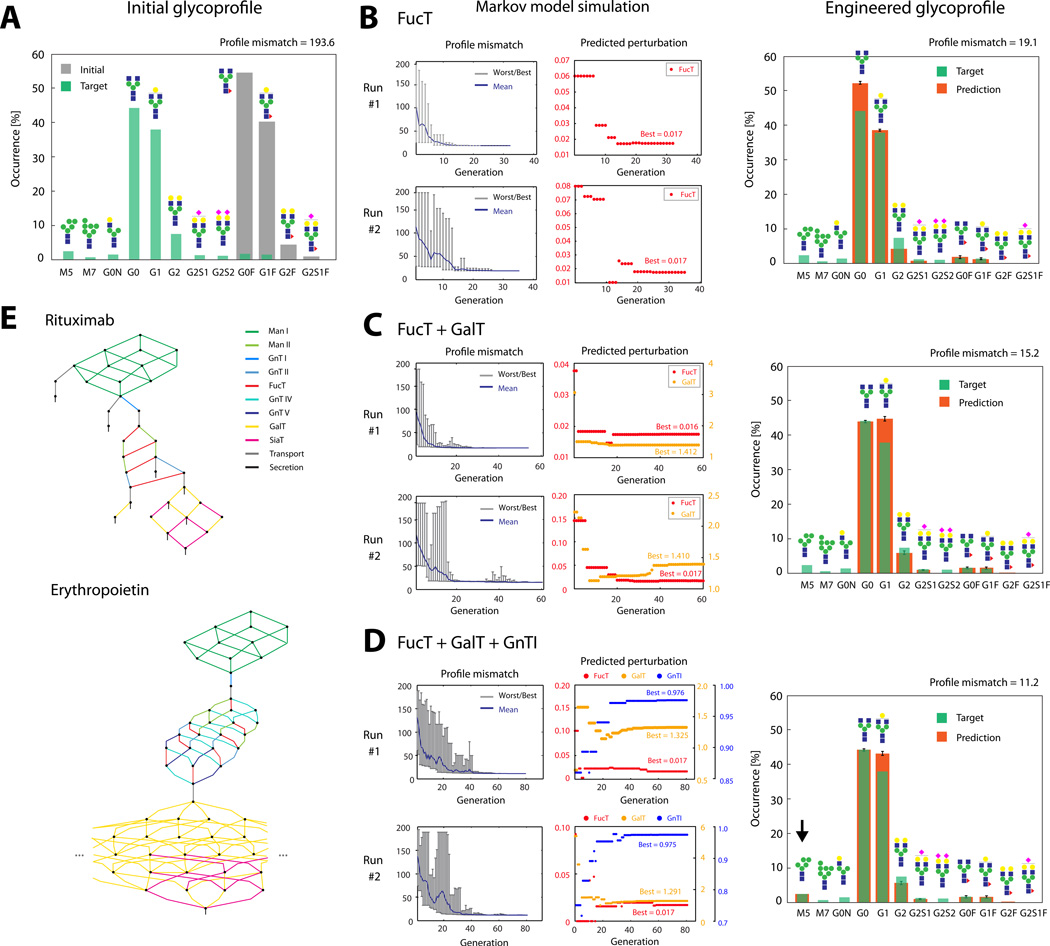
We demonstrate the workflow with Rituxmab, a monoclonal antibody of high commercial interest (Daniel et al., 2015; Turner, 2015; Visser et al., 2013). The glycoprofile of the innovator drug shows the typical pattern of an IgG1 antibody featuring G0, G1 and G2 glycoforms as well as a low level of fucosylation (Visser et al., 2013) (Fig. 1A). Achieving this glycoprofile usually requires a considerable degree of glycoengineering since CHO cell lines used for production typically display profiles that show higher degrees of fucosylation. When produced in a CHO-S line, the Rituximab glycoprofile has all glycoforms being fucosylated (Fig. 1A). Since the target profile on the innovator drug shows only minor fractions of fucosylation, the FucT reaction set is an obvious target for downregulation through glycoengineering, but due to the intertwined nature of the glycosylation reaction network (Fig. 1E) the magnitude of downregulation of FucT activity is non-obvious. Running the Markov-model based simulation, a downregulation to ~2% is predicted to give the closest possible match to the desired target profile, if only fucosylation is allowed to be altered (Fig. 1B). However, as evident from the frequency mismatches in the predicted profile (glycans G0-G2), the increased levels of galactosylated glycoforms present on the innovator profile require further modification, so it is reasonable to alter the GalT reaction set as well. By allowing perturbation of both reaction sets, an optimal perturbation that matches the degree of both galactosylation and fucosylation is computed to be at 41% upregulation of the GalT reaction set and downregulation to ~2% of the FucT reaction set (Fig. 1C). As mentioned above, custom weights could be applied for individual glycans, e.g. if a match in G1 (rather than G0) is to be given higher priority because stability concerns are not an issue (Raju and Scallon, 2007). Finally, the target profile also includes a fraction of high-mannose glycoforms, so GnTI can be added to the list of reaction sets to be altered because failure to add the respective GlcNAc at this point in the network will cause secretion of unprocessed (high-mannose) glycoforms. As predicted by the simulation, an excellent match to the target profile can be obtained by reducing GnTI to ~98% while increasing galactosylation by ~30% and decreasing fucosylation to ~2% since also the M5 fraction will be successfully reproduced (Fig. 1D, arrow). Thus, our model-based simulation can give helpful clues into how close a certain target profile can be matched with a given initial cell line, and also provides the predicted reaction rate perturbations that will transform the initial profile accordingly.
Figure 1.
(A): Glycoprofiles on Rituximab, expressed in CHO-S cell line before any glycoengineering (“Initial”). The target profile shows the glycoprofile of the innovator drug as reported previously (Visser et al., 2013). (B)–(D): Results from the Markov-model based glycoengineering simulation (two representative runs shown) considering only FucT (B), FucT+GalT (C) or FucT+GalT+GnTI (D) reaction sets for perturbation. Profile mismatch evolution plots (left) show the worst, best and mean profile mismatches in each generation of the genetic algorithm. Perturbation plots (right) show the quantity of predicted perturbation yielding the best match in each generation. Glycoprofiles (far right) are calculated with the averages of perturbations throughout all independent simulation runs. Error bars are standard deviation. (E): Reaction network for the glycoprofiles on Rituximab and partial reaction network reconstructed for the glycoprofile on erythropoietin. Reactions are color-coded according to the glycosylation enzyme involved.
3.3 Validation based on glycosyltransferase knock-out cell lines
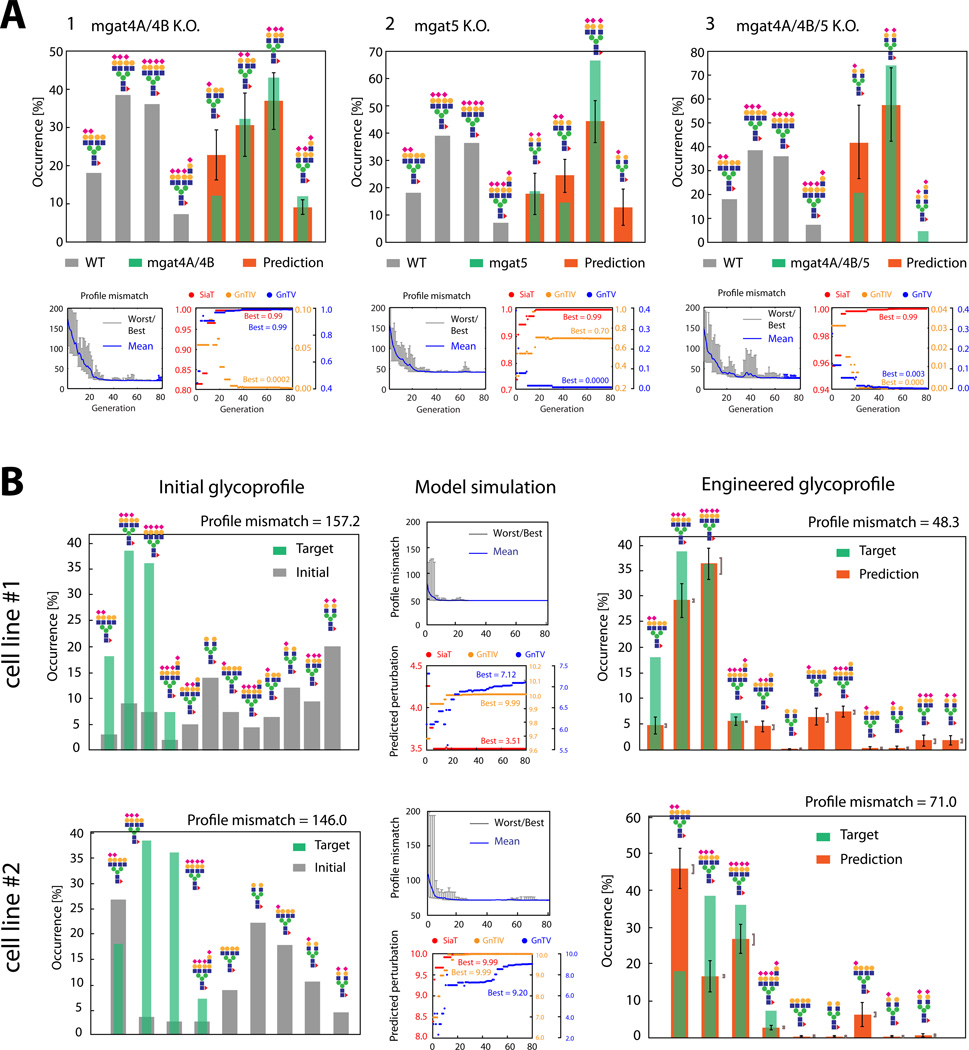
In order to validate that our simulation framework can infer perturbations based on glycoprofiles, we tested it on glycosyltransferase knock-out cell lines in CHO, published previously (Yang et al., 2015). A starting glycoprofile was obtained for EPO, expressed in a wildtype CHO-GS cell line. The model was used to predict the modifications required to match target profiles for mgat4A/4B, mgat5 and mgat4A/4B/5 knock-out cell lines, and the simulation correctly inferred the complete abrogation of GnTIV, GnTV and GnTIV + GnTV, respectively (Fig. 2A, 1–3). Interestingly, a slight perturbation to GnTIV was also seen for the GnTV knock-out, suggesting that those reactions were also moderately affected. However, the results here demonstrated that even for more complex multi-gene strategies, the model can accurately predict the strategy to perturb glycosylation to achieve a desired glycoprofile.
Figure 2.
(A): Glycoprofiles on EPO, expressed in CHO-GS (Wildtype and knock-out data from (Yang et al., 2015)). The simulation infers the required perturbations in SiaT, GnTIV and GnTV that transform the wildtype profile into the observed knock-out profiles (one representative run shown). Error bars are standard deviation. (B): Glycoengineering of two EPO-producing CHO lines with varying degrees of branching and sialylation. The reaction sets dependent on the branching enzymes GnTIV & GnTV and sialyltransferase (SiaT) are subject to perturbation in simulations (one representative run shown). Glycoprofiles are calculated with the averages of perturbations throughout all independent simulation runs. Error bars are standard deviation. Brackets indicate the range of the predicted averages when optimal predicted perturbations are varied by 10% (Methods).
3.4 Clones can be screened to test feasibility of engineering strategies
The approach presented here allows one to predict how to engineer a cell line to obtain glycoprofiles that approximate a desired innovator drug profile. However, different cell lines typically produce glycoprofiles that differ in their consistency with the target profile. Thus, it can be difficult to select the best cell line for glycoengineering efforts because it could be unclear which one requires less complex genetic or processing-related adjustments. Furthermore, it is often unclear which cell lines can be brought closest to the target profile. Our approach could therefore be valuable in choosing an optimal production line as it can give a quick guidance into the complexity of engineering strategies and assess the degree of match achievable for each cell line. We exemplify this application with two glycoprofiles on EPO produced in CHO lines, and compare the optimal strategies to achieve a profile from the literature (Yang et al., 2015). As shown by the simulation, both cell lines can approach the target profile through perturbation of SiaT, GnTIV and GnTV-dependent reactions. However, cell line #1 attains a considerably better match after glycoengineering, despite having an initial glycoprofile that differs more substantially from the innovator drug than cell line #2 (Fig. 2B). Indeed, the simulation indicates that even an optimal glycoengineering strategy can only obtain a mediocre match to the target profile for cell line # 2 (Fig. 2B). Consequently, the optimal cell line for glycoengineering is not necessarily the one showing the best initial match to the target profile. Computational analyses like these could be carried out even on a large collection of cell line candidates and help in prioritization and proper selection of glycoengineering methods.
4 Discussion
Here we have presented a novel approach that allows the prediction of engineering strategies to obtain a target glycoprofile, such as a profile from an innovator drug. It is important to note that the strategies predicted by our model represent quantities by which glycosylation reaction rates need to be perturbed. As such, our model makes no restriction on how these perturbations could be carried out experimentally. In fact, finding the right experimental tool for implementing these predicted quantitative perturbations is a challenge to the cell engineer; fortunately, recent progress in genetic and metabolic cell culture engineering have made a wide range of options available in this field the choice of which will largely depend on the specific perturbation to be obtained (Brühlman et al., 2015).
For example, if the model suggests a reduction in reaction rates, small-interfering RNA represents an inexpensive and effective option since downregulation of glycosylation genes can effectively change the glycoprofile (Imai-Nishiya et al., 2007). Alternatively, small molecules, such as sugar analogs or chemical inhibitors, can be used to reduce the rates of certain glycosylation reactions (Gardai et al., 2015; Okeley et al., 2013; Pande et al., 2015; Rillahan et al., 2012; Surve and Gadgil, 2015); also, the reduction of glucose has been reported as a way to reduce galactosylation levels (Villacres et al., 2015). Future work will aim to link to various media and bioprocesses treatments to other enzymatic reactions, thus further expanding the means by which such interventions can be used in a predictable manner. Since these non-genetic methods only require modification of the growth medium, they would be preferable in cases where manipulation of the cell line in use is not desired. Since the compounds used (siRNA or small molecules, respectively) can be titrated, these methods are particularly attractive for partial reduction of reaction rates where the quantitative level of reaction activity is supposed to match the level predicted by the model. In contrast, if total abrogation (i.e. a reduction to nearly 0%) is indicated by the model, genetic knock-outs of one or more glycosylation genes through zinc-finger nucleases, TALENS or CRISPR-Cas9 technology (Gaj et al., 2013; Kim and Kim, 2014; Lee et al., 2015) may be more attractive options since these methods provide a more effective way to permanently eliminate gene activity.
In cases where the model suggests increases in glycosylation reaction rates (see Fig. 2B), both genetic and metabolic perturbation strategies could be employed as well. Genetic approaches would, for example, include ectopic expression of glycosyltransferases or CRISPR-mediated gene activation in order to increase certain glycosylation reaction rates (Lin et al., 2015; Perez-Pinera et al., 2013; Weikert et al., 1999; Zalatan et al., 2015). Such ectopic expression could be combined with inducible promoters (Gossen and Bujard, 1992; Weber et al., 2004) to quantitatively control the increase of transgene expression, and thus reaction rates. Alternatively, supplementation of the growth medium with sugars, sugar-nucleotides or cofactors (e.g. manganese) provides a powerful method to increase certain glycosylation reactions as exemplified in several studies (Grainger and James, 2013; Gramer et al., 2011; St Amand et al., 2014). In addition to modifications of either the growth media or the host genome or transcriptome, protein yield can be another cell culture parameter affecting the glycoprofile. For instance, increases in protein yield can indirectly downregulate glycosylation reaction rates since under high protein production sugar substrates can become limiting. Also, at high protein production rates, glycosylation enzymes might not be capable of processing the entire bulk of Golgi-transiting proteins, thus leaving a higher fraction to remain in their unmodified, high-mannose glycoform (del Val et al., 2016).
Thus, multiple different approaches could be taken to fine-tune the activity of specific biosynthetic reactions in glycosylation. However, the ease of supplementing media with either inhibitory or stimulating agents makes it the most convenient strategy for implementing the model-predicted down- or upregulations, respectively. Genetic knock-outs might be the preferred method if a reaction set needs to be shut down completely and if genetic engineering of the production cell line is an option.
As demonstrated by the increasing profile similarity (Fig. 1 B–D), inclusion of multiple reaction sets for perturbation should typically obtain a closer match to the target profile, but it also increases complexity of the optimization problem resulting in slower convergence (i.e. increased computation time). More practically, however, the inclusion of more enzymes will require more complex experiments to fine-tune the engineering strategies. Also, glycosylation reactions are not perfectly independent since they all rely upon a shared sugar nucleotide metabolic network. Future advanced models could take this into account by integrating sugar metabolism into the prediction of glycoforms, as already demonstrated for kinetic glycosylation models (Jedrzejewski et al., 2014; Villiger et al., 2016). Thus, efforts that employ the simplest strategy resulting in a glycoprofile that meets requirements of safety, efficacy, and similarity are preferred.
In summary, while this approach contributes to a growing body of work on modeling glycosylation with a variety of computational methods (Hossler, 2012; Neelamegham and Liu, 2011; Spahn and Lewis, 2014), our method is the first that directly addresses model-based prediction of biosimilar design. The probabilistic foundation of it makes it especially valuable for rapid in silico screens as running the simulation requires no input other than the initial and the target glycoprofiles and the specification of the reaction sets to be perturbed. The method is fully flexible and can be used for any cell line and any recombinant glycoprotein. It can, thus, be a helpful asset in QbD-oriented glycoengineering efforts.
Acknowledgments
We acknowledge funding from the National Institutes of Health (grants R21 HD080682-01A1 and R35 GM119850) and generous funding through the Novo Nordisk Foundation Center for Biosustainability at the Technical University of Denmark (NNF16CC0021858). In addition, we thank Julio Baez for insightful discussion. This work is covered in a pending patent. The authors would like to thank Helle Munck Petersen for excellent technical assistance.
Abbreviations
- mAb
monoclonal antibody
- EPO
erythropoietin
- CHO
Chinese hamster ovary
- QbD
Quality by Design
Footnotes
Conflict of interest
We declare that there is no conflict of interest.
The authors declare no financial or commercial conflict of interest.
References
- Brühlman D, Jordan M, Hemberger J, Sauer M, Stettler M, Broly H. Tailoring recombinant protein quality by rational media design. Biotechnol. Prog. 2015;31:615–629. doi: 10.1002/btpr.2089. [DOI] [PubMed] [Google Scholar]
- Chiang AW, Li S, Spahn PN, Richelle A, Kuo C-C, Samoudi M, Lewis NE. Modulating carbohydrate–protein interactions through glycoengineering of monoclonal antibodies to impact cancer physiology. Curr. Opin. Struct. Biol. 2016;40:104–111. doi: 10.1016/j.sbi.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel M, Crispin M, Scanlan CN, Zitzmann N, Dwek Ra. Emerging principles for the therapeutic exploitation of glycosylation. Science. 2014;343:1235681. doi: 10.1126/science.1235681. [DOI] [PubMed] [Google Scholar]
- Daniel MG, Pawlik TM, Fader AN, Esnaola NF, Makary MA. The Orphan Drug Act. Am. J. Clin. Oncol. 2015;39:210–213. doi: 10.1097/COC.0000000000000251. [DOI] [PubMed] [Google Scholar]
- del Val IJ, Fan Y, Weilguny D. Dynamics of immature mAb glycoform secretion during CHO cell culture: An integrated modelling framework. Biotechnol. J. 2016:1–34. doi: 10.1002/biot.201400663. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach Ca, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardai S, Epp A, Linares G, Westerndorf L, Sutherland M, Neff-LaFord H, Peng S, Law C-L. SEA-CD40, a sugar engineered non-fucosylated anti-CD40 antibody with improved immune activating capabilities, in: AACR 106th Annual Meeting 2015. Cancer Res. 2015:75. 2015; Abstract nr 2472. [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U. S. A. 1992;89:5547–551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger RK, James DC. CHO cell line specific prediction and control of recombinant monoclonal antibody N-glycosylation. Biotechnol. Bioeng. 2013;110:2970–2983. doi: 10.1002/bit.24959. [DOI] [PubMed] [Google Scholar]
- Gramer MJ, Eckblad JJ, Donahue R, Brown J, Shultz C, Vickerman K, Priem P, van den Bremer ETJ, Gerritsen J, van Berkel PHC. Modulation of antibody galactosylation through feeding of uridine, manganese chloride, and galactose. Biotechnol. Bioeng. 2011;108:1591–1602. doi: 10.1002/bit.23075. [DOI] [PubMed] [Google Scholar]
- Grav LM, Lee JS, Gerling S, Beuchert Kallehauge T, Holmgaard Hansen A, Kol S, Lee GM, Ebdrup Pedersen L, Faustrup Kildegaard H. One-step generation of triple knockout CHO cell lines using CRISPR Cas9 and fluorescent enrichment. Biotechnol. J. 2015;10:1446–1456. doi: 10.1002/biot.201500027. [DOI] [PubMed] [Google Scholar]
- Griebenow KAI, Sola RJ. Effects of Glycosylation on the Stability of Protein Pharmaceuticals. J. Pharm. Sci. 2009;98:1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossler P. Protein Glycosylation Control in Mammalian Cell Culture : Past Precedents and Contemporary Prospects. Genomics and Systems Biology of Mammalian Cell Culture. 2012:187–219. doi: 10.1007/10_2011_113. [DOI] [PubMed] [Google Scholar]
- Imai-Nishiya H, Mori K, Inoue M, Wakitani M, Iida S, Shitara K, Satoh M. Double knockdown of alpha1,6-fucosyltransferase (FUT8) and GDP-mannose 4,6-dehydratase (GMD) in antibody-producing cells: a new strategy for generating fully non-fucosylated therapeutic antibodies with enhanced ADCC. BMC Biotechnol. 2007;7:84. doi: 10.1186/1472-6750-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrzejewski PM, del Val IJ, Constantinou A, Dell A, Haslam SM, Polizzi KM, Kontoravdi C. Towards controlling the glycoform: a model framework linking extracellular metabolites to antibody glycosylation. Int. J. Mol. Sci. 2014;15:4492–4522. doi: 10.3390/ijms15034492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat. Rev. Drug Discov. 2009;8:226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Lee JS, Grav LM, Lewis NE, Kildegaard HF. CRISPR/Cas9-mediated genome engineering of CHO cell factories: Application and perspectives. Biotechnol. J. 2015;10:979–994. doi: 10.1002/biot.201500082. [DOI] [PubMed] [Google Scholar]
- Li H, d’Anjou M. Pharmacological significance of glycosylation in therapeutic proteins. Curr. Opin. Biotechnol. 2009;20:678–684. doi: 10.1016/j.copbio.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Lin N, Mascarenhas J, Sealover NR, George HJ, Brooks J, Kayser KJ, Gau B, Yasa I, Azadi P, Archer-Hartmann S. Chinese hamster ovary (CHO) host cell engineering to increase sialylation of recombinant therapeutic proteins by modulating sialyltransferase expression. Biotechnol. Prog. 2015;31:334–346. doi: 10.1002/btpr.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M. An introduction to Genetic Algorithms. MIT Press; 1998. [Google Scholar]
- Neelamegham S, Liu G. Systems glycobiology: biochemical reaction networks regulating glycan structure and function. Glycobiology. 2011;21:1541–1553. doi: 10.1093/glycob/cwr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R, Satoh M. The Current Status and Prospects of Antibody Engineering for Therapeutic Use : Focus on Glycoengineering Technology. J. Pharm. Sci. 2015;104:930–941. doi: 10.1002/jps.24316. [DOI] [PubMed] [Google Scholar]
- Okeley NM, Toki BE, Zhang X, Jeffrey SC, Burke PJ, Alley SC, Senter PD. Metabolic engineering of monoclonal antibody carbohydrates for antibody-drug conjugation. Bioconjug. Chem. 2013;24:1650–1655. doi: 10.1021/bc4002695. [DOI] [PubMed] [Google Scholar]
- Pande S, Rahardjo A, Livingston B, Mujacic M. Monensin, a small molecule ionophore, can be used to increase high mannose levels on monoclonal antibodies generated by Chinese hamster ovary production cell-lines. Biotechnol. Bioeng. 2015;112:1383–1394. doi: 10.1002/bit.25551. [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju TS. Terminal sugars of Fc glycans influence antibody effector functions of IgGs. Curr. Opin. Immunol. 2008;20:471–478. doi: 10.1016/j.coi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Raju TS, Scallon B. Fc glycans terminated with N-acetylglucosamine residues increase antibody resistance to papain. Biotechnol. Prog. 2007;23:964–971. doi: 10.1021/bp070118k. [DOI] [PubMed] [Google Scholar]
- Rillahan CD, Antonopoulos A, Lefort CT, Sonon R, Azadi P, Ley K, Dell A, Haslam SM, Paulson JC. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat. Chem. Biol. 2012;8:661–668. doi: 10.1038/nchembio.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solá RJ, Griebenow K. Glycosylation of Therapeutic Proteins: An Effective Strategy to Optimiza Efficacy. BioDrugs. 2011;24:9–21. doi: 10.2165/11530550-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn PN, Hansen AH, Hansen HG, Arnsdorf J, Kildegaard HF, Lewis NE. A Markov chain model for N-linked protein glycosylation – towards a low-parameter tool for model-driven glycoengineering. Metab. Eng. 2016;33:52–66. doi: 10.1016/j.ymben.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn PN, Lewis NE. Systems glycobiology for glycoengineering. Curr. Opin. Biotechnol. 2014;30C:218–224. doi: 10.1016/j.copbio.2014.08.004. [DOI] [PubMed] [Google Scholar]
- St Amand MM, Radhakrishnan D, Robinson AS, Ogunnaike Ba. Identification of manipulated variables for a glycosylation control strategy. Biotechnol. Bioeng. 2014;111:1957–1970. doi: 10.1002/bit.25251. [DOI] [PubMed] [Google Scholar]
- Surve T, Gadgil M. Manganese increases high mannose glycoform on monoclonal antibody expressed in CHO when glucose is absent or limiting: Implications for use of alternate sugars. Biotechnol. Prog. 2015;31:460–467. doi: 10.1002/btpr.2029. [DOI] [PubMed] [Google Scholar]
- Tsuruta LR, Lopes dos Santos M, Moro AM. Biosimilar advancements: moving on to the future. Biotechnol. Prog. 2015;31:1139–1149. doi: 10.1002/btpr.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M. Magic bullets to blockbusters. Nature. 2015;523:34. [Google Scholar]
- Villacres C, Tayi VS, Lattova E, Perreault H, Butler M. Low glucose depletes glycan precursors, reduces site occupancy and galactosylation of a monoclonal antibody in CHO culture. Biotechnol. J. 2015;10:1051–1066. doi: 10.1002/biot.201400662. [DOI] [PubMed] [Google Scholar]
- Villiger TK, Roulet A, Périlleux A, Stettler M, Broly H, Morbidelli M, Soos M. Controlling the time evolution of mAb N-linked glycosylation - Part I: micro-bioreactor experiments. Biotechnol. Prog. 2016;32(5):1135–1148. doi: 10.1002/btpr.2315. [DOI] [PubMed] [Google Scholar]
- Visser J, Feuerstein I, Stangler T, Schmiederer T, Fritsch C, Schiestl M. Physicochemical and functional comparability between the proposed biosimilar rituximab GP2013 and originator rituximab. BioDrugs. 2013;27:495–507. doi: 10.1007/s40259-013-0036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber W, Rimann M, Spielmann M, Keller B, Daoud-El Baba M, Aubel D, Weber CC, Fussenegger M. Gas-inducible transgene expression in mammalian cells and mice. Nat. Biotechnol. 2004;22:1440–1444. doi: 10.1038/nbt1021. [DOI] [PubMed] [Google Scholar]
- Weikert S, Papac D, Briggs J, Cowfer D, Tom S, Gawlitzek M, Lofgren J, Mehta S, Chisholm V, Modi N, Eppler S, Carroll K, Chamow S, Peers D, Berman P, Krummen L. Engineering Chinese hamster ovary cells to maximize sialic acid content of recombinant glycoproteins. Nat. Biotechnol. 1999;17:1116–1121. doi: 10.1038/15104. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wang S, Halim A, Schulz MA, Frodin M, Rahman SH, Vester-Christensen MB, Behrens C, Kristensen C, Vakhrushev SY, Bennett EP, Wandall HH, Clausen H. Engineered CHO cells for production of diverse, homogeneous glycoproteins. Nat. Biotechnol. 2015;33:842–844. doi: 10.1038/nbt.3280. [DOI] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, Lim WA. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Woen S, Wang T, Liau B, Zhao S, Chen C, Yang Y, Song Z, Wormald MR, Yu C, Rudd PM. Challenges of glycosylation analysis and control: An integrated approach to producing optimal and consistent therapeutic drugs. Drug Discov. Today. 2016;21:740–765. doi: 10.1016/j.drudis.2016.01.006. [DOI] [PubMed] [Google Scholar]