Abstract
Previous research has shown a connection between metabolic abnormalities in the methionine cycle and transsulfuration pathway and autism spectrum disorder. Using clinical data from a case-control study investigating measurements of transmethylation and transsulfuration metabolites, a steady-state model of these metabolites in liver cells was developed and participant-specific parameters were identified. Comparison of mean parameter values and parameter distributions between neurotypical study participants and those on the autism spectrum revealed significant differences for four model parameters. Sensitivity analysis identified the parameter describing the rate of glutamylcysteine synthesis, the rate-limiting step in glutathione production, to be particularly important in determining steady-state metabolite concentrations. These results may provide insight into key reactions to target for potential intervention strategies relating to autism spectrum disorder.
Keywords: Personalized model, transmethylation, steady-state analysis, parameter estimation, sensitivity analysis
1. Introduction
Autism spectrum disorder (ASD) is a general diagnosis for a group of neurodevelopmental disabilities that appear in the early years of childhood, typically before the age of two (American Psychiatric Association, 2013). Although the disorder can be associated with a large array of symptoms, the two primary criteria for diagnosis of ASD are difficulties with social interaction and communication and the display of restricted, repetitive behaviors (American Psychiatric Association, 2013). The Center for Disease Control’s most recent estimate of ASD prevalence among children in the United States is 1 in 68 (Christensen et al., 2016). This is a substantial increase from its 1996 estimate of 1 in 294 (Yeargin-Allsopp et al., 2003), and even more so from international estimates in the early 1970s of approximately 1 in 2300 (Gillberg and Wing, 1999).
The significant rise in ASD prevalence has prompted research into certain risk factors for the disorder. While there is clearly a genetic component involved in the development of ASD (Abrahams and Geschwind, 2008; Bailey et al., 1995; Rai, 2016), recent twin studies indicate that heritability of the disorder is potentially lower than previously estimated (Gaugler et al., 2014; Hallmayer et al., 2011). Environmental factors are also suggested to contribute to increased ASD susceptibility through a variety of mechanisms (Rossignol et al., 2014). For example, a recent study found significant correlations between the concentrations of organic pollutants, such as pesticides, in the blood of children and the severity of ASD-associated behaviors in those children with the disorder potentially also affected by a child’s genetic susceptibilities (Boggess et al., 2016). The results of another study indicated a significant association between maternal antidepressant treatment before pregnancy and ASD risk in children (Castro et al., 2016). Correlations have also been found between concentrations of certain toxic metals in blood and urine and ASD severity (Adams et al., 2012; Adams et al., 2016). While there is an ongoing debate on what factors contribute to ASD (and in what capacity), these findings provide evidence that the factors involved in ASD risk are much more complex than just genetic predisposition alone.
Recent research has also pointed to a critical connection between incidence of ASD and irregularities in folate-dependent one-carbon metabolism and transsulfuration. Several studies have found evidence for reduced methylation capacity and increased oxidative stress in people with ASD compared to age-matched controls (Adams et al., 2011; James et al., 2006; Melnyk et al., 2012). Since the metabolic pathways directly responsible for these abnormalities are the methionine cycle and transsulfuration pathway, these results suggest that some dysfunction in these pathways might be associated with ASD. While the methionine cycle is found in all cells in the body, transsulfuration is limited to the liver, pancreas, small intestine, kidney, and brain (Finkelstein and Martin, 2000; Vitvitsky et al., 2006). Combined, these pathways have many diverse functions in the human body, including the regulation of gene expression through addition of methyl groups to DNA (Ulrey et al., 2005), myelin protein stabilization in nerve cells (Miller, 2003), and synthesis of glutathione, one of the body’s major antioxidants (Wu et al., 2004). Glutathione plays an important role in detoxification and removal of reactive oxygen species in the body. In mammals, glutathione is found in all tissue types, with high concentrations found in the liver (Lu, 1999) where it is primarily synthesized. The main function of glutathione is antioxidant defense against reactive oxygen species, including free radicals (Fang et al., 2002), and detoxification of environmental toxins, including heavy metals (Adams et al., 2011). Metabolic abnormalities and deficiency of glutathione can result in increased intracellular oxidative stress. Studies suggest that elevated levels of oxidative stress are associated with the pathophysiology of a number of diseases, including Alzheimer’s disease (Markesbery, 1997), diabetes (Giugliano et al., 1996), and cystic fibrosis (Roum et al., 1993), as well as ASD (James et al., 2006). Although the link between glutathione, oxidative stress, and disease has been well-studied, exact explanations for why these relationships exist have yet to be found (Ballatori et al., 2009).
This paper seeks to contribute to our understanding of how metabolites of the methionine cycle and transsulfuration pathway interact by creating a mathematical model where the probability density functions (PDFs) of the parameter values are determined from clinical data. Detailed models of the methionine cycle and transsulfuration pathway exist (Duncan et al., 2013a, 2013b; Reed et al., 2008; Reed et al., 2004) that include complex nonlinear formulas for the rates of each of the reactions, as well as inhibitory and excitatory effects of metabolites on enzymes. These models have a large number of parameters which cannot all be estimated by measuring only a small number of metabolites in the blood. Instead, we chose to develop a smaller model of the pathways with unidirectional linear kinetics. This model does not have the biological detail of the larger models, but it has the advantage of having only 8 parameters (the kinetic rate constants), which can be determined from clinical data. Estimation of the PDFs of these parameters for both neurotypical study participants and participants with ASD, as well as performing sensitivity analysis on the model, allows for identification of important reactions that could potentially be manipulated for future intervention strategies for ASD.
2. Materials and Methods
2.1. Plasma metabolite data
The clinical data used in this model come from the Integrated Metabolic and Genomic Endeavor (IMAGE) study at Arkansas Children’s Hospital Research Institute (Melnyk et al., 2012). The IMAGE study protocol was approved by the University of Arkansas for Medical Sciences’ Institutional Review Board, and parents provided written informed consent. Data for 82 neurotypical study participants (control) and 93 participants on the autism spectrum (case) were used for the model in this work. These data reflect plasma concentrations of transmethylation and transsulfuration metabolites, as well as those of tyrosine, nitrotyrosine, and chlorotyrosine, quantified with high performance liquid chromatography (HPLC) (Melnyk et al., 1999; Melnyk et al., 2000). Amounts of cytosine, 5-methylcytosine, and 8-oxo-deoxyguanosine in DNA were also quantified using a HPLC-ultraviolet system and HPLC electrochemical detection (Helbock et al., 1998). The interested reader is referred to (Melnyk et al., 2012) for further information regarding data collection and experimental procedures.
2.2. Model development
The components and structure of the methionine cycle and transsulfuration pathway can be found in the public literature (Reed et al., 2008). The mathematical model developed here is based upon the structure of these metabolic pathways in liver cells, where 8 metabolites measured in the clinical study are modeled. The model consists of the component balances of the individual metabolites at steady state, is based on mass action kinetics, and can be mathematically described as a set of linear algebraic equations. Molar concentrations of metabolites (c), participant-specific rate parameters (p), and efflux rate constants (f) represent the inputs to the system.
A compartmental model, where the concentrations of the metabolites within each compartment are considered to be well-mixed, is used. There are potentially fluxes of components into and out of each compartment, resulting in a net flux rate. Similarly, reactions within each compartment can result in the concentration of a metabolite being increased or decreased. This is represented by the general component balance:
| (1) |
The structure of the metabolic reactions described by the mathematical model is shown in Figure 1. Methionine, S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH), and homocysteine make up the methionine cycle. Cysteine, glutamylcysteine, glutathione (GSH), and glutathione disulfide (GSSG) are the modeled components of the transsulfuration pathway. Cystathionine, an intermediate in the reaction of homocysteine converting to cysteine, was omitted from the transsulfuration pathway due to lack of available measurements for it.
Figure 1.
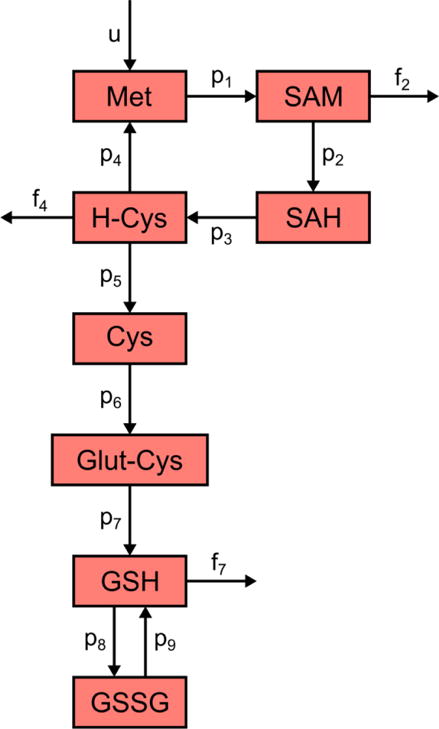
The structure of the methionine cycle and transsulfuration pathway as described by the mathematical model. Met: methionine, SAM: S-adenosylmethionine, SAH: S-adenosylhomocysteine, H-Cys: homocysteine, Cys: cysteine, Glut-Cys: glutamylcysteine, GSH: glutathione, GSSG: glutathione disulfide. The p variables represent participant-specific rate parameters, while the f variables correspond to flux rate constants. Variable u is a zero-order, participant-dependent methionine influx used to keep the system at steady state.
The model consists of 8 equations (Table 1) and contains 9 participant-specific parameters represented by variables p1 through p9, each having units of hr−1. Included are also 3 global efflux rate constants representing the removal rates of SAM, homocysteine, and GSH from the compartment. These flux rates are represented by variables f2 = 9 hr−1 (Duncan et al., 2013a; Stabler and Allen, 2004), f4 = 0.001 hr−1 (Chwatko and Jakubowski, 2005; Duncan et al., 2013a; Refsum et al., 1985), and f7 = 3.8 hr−1 (Hong et al., 2005). To maintain the steady state properties of the model, a zero-order influx rate constant u is used to supply methionine to the system. This variable has units of μM/hr and is equal to the sum of a participant’s three efflux rates to achieve a steady state balance:
| (2) |
with f2, f4, and f7 being the removal rates (in hr−1) of SAM, homocysteine, and GSH, respectively, and c2, c4, and c7 being the concentrations (in μM) of SAM, homocysteine, and GSH, respectively.
Table 1.
Metabolites included in and linear algebraic equations of the model.
| Metabolite | Concentration Variable | Model Equation |
|---|---|---|
| Methionine | c1 | 0 = p4c4 − p1c1 + f2c2 + f4c4 + f7c7 |
| S-adenosylmethionine (SAM) | c2 | 0 = p1c1 − p2c2 − f2c2 |
| S-adenosylhomocysteine (SAH) | c3 | 0 = p2c2 −p3c3 |
| Homocysteine | c4 | 0 = p3c3 − p4c4 − p5c4 − f4c4 |
| Cysteine | c5 | 0 = p5c4 −p6c5 |
| Glutamylcysteine | c6 | 0 = p6c5 − p7c6 |
| Glutathione (GSH) | c7 | 0 = p7c6 − p8c7 + p9c8 − f7c7 |
| Glutathione disulfide (GSSG) | c8 | 0 = P8c7 − p9c8 |
In this model, methionine enters the system at the rate defined in Equation 2, and is converted into SAM with rate constant p1. SAM is then converted to SAH in a DNA methylation reaction with rate constant p2, while some of it is also lost via breakdown or excretion (represented by efflux rate constant f2). SAH turns into homocysteine according to the rate constant p3. Homocysteine then follows several paths: it is re-methylated to form methionine with rate constant p4, contributes to the transsulfuration pathway by converting to cysteine with rate constant p5, and leaves the system with rate constant f4. Cysteine is converted to glutamylcysteine according to rate constant p6, with glutamylcysteine subsequently being depleted to form GSH with rate constant p7. GSH is transported out of liver cells or degraded (represented by flux rate constant f7) or is oxidized to GSSG at a rate defined by p8. GSSG is reduced back to GSH according to rate constant p9.
Each participant-specific rate parameter in the model represents the activity of an enzyme associated with the methionine cycle or transsulfuration pathway. For example, p1 corresponds to the combined activities of methionine adenosyltransferase I (MAT-I) and methionine adenosyltransferase III (MAT-III), two enzymes responsible for the conversion of methionine to SAM. p2 describes the combined rate of the enzymes DNA methyltransferase (DNMT), glycine N-methyltransferase (GNMT), and other methyltransferases, while p3 characterizes the activity of S-adenosylhomocysteine hydrolase (SAHH) and p4 captures the combined effects of methionine synthase (MS) and betaine-homocysteine methyltransferase (BHMT). p5 describes a lumped reaction of homocysteine being converted directly to cysteine with no cystathionine intermediate; this parameter is taken to represent solely the activity of cystathionine β-synthase (CBS), which is responsible for the conversion of homocysteine to cystathionine. Parameter p6 corresponds to glutamate-cysteine ligase (GCL), while p7 describes the activity of glutathione synthetase (GS). p8 captures the rate of glutathione peroxidase (GPX), and p9 describes the action of glutathione reductase (GR). These model enzymes are summarized in Table 2.
Table 2.
Enzymes and fluxes represented by parameters used in the mathematical model.
| Parameter | Representative Enzyme(s) or Flux | Abbreviation |
|---|---|---|
| p1 | methionine adenosyltransferase I | MAT-I |
| methionine adenosyltransferase III | MAT-III | |
| p2 | DNA methyltransferases | DNMT |
| glycine N-methyltransferase | GNMT | |
| p3 | S-adenosylhomocysteine hydrolase | SAHH |
| p4 | methionine synthase | MS |
| betaine-homocysteine methyltransferase | BHMT | |
| p5 | cystathionine β-synthase | CBS |
| p6 | glutamate-cysteine ligase | GCL |
| p7 | glutathione synthetase | GS |
| p8 | glutathione peroxidase | GPX |
| p9 | glutathione reductase | GR |
| f2 | SAM efflux | – |
| f4 | homocysteine efflux | – |
| f7 | GSH efflux | – |
2.3. Parameter estimation
When estimating parameters for each study participant, the values of c and f as given in Table 1 are known; these are the measured metabolite concentrations and defined efflux rates, respectively. The values of p represent the unknown participant-specific rate parameters and need to be computed. Since the relationship between GSH and GSSG is underdetermined in this system, however, the individual values of p8 and p9 cannot be computed; instead, only the ratio of p8 to p9 (expressed p8:p9 or GPX:GR) can be calculated. This ratio was thus considered to be one combined term describing the relative values of p8 and p9. To account for this, the component balance equation for GSSG (Table 1) was rewritten as:
| (3) |
This effectively reduces the model to 8 parameters. Given that the model is still linear, it can be written such that the c values are in an 8×8 coefficient matrix multiplied by an 8×1 vector of unknown p variables. This product is then summed with an 8×1 vector of the f·c efflux terms, and this sum is set equal to the 8×1 zero vector. Formulating the model in this manner allows for estimation of the 8 unknown p values by solving a system of linear equations. Estimation of model parameters was performed for each study participant in the neurotypical and ASD groups using their individual measured values of c from the clinical data.
2.4. Model assumptions
Certain assumptions needed to be made for this work to develop the mathematical structure of the model and explain how it is affected by the availability of data:
The derived model has a linear structure and includes 8 parameters to allow for the distributions of the parameter values to be estimated from the clinical data. While a more detailed model of the pathway may allow for a more accurate description, it is assumed that a model of the chosen complexity can reflect the pathway behavior reasonably well.
The model system is at steady state, so the concentrations of metabolites are not changing over time. This has to be assumed because each study participant only had metabolite measurements taken at one point in time. Along with this, it is assumed that the combined contributions of an enzyme’s concentration and activity do not change over time.
Changes in concentrations of metabolites in plasma are reflected by proportional changes in concentrations of metabolites in liver cells, although this is not always true in some cases (Duncan et al., 2013a). However, this assumption must be made here because the clinical data consist of measurements taken in plasma while the model structure describes the metabolic pathways in liver cells.
Other assumptions were made to explain the biological implications of each reaction in the model and how these were also affected by the available data:
-
4
The conversion of methionine to SAM is due to the combined effects of MAT-I and MAT-III. Having only a single measurement in time for each metabolite does not allow for understanding the effects of individual enzymes acting in parallel, so only the combined effect is modeled.
-
5
The conversion of SAM to SAH is due to the combined contributions of DNMT enzymes, GNMT, and approximately 150 other methyltransferases. For reasons given in Assumption 4, only the combined effect of these enzymes is modeled. Additionally, GNMT uses glycine as a second substrate, but the associated reaction is treated as pseudo first order so only the effects of SAM concentration are considered.
-
6
While the reaction converting SAH to homocysteine by the enzyme SAHH is reversible, it is represented by one net forward rate constant in the model. At steady state, it is not possible to determine the individual rate constants of the forward and reverse reactions. However, the rate of the forward reaction must be greater than the rate of the reverse reaction for the system to remain at steady state.
-
7
The conversion of homocysteine to methionine is due to the combined contributions of MS and BHMT. This assumption is made for reasons similar to Assumption 4, so again only the combined contribution of these enzymes is modeled. Additionally, BHMT uses betaine as a second substrate, but the associated reaction is treated as pseudo first order so only the effects of homocysteine concentration are considered.
-
8
The reactions involving the conversion of homocysteine to cystathionine and the conversion of cystathionine to cysteine are represented by a single lumped reaction where homocysteine is directly converted to cysteine. This is due to no measurements being taken for cystathionine.
Furthermore, an assumption was made to focus the analyses on just the model system of interest:
-
9
There are no indirect inhibitory or regulatory effects between metabolites in the described pathways. In addition, it is assumed that there are no interaction effects from other metabolic pathways. While it is known that other pathways and metabolites, such as folate, have long-range regulatory effects (Reed et al., 2008), these mechanisms are ignored in this model so that parameters can be estimated as stated in Section 2.3.
The final assumption was needed to reflect that certain information is not readily available for subsets of participants:
-
10
The rates of SAM removal, homocysteine excretion, and GSH degradation are identical in neurotypical and ASD study participants. No reported numbers for the values of these rates are available for participants on the autism spectrum. Therefore, the reported values of these rates for neurotypical individuals were assumed to be identical to those for participants on the autism spectrum.
2.5. Sensitivity analysis
The sensitivity of a model to a specific parameter is commonly measured as the change in the model’s output due to a perturbation of that parameter (Frey and Patil, 2002). Local sensitivity analysis can be performed by individually perturbing each parameter from its baseline value while keeping the values of all other parameters constant. This is also referred to as a one-factor-at-a-time approach (Hamby, 1994; Saltelli and Annoni, 2010). A simple method for perturbing a chosen parameter is to add or subtract a percentage of its base value (Hamby, 1994).
The baseline value of each model parameter was taken to be the mean value of that parameter among all neurotypical participants. The resulting set of baseline parameters, which were then known, corresponded to a particular unknown set of metabolite concentrations, which were treated as the outputs of the system. However, calculating the output’s sensitivity to changes in parameters required knowledge of the baseline concentrations that would serve as a point of reference for the model output. This required calculating the metabolite concentrations that corresponded to the baseline parameter values.
To solve for these unknown baseline concentrations, an approach similar to that described in Section 2.3 was taken. Using the model equations in Table 1, an 8×8 matrix of average p and f terms was constructed and multiplied by an 8×1 vector of unknown baseline c values. Their product was then set equal to an 8×1 zero vector to form a homogeneous system of linear equations. Given that any row of the matrix is equal to the negative sum of all other rows, all rows are not linearly independent; the calculated rank of the matrix was then found to be 7. Since the largest potential rank of this matrix is 8, the rank-nullity theorem states there exists one solution for the right null space of this matrix. The right null space was then solved for by using the null command in MATLAB. The absolute values of these vector elements were taken to be the baseline metabolite concentrations for the purposes of sensitivity analysis.
Each model parameter, as well as each efflux rate constant, was perturbed according to the one-factor-at-a-time approach. To observe the output’s response to small perturbations, parameters were increased and decreased by 1% of their baseline value. The metabolite concentrations corresponding to each new perturbation were then recalculated by finding the right null space of the parameter matrix as described above. Each new metabolite concentration resulting from a given perturbation was compared to its baseline value using the relative change, which was used to account for large differences in magnitude between metabolite concentrations. The sensitivity measure for a particular perturbation was taken to be the Euclidean norm of the vector containing these 8 relative changes (one for each metabolite). Total sensitivity of the output to changes in a certain parameter was calculated as the average of this sensitivity measure for the negative and positive perturbations of that parameter.
2.5.1 Sensitivity analysis and cross validation
The many model assumptions and the uncertainties in model parameter estimates may limit the confidence with which the results of sensitivity analysis can be interpreted. Thus, these results were validated using a leave-one-out cross validation approach (Kohavi, 1995). The cross-validation procedure was applied by removing the first neurotypical participant’s set of parameters from the data – this participant was treated as the test set, while the remaining 81 participants were treated as the training set. Sensitivity analysis was performed, as described above, independently for the training set and the test set. The obtained sensitivity measures were recorded, the first participant’s data were replaced, and the process was repeated one at a time for each of the other neurotypical participants. Thus, sensitivity analysis was performed for 82 training sets and 82 test sets. The averages of the sensitivity measures between the training sets and test sets were then compared. Performing the analysis in this manner helped to ensure that the sensitivity results were robust to changes in the available sample set. Efflux rates were also considered in this cross-validation.
3. Results
3.1. Parameter estimation
The 8 participant-specific parameters were estimated for the 82 neurotypical participants and 93 participants with ASD for who concentrations of all 8 model metabolites were available in the clinical data. Means and standard deviations for metabolite concentrations among the participants included in this study are provided in Table 3. Significant statistical differences were observed between the neurotypical and ASD groups for all metabolites except homocysteine (α = 0.01).
Table 3.
Means and standard deviations of metabolite concentrations for the neurotypical and ASD groups.
| Metabolite | Concentration Mean ± SD (μM)
|
p-value | |
|---|---|---|---|
| Neurotypical (n = 82) | ASD (n = 93) | ||
| Methionine | 23.1 ± 3.6 | 19.0 ± 2.9 | < 0.0001 |
| SAM | 0.0696 ± 0.0100 | 0.0600 ± 0.0104 | < 0.0001 |
| SAH | 0.0152 ± 0.0036 | 0.0186 ± 0.0047 | < 0.0001 |
| Homocysteine | 4.74 ± 1.01 | 5.10 ± 1.20 | 0.0323 |
| Cysteine | 209 ± 17 | 190. ± 18 | < 0.0001 |
| Glutamylcysteine | 2.33 ± 0.59 | 1.85 ± 0.48 | < 0.0001 |
| GSH | 2.31 ± 0.70 | 1.71 ± 0.36 | < 0.0001 |
| GSSG | 0.147 ± 0.058 | 0.236 ± 0.100 | < 0.0001 |
Following the methodology outlined in Section 2.3, a system of linear equations was constructed using a coefficient matrix of c values, a vector of unknown p values, and a vector of efflux terms f·c. However, the matrix of c values was found to be rank deficient (for reasons identical to those given in Section 2.5 for a matrix of p and f terms). The four components describing the methionine cycle were specifically responsible for this condition. Finding a unique solution for the associated parameters required that a constraint be placed on one of the four parameters in the methionine cycle. To determine the optimal parameter to constrain, an equality constraint was applied to one parameter in the cycle, and values of the remaining parameters were estimated. The constraint was then removed and placed on another parameter in the methionine cycle, and the new group of unconstrained parameters was estimated. This process was repeated for all four parameters in the methionine cycle. Estimates of all model parameters resulting from each of the constraints for neurotypical participants were compared with approximated biological values of these rate constants from the literature. The lsqlin routine in MATLAB was used to define each constraint and estimate the parameters such that the constraints were satisfied.
The equality constraint used to estimate parameters relates the rate of each reaction in the methionine cycle to a nominal rate, given in μM/hr. Each constraint has the form:
| (4) |
where pi is a participant’s estimated value of the i-th rate constant (to be solved), ci is that participant’s concentration of the i-th metabolite, pnom is a nominal value for the rate parameter being estimated (taken from the literature), and is the average concentration of the i-th metabolite for the group that the participant belongs to (neurotypical or ASD). Each constraint is only applied to a reaction within the methionine cycle, i.e., i ranges from 1 to 4.
During the process of determining the optimal constraint, only data for neurotypical participants were considered. When the constraint described by Equation 4 was applied to each methionine cycle parameter in turn, the parameter estimates in Table 4 were obtained. Approximate nominal values from the literature are also provided. The linear structure of the transsulfuration pathway results in the estimates for parameters p5 through p8:p9 remaining unchanged across the different constraints. Placing a constraint on p1 produces very large values for the other methionine cycle parameters. Constraining p2 gives much smaller estimates of p1 and p3, with unacceptable negative values for p4. The constraint on p3, however, produces parameter estimates that are most similar to the values from the literature. Values for p2 and p3 resulting from the constraint on p4 are too large to consider acceptable. Thus, constraining p3 is the preferred approach to estimating parameters for the methionine cycle.
Table 4.
Estimates of control parameters when p1 through p4 are individually constrained. All parameters have units of hr−1 except for p8:p9, which is dimensionless.
| Mean Parameter Value ± SD
|
Literature value | |||||
|---|---|---|---|---|---|---|
| Parameter |
p1 constrained |
p2 constrained |
P3 constrained |
P4 constrained |
Reference | |
| p1 | 4.90 ± 0.78 | 0.412 ± 0.064 | 1.90 ± 0.30 | 2.97 ± 0.46 | 4.78 | (Broxson et al., 1989; del Pino et al., 2000; Nijhout et al., 2006; Sullivan and Hoffman, 1983) |
| p2 | 1610 ± 218 | 127 ± 17 | 620. ± 84 | 976 ± 140. | 125 | (Castro et al., 2003; Flynn and Reich, 1998; Luka and Wagner, 2003; Nijhout et al., 2006) |
| p3 | 7720 ± 2215 | 610. ± 175 | 2970 ± 852 | 4680 ± 1410 | 2780 | (Castro et al., 2003; Døskeland and Ueland, 1982; Nakanishi et al., 2001) |
| p4 | 22.3 ± 4.6 | −0.0210 ± 0.5858 | 7.38 ± 1.62 | 12.7 ± 2.6 | 12.2 | (Castro et al., 2003; Finkelstein and Martin, 1984) |
| p5 | 1.93 ± 0.70 | 1.93 ± 0.70 | 1.93 ± 0.70 | 1.93 ± 0.70 | 10.4 | (Castro et al., 2003; Finkelstein and Martin, 1984) |
| p6 | 0.0421 ± 0.0125 | 0.0421 ± 0.0125 | 0.0421 ± 0.0125 | 0.0421 ± 0.0125 | 0.0276 | (Davis et al., 2006; Huang et al., 1988; Toroser et al., 2006) |
| p7 | 3.82 ± 0.77 | 3.82 ± 0.77 | 3.82 ± 0.77 | 3.82 ± 0.77 | 10.0 | (Lyons et al., 2000; Martensson, 1987) |
| p8:p9 | 0.0660 ± 0.0244 | 0.0660 ± 0.0244 | 0.0660 ± 0.0244 | 0.0660 ± 0.0244 | 0.0118 | (Blanco et al., 2007; López-Barea and Lee, 1979; Maddipati and Marnett, 1987) |
Using the optimal constraint on p3, parameter values for ASD participants were then estimated. A comparison of the estimates for control and ASD participants is provided in Table 5. Significant statistical differences can be seen between the neurotypical and ASD groups for all parameters except p3 and p7 (α = 0.01). Additional insight into these differences can be gained from observing the respective PDFs of each parameter for the control and ASD groups. Using kernel density estimation with a normal kernel (using the ksdensity command in MATLAB), PDFs were constructed for each parameter (Figure 2). The most apparent differences between neurotypical and ASD distributions are seen for parameters p1 (Figure 2A), p2 (Figure 2B), p4 (Figure 2D), and p8:p9 (Figure 2H). Although p5 and p6 (Figure 2E and 2F, respectively) displayed significant statistical differences, there are not observable differences in the distributions for the two groups of participants. The two distributions for p3 (Figure 2C) are nearly identical, which is a consequence of the constraint placed upon the parameter.
Table 5.
Comparison of model parameter estimates for control and ASD participants. All parameters have units of hr−1 except for p8:p9, which is dimensionless.
| Parameter | Parameter Mean ± SD
|
p-value | |
|---|---|---|---|
| Neurotypical (n = 82) | ASD (n = 93) | ||
| p1 | 1.90 ± 0.30 | 2.81 ± 0.42 | < 0.0001 |
| p2 | 620. ± 84 | 889 ± 154 | < 0.0001 |
| p3 | 2970 ± 852 | 3020 ± 1020 | 0.726 |
| p4 | 7.38 ± 1.62 | 9.32 ± 2.00 | < 0.0001 |
| p5 | 1.93 ± 0.70 | 1.35 ± 0.47 | < 0.0001 |
| p6 | 0.0421 ± 0.0125 | 0.0346 ± 0.0088 | < 0.0001 |
| p7 | 3.82 ± 0.77 | 3.59 ± 0.63 | 0.031 |
| p8:p9 | 0.0660 ± 0.0244 | 0.144 ± 0.065 | < 0.0001 |
Figure 2.
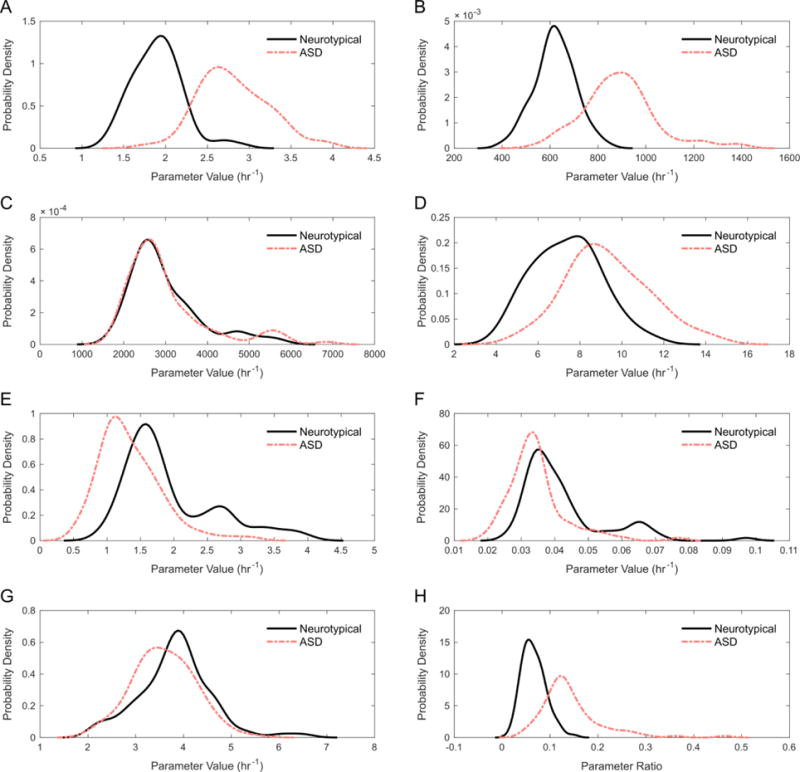
PDFs of the eight participant-specific model parameters for the neurotypical and ASD participant groups. (A) Rate parameter p1 for MAT-I + MAT-III. (B) Rate parameter p2 for DNMT + GNMT. (C) Rate parameter p3 for SAHH. (D) Rate parameter p4 for MS + BHMT. (E) Rate parameter p5 for CBS. (F) Rate parameter p6 for GCL. (G) Rate parameter p7 for GS. (H) Ratio of rate parameters p8 and p9 for GPX:GR.
3.3. Sensitivity analysis
Metabolite concentrations were considered to be the outputs of the system for local sensitivity analysis. Since the system is linear, local sensitivity analysis results for positive and negative 1% perturbations should be identical. However, both perturbations were performed for each parameter to ensure the results were not affected by numerical inaccuracies. The results of both perturbations were indeed found to be almost identical, and the average results are reported in Table 6.
Table 6.
Results of sensitivity analysis from perturbing each parameter and efflux rate constant by 1%. Also shown are the results of leave-one-out cross validation for this analysis.
| Parameter or Efflux Rate Constant | Sensitivity (all samples) | Mean Training Sensitivity ± SD | Mean Test Sensitivity ± SD |
|---|---|---|---|
| p1 | 0.0099 | 0.0099 ± 0.0000 | 0.0099 ± 0.0000 |
| p2 | 0.0100 | 0.0100 ± 0.0000 | 0.0100 ± 0.0000 |
| p3 | 0.0100 | 0.0100 ± 0.0000 | 0.0100 ± 0.0000 |
| p4 | 0.0136 | 0.0136 ± 0.0000 | 0.0136 ± 0.0011 |
| p5 | 0.0168 | 0.0168 ± 0.0000 | 0.0168 ± 0.0008 |
| p6 | 0.0261 | 0.0261 ± 0.0000 | 0.0261 ± 0.0001 |
| p7 | 0.0100 | 0.0100 ± 0.0000 | 0.0100 ± 0.0000 |
| p8: p9 | 0.0100 | 0.0100 ± 0.0000 | 0.0100 ± 0.0000 |
| f2 | 0.0001 | 0.0001 ± 0.0000 | 0.0001 ± 0.0000 |
| f4 | 0.0000 | 0.0000 ± 0.0000 | 0.0000 ± 0.0000 |
| f7 | 0.0141 | 0.0141 ± 0.0000 | 0.0141 ± 0.0000 |
The metabolite concentrations were found to be most sensitive to changes in parameter p6, which describes the rate at which cysteine is converted to glutamylcysteine. In other words, altering the value of p6 causes the largest cumulative relative change in the steady state metabolite concentrations. It is thus an important parameter and has the most influence on the model output. After p6, the output was most sensitive to changes in parameter p5, which defines the rate of production of cysteine from homocysteine, and to changes in efflux rate constant f7, which represents the removal rate of GSH from the system. The model output also showed relatively high sensitivity to changes in parameter p4, which describes the rate at which homocysteine is converted to methionine. These results indicate that these parameters also have a greater effect on the model concentrations. The remaining participant-specific parameters, having equally low measures of sensitivity, were indicated to have less effect on steady state metabolite concentrations. Efflux rate constants f2 and f4 appeared to have minimal influence on the concentrations of metabolites. Additionally, the cross-validation results (Table 6) revealed no effective differences between the parameter sensitivity measures as calculated from all samples, from the averaged training results, and from the averaged test results.
4. Discussion
This work developed a model of the methionine cycle and transsulfuration pathway and this is the first effort which estimates parameters for such a model using clinical data from both neurotypical and ASD study participants. Furthermore, the distributions of the parameters were estimated and compared between the groups of study participants, as each participant had different metabolite concentrations, resulting in different model parameters.
Development of the presented model was based on a number of assumptions. Some of these assumptions may limit the accuracy of the model, while others may have little or no bearing on the model’s ability to describe true metabolic behavior. The assumption of a steady state system can be particularly limiting since biological systems are rarely, if ever, in this state of equilibrium. That being said, few physiological measurements, especially if they involve metabolites in clinical studies, are taken over time and as such this is a reasonable assumption for this particular application. Additionally, assuming linear interactions between enzymes and substrates may neglect to capture the true behavior of these reactions. Michaelis-Menten equations are often used to describe the nonlinear kinetics of enzymes, but the kinetic parameters of these equations are difficult to estimate without available time-series data. In general, the assumptions made are based upon the state-of-the-art of what can be done in a clinical research trial. Regardless of the implications of these assumptions, the overall purpose of this study was to characterize and model metabolic differences between neurotypical and ASD participants. The current model provides a starting point for describing the interactions of the studied metabolic pathways in ASD.
The most significant limitation of the model is that the efflux rate constants are not necessarily specific to each participant. Given the variability in metabolite concentrations between participants, it is unlikely that these efflux rates are identical for participants within the same group, let alone identical between the neurotypical and ASD participant groups. Differences in these efflux rates could explain the differences observed for certain participant-specific rate parameters; using a one-size-fits-all approach could result in forced parameter differences that would not be present if the efflux rates had been specific to each participant. However, the lack of data describing these individualized effluxes makes this problem difficult to address. There are also very limited studies investigating these processes in individuals with ASD, so it is challenging to characterize any flux differences for those participants compared to the neurotypical group. Thus, the efflux rate constants must be assumed to be the same for every participant until more data regarding the rates of these processes become available.
The mean values for many of the estimated parameters in the neurotypical group are close to the biological activity rates of their corresponding enzymes that are found in the literature. Referring to the literature values in Table 4, it can be seen that the approximate rate constant for MAT-I + MAT-III activity is fairly close to the mean estimated value for control participants. The estimates for the DNMT+GNMT and MS+BHMT rate constants are also within acceptable ranges of their literature values. Additionally, the mean estimate of the rate constant for SAHH is very close to the true rate constant, although this is expected due to the constraint applied during estimation. The estimated rate constants for GCL, GS, and the GPX:GR ratio are close to their approximate nominal values as well. However, the mean estimate for CBS is significantly different from its nominal value. It is unlikely that this discrepancy is a consequence of the lumped homocysteine-cystathionine-cysteine reaction used in the model. At steady state, the theoretical metabolic flux from homocysteine to cystathionine will be equivalent to the flux from cystathionine to cysteine; since the flux throughout the transsulfuration pathway needs to be the same at every reaction step, the lumped reaction rate from homocysteine to cysteine will also be equal to either of these two individual reaction rates. Therefore, the product of the homocysteine concentration and rate constant p5 still reflects the actual conversion rate of homocysteine to cystathionine, and p5 should be a valid approximation of CBS activity. A more likely reason for the discrepancy is the fact that cysteine is used in many other metabolic processes besides the transsulfuration pathway; it is also involved in the synthesis of taurine, pyruvates, sulfates, and sulfites, for example (Stipanuk et al., 2006). The model does not describe these alternative pathways, and thus does not account for the higher levels of cysteine that would be required to carry out all of these reactions. This may explain why the rate of cysteine synthesis is underestimated in the model.
Parameter estimation revealed observable differences in parameters p1, p2, p4, and p8:p9 between neurotypical and ASD participants. This suggests some degree of difference in activity rate for MAT-I+MAT-III, DNMT+GNMT, MS+BHMT, and GPX:GR in participants with ASD as compared to neurotypical participants. The GPX:GR difference is expected; the ratio of reduced to oxidized glutathione as a marker of oxidative stress has been well-documented (Asensi et al., 1999; James et al., 2006) and there is evidence to suggest that a decreased ratio is present in participants with ASD (Adams et al., 2011; James et al., 2006; Melnyk et al., 2012). The DNMT+GNMT parameter represents the rate at which SAM is converted into SAH, and reflects the SAM/SAH ratio, which is an indicator of DNA methylation capacity (Yi et al., 2000). In participants with ASD, the SAM/SAH ratio has been found to be significantly reduced (Adams et al., 2011; James et al., 2006; Melnyk et al., 2012). With regards to the differences in MAT-I + MAT-III, oxidative stress has been linked to a down-regulation of the methionine adenosyltransferase enzymes (Avila et al., 1998). Given that increased oxidative stress is characteristic of ASD, it is possible that there could be altered MAT rates in participants on the spectrum. Finally, it has been found that increased oxidative stress can be responsible for reduced MS activity (Muratore et al., 2013). This mechanism leaves more homocysteine available to be eventually converted to glutathione, which can then be used to lower oxidative stress. These observations are in line with the finding that the participants with ASD had different values of the p4 parameter as compared to their neurotypical counterparts.
Model metabolite concentrations were found to be most sensitive to changes in parameter p6, followed by parameter p5, efflux rate constant f7, and parameter p4. These correspond to the first-order rate constants of the enzymes GCL and CBS, the removal of GSH, and the enzymes MS+BHMT, respectively. GCL is responsible for the conversion of cysteine to glutamylcysteine, so the sensitivity results suggest that the formation of glutamylcysteine has an important role in the model. This is somewhat surprising because the formation of GSH was expected to have a significant impact on the model predictions; instead the synthesis of its precursor was identified as more important. That being said, the synthesis of a GSH precursor will ultimately affect GSH formation, and formation of glutamylcysteine is considered to be the rate-limiting step of GSH synthesis (Hamilton et al., 2003; Lu, 2009; Shi et al., 1994). In addition, the relatively high sensitivity to the removal of GSH does suggest that levels of GSH have a significant effect on other metabolites in the methionine cycle and transsulfuration pathway; this could be supported by findings of redox levels (which are influenced by GSH) affecting synthesis of certain metabolites in these pathways (Mosharov et al., 2000; Richman and Meister, 1975). The other important model enzymes, CBS and MS+BHMT, are involved in the synthesis of cysteine and methionine, respectively, both from homocysteine. Given that these enzymes form the juncture between the methionine cycle and transsulfuration pathway, and determine the fate of homocysteine, it is not as surprising that these are among the most important parameters in the model.
The robustness of the most important sensitivity analysis results to changes in the study participants was verified by leave-one-out cross validation, where no substantial differences in sensitivity measures were observed between the averaged training and test set results (Table 6). The sensitivity results were nearly identical across training sets since each training set’s baseline parameter values were averaged from the parameters of all neurotypical participants except one, and removing one different participant from the average of 81 participants in each training set is unlikely to have a significant effect on the average parameter values. This explains why the variations in the sensitivity measures between training sets are below the used cutoff. Unlike the training set, however, the test sets did show some variation in their sensitivity measures for certain parameters. Despite this difference, the test sets’ averaged results were very close to those from the averaged training results, suggesting that the parameters’ sensitivity measures are much less dependent on individual parameter values than on the structure of the model itself.
Model robustness is further suggested by the small values of the sensitivity measures for each parameter. However, a potential drawback of these small sensitivities is poor observability of the parameters in the experimental data. One possible way to address this issue could be to implement a different measure of sensitivity, perhaps by considering a relationship among the metabolites’ relative changes other than the Euclidean norm, or by replacing the relative change of a metabolite with a different calculation of change altogether. Another solution is to employ regularization methods, such as Tikhonov regularization (Engl et al., 1996), that can deal with poor observability by providing more stable estimates of model parameters (Golub et al., 1999). Experimentation with Tikhonov regularization (using neurotypical participants and values of the regularization parameter up to 0.1) yielded parameter values that, on average, varied by less than 6% of their current solutions. This was the case for all parameters except p8:p9, which had an average percent change of 89.1%. These results indicate that the parameters are robust with respect to the parameter observability, with the exception of p8:p9. However, the p8:p9 ratio is not one of the key variables in the model as determined by sensitivity analysis, and thus the conclusions do not change as a result of this analysis.
Analysis of the raw plasma concentration data in (Melnyk et al., 2012) yielded many important observations of metabolic abnormalities in study participants with ASD. Among the primary outcomes of that study were significant measured differences for methionine, SAM, SAH, total cysteine, GSH, and GSSG concentrations in control and case participants. The study concluded that participants with ASD have genome-wide reduced DNA methylation, in addition to finding evidence for increased oxidative stress in participants with ASD. While the current model does not provide any insight into genome-wide trends, it does offer some results similar to those obtained by analysis of the raw data. Mainly, observed differences in the p1, p2, p4, and p8:p9 rate parameters point to equivalent metabolic differences identified in (Melnyk et al., 2012). An important distinction to make, however, is that those analyses focused directly on the metabolite concentrations, while the model analyses here are aimed toward the rates at which the metabolic reactions take place. With this in mind, the model also produced some novel results that were not previously extracted from the data. Sensitivity analysis found two parameters relating to the fate of homocysteine to be very important, while no significant difference in homocysteine concentration was found in the raw data. While this could be an important finding, it could also be an artifact of the model; the idea of homocysteine metabolism being altered in response to oxidative stress (Muratore et al., 2013) suggests the former, however. The parameter pertaining to glutamylcysteine synthesis (by GCL) was indicated to be of even greater importance, but no analysis of glutamylcysteine concentrations in the raw data was performed. Future investigations should explore in more detail the importance of GCL, and its rate-limiting reaction, in GSH synthesis.
5. Conclusions
This study developed a model for two pathways of potential importance to ASD and identified model parameters, as well as their PDFs, from clinical data from neurotypical participants and those on the spectrum. Significant statistical differences can be seen in four of the parameters between the two groups through comparison of their mean parameter values and their parameter distributions. Almost all estimated rate parameters were found to match with corresponding experimental values from the literature, with the exception of the rate constant for cystathionine β-synthase. Sensitivity analysis revealed parameter p6, which reflects the activity of the enzyme glutamate-cysteine ligase, to be especially important to the model’s predictions. This finding aligns with the observation that the reaction the enzyme is responsible for, i.e., the formation of glutamylcysteine, is the rate-limiting step in glutathione synthesis. Development of this model serves as the first step for identifying targets for potential future intervention strategies. Reactions with significant differences among the two participant groups can serve as a potential starting point for future investigations.
Highlights.
Previous studies linked autism spectrum disorder (ASD) to metabolic irregularities.
A model describing transmethylation and transsulfuration pathways was developed.
Case-control clinical data were used to estimate distribution of model parameters.
Sensitivity analysis identified potential metabolic reactions of interest.
Results of this study hint at potential intervention strategies for ASD.
Acknowledgments
The authors gratefully acknowledge partial financial support from the National Institutes of Health (Grant R01AI110642).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Author Contributions
TV participated in the design of the study, performed the analysis, interpreted results, and drafted the manuscript; DPH participated in the design of the study, and helped to interpret results and draft the manuscript; SM and SJJ developed the methodology for collection of the clinical data, and collected and interpreted this data; JH conceived of the study and participated in its design and coordination, and helped to interpret results and draft the manuscript. All authors read and approved the final manuscript.
References
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nature Reviews Genetics. 2008;9(5):341–355. doi: 10.1038/nrg2346. https://doi.org/10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JB, Audhya T, McDonough-Means S, Rubin RA, Quig D, Geis E, Lee W. Nutritional and metabolic status of children with autism vs. neurotypical children, and the association with autism severity. Nutrition & Metabolism. 2011;8:34. doi: 10.1186/1743-7075-8-34. https://doi.org/10.1186/1743-7075-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JB, Audhya T, McDonough-Means S, Rubin RA, Quig D, Geis E, Lee W. Toxicological status of children with autism vs. neurotypical children and the association with autism severity. Biological Trace Element Research. 2012;151(2):171–180. doi: 10.1007/s12011-012-9551-1. https://doi.org/10.1007/s12011-012-9551-1. [DOI] [PubMed] [Google Scholar]
- Adams JB, Howsmon DP, Kruger U, Geis E, Gehn E, Fimbres V, et al. Significant Association of Urinary Toxic Metals and Autism-Related Symptoms—A Nonlinear Statistical Analysis with Cross Validation. PLoS ONE. 2017;12(1):e0169526. doi: 10.1371/journal.pone.0169526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 5th. Washington, DC: American Psychiatric Publishing; 2013. [Google Scholar]
- Asensi M, Sastre J, Pallardo FV, Lloret A, Lehner M, Garcia-de-la Asuncion J, Viña J. Ratio of reduced to oxidized glutathione as indicator of oxidative stress status and DNA damage. Methods in Enzymology. 1999;299:267–276. doi: 10.1016/s0076-6879(99)99026-2. [DOI] [PubMed] [Google Scholar]
- Avila MA, Carretero MV, Rodriguez EN, Mato JM. Regulation by hypoxia of methionine adenosyltransferase activity and gene expression in rat hepatocytes. Gastroenterology. 1998;114(2):364–371. doi: 10.1016/s0016-5085(98)70489-5. https://doi.org/10.1016/S0016-5085(98)70489-5. [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychological Medicine. 1995;25(1):63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Ballatori N, Krance SM, Notenboom S, Shi S, Tieu K, Hammond CL. Glutathione dysregulation and the etiology and progression of human diseases. Biological Chemistry. 2009;390(3):191–214. doi: 10.1515/BC.2009.033. https://doi.org/10.1515/BC.2009.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco RA, Ziegler TR, Carlson BA, Cheng PY, Park Y, Cotsonis GA, Jones DP. Diurnal variation in glutathione and cysteine redox states in human plasma. The American Journal of Clinical Nutrition. 2007;86(4):1016–1023. doi: 10.1093/ajcn/86.4.1016. [DOI] [PubMed] [Google Scholar]
- Boggess A, Faber S, Kern J, Kingston HMS. Mean serum-level of common organic pollutants is predictive of behavioral severity in children with autism spectrum disorders. Scientific Reports. 2016;6:26185. doi: 10.1038/srep26185. https://doi.org/10.1038/srep26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxson EH, Stork LC, Allen RH, Stabler SP, Kolhouse JF. Changes in plasma methionine and total homocysteine levels in patients receiving methotrexate infusions. Cancer Research. 1989;49(21):5879–5883. [PubMed] [Google Scholar]
- Castro R, Rivera I, Struys EA, Jansen EEW, Ravasco P, Camilo ME, Tavares de Almeida I. Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clinical Chemistry. 2003;49(8):1292–1296. doi: 10.1373/49.8.1292. https://doi.org/10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- Castro VM, Kong SW, Clements CC, Brady R, Kaimal AJ, Doyle AE, Perlis RH. Absence of evidence for increase in risk for autism or attention-deficit hyperactivity disorder following antidepressant exposure during pregnancy: a replication study. Translational Psychiatry. 2016;6(1):e708. doi: 10.1038/tp.2015.190. https://doi.org/10.1038/tp.2015.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DL, Baio J, Braun KVN, Bilder D, Charles J, Constantino JN, Yeargin-Allsopp M. Prevalence and characteristics of autism spectrum disorder among children aged 8 years — Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2012. Morbidity and Mortality Weekly Report. 2016;65(3):1–23. doi: 10.15585/mmwr.ss6503a1. https://doi.org/10.15585/mmwr.ss6503a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwatko G, Jakubowski H. Urinary excretion of homocysteine-thiolactone in humans. Clinical Chemistry. 2005;51(2):408–415. doi: 10.1373/clinchem.2004.042531. https://doi.org/10.1373/clinchem.2004.042531. [DOI] [PubMed] [Google Scholar]
- Davis SR, Quinlivan EP, Stacpoole PW, Gregory JF. Plasma glutathione and cystathionine concentrations are elevated but cysteine flux is cnchanged by dietary vitamin B-6 restriction in young men and women. The Journal of Nutrition. 2006;136(2):373–378. doi: 10.1093/jn/136.2.373. [DOI] [PubMed] [Google Scholar]
- del Pino MMS, Corrales FJ, Mato JM. Hysteretic behavior of methionine adenosyltransferase III. Methionine switches between two conformations of the enzyme with different specific activity. Journal of Biological Chemistry. 2000;275(31):23476–23482. doi: 10.1074/jbc.M002730200. https://doi.org/10.1074/jbc.M002730200. [DOI] [PubMed] [Google Scholar]
- Døskeland SO, Ueland PM. Comparison of some physicochemical and kinetic properties of S-adenosylhomocysteine hydrolase from bovine liver, bovine adrenal cortex and mouse liver. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1982;708(2):185–193. doi: 10.1016/0167-4838(82)90219-9. https://doi.org/10.1016/0167-4838(82)90219-9. [DOI] [PubMed] [Google Scholar]
- Duncan TM, Reed MC, Nijhout HF. A population model of folate-mediated one-carbon metabolism. Nutrients. 2013a;5(7):2457–2474. doi: 10.3390/nu5072457. https://doi.org/10.3390/nu5072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan TM, Reed MC, Nijhout HF. The relationship between intracellular and plasma levels of folate and metabolites in the methionine cycle: A model. Molecular Nutrition & Food Research. 2013b;57(4):628–636. doi: 10.1002/mnfr.201200125. https://doi.org/10.1002/mnfr.201200125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engl HW, Hanke M, Neubauer A. Regularization of Inverse Problems. Springer Science & Business Media; 1996. [Google Scholar]
- Fang YZ, Yang S, Wu G. Free radicals, antioxidants, and nutrition. Nutrition. 2002;18(10):872–879. doi: 10.1016/s0899-9007(02)00916-4. https://doi.org/10.1016/S0899-9007(02)00916-4. [DOI] [PubMed] [Google Scholar]
- Finkelstein JD, Martin JJ. Methionine metabolism in mammals. Distribution of homocysteine between competing pathways. Journal of Biological Chemistry. 1984;259(15):9508–9513. [PubMed] [Google Scholar]
- Finkelstein JD, Martin JJ. Homocysteine. The International Journal of Biochemistry & Cell Biology. 2000;32(4):385–389. doi: 10.1016/s1357-2725(99)00138-7. https://doi.org/10.1016/S1357-2725(99)00138-7. [DOI] [PubMed] [Google Scholar]
- Flynn J, Reich N. Murine DNA (Cytosine-5-)-methyltransferase: Steady-state and substrate trapping analyses of the kinetic mechanism. Biochemistry. 1998;37(43):15162–15169. doi: 10.1021/bi9810609. https://doi.org/10.1021/bi9810609. [DOI] [PubMed] [Google Scholar]
- Frey HC, Patil SR. Identification and review of sensitivity analysis methods. Risk Analysis. 2002;22(3):553–578. https://doi.org/10.1111/0272-4332.00039. [PubMed] [Google Scholar]
- Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, Buxbaum JD. Most genetic risk for autism resides with common variation. Nature Genetics. 2014;46(8):881–885. doi: 10.1038/ng.3039. https://doi.org/10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillberg C, Wing L. Autism: not an extremely rare disorder. Acta Psychiatrica Scandinavica. 1999;99(6):399–406. doi: 10.1111/j.1600-0447.1999.tb00984.x. https://doi.org/10.1111/j.1600-0447.1999.tb00984.x. [DOI] [PubMed] [Google Scholar]
- Giugliano D, Ceriello A, Paolisso G. Oxidative stress and diabetic vascular complications. Diabetes Care. 1996;19(3):257–267. doi: 10.2337/diacare.19.3.257. [DOI] [PubMed] [Google Scholar]
- Golub G, Hansen P, O’Leary D. Tikhonov Regularization and Total Least Squares. SIAM Journal on Matrix Analysis and Applications. 1999;21(1):185–194. https://doi.org/10.1137/S0895479897326432. [Google Scholar]
- Hallmayer J, Cleveland S, Torres A, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Archives of General Psychiatry. 2011;68(11):1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. https://doi.org/10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamby DM. A review of techniques for parameter sensitivity analysis of environmental models. Environmental Monitoring and Assessment. 1994;32(2):135–154. doi: 10.1007/BF00547132. https://doi.org/10.1007/BF00547132. [DOI] [PubMed] [Google Scholar]
- Hamilton D, Wu JH, Alaoui-Jamali M, Batist G. A novel missense mutation in the γ-glutamylcysteine synthetase catalytic subunit gene causes both decreased enzymatic activity and glutathione production. Blood. 2003;102(2):725–730. doi: 10.1182/blood-2002-11-3622. https://doi.org/10.1182/blood-2002-11-3622. [DOI] [PubMed] [Google Scholar]
- Helbock HJ, Beckman KB, Shigenaga MK, Walter PB, Woodall AA, Yeo HC, Ames BN. DNA oxidation matters: The HPLC–electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(1):288–293. doi: 10.1073/pnas.95.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SY, Gil HW, Yang JO, Lee EY, Kim HK, Kim SH, Lee Z-W. Pharmacokinetics of glutathione and its metabolites in normal subjects. Journal of Korean Medical Science. 2005;20(5):721–726. doi: 10.3346/jkms.2005.20.5.721. https://doi.org/10.3346/jkms.2005.20.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CS, Moore WR, Meister A. On the active site thiol of gamma-glutamylcysteine synthetase: relationships to catalysis, inhibition, and regulation. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(8):2464–2468. doi: 10.1073/pnas.85.8.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SJ, Melnyk S, Jernigan S, Cleves MA, Halsted CH, Wong DH, Gaylor DW. Metabolic endophenotype and related genotypes are associated with oxidative stress in children with autism. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2006;141B(8):947–956. doi: 10.1002/ajmg.b.30366. https://doi.org/10.1002/ajmg.b.30366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohavi R. Proceedings of the 14th International Joint Conference on Artificial Intelligence - Volume 2. San Francisco, CA, USA: Morgan Kaufmann Publishers Inc; 1995. A Study of Cross-validation and Bootstrap for Accuracy Estimation and Model Selection; pp. 1137–1143. Retrieved from http://dl.acm.org/citation.cfm?id=1643031.1643047. [Google Scholar]
- López-Barea J, Lee CY. Mouse-liver glutathione reductase. European Journal of Biochemistry. 1979;98(2):487–499. doi: 10.1111/j.1432-1033.1979.tb13210.x. https://doi.org/10.1111/j.1432-1033.1979.tb13210.x. [DOI] [PubMed] [Google Scholar]
- Lu SC. Regulation of hepatic glutathione synthesis: current concepts and controversies. The FASEB Journal. 1999;13(10):1169–1183. [PubMed] [Google Scholar]
- Lu SC. Regulation of glutathione synthesis. Molecular Aspects of Medicine. 2009;30(1–2):42–59. doi: 10.1016/j.mam.2008.05.005. https://doi.org/10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luka Z, Wagner C. Effect of naturally occurring mutations in human glycine N-methyltransferase on activity and conformation. Biochemical and Biophysical Research Communications. 2003;312(4):1067–1072. doi: 10.1016/j.bbrc.2003.11.037. https://doi.org/10.1016/j.bbrc.2003.11.037. [DOI] [PubMed] [Google Scholar]
- Lyons J, Rauh-Pfeiffer A, Yu YM, Lu XM, Zurakowski D, Tompkins RG, Castillo L. Blood glutathione synthesis rates in healthy adults receiving a sulfur amino acid-free diet. Proceedings of the National Academy of Sciences. 2000;97(10):5071–5076. doi: 10.1073/pnas.090083297. https://doi.org/10.1073/pnas.090083297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddipati KR, Marnett LJ. Characterization of the major hydroperoxide-reducing activity of human plasma. Purification and properties of a selenium-dependent glutathione peroxidase. Journal of Biological Chemistry. 1987;262(36):17398–17403. [PubMed] [Google Scholar]
- Markesbery WR. Oxidative Stress Hypothesis in Alzheimer’s Disease. Free Radical Biology and Medicine. 1997;23(1):134–147. doi: 10.1016/s0891-5849(96)00629-6. https://doi.org/10.1016/S0891-5849(96)00629-6. [DOI] [PubMed] [Google Scholar]
- Martensson J. Method for determination of free and total glutathione and gamma-glutamylcysteine concentrations in human leukocytes and plasma. Journal of Chromatography B: Biomedical Sciences and Applications. 1987;420:152–157. doi: 10.1016/0378-4347(87)80166-4. https://doi.org/10.1016/0378-4347(87)80166-4. [DOI] [PubMed] [Google Scholar]
- Melnyk S, Fuchs GJ, Schulz E, Lopez M, Kahler SG, Fussell JJ, James SJ. Metabolic imbalance associated with methylation dysregulation and oxidative damage in children with autism. Journal of Autism and Developmental Disorders. 2012;42(3):367–377. doi: 10.1007/s10803-011-1260-7. https://doi.org/10.1007/s10803-011-1260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk S, Pogribna M, Pogribny I, Hine RJ, James SJ. A new HPLC method for the simultaneous determination of oxidized and reduced plasma aminothiols using coulometric electrochemical detection1. The Journal of Nutritional Biochemistry. 1999;10(8):490–497. doi: 10.1016/s0955-2863(99)00033-9. https://doi.org/10.1016/S0955-2863(99)00033-9. [DOI] [PubMed] [Google Scholar]
- Melnyk S, Pogribna M, Pogribny IP, Yi P, James SJ. Measurement of Plasma and Intracellular S-Adenosylmethionine and S-Adenosylhomocysteine Utilizing Coulometric Electrochemical Detection: Alterations with Plasma Homocysteine and Pyridoxal 5′-Phosphate Concentrations. Clinical Chemistry. 2000;46(2):265–272. [PubMed] [Google Scholar]
- Miller AL. The methionine-homocysteine cycle and its effects on cognitive diseases. Alternative Medicine Review. 2003;8(1):7–19. [PubMed] [Google Scholar]
- Mosharov E, Cranford MR, Banerjee R. The quantitatively important relationship between homocysteine metabolism and glutathione synthesis by the transsulfuration pathway and its regulation by redox changes. Biochemistry. 2000;39(42):13005–13011. doi: 10.1021/bi001088w. [DOI] [PubMed] [Google Scholar]
- Muratore CR, Hodgson NW, Trivedi MS, Abdolmaleky HM, Persico AM, Lintas C, Deth RC. Age-dependent decrease and alternative splicing of methionine synthase mRNA in human cerebral cortex and an accelerated decrease in autism. PLOS ONE. 2013;8(2):e56927. doi: 10.1371/journal.pone.0056927. https://doi.org/10.1371/journal.pone.0056927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi M, Iwata A, Yatome C, Kitade Y. Purification and properties of recombinant Plasmodium falciparum S-adenosyl-L-homocysteine hydrolase. Journal of Biochemistry. 2001;129(1):101–105. doi: 10.1093/oxfordjournals.jbchem.a002819. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Reed MC, Anderson DF, Mattingly JC, James SJ, Ulrich CM. Long-range allosteric interactions between the folate and methionine cycles stabilize DNA methylation reaction rate. Epigenetics. 2006;1(2):81–87. doi: 10.4161/epi.1.2.2677. [DOI] [PubMed] [Google Scholar]
- Rai V. Association of methylenetetrahydrofolate reductase (MTHFR) gene C677T polymorphism with autism: evidence of genetic susceptibility. Metabolic Brain Disease. 2016;31(4):727–735. doi: 10.1007/s11011-016-9815-0. https://doi.org/10.1007/s11011-016-9815-0. [DOI] [PubMed] [Google Scholar]
- Reed MC, Nijhout HF, Sparks R, Ulrich CM. A mathematical model of the methionine cycle. Journal of Theoretical Biology. 2004;226(1):33–43. doi: 10.1016/j.jtbi.2003.08.001. https://doi.org/10.1016/j.jtbi.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Reed MC, Thomas RL, Pavisic J, James SJ, Ulrich CM, Nijhout HF. A mathematical model of glutathione metabolism. Theoretical Biology and Medical Modelling. 2008;5:8. doi: 10.1186/1742-4682-5-8. https://doi.org/10.1186/1742-4682-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsum H, Helland S, Ueland PM. Radioenzymic determination of homocysteine in plasma and urine. Clinical Chemistry. 1985;31(4):624–628. [PubMed] [Google Scholar]
- Richman PG, Meister A. Regulation of gamma-glutamyl-cysteine synthetase by nonallosteric feedback inhibition by glutathione. Journal of Biological Chemistry. 1975;250(4):1422–1426. [PubMed] [Google Scholar]
- Rossignol DA, Genuis SJ, Frye RE. Environmental toxicants and autism spectrum disorders: a systematic review. Translational Psychiatry. 2014;4(2):e360. doi: 10.1038/tp.2014.4. https://doi.org/10.1038/tp.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roum JH, Buhl R, McElvaney NG, Borok Z, Crystal RG. Systemic deficiency of glutathione in cystic fibrosis. Journal of Applied Physiology. 1993;75(6):2419–2424. doi: 10.1152/jappl.1993.75.6.2419. [DOI] [PubMed] [Google Scholar]
- Saltelli A, Annoni P. How to avoid a perfunctory sensitivity analysis. Environmental Modelling & Software. 2010;25(12):1508–1517. https://doi.org/10.1016/j.envsoft.2010.04.012. [Google Scholar]
- Shi MM, Kugelman A, Iwamoto T, Tian L, Forman HJ. Quinone-induced oxidative stress elevates glutathione and induces gamma-glutamylcysteine synthetase activity in rat lung epithelial L2 cells. Journal of Biological Chemistry. 1994;269(42):26512–26517. [PubMed] [Google Scholar]
- Stabler SP, Allen RH. Quantification of serum and urinary S-adenosylmethionine and S-adenosylhomocysteine by stable-isotope-dilution liquid chromatography-mass spectrometry. Clinical Chemistry. 2004;50(2):365–372. doi: 10.1373/clinchem.2003.026252. https://doi.org/10.1373/clinchem.2003.026252. [DOI] [PubMed] [Google Scholar]
- Stipanuk MH, Dominy JE, Lee JI, Coloso RM. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. The Journal of Nutrition. 2006;136(6):1652S–1659S. doi: 10.1093/jn/136.6.1652S. [DOI] [PubMed] [Google Scholar]
- Sullivan DM, Hoffman JL. Fractionation and kinetic properties of rat liver and kidney methionine adenosyltransferase isozymes. Biochemistry. 1983;22(7):1636–1641. doi: 10.1021/bi00276a017. [DOI] [PubMed] [Google Scholar]
- Toroser D, Yarian CS, Orr WC, Sohal RS. Mechanisms of γ-glutamylcysteine ligase regulation. Biochimica et Biophysica Acta. 2006;1760(2):233–244. doi: 10.1016/j.bbagen.2005.10.010. https://doi.org/10.1016/j.bbagen.2005.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrey CL, Liu L, Andrews LG, Tollefsbol TO. The impact of metabolism on DNA methylation. Human Molecular Genetics. 2005;14(suppl 1):R139–R147. doi: 10.1093/hmg/ddi100. https://doi.org/10.1093/hmg/ddi100. [DOI] [PubMed] [Google Scholar]
- Vitvitsky V, Thomas M, Ghorpade A, Gendelman HE, Banerjee R. A Functional Transsulfuration Pathway in the Brain Links to Glutathione Homeostasis. Journal of Biological Chemistry. 2006;281(47):35785–35793. doi: 10.1074/jbc.M602799200. https://doi.org/10.1074/jbc.M602799200. [DOI] [PubMed] [Google Scholar]
- Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. The Journal of Nutrition. 2004;134(3):489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- Yeargin-Allsopp M, Rice C, Karapurkar T, Doernberg N, Boyle C, Murphy C. Prevalence of autism in a US metropolitan area. Journal of the American Medical Association. 2003;289(1):49–55. doi: 10.1001/jama.289.1.49. [DOI] [PubMed] [Google Scholar]
- Yi P, Melnyk S, Pogribna M, Pogribny IP, Hine RJ, James SJ. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. Journal of Biological Chemistry. 2000;275(38):29318–29323. doi: 10.1074/jbc.M002725200. https://doi.org/10.1074/jbc.M002725200. [DOI] [PubMed] [Google Scholar]


