Abstract
Characterization of carotid plaque composition, more specifically the amount of lipid core, fibrous tissue, and calcified tissue, is an important task for the identification of plaques that are prone to rupture, and thus for early risk estimation of cardiovascular and cerebrovascular events. Due to its low costs and wide availability, carotid ultrasound has the potential to become the modality of choice for plaque characterization in clinical practice. However, its significant image noise, coupled with the small size of the plaques and their complex appearance, makes it difficult for automated techniques to discriminate between the different plaque constituents. In this paper, we propose to address this challenging problem by exploiting the unique capabilities of the emerging deep learning framework. More specifically, and unlike existing works which require a priori definition of specific imaging features or thresholding values, we propose to build a convolutional neural network that will automatically extract from the images the information that is optimal for the identification of the different plaque constituents. We used approximately 90,000 patches extracted from a database of images and corresponding expert plaque characterizations to train and to validate the proposed convolutional neural network (CNN). The results of cross-validation experiments show a correlation of about 0.90 with the clinical assessment for the estimation of lipid core, fibrous cap, and calcified tissue areas, indicating the potential of deep learning for the challenging task of automatic characterization of plaque composition in carotid ultrasound
Index Terms: Atherosclerosis, carotid artery, ultrasound, plaque composition, convolutional neural networks
I. Introduction
CARDIOVASCULAR AND CEREBROVASCULAR events, otherwise referred to as myocardial infarction and stroke, are major causes of mortality and morbidity in the developed world [1], [2]. These serious accidents occur when atherosclerotic plaques in the arteries suddenly rupture, leading to the obstruction of the blood flow to the heart or to the brain. Early and accurate prediction of individuals at high risk of myocardial infarction and stroke would allow for preventive (e.g. diet modifications), therapeutic (e.g. lipid lowering therapy), or surgical (e.g. stenting) measures to be applied to the patient before any of these life threatening events take place.
In clinical practice, the identification of high-risk individuals is carried out thus far by using risk prediction calculators, which combine standard risk factors such as age, smoking, blood pressure, family history, diabetes, and body mass index [3], [4]. However, these risk predictions do not take into account patient-specific information describing the presence and the type of any existing atherosclerotic plaques, resulting in estimations that are approximate averages over population risk factors. In contrast, in the era of personalized medicine, plaque image analysis [5] has the potential to extract valuable information about the plaques and thus to identify more accurately the patients at risk of plaque rupture. It is now well established that the tissue composition plays a central role for the stability or vulnerability of atherosclerotic plaques [6], [7]. More specifically, plaques with large lipid cores and thin fibrous caps are more prone to rupture, while plaques that contain calcified tissue tending to be more stable [8]. In contrast, several studies have shown that morphological measures used in current practice, such as the intima-media thickness (IMT), have limited prediction powers [9].
It is therefore important to develop computational techniques that can automatically and objectively determine the plaque constituents of atherosclerotic plaques from image data. But the task remains challenging due to the small size of the plaques and the complexity of the tissue appearance. So far, most of the computational techniques for plaque tissue characterization have been developed for multi-contrast MRI [10]–[19], which provides relatively high resolution and good quality images of the plaques. However, multi-contrast MRI has cost and scanning limitations that make its use in daily clinical practice limited, thus remaining largely dedicated for research purposes. In contrast, B-mode ultrasound is widely used for the assessment of atherosclerosis in the carotid artery due to its ease of use, very low costs, and wide availability [20]. Histology-based studies have shown the capability of ultrasound to separate the lipid, fibrous, and calcified tissues in the carotid [21]–[23]. Due to these benefits, carotid ultrasound has significant potential as the modality of choice for atherosclerosis assessment in clinical practice [20].
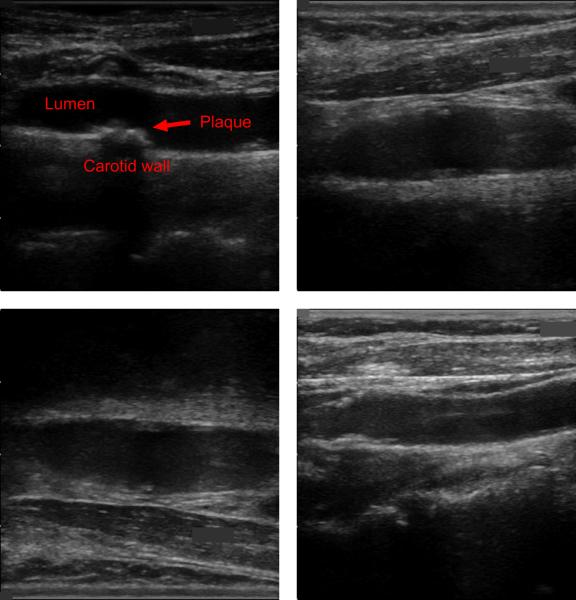
As illustrated by the examples of Fig. 1, ultrasound images of the carotid bifurcation have typically lower image quality [24], incorporating significant noise, artifacts, shadowing, and reverberation. Under these conditions, the detection and characterization of the plaques becomes tedious and inconsistent, even for an expert clinician.
Fig. 1.
Four examples of carotid ultrasound images, incorporating noise, artifacts, shadowing, and reverberation. All of these examples contain plaques, but their detection and characterization is challenging even for an expert clinician, resulting in tedious and inconsistent assessments.
An automated technique is thus required to obtain fast, robust, and user-independent analyses of the plaque composition. Thus far, this has been achieved in the literature largely based on thresholding of the imaging intensities inside the plaque [21], [23], [25], with each of these papers using different threshold values for the plaque constituents. A recent study by Pazinato et al. [26] has shown that there is significant overlap between the intensity distributions of the different constituents in ultrasound, thus leading to accuracy results just over 50% even with the choice of optimal threshold values [26]. The authors proposed instead to estimate imaging features from patches around each pixel before classification, improving the identification of the plaque constituents to over 70% accuracy. However, this technique required the use of predefined standard imaging features such as statistical moments or histograms of oriented gradients. Furthermore, the validation was carried out with a small sample size of only six real datasets, thus with limited variability in the type and appearance of the plaques.
In this paper, we investigate a deep learning approach in order to address the challenging task of automatically characterizing plaque composition in carotid ultrasound. The proposed technique does not require the pre-definition of intensity thresholds or imaging features. Instead, we built a convolutional neural network that can extract automatically from a training sample of imaging patches around each pixel position the image information in the ultrasound data that are optimal for the discrimination of the different plaque constituents. We trained and validated our deep learning architecture using a total of 56 in vivo cases acquired from clinical practice and representing different plaque characteristics and types.
II. Methods
A. Deep Learning
Deep learning is a promising machine learning tools for the automatic classification and interpretation of medical image data. In the last year, the paradigm has been applied extensively in several medical imaging applications, such as for brain [27], [28], lung [29], [30], and breast [31], [32] imaging. Its application to arterial structures such as the carotid artery and for noisy image data as those found in ultrasound of the carotid bifurcation has not been reported yet.
Essentially, deep learning attempts to extract high level representations of the data that are most relevant to a specific learning task by using a deep graph composed of multiple processing layers. One key attribute of deep learning is the ability to build multiple layers encoding different levels of abstractions, each contributing to the discriminative or prediction power of the whole [33]. For the analysis of images, deep learning can be used to transform an image into a set of hierarchical nonlinear processing units representing visual or statistical information (e.g., edges/shapes, intensity profiles, statistical patterns), such that this information is optimal for a given image analysis task (e.g., object recognition, image classification, image quantification).
One of the promises of deep learning is thus to replace generic imaging features (e.g. wavelets, spatial textures, statistical moments) with processing layers that are more complex as well as more specific to the data and task in question, leading to an optimal use of information and improved prediction power. In the case of plaque characterization, our hypothesis is that the complex and overlapping appearance of the plaque constituents in noisy and low resolution ultrasound data cannot be represented with standard imaging features and that deep learning can extract new discriminative features combining both global and local imaging information of the plaque.
B. Plaque Image Patches
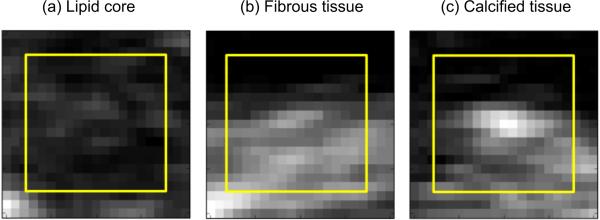
The characterization of plaque composition at each pixel position based solely on the intensity value at that pixel, as proposed in several papers [21], [23], [25], is challenging due to the significant overlap between the intensity distributions of the different plaque constituents, as demonstrated in this recent study [26]. Instead, using patches around each pixel position (see examples of Fig. 2) would enable to take context into account in the classification, by learning the relative appearance of each pixel with respect to other tissue components, as illustrated in the example of Fig. 3. In [26], the authors considered patches around the pixel position but only to calculate predefined imaging features such as statistical moments and histograms of oriented gradients before the learning and classification stages. This means not all the information contained in the patches was used for the plaque discrimination. In the proposed approach, the idea is to use the entire patch as the input of the deep learning, which will then choose automatically the information that is most relevant and optimal to the discrimination of the different plaque constituents. With this method, the imaging features are not chosen empirically before the learning stage but instead are optimally extracted as part of the learning and classification tasks. In this work, we empirically selected an image patch size 15 × 15 pixels for our CNN model, which we found to be sufficient to adequately encode inter-variability between neighboring constituents.
Fig. 2.
Examples of input image patches for different plaque tissue classes. The yellow frames outline the input to the CNN.
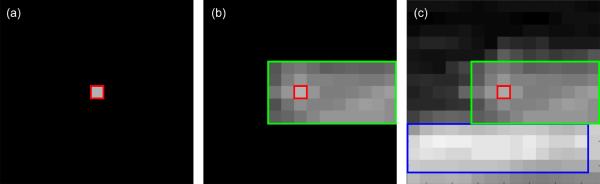
Fig. 3.
Illustration of the advantage of using image patches around each pixel position in the plaque for the characterization of the constituents. In this example, by looking only at the central intensity value in (a), the corresponding tissue (green rectangle) in (b) can be potentially mistaken for an early stage classified tissue, instead of a fibrous tissue. However, by looking at the entire intensity patch as shown in (c), the presence of a brighter component (blue rectangle) can be exploited by the deep learning model to eliminate potential ambiguities, thus correctly classifying the tissue of interest into fibrous tissue.
C. Plaque Intensity Normalization
One of the limitations of ultrasound data is the variability in the appearance of tissues between different image acquisitions depending on the equipment, operator, patient, and imaging settings. Consequently, it is important to develop methods that can address the variability in the appearance of ultrasound images and of their tissues. Traditionally, this has been achieved using image normalization, i.e. by transforming the image data such that same tissues have approximatively similar intensity values.
For carotid ultrasound imaging, normalization has been done through manual interaction, i.e. through an operator who marks two corresponding regions in different images, which are then used as the references for intensity normalization of the dynamic ranges of the images. In most works, such as the recent study using SVM [26], the operator is required to draw manually two regions corresponding to the blood and to the adventitia wall. Such an approach, however, is dependent on the user input, which can change significantly due to the variability in the appearance of both the blood and the adventitia in the same image that result from the presence of shadowing, reverberations, and inconsistencies within same tissues.
In this work, our hypothesis is that the deep learning paradigm can extract high-level features from the imaging patches that take into account relative values of the constituents and therefore that are less sensitive to image normalization. Accordingly, we have applied a linear scaling between the minimum and maximum values of the full image as a standard method for normalization, without the use of any user interaction.
D. Plaque CNN Architecture
Convolutional neural networks (CNNs) are types of deep learning architectures that use convolutional filters (i.e. kernels of varying sizes) at each layer to extract new features from training images that are relevant for the image interpretation task. They have shown their promise by successfully addressing a number of challenging image recognition tasks [34].
Mathematically, given a training dataset of images {Ii} and the corresponding class labels {ci}, the goal of CNNs is to learn an optimal mapping F(I) = c in the form of an L-level composition F(I) = fL ∘…∘ f1(I). Each of the functions fl define the l-th layer of the CNN, where the 0-th layer correspond to the input data, i.e. f0 = I. In this notation, the l-th layer is a convolution of the previous layer with a kernel Wl with added bias bl, i.e. fl = f(Wl * fl−1 + bi), where f is a non-linearity function, e.g. f(.) = max(0,.) + α min(0,.). The kernels Wl and bias terms bl representing the new discriminative features are learned at the training phase, while the number of layers L and the kernel sizes are defined by the architecture design depending on the image analysis problem.
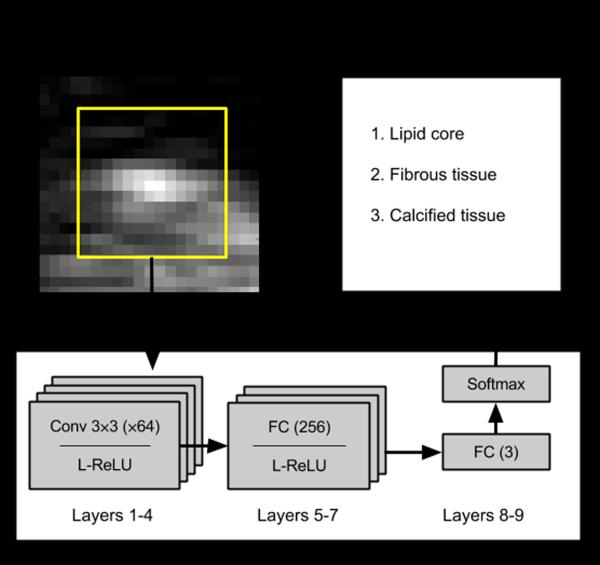
For our plaque characterization problem, as it can be seen from Figs. 2 and 3, the composition of the plaque is distinguishable at a neighborhood close to 15 × 15 and therefore we adapted a CNN to describe such context. Specifically, we implemented a design consisting of four convolutional layers with kernel sizes of 3×3 and three fully-connected layers each followed by leaky rectified linear units (ReLU) as illustrated schematically in Fig. 4. We used the cross-entropy loss function with L2 regularization penalty on the weights, as well as dropout layers [35] after the convolutional and fully connected layers at training phase. We solved the optimization problem using the Adam method [36].
Fig. 4.
Schematic diagram of the CNN architecture used for automatic characterization of plaque constituents in carotid ultrasound images. The architecture consisted of four convolutional (Conv) and three fully connected (FC) layers with leaky rectified linear units (L-ReLU) non-linearity functions, and an FC layer with class scores followed by the softmax function.
Furthermore, we used a batch size of 1000 for training and the recommended fixed hyper-parameters for the leaky ReLU slope (0.01). Hyper-parameter optimization was performed to select the weight decay (0.005) and the initial learning rate (0.001) in an inverse annealing scheme.
III. Evaluation
A. Datasets
The evaluation of the proposed deep learning approach for the characterization of the plaque constituents was achieved based on a total of 56 in vivo cases obtained from the University Hospital Arnau de Vilanova, Lleida, Spain. We selected only 56 cases for this study due to the highly tedious and time consuming work required to obtain ground truth in clinical practice for plaque composition. However, this sample translates into about 90,000 imaging patches, which are sufficient to train the CNN architecture described in Section II-D. Furthermore, as shown in Table I, the validation sample displays significant variability in the properties of the plaque and images. In particular, the plaque composition varies highly between the 56 cases (see the variation in the percentages of the different constituents in Table II), thus enabling the validation of the proposed technique with respect to various plaque types.
TABLE I.
Metadata of the Plaques Used for the Validation of the Deep Learning Approach
| Variability: | From | To |
|---|---|---|
|
| ||
| Size of plaque (mm2) | 5.6 | 196.3 |
| Percentage of lipid core (%) | 0.4 | 82.9 |
| Percentage of fibrous tissue (%) | 13.2 | 69.5 |
| Percentage of calcified tissue (%) | 0.9 | 86.1 |
| Maximal intensity | 180 | 255 |
TABLE II.
Summary of the Pixel-Based Accuracy Obtained by the Proposed and Existing SVM Techniques
| Median | Mean | Std. | Min | Max | % best result | |
|---|---|---|---|---|---|---|
|
| ||||||
| Single-scale SVM | 0.66 | 0.61 | 0.17 | 0.21 | 0.87 | 7.2 |
| Multi-scale SVM | 0.70 | 0.69 | 0.16 | 0.20 | 0.94 | 14.3 |
| Proposed CNN | 0.80 | 0.75 | 0.16 | 0.26 | 0.97 | 78.5 |
The images of the carotid bifurcation were acquired in B-mode using a General Electric Vivid-i system equipped with a 4–7 MHz transducer. An image resolution of 8.5 pixels per mm was obtained. Note that all the carotid ultrasound scans were performed between 2009 and 2013.
B. Clinical Assessment
As ground truth, we used in this study the clinical results obtained from the Unit for the Detection and Treatment of Athero-thrombotic Diseases (UDETMA), Lleida, Spain. This specialized and accredited center has decades of experience in the assessment of atherosclerosis using carotid ultrasound data. The expert observer for this study was our co-author Dr. Angels Betriu, who firstly delineated for each image the boundaries of the plaque. Subsequently, each plaque constituent was defined inside the plaque (lipid core in red, fibrous tissue in yellow, and calcified tissue in green, see examples in Fig. 8). Finally, for each plaque constituent, the area in mm2 was calculated and was used as the basis to assess the potential vulnerability or stability of the plaque.
Fig. 8.
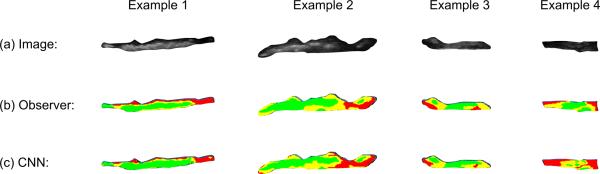
Visual examples of the classification results obtained by the CNN model for varying degrees of accuracy (red: lipid core, yellow: fibrous tissue, green: calcified tissue). The accuracy values for these examples (1 to 4) are 0.96, 0.82, 0.67, and 0.48, in this order.
Our aim is not to define accurately every pixel but rather to assess whether our CNN can automatically reproduce similar results than those obtained in clinical practice.
C. Pixel-Based Accuracy Results
In this section, we assessed the strength of the proposed classification technique for plaque tissue characterization by computing accuracy as the percentage of correctly classified pixels.
Although we acknowledge that the ground truth provided by the expert observer cannot be consistently accurate for each individual pixel within the plaque, in particular at the boundaries between the different constituents, this evaluation provides an indication of the improvement achieved by the proposed deep learning approach.
For comparison, we implemented the technique based on support vector classification and statistical moment descriptors (SVM-SMD) recently proposed in [26]. We used the optimal implementation as reported by the authors, i.e. by calculating seven statistical moment descriptors (mean, standard deviation, skewness, kurtosis, median, entropy, range) from 13 × 13 windows as the underlying imaging features for the SVM classification. We also implemented the multi-scale SVM technique by considering 9×9, 11×11, and 13×13 image patches as recommended by the authors in [26]. We used the dot product kernel function for the SVM classification.
Note that all experiments were carried out using 5-fold cross-validation, i.e. one fifth of the cases were used for testing and the rest for training and optimizing the CNN architecture. For a fair comparison, both the proposed and existing methods were trained and validated with the exact same datasets.
The accuracy results are summarized in Table III, where it can be seen that the CNN approach improves all the accuracy statistics (median, mean, standard deviation, minimum and maximum) in comparison to the SVM approaches. Specifically, the CNN approach outputs the best classification in 78.5% of the cases, versus 14.3% for the multi-scale SVM and 7.2% of the cases for the single-scale SVM. The CNN obtains a mean accuracy of 0.75 ± 0.16 (median 0.80), which is an improvement over the results of the single-scale SVM (mean: 0.63 ± 0.14), as well as the multi-scale SVM (mean: 0.69 ± 0.16). This translates into an average improvement of 21.4% over the single-scale SVM and 14.2% over the multi-scale SVM approaches. These results clearly indicate the benefits of minimal loss of information achieved with deep learning by extracting optimal features for tissue classification, instead of using pre-defined features such as the statistical moment descriptors.
TABLE III.
Sensitivity and Specificity of the Deep Learning Technique for the Three Tissue Components
| Sensitivity | Specificity | |
|---|---|---|
|
| ||
| Lipid core | 0.83 ± 0.12 | 0.90 ± 0.13 |
| Fibrous cap | 0.70 ± 0.16 | 0.80 ± 0.14 |
| Calcified tissue | 0.76 ± 0.15 | 0.89 ± 0.12 |
In Table IV, the sensitivity and specificity of the deep learning technique for the three tissue constituents are summarized. It can be seen that the classification performance is higher for the lipid core as it appears bright in the images. On the contrary, the fibrous cap is the tissue classified with the lowest performance due to its generally more variable appearance and small size. All tissues were classified with sensitivity over 0.70 and specificity over 0.80.
TABLE IV.
Confusion Matrix of the Deep Learning Technique for the Three Plaque Constituents (in %)
| Predicted lipids | Predicted fibrous | Predicted calcium | |
|---|---|---|---|
|
| |||
| Actual lipids | 83.4 | 16.2 | 0.4 |
| Actual fibrous | 12.6 | 70.2 | 17.2 |
| Actual calcium | 2.4 | 21.0 | 76.6 |
Additionally, the derived confusion matrix for the deep learning technique is given in Table IV. It can be noted that the highest confusion is obtained between the fibrous and calcified tissues due to similar appearances depending on the quality of the ultrasound scan. Nevertheless, the classification performance remains above the 70% mark for all constituents, and is particularly high for the lipid core, which plays an important role in defining the vulnerability of the plaques.
D. Area-Based Quantification Results
In clinical practice, the important quantification to be used for the assessment of plaque composition is not the classification of each individual pixel, which can be difficult to determine consistently for all pixels and which can furthermore vary between expert observers. Instead, clinicians estimate the amount of tissue for each of the constituents, which then give an indication of the type of plaque under investigation. For example, a plaque with a large lipid core applies higher pressure forces towards its walls and is thus more prone to rupture. On the other hand, plaques with large calcified tissue tend to be more stable and less at risk of rupture.
Thus, after having established the superiority of the deep learning approach for tissue classification, we calculated the areas in mm2 of the different constituents as obtained by the expert clinician and the proposed automatic technique. The agreement between the two methods is visually assessed in Figs. 5, 6, and 7, for the lipid core, fibrous tissue, and calcified tissue, resp7ectively.
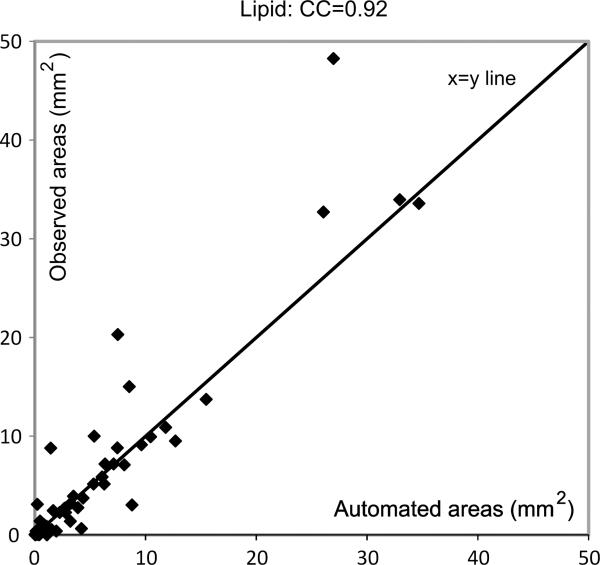
Fig. 5.
Figure showing degree of agreement between expert and automated area estimations for the lipid core.
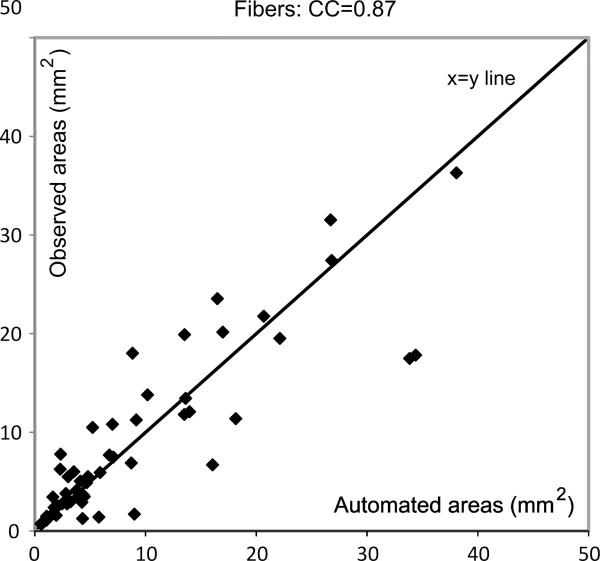
Fig. 6.
Figure showing degree of agreement between expert and automated area estimations for the fibrous cap.
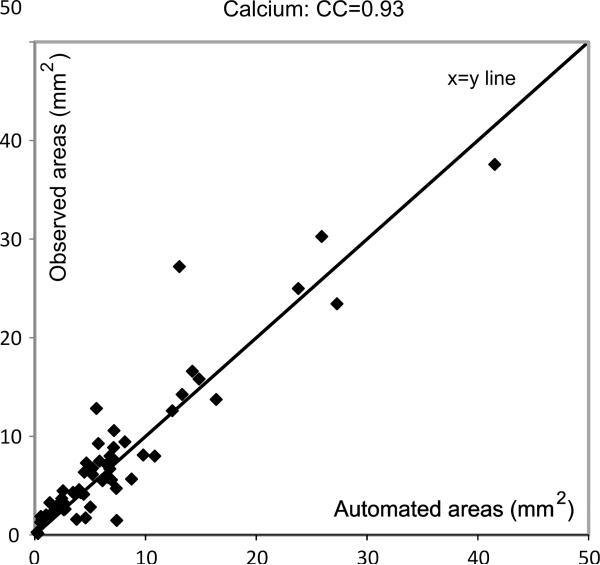
Fig. 7.
Figure showing degree of agreement between expert and automated area estimations for the calcified tissue.
It can be seen from these figures that there is good agreement between the automatic and expert assessments of the three tissue components (except for a few minor outliers), with Pearson's correlation coefficients equal to 0.92, 0.87, and 0.93, for the lipid core, fibrous tissue, and calcified tissues, respectively. This means the automatic measurements can be used to make clinical predictions on the type of the plaques that are under investigation. Note that the correlation coefficients obtained with the SVM-based techniques were 0.88, 0.82 and 0.86, in the same order.
E. Visual Illustrations
Fig. 8 shows four examples of visual results with varying degrees of accuracy by the CNN technique (red: lipid core, yellow: fibrous tissue, green: calcified tissue). In the examples 1 and 2, good separation of the plaque components is obtained by the CNN classification, with only minor localized errors (the classification accuracy equals 0.96 and 0.82, respectively). The examples 3 and 4 are associated with higher misclassifications by the proposed technique due to a particularly limited contrast inside the plaque as compared to the general quality of the images in the sample, which can be due to ultrasound suboptimal scanning by the operator (the classification accuracy equals 0.67 and 0.48, respectively). In this case, the CNN classification still manages to identify all the main constituents but with under-estimated lipid and calcified tissues respectively for the examples 3 and 4.
F. Effect of Training Size
Finally, we evaluated the effect of the training size on the accuracy of the CNN classifier, thus to estimate whether the performance is near an optimal classification based on the 56 cases, or whether improvement of the classification accuracy is possible by adding more data to the training. To continue with the 5-fold validation used in this paper, we subdivided in each test randomly the 56 cases as follows:
Testing subset: 1/5th of the sample (12 cases).
Validation subset: 1/5th (11 cases).
Training subset: 1/5th, then 2/5th, and 3/5th of the sample (i.e. 11 cases, then 22, then 33).
The obtained results in Table V indicate that all statistics (mean, standard deviation, minimum and maximum) continue to improve progressively as the training size increases, though at a relatively modest rate. This increase is indicative of the method's potential to further improve the result by training on larger datasets when possible.
TABLE V.
Effect of the Training Sample Size on the Accuracy of the Tissue Components Classification (in %)
| Training size | Mean | Std. | Min | Max. |
|---|---|---|---|---|
|
| ||||
| 1/5th (11 cases) | 0.70 | 0.16 | 0.21 | 0.94 |
| 2/5th (22 cases) | 0.72 | 0.16 | 0.21 | 0.96 |
| 3/5th (33 cases) | 0.75 | 0.14 | 0.23 | 0.98 |
IV. Discussion & Conclusions
In this paper, we presented a deep learning approach for automatic characterization of plaque composition in carotid ultrasound images. The validation with a highly variable sample of 56 in vivo cases shows that the proposed technique improves upon the results obtained with the most advanced recent method based on support vector machine and pre-defined imaging features, with an improvement in 78.5% of the cases. Furthermore, the quantitative results indicate good agreement between the expert and automatic measurements for the estimation of the amount of the different constituents, with about 0.90 correlations between the two methods. This means the automatic measurements can be used to make clinical predictions on the type of the plaques that are under investigation.
In terms of time complexity, while the duration of the training is relatively long (an average of 1.5 hours ± 0.6 hours), the testing stage was achieved in near real time, i.e. 52 ms ± 13ms for each individual image. These running times were obtained using a GPU server on NVIDIA K20 graphics card.
This present study has some limitations that are important to mention. Firstly, we used only 56 cases for the training and validation of the CNN model. This is because it is very tedious and time consuming for an expert observer to delineate the different plaque constituents in such small and complex atherosclerotic plaques. However, due to the use of a patch-based approach, our sample translates into about 90,000 input imaging patches for the CNN model. Furthermore, this sample displays significant variability in their characteristics as shown in Table I. In comparison, the most recent technique on the topic was validated with only six real datasets [26].
Secondly, the ground truth for this study was provided by a single expert clinician, though with decades of experience working with carotid ultrasound data. Nevertheless, the CNN architecture is precisely designed to imitate and reproduce the assessment of such an expert clinician automatically, with the goal to obtain consistent and fast measurements in daily clinical practice, such as to measure response to drug therapy.
In previous works, such as in Moeskops et al. [37], it was found that patch size can play an important role in the performance of the CNN classification. However, in our application, by varying the patches between 9×9, 11×11, 13×13, and 15×15 sizes, as well as by fusing these multiple scales in a multi-path CNN framework, the change in accuracy is minimal (around 0.003). This can be explained by the fact that the constituents and plaques are generally very small compared to the rest of the image and artery, so increasing the patch size does not add much information to the classification in our case. In contrast, in the work of Moeskops et al., the entire brain was segmented and the various brain structures have significantly different sizes, thus the influence of patch size is more evident.
Similarly, data augmentation [34], such as using image flipping, rotation, and translation, has been used to boost the classification in many medical imaging applications. In our work, we have augmented our training data by flipping the images in both axis of the plane (2 flips), and small rotations of the original and flipped patches around the ground truth position (± pi/8), which resulted in a 9-fold boost of the number of training patches. However, this resulted in very similar results (less than 1% differences). This is because data augmentation does not change the plaque composition itself but only the position of the plaque in the image, which explains the limited impact for our specific application.
Based on the results of the experiments in Section III-E, showing steady improvement with increase training size, our goal is to progressively build a larger database of training cases together with our clinical partner to further enrich the plaque CNN classification over time as new datasets become available and to make it more robust to larger variability.
Acknowledgment
The work of K. Lekadir was funded by a Marie-Curie research fellowship from the European Commission.
The research of K. Lekadir received funding from the Marie-Curie Actions Program of the European Union (FP7/2007–2013) under REA grant agreement n° 600388 (TECNIOspring program) and from ACCIÓ. The work of A. Galimzianova and D. L. Rubin was funded through NIH U01CA142555 and 1U01CA190214 grants, and through a hardware grant from NVIDIA. M. Vila was supported through the FIS CP12/03287 and RD12/0042/0061 grants from the Instituto de Salud Carlos III and the European Regions Development Fund. The work of L. Igual and P. Radeva was partially funded by the TIN2012-38187-C03-01 and SGR 1219 grants. P. Radeva was partly supported by an ICREA Academia 2014 grant. The work of S. Napel was supported by the NIH R01 CA160251 and U01 CA187947 grants.
References
- [1].Paiva L, Providencia R, Barra S, Dinis P, Faustino AC, Goncalves L. Universal definition of myocardial infarction: Clinical insights. Cardiol. 2015;131(1):13–21. doi: 10.1159/000371739. [DOI] [PubMed] [Google Scholar]
- [2].W. H. Organisation WHO: Stroke, Cerebrovascular accident. Stroke. 2011 [Online]. Available: http://www.who.int/topics/cerebrovascular_accident/en/
- [3].Hippisley-Cox J, Coupland C, Vinogradova Y, Robson J, Brindle P. Performance of the QRISK cardiovascular risk prediction algorithm in an independent UK sample of patients from general practice: a validation study. Heart. 2008;94(1):34–39. doi: 10.1136/hrt.2007.134890. [DOI] [PubMed] [Google Scholar]
- [4].Wolf PA, D'Agostino RB, Belanger AJ, Kannel WB. Probability of stroke: a risk profile from the Framingham Study. Stroke. 1991;22(3):312–318. doi: 10.1161/01.str.22.3.312. [DOI] [PubMed] [Google Scholar]
- [5].Vancraeynest D, Pasquet A, Roelants V, Gerber BL, Vanoverschelde JLJ. Imaging the vulnerable plaque. J. Am. Coll. Cardiol. 2011;57(20):1961–1979. doi: 10.1016/j.jacc.2011.02.018. [DOI] [PubMed] [Google Scholar]
- [6].Alsheikh-Ali AA, Kitsios GD, Balk EM, Lau J, Ip S. The vulnerable atherosclerotic plaque: Scope of the literature. Ann. Intern. Med. 2010;153(6):387–395. doi: 10.7326/0003-4819-153-6-201009210-00272. [DOI] [PubMed] [Google Scholar]
- [7].Finn AV, Nakano M, Narula J, Kolodgie FD, Virmani R. Concept of vulnerable/unstable plaque. Arteriosclerosis, Thrombosis, and Vascular Biology. 2010;30(7):1282–1292. doi: 10.1161/ATVBAHA.108.179739. [DOI] [PubMed] [Google Scholar]
- [8].Moreno PR. Vulnerable Plaque: Definition, Diagnosis, and Treatment. Cardiology Clinics. 2010;28(1):1–30. doi: 10.1016/j.ccl.2009.09.008. [DOI] [PubMed] [Google Scholar]
- [9].Lorenz MW, Schaefer C, Steinmetz H, Sitzer M. Is Carotid intima media thickness useful for individual prediction of cardiovascular risk? Ten-year results from the Carotid Atherosclerosis Progression Study (CAPS) Eur. Heart J. 2010;31(16):2041–2048. doi: 10.1093/eurheartj/ehq189. [DOI] [PubMed] [Google Scholar]
- [10].Yoneyama T, Sun J, Hippe DS, Balu N, Xu D, Kerwin WS, Hatsukami TS, Yuan C. In vivo semi-automatic segmentation of multicontrast cardiovascular magnetic resonance for prospective cohort studies on plaque tissue composition: initial experience. Int. J. Cardiovasc. Imaging. 2016;32(1):73–81. doi: 10.1007/s10554-015-0704-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Clarke SE, Beletsky V, Hammond RR, Hegele RA, Rutt BK. Validation of automatically classified magnetic resonance images for carotid plaque compositional analysis. Stroke. 2006;37(1):93–97. doi: 10.1161/01.STR.0000196985.38701.0c. [DOI] [PubMed] [Google Scholar]
- [12].Hofman JMA, Branderhorst WJ, Ten Eikelder HMM, Cappendijk VC, Heeneman S, Kooi ME, Hilbers PAJ, Ter Haar Romeny BM. Quantification of atherosclerotic plaque components using in vivo MRI and supervised classifiers. Magn. Reson. Med. 2006;55(4):790–799. doi: 10.1002/mrm.20828. [DOI] [PubMed] [Google Scholar]
- [13].Liu W, Balu N, Sun J, Zhao X, Chen H, Yuan C, Zhao H, Xu J, Wang G, Kerwin WS. Segmentation of carotid plaque using multicontrast 3D gradient echo MRI. J. Magn. Reson. Imaging. 2012;35(4):812–819. doi: 10.1002/jmri.22886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Van 't Klooster R, Naggara O, Marsico R, Reiber JHC, Meder JF, Van Der Geest RJ, Touzé E, Oppenheim C. Automated versus manual in vivo segmentation of carotid plaque MRI. Am. J. Neuroradiol. 2012;33(8):1621–1627. doi: 10.3174/ajnr.A3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gao S, van 't Klooster R, van Wijk DF, Nederveen AJ, Lelieveldt BPF, van der Geest RJ. Repeatability of in vivo quantification of atherosclerotic carotid artery plaque components by supervised multispectral classification. Magn. Reson. Mater. Physics, Biol. Med. 2015;28(6):535–545. doi: 10.1007/s10334-015-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Van Engelen A, Niessen WJ, Klein S, Groen HC, Verhagen HJM, Wentzel JJ, Van Der Lugt A, De Bruijne M. Atherosclerotic plaque component segmentation in combined carotid MRI and CTA data incorporating class label uncertainty. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0094840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sakellarios AI, Stefanou K, Siogkas P, Tsakanikas VD, Bourantas CV, Athanasiou L, Exarchos TP, Fotiou E, Naka KK, Papafaklis MI, Patterson AJ, Young VEL, Gillard JH, Michalis LK, Fotiadis DI. Novel methodology for 3D reconstruction of carotid arteries and plaque characterization based upon magnetic resonance imaging carotid angiography data. Magn. Reson. Imaging. 2012;30(8):1068–1082. doi: 10.1016/j.mri.2012.03.004. [DOI] [PubMed] [Google Scholar]
- [18].Itskovich VV, Samber DD, Mani V, Aguinaldo JGS, Fallon JT, Tang CY, Fuster V, Fayad ZA. Quantification of human atherosclerotic plaques using spatially enhanced cluster analysis of multicontrast-weighted magnetic resonance images. Magn. Reson. Med. 2004;52(3):515–23. doi: 10.1002/mrm.20154. [DOI] [PubMed] [Google Scholar]
- [19].Fan Z, Yu W, Xie Y, Dong L, Yang L, Wang Z, Conte AH, Bi X, An J, Zhang T, Laub G, Shah PK, Zhang Z, Li D. Multi-contrast atherosclerosis characterization (MATCH) of carotid plaque with a single 5-min scan: technical development and clinical feasibility. J. Cardiovasc. Magn. Reson. 2014;16:53. doi: 10.1186/s12968-014-0053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Crişan S. Carotid ultrasound. Med. Ultrason. 2011;13(4):326–330. [PubMed] [Google Scholar]
- [21].Lal BK, Hobson RW, Pappas PJ, Kubicka R, Hameed M, Chakhtura EY, Jamil Z, Padberg FT, Haser PB, Duran WN. Pixel distribution analysis of B-mode ultrasound scan images predicts histologic features of atherosclerotic carotid plaques. J. Vasc. Surg. 2002;35(6):1210–1217. doi: 10.1067/mva.2002.122888. [DOI] [PubMed] [Google Scholar]
- [22].Urbani MP, Picano E, Parenti G, Mazzarisi a, Fiori L, Paterni M, Pelosi G, Landini L. In vivo radiofrequency-based ultrasonic tissue characterization of the atherosclerotic plaque. Stroke. 1993;24(10):1507–12. doi: 10.1161/01.str.24.10.1507. [DOI] [PubMed] [Google Scholar]
- [23].Hashimoto H, Tagaya M, Niki H, Etani H. Computer-assisted analysis of heterogeneity on B-mode imaging predicts instability of asymptomatic carotid plaque. Cerebrovasc. Dis. 2009;28(4):357–364. doi: 10.1159/000229554. [DOI] [PubMed] [Google Scholar]
- [24].Hashimoto BE. Pitfalls in carotid ultrasound diagnosis. Ultrasound Clin. 2011;6(4):463–476. [Google Scholar]
- [25].Madycki G, Staszkiewicz W, Gabrusiewicz A. Carotid plaque texture analysis can predict the incidence of silent brain infarcts among patients undergoing carotid endarterectomy. Eur. J. Vasc. Endovasc. Surg. 2006;31(4):373–380. doi: 10.1016/j.ejvs.2005.10.010. [DOI] [PubMed] [Google Scholar]
- [26].Pazinato DV, Stein BV, de Almeida WR, de O Werneck R, Junior PRM, Penatti OAB, da S. Torres R, Menezes FH, Rocha A. Pixel-Level Tissue Classification for Ultrasound Images. IEEE J. Biomed. Heal. informatics. 2016;20(1):256–67. doi: 10.1109/JBHI.2014.2386796. [DOI] [PubMed] [Google Scholar]
- [27].Pereira S, Pinto A, Alves V, Silva CA. Brain Tumor Segmentation using Convolutional Neural Networks in MRI Images. IEEE Trans. Med. Imaging. 2016;PP(99):1. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- [28].Zhang W, Li R, Deng H, Wang L, Lin W, Ji S, Shen D. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. Neuroimage. 2015;108:214–224. doi: 10.1016/j.neuroimage.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S. Lung Pattern Classification for Interstitial Lung Diseases Using a Deep Convolutional Neural Network. IEEE Trans. Med. Imaging. 2016;62(c):1–1. doi: 10.1109/TMI.2016.2535865. [DOI] [PubMed] [Google Scholar]
- [30].Ciompi F, de Hoop B, van Riel SJ, Chung K, Scholten ET, Oudkerk M, de Jong PA, Prokop M, van Ginneken B. Automatic classification of pulmonary peri-fissural nodules in computed tomography using an ensemble of 2D views and a convolutional neural network out-of-the-box. Med. Image Anal. 2015;26(1):195–202. doi: 10.1016/j.media.2015.08.001. [DOI] [PubMed] [Google Scholar]
- [31].Albarqouni S, Baur C, Achilles F, Belagiannis V, Demirci S, Navab N. AggNet: Deep Learning from Crowds for Mitosis Detection in Breast Cancer Histology Images. IEEE Trans. Med. Imaging. 2016;35(5):1–1. doi: 10.1109/TMI.2016.2528120. [DOI] [PubMed] [Google Scholar]
- [32].Kallenberg M, Petersen K, Nielsen M, Ng A, Diao P, Igel C, Vachon C, Holland K, Karssemeijer N, Lillholm M. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans. Med. Imaging. 2016;62(c):1–10. doi: 10.1109/TMI.2016.2532122. [DOI] [PubMed] [Google Scholar]
- [33].LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- [34].Krizhevsky A, Sutskever I, Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks. Adv. Neural Inf. Process. Syst. 2012:1–9. [Google Scholar]
- [35].Srivastava N, Hinton GE, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout : A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014;15:1929–1958. [Google Scholar]
- [36].Kingma D, Ba J. Adam: A Method for Stochastic Optimization. Int. Conf. Learn. Represent. 2014:1–13. [Google Scholar]
- [37].Moeskops P, Viergever MA, Mendrik AM, De Vries LS, Benders MJNL, Isgum I. Automatic Segmentation of MR Brain Images with a Convolutional Neural Network. IEEE Trans. Med. Imaging. 2016;35(5):1252–1261. doi: 10.1109/TMI.2016.2548501. [DOI] [PubMed] [Google Scholar]