Abstract
Viral diseases are perpetual threats to human and animal health. Detection and characterization of viral pathogens require accurate, sensitive and rapid diagnostic assays. For field and clinical samples, the sample preparation procedures limit the ultimate performance and utility of the overall virus diagnostic protocols. Here, we presented the development of a microfluidic device embedded with porous silicon nanowire (pSiNW) forest for label-free size-based point-of-care virus capture in a continuous curved flow design. The pSiNW forests with specific inter-wire spacing were synthesized in situ on both bottom and sidewalls of the microchannels in a batch process. With the enhancement effect of Dean flow, we demonstrated ~50% H5N2 avian influenza viruses were physically trapped without device clogging. A unique feature of the device is that captured viruses can be released by inducing self-degradation of the pSiNWs in physiological aqueous environment. About 60% of captured viruses can be released within 24 hours for virus culture, subsequent molecular diagnosis and other virus characterization and analyses. This device performs viable, unbiased and label-free virus isolation and release. It has great potentials for virus discovery, virus isolation and culture, functional studies of virus pathogenicity, transmission, drug screening, and vaccine development.
Keywords: Porous silicon nanowire, point of care, label free, Dean flow, virus capture
1. Introduction
Viruses are infectious agents at sub-micrometer scale, which can infect all types of life forms, including animals, plants and bacteria. The spread of highly pathogenic viral infections caused some of the deadliest pandemics in recorded human history and had significant negative impact on global economy.[1–7] Moreover, high mutation rate and rapid adaptation of viruses, human demographics and behavior, environmental changes, technology and economic development, international travels, and global trades facilitate the rapid spread of viral infectious diseases global.[8, 9] There is an urgent need for the development of new techniques that can rapidly detect viruses and perform surveillance of viral infectious diseases at any location. A number of methods have been developed for virus detection. The viral antigens, nucleic acids and serological antibodies are the core repertoire of techniques used for laboratory diagnosis of viral infection.[10–13] However, in clinical and field applications, the low virus titer and the high level of contaminants from host and environment often severely limit the performance of these detection methods. Thus the enrichment and concentration of viruses from practical samples, or more generally referred as virus sample preparation, become critical in the overall virus diagnosis protocols.
Virus enrichment methods fall into two categories of biological methods and physical methods.[11, 14, 15] Biological methods exploit bioaffinities using antibodies to identify virus surface antigens to isolate viruses, in which the viral antigens need to express on the surface and the antibodies have to be available. On the other hand, physical methods, including differential ultracentrifugation, dielectrophoresis, and filtration, mainly utilize differences in density, electrical affinity, and size between viruses and impurities.[16–20] Physical methods are label-free and can be operated easily and offer excellent flexibility in subsequent molecular analyses and virus culture without affecting virus’s capability in pathogenesis and transmission.[21] Among the available methods, size-based filtration is frequently used.[22–24] As most of viruses have unique spectra of size distribution, ranging from 20 nm to 400 nm (Fig. 1A),[17, 18, 25–28] size-based isolation can differentiate viruses from impurities, such as proteins and protein complexes of 10 nm and below in size, bacteria and mammalian cells of 1 µm and above in size. So it can be used for unknown or unidentified viruses. Nanostructures and nanomaterials like nanowires, graphene are frequently used to control cell capture and release.[29–38] The unique characteristics of nanomaterials, such as large surface-to-volume ratio, lightweight, high tensile strength, biocompatibility, and tunable pore size at nanoscale, are particularly appealing for virus isolation and analysis.[25, 28] In our recent study, we successfully demonstrated that the vertically aligned carbon nanotube (CNT) forests with tunable inter-tubular distance could be integrated into a microfluidic virus isolation device in a dead-end filtration configuration for virus capture.[24] Isolated viruses are available for subsequent molecular analyses, virus culture or other virus characterization and studies. Although the device was successful in isolating emerging viruses from field samples for virus identification, there are still opportunities for new developments, including the small batch fabrication, custom-made nanomaterial synthesis system, and limited sample capacity. Most of these size-based filtration systems, including CNT microdevice mentioned above, are configured as dead-end filtration. Unlike cross-flow filtration, impurities especially large particles can quickly block pores,[39] leading to low sample capacity and slow processing speed. Moreover, as captured viruses are buried firmly inside the stable nanomaterial of the filters, it is difficult to recovery the viruses alive and intactly from the devices for further analyses.
Figure 1.
The design and operation of the pSiNWs forest based device for viral isolation. (A) Size of various biological molecules in the micro and nano scale; (B) Illustrations of pSiNWs forest based microfluidic device showing the overall microfluidic design in 3D, the location of the pSiNWs forests (top inset, cross-sectional view), and nanoparticles captured inside the pSiNWs forest (bottom inset); (C) Photo of the fabricated device.
In this report, we describe a new continuous flow microfluidic point-of-care (POC) device integrated with pSiNWs forest by in situ synthesis for high efficient virus isolation and release. As a nanoscale material with unique properties[40–43] and a variety of morphologies and tunable fabrication methods,[44–47] silicon nanowires have been studied extensively for their applications in electronics,[48, 49] photonics,[49–51] energy generation and storage,[52–54] and biomedicine.[55–57] In virology, silicon nanowires have been used in field effect transistors for label-free viral detection.[25, 58, 59] However, to the best of our knowledge, they have never been explored for viral isolation. The device adopts a continuously flow design similar to cross-flow filtration configuration to achieve large sample capacity and high processing speed. With judiciously optimized the curvature and dimensions of channels, our results show that approximately 50% of influenza viruses can be isolated in 30 minutes with clinical relevant sample volume from microliters to milliliters. Moreover, our pSiNWs forest is biodegradable in physiological conditions attributed to the extensive porous surfaces,[60, 61] enabling the release and harvest of trapped viruses in 24 hours for further virus culture and molecular analysis. Together, combined with the portability, high isolation efficiency, large sample capacity, and unique viral release mechanism, this POC devices can provide much faster access to results at or near the sites of the patient care, discover unknown or emerging virus, and monitor outbreaks of viral infectious diseases.
2. Results and Discussion
2.1. Design of the porous silicon nanowires forest-embedded microfluidic device
Overall the microfluidic device adopts a continuous flow design and the virus capture is configured similar to cross-flow filtration. The pSiNWs forests used to capture virus were on bottom and sidewalls, perpendicular to the flow direction. Meanwhile, the channel curvature introduces circular flow pattern within the transverse plane of the channel,[62–64] which spontaneously brings viruses and other sub-micrometer particles to the pSiNWs forest on the bottom and sidewalls (Fig. 1B). The pSiNWs forest with well-controlled inter-wire space was prepared by metal-assisted wet etching within the curved channels (Fig. S1–S2). Viruses within a certain size range can enter the gaps between pSiNWs and physically trapped inside the forest. Meanwhile, larger impurities can be directly excluded, while smaller ones may escape from the pSiNWs forest during continuous fluidic flow of sample loading and the subsequent washing step. Moreover, this design allows uncaptured particles removed from the outlet quickly, minimizing pore blockage by impurities.
2.2. Tuning the inter-wire spacing of the pSiNWs forest for size-based capture of sub-micrometer particle
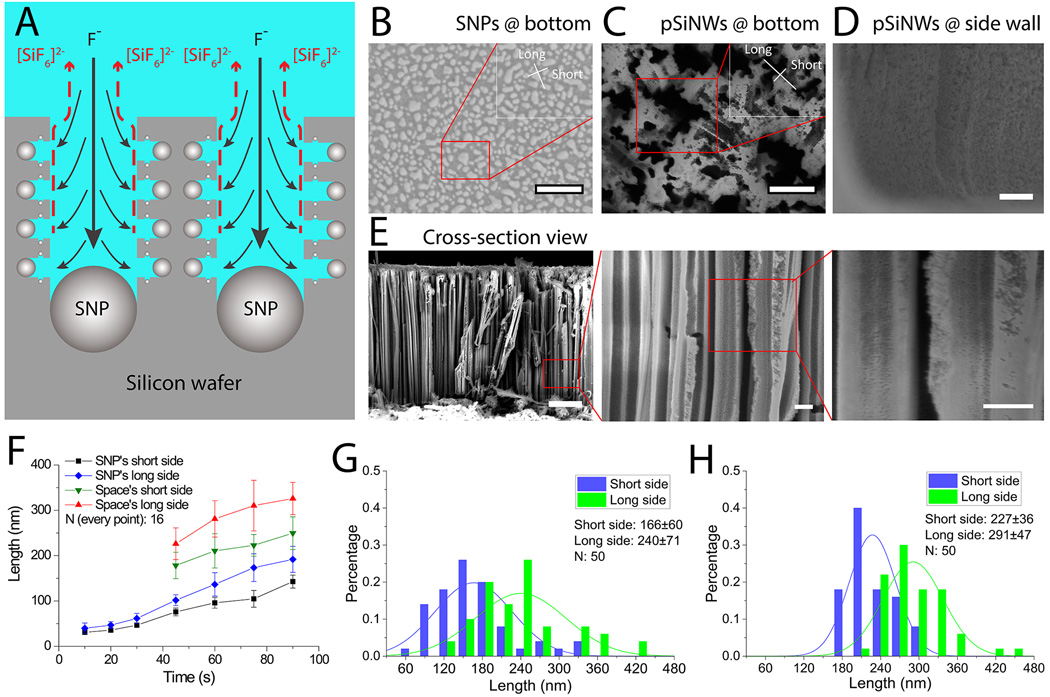
For label-free size-based isolation of nanoparticles in the pSiNWs continuous flow microfluidic device, the inter-wire spacing between pSiNWs needs to be slightly larger than the nanoparticles to allow them trapped inside the pSiNWs forest. Therefore, the ideal inter-wire spacing was determined first. In metal-assist etching, silver nanoparticles (SNPs) serve as the catalyst and only silicon beneath the SNPs are effectively etched (Fig. 2A), thus the size of SNPs after deposition directly determines the inter-wire spacing (Fig. 2B–D) and the diffusion of small SNPs determines the size of the mesopores on a single nanowire (Fig. 2E). By adjusting the SNP deposition time from 45 s to 90 s, We found that the average size of the SNPs clusters increases from approximately 75 nm × 100 nm to 140 nm × 190 nm (Fig. 2B and 2F). Accordingly, after wet etching the average size of inter-wire spacing increased from about 180 nm × 230 nm to 250 nm × 330 nm (Fig. 2C and 2F). As the size range of influenza viruses used in this study is approximately from 80 to 120 nm,[65] we need to generate the inter-wire spacing slightly larger than 120 nm, which would require 45–60 s for deposition of SNPs (Fig. 2F). To determine the ideal deposition time, 50 locations were randomly selected to measure the inter-wire spacing for pSiNW synthesis after 45 and 60 seconds SNP deposition (Fig. 2G and 2H). It clearly shows that over 30% of the inter-wire spacing is smaller than 120 nm if the deposition time is reduced to 45 s. In contrast, with 60 s deposition, almost all of inter-wire spacing is larger than the threshold value of 120 nm (Fig. 2H), and the average inter-wire spacing is about 227 nm × 291 nm (Fig. 2G). Such inter-wire spacing distribution would allow viruses and other sub-micrometer particles around 100 nm in size to enter into pSiNWs forest and trap them within the forest. On the other hand, as less than 10% of pores (short side) are larger than 300 nm, particles in micrometer size range, such as bacteria, eukaryote cells and large cell organelles (e.g. nuclei, mitochondria, chloroplasts), are unable to penetrate the relatively narrow gap and enter into pSiNWs forest; while smaller nanoparticles and macrobiomolecules, e.g. serum proteins, protein complexes and ribosomes, are much smaller and can enter and escape from the pSiNWs forest under continuous flow. Thus, deposition time of 60 seconds was used to prepare pSiNWs forest to isolate influenza virus.
Figure 2.
Synthesis and characterization of pSiNWs. (A) Sketch of synthesizing pSiNWs showing larger SNPs defining the inter-wire spacing between silicon nanowires and tiny SNPs generating porous structures on individual silicon nanowire. Black solid arrows indicate reactant fluoride ions coming to the surface. Red dash lines indicate product silicon hexafluoriode anions leaving the surface into the bulk. (B) Scanning Electron Microscope (SEM) of SNPs formed on the bottom of the channel (bar: 500 nm, insert bar: 200 nm). (C) SEM images of pSiNWs on channel bottom (scale bars: 500 nm, 200 nm for the inset). (D) SEM images of pSiNWs formed on the channel sidewall (scale bar: 2 µm). (E) SEM images of cross-section view of the pSiNWs (scale bars: 2 µm, 500 nm for the two insets). (F) Sizes of SNPs and inter-wire spacing of pSiNWs forests versus the SNP deposition time. (G) & (H) Distribution of the inter-wire spacing of pSiNWs with silver nanoparticle deposition time of 45 s (G) and 60 s (H), respectively.
2.3. Optimization of the microchannel geometry and flow rate for efficient capture of sub-micrometer particles
In our design of the curved channel geometry, the channel curvature amplifies the lateral instability that drives the Dean flow, which is a secondary flow field in the transverse plane. When the channel height is comparable to the channel width, the secondary flow field in the transverse plan is characterized by the presence of two main counter-rotating vortices located above and below the horizontal plane of symmetry of the channel.[66] Inertial focusing of spherical microparticles with diameters ranging between 5 and 20 µm has demonstrated the promise of efficient separation of micrometer-sized particles at relatively high Reynolds number for increased throughput.[62–64] To date, the Dean flow-based technique has not been reported for nanoparticles. We tested the idea of employing the Dean flow to improve particle capture and separation at the sub-micrometer scale.
Our idea of employing Dean flow for sub-micrometer particle separation is different from those of microparticle separation. In separation of particles in the micrometer scale, microparticles obtain their balanced locations at the transverse plane and are thus separated. For efficient virus capture by the pSiNWs on the microchannel surface, the viruses (~100 nm in size) inside the flow need to be brought to the pSiNWs surface. In our design, we achieve this goal by designing microchannel curvature and adjusting flow rate to generate vortices in the transverse plane of the flow. Moreover, in the flow, the Dean force pushing virus particles toward the microchannel walls is expected to be larger than the wall lift force in the opposite direction by several orders of magnitude (Fig. S3). Thus it can guide viruses into the inter-wise spacing of the pSiNWs forest, and make them trapped inside.
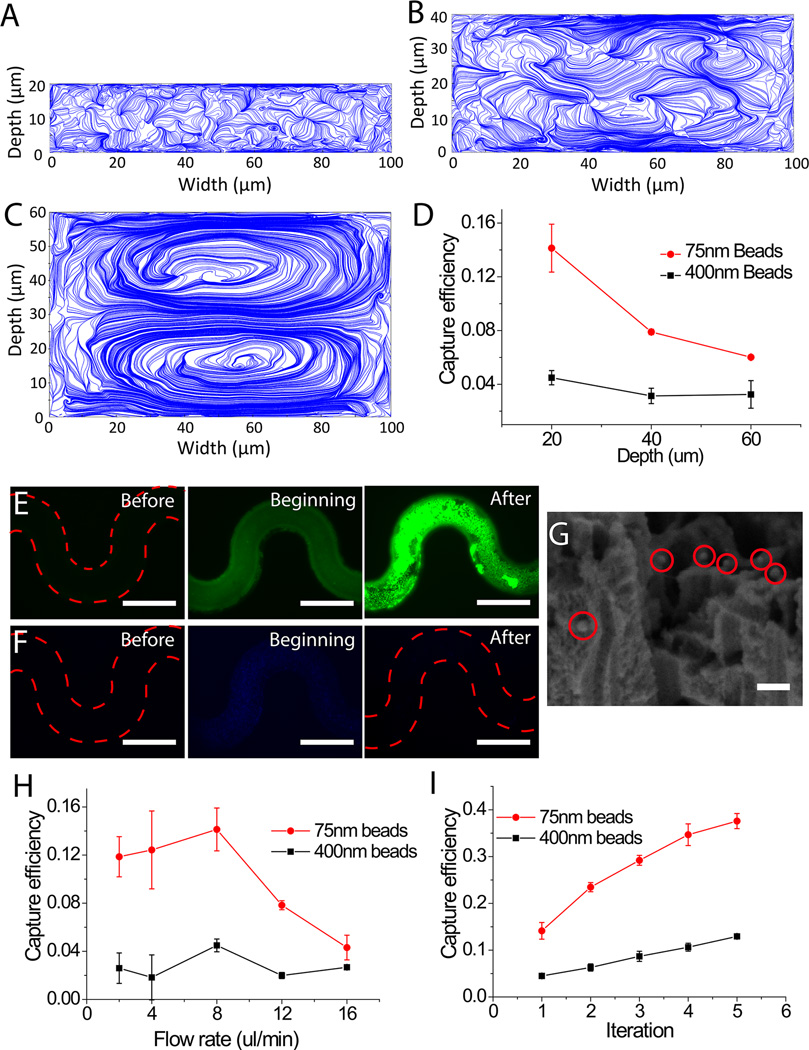
Given the curvature of channels is fixed in photomask design, the vortices in the transverse plane could be optimized by changing the channel height during device fabrication and flow rate during device operation. To study the effect of channel height on the Dean flow in the transverse plane, a finite element method (FEM) model was established to simulate the fluidic field inside the channel. We found the intensity (Fig. S4–S6) and position of local vortices (Fig. 3A–3C) changed in the three designs of different channel heights of 20 µm, 40 µm and 60 µm. Relatively more local vortices near the channel wall were observed in the curved channels of 20 µm height in comparison with those in the rest two groups, and thus we conclude more viruses would be brought into the pSiNWs forest in channels with relatively low channel height. Further, we experimentally investigated the height effect on capture efficiency using fluorescent nanobeads. Green or blue fluorescent nanobeads of 75 nm and 400 nm in size mimic the influenza virus particles and larger impurities in body fluid, respectively. In consistent with the design principle and simulation results, when the flow rate was kept at constant 8 µl/min and the channel height decreased from 60 µm to 20 µm, the capture efficiency of 75 nm nanobeads increased from 6.4% to 14.1%, while that of the 400 nm nanobeads was approximately 4% in all groups (Fig. 3D). Thus, the channels with 20 µm height were prepared for the following optimization.
Figure 3.
Capture of nanoscale particles inside the pSiNWs microfluidic devices with the effect of the Dean flow. (A)–(C) FEM simulations showing velocity field on the cross-sectional planes of meandering flow channels with channel depths of 20, 40 and 60 µm, respectively. 0 and 100 µm point the outer and inner rim of channel, respectively. (D) Capture efficiency of 75 nm and 400 nm nanobeads in channels with 20, 40 and 60 µm height, respectively. (E) Capture of 75 nm green nanobeads showing top views of the pSiNWs flow channels before, at the beginning, and after the introduction of the nanobeads (bar: 200 µm); (F) Capture of 400 nm blue nanobeads showing top views of the pSiNWs flow channels before, at the beginning and after the introduction of the nanobeads (bar: 200 µm); (G) SEM image of captured 75 nm nanobeads (bar: 150 nm). (H) Capture efficiency of 75 nm and 400 nm nanobeads under different flow rates. (I) Capture efficiency of 75 nm and 400 nm nanobeads versus number of run times of the same sample in the same device.
In addition to channel height, flow rate can also affect the capture efficiency of viruses. In general, high flow rate is preferred as it can shorten sample-processing time and increase sample capacity. In addition, to generate strong local vortices in the transverse plane, relatively high flow velocity is needed. However, our designed extremely high flow rate limit the interaction time between the nanoparticles and the pSiNWs, resulting in lower capture efficiency. To study the effect of flow rate on the capture efficiency, we tested flow rates ranging from 2 µl/min to 16 µl/min using 75 nm and 400 nm nanobeads, respectively. As shown in Figure 3H, the capture efficiency of 75 nm nanobeads reaches the maximal value of 14.1% at 8 µl/min. In the regime of lower flow rate, capture efficiency of 75 nm nanobeads is gradually improved by increasing flow rate. This can be explained by strengthening the local vortices by increasing the flow rate, which effectively brings nanobeads to the surface of the pSiNWs forest. However, once the flow rate reaches or beyond 12 µl/min the capture efficiency of 75 nm nanobeads decreases significantly. We believe that at this high flow rate regime, nanobeads might not have sufficient time to interact with the pSiNWs forest and are flushed out of the device. In the control group using 400 nm nanobeads, which are expected to be barely captured inside the pSiNWs forest based on their size compared with the inter-wire spacing of the pSiNWs forest, the capture efficiency slightly fluctuates between 2% and 4.5% (Fig. 3H). Thus flow rate of 8 µl/min is used as optimized flow rate of the pSiNW microdevice for capture of sub-micrometer particles.
2.4. Study the size selectivity of the pSiNW microdevice
To study the capture selectivity, 40 µl suspension of mixed nanobeads of 75 nm and 400 nm in diameter were introduced into the pSiNW device with 20 µm channel height at flow rate of 8 µl/min. As the inter-wire spacing of the pSiNWs was approximately 227 nm × 291 nm, we expected 75 nm nanobeads (mimicking viruses) would be trapped, while 400 nm nanobeads (mimicking large impurities) would be excluded. After thoroughly rinsing with DI water, the green fluorescence derived from the 75 nm nanobeads inside the microfluidic channel was more than 3 times stronger than that of the original beads suspension in pSiNW device at the beginning of the experiment (Fig. 3E and Fig. S7A), and the capture efficiency was ~14.1%, indicating that 75 nm nanobeads were efficiently captured and enriched inside the pSiNWs. On the contrary, the blue fluorescence from the 400 nm nanobeads was extremely weak (~1/8 of the original beads suspension) and very close to the pSiNW microchannel background after capture and rinse (Fig. 3F and Fig. S7B). The capture efficiency was ~4.5%, suggesting the pSiNW forests could barely capture the 400 nm nanobeads. The SEM image further validated that many 75 nm nanobeads and very few 400 nm nanobeads were captured inside the pSiNW forests (Fig. 3G, nanobeads labeled by red circles). These experimental results obtained in our study support that the pSiNW forest with ~250 nm inter-wires spacing in the curved microchannels of 20 µm channel height can effectively and specifically isolate and enrich 75 nm nanobeads, but reject nanobeads of 400 nm in diameter. So by choosing the right inter-wire space of pSiNWs, this device can capture particles with smaller size and exclude larger impurities.
2.5. Improvement of capture efficiency by cyclic iteration
In the cyclic iteration mode of sample introduction, the same sample run for multiple times inside the same device to increase the capture efficiency. This strategy has been used previously to isolate rare molecules.[67] As shown in Fig. 3D, after a single run of the 75 nm green fluorescent nanobeads in the curved channels of 20 µm channel height, 14.1% capture efficiency can be achieved. However, more careful observation found the fluorescence distribution was random and non-uniform (Fig. 3D), and there were still many empty pores and gaps beneath nanowires’ top surface (Fig. 3G), indicating there was still considerable empty spacing inside the pSiNW forest for trapping 75 nm nanobeads. Hence, we hypothesized that the capture efficiency of 75 nm nanobeads could be significantly improved by re-introducing the sample into the same device as a cyclic iteration operation. In the following experiment, we recollected the nanobeads suspension from the device outlet and re-introduced them into the same devices for up to 5 times. The capture efficiency of 75 nm nanobeads significantly increased from 14.1% to 37.6 % after the five iterations. In contrast, the capture efficiency of 400 nm nanobeads was increased from 4.5% to 12.9 % (Fig. 3I). After five iterations, the pSiNW device still captures much more 75 nm nanobeads than 400 nm nanobeads. However, the ratio of the captured beads of 75 nm in diameter to those of 400 nm maintains roughly the same of ~3, suggesting the cyclic iteration runs do not change the size selectivity of the pSiNW device. Although cyclic iteration cannot increase the purity of captured nanobeads, it can further enrich the targets by ~2.7 times. It is important especially when the concentration or quantity of the target in the original samples is low.
2.6. Degradation of the pSiNW forests inside the microdevice
Although sub-micrometer particles like viruses can be trapped and enriched in the device, without releasing and collecting, they cannot be used for either further characterization (e.g. virus isolation/culture, genomic sequencing) or functional studies (e.g. virus pathogenicity, transmission, drug screening and vaccine development). Different from solid silicon nanowires, pSiNWs have been demonstrated to be degradable in various aqueous solutions including phosphate buffered saline (PBS) due to its mesopores at nanoscale (Equation 1):[60]
| (Equation 1) |
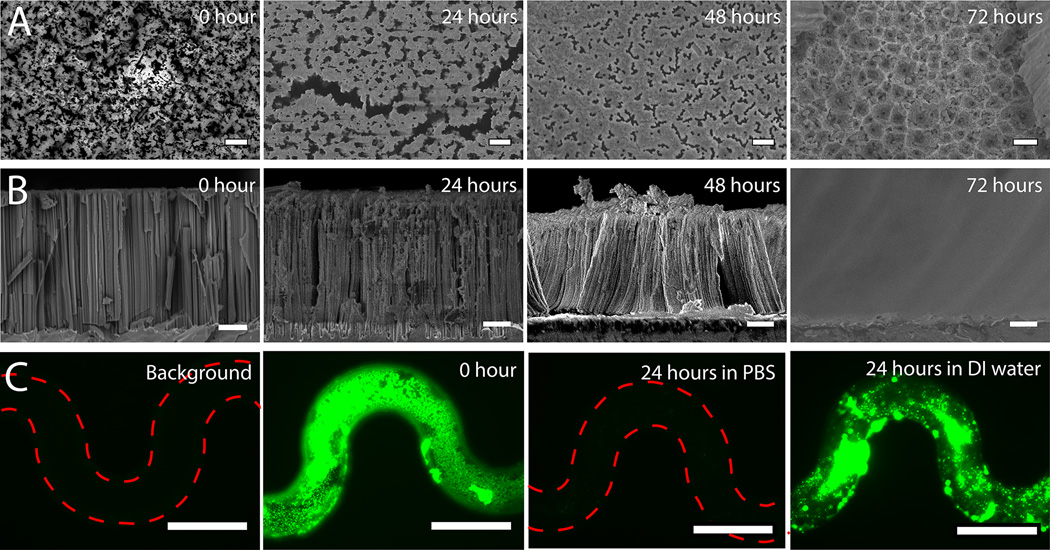
However, they cannot be simply degraded in deionized (DI) water.[60] The degradation of pSiNWs will allow captured particles to escape from the pSiNW forest. We tested in situ degradation of the pSiNWs by flowing 1× PBS continuously through the pSiNW integrated microfluidic device for 24, 48 and 72 hours at RT, respectively. The appearance of pSiNW forest at each time point was imaged by SEM in Figure 4A and 4B. The images reveal that the degree of degradation is time-dependent, and the pSiNW forest can totally degrade within 72 hours.
Figure 4.
Degradation of pSiNWs s and release of captured nanobeads. (A) SEM images of pSiNWs s from the top view after soaking in PBS for 0, 24, 48 and 72 hours (bar: 1 µm). (B) SEM images of pSiNWs from the side view after soaking in PBS for 0, 24, 48 and 72 hours (bar: 2 µm). (C) Releasing nanobeads by degrading pSiNWs forests for 24 hours in PBS (bar: 200 µm).
The feasibility of releasing captured 75 nm nanobeads was further tested. As captured nanobeads will not be buried too deep in pSiNW forest, degradation time of 24 hours is initially chosen to release them as longer degradation time might decrease the viability of viruses. After continuous perfusion with 1× PBS for 24 hours under flow rate of 4–15 µl/hr, the green fluorescence (from the 75 nm nanobeads) was only ~2% of that before degradation within the location of the microchannels (Fig. 4C and Fig. S8A), indicating trapped 75 nm nanobeads were released from forest. Extended perfusion would not significantly improve the release efficiency since only ~2% of captured beads were left after 24-hour perfusion, thus 24 hours are long enough for releasing captured particles. However, as expected, after perfusion with DI water, the green fluorescence within the microchannel remains ~85% compared with that before perfusion (Fig. 5C and Fig. S8A), indicating the pSiNW forest did not degrade in DI water and only a small percentage of nanobeads could be released by extensive perfusion only. Compared with the flow rate during the capture of sub-micrometer particles, the flow rate during particle release was much lower for two purposes. First, it limits the total volume after particle release, thus particle concentration could be maintained at relatively high level to facilitate their detection. Second, the slow flow rate induces small shear stress on the particles, about one order of magnitude lower than the shear stress in arterioles and capillaries,[68] which is expected to maintain the integrity of the fragile biological particles (e.g. viruses) and not affect their viability.
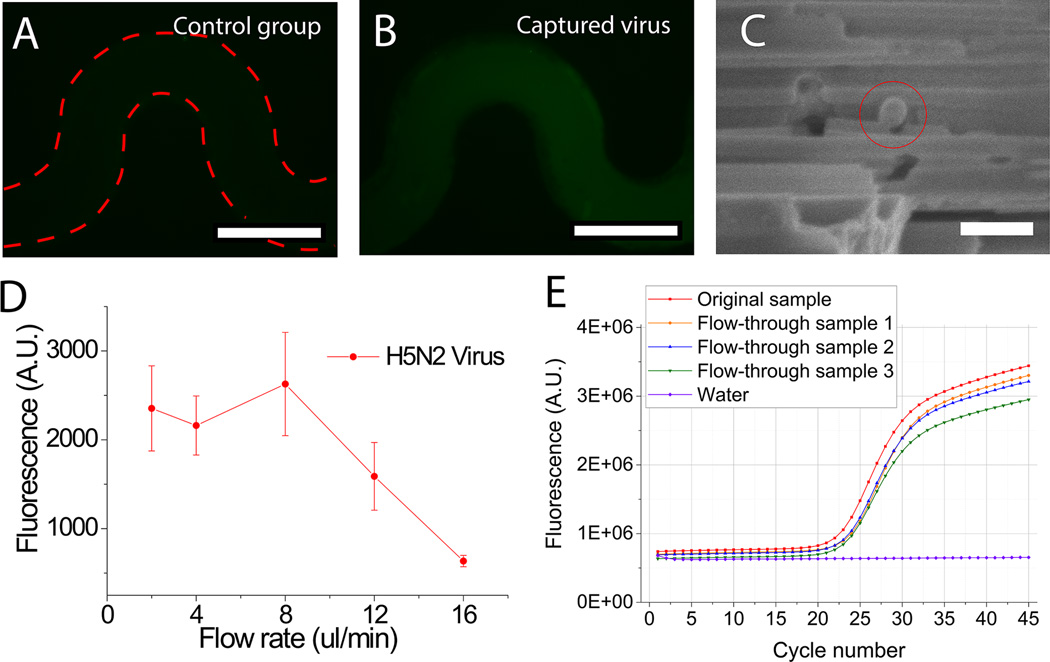
Figure 5.
Capture of viruses inside the pSiNW microfluidic devices. (A) Control group using PBS without viruses (scale bar: 200 µm). (B) Capture of virus-containing sample by the pSiNW device followed by in situ immunofluorescent staining with antibody against H5 antigen on the H5N2 AIV surface (scale bar: 200 µm). (C) SEM image of a captured virus (scale bar: 150 nm). (D) The fluorescence intensity of the microchannels after in situ immunofluorescent staining versus the flow rate during virus capture. (E) RT-qPCR profiles of the original sample (only one is shown here) and the flow-through of three separate virus-containing samples.
2.7. Capture of influenza virus using pSiNW forest embedded microfluidic device
After optimizing operation parameters using nanobeads, the pSiNW forest embedded microfluidic device was used to capture H5N2 avian influenza virus (AIV). For the experimental group, 40 µl of H5N2 AIV suspension with hemagglutination (HA) titer of 1:512 was introduced into the device under various flow rates ranging from 2 µl/min to 16 µl/min with 3 cyclic iterations. The captured viruses were then detected by indirect in situ immunofluorescence assay using H5 specific monoclonal antibody (mAb) to react with the H5N2 AIV first. We measured the fluorescence intensity after the in situ immunofluorescence assay and found that the fluorescence from the device processed with the virus-containing sample was approximately 5.7 times higher than that in negative control group that contained no viruses (Fig. 5A–5B, Fig. S8B). SEM images also demonstrated H5N2 AIVs were captured inside the pSiNW forest (Fig. 5C and Fig. S9). Similar to the measurement of capture efficiency of the 75 nm nanobeads, the highest capture efficiency of H5N2 AIV was also achieved at 8 µl/min (Fig. 5D). Since most POC applications for virus diagnosis use sample volume in the range of 1–100 µl,[11, 14, 69–73] the high flow rate for optimal capture efficiency translates to a processing time of less 13 minutes for a 100 µl sample in a single pass of a single device. If we scale-up the device size to standard 1 cm × 1 cm, we will be able to process a 100 µl sample in ~2.5 minutes for a single run. The time-scale of pSiNW degradation in 1× PBS solution is in hours to days, while the whole capture process takes only 30 min, therefore it is safe to assume the pSiNW forest structure will be stable during the capture process.[61, 74]
To further quantitatively determine the capture efficiency of H5N2 AIV, we collected the flow-through of each sample, measured the virus concentration inside the flow-through using quantitative reverse transcription PCR (RT-qPCR) and compared it with the original sample (Fig, 5E). Based on the Ct value of the flow-through of each group, we determined virus capture efficiency using our device as 48±4% (Table 1).
Table 1.
Capture efficiency of the pSiNW device for H5N2 AIV measured by RT-qPCR
| H5N2 AIV | Ct value |
|---|---|
| Original sample 1 | 19.58 |
| Original sample 2 | 19.25 |
| Original sample 3 | 19.50 |
| Flow-through sample 1 | 20.47 |
| Flow-through sample 2 | 20.25 |
| Flow-through sample 3 | 20.40 |
2.8. Release of influenza virus captured in the pSiNW forest embedded microfluidic device
To lease the captured H5N2 AIV, the microchannel was continuously perfused with 1× PBS for 24 hours after immunofluorescence staining. The green fluorescence intensity of the pSiNW microchannel was only ~6% after virus release (Fig. S8B). While when perfused with DI water, the green fluorescence intensity only decreased to ~65% after 24 hours (Fig. 6A and Fig. S8B). This result indicated that captured viruses were released by degradation of pSiNWs forest with PBS and flowed out in the PBS flow. In contrast, simply perfusing using DI water, which has insignificant effect on degradation of pSiNWs, is inefficient to directly release trapped viruses from the pSiNW forest.
Figure 6.
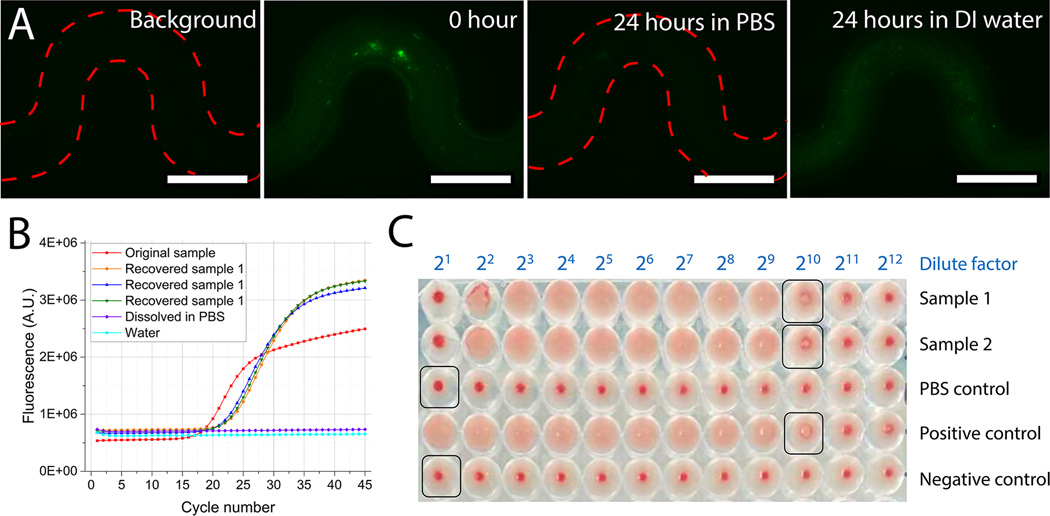
Release, recovery and propagation of captured virus. (A) Releasing virus by degrading pSiNW forests for 24 hours in PBS (scale bar: 200 µm). (B) RT-qPCR profiles of the original sample and recovered samples to measure the recovery efficiency of virus release. “Dissolved in PBS” is a negative control of pSiNW degradation in PBS solution without viruses. “Water” is a negative control of RT-qPCR using DI water as sample surrogate. (C) HA tests on two samples (Sample 1 and Sample 2) after virus release and propagation in embryonated chicken eggs. “PBS control” is a negative control sample of pSiNW degradation in PBS solution without viruses. “Positive control” is a virus suspension with a titer of 1:29. “Negative control” is DI water.
The released viruses were collected for subsequent identification and culture. First, a sandwich enzyme-linked immunosorbent assay (ELISA) was performed on the surface of magnetic beads for released virus identification (Fig. S10). The red fluorescence intensity in the experimental group was more than twice higher than that of the negative control group containing pSiNW degradation solution only without any viruses (Fig. S10D), indicating released viruses can be successfully collected for identification tests.
To quantify the virus recovery efficiency, we measured the virus concentration in the releasing buffer by using RT-qPCR (Fig 6B, and Table 2)., The overall recovery efficiency in Ct values was 29±7% in virus-releasing solutions. The range of the solution volume spans from 60 µl to 1000 µl. An estimated amount of ~60% of the captured viruses could be released and collected from the pSiNW microdevice in considering the capture efficiency.
Table 2.
The release and recovery of captured H5N2 AIV from the pSiNW microdevice
| Initial volume | Diluted volume for RT- qPCR |
CT value of RT-qPCR |
Overall recovery efficiency |
|
|---|---|---|---|---|
| Original sample A | 120 µl | 120 µl | 15.92 | - |
| Recovered sample A1 | 60 µl | 60 µl | 17.43 | 17.6% |
| Recovered sample A2 | 60 µl | 60 µl | 16.33 | 37.6% |
| Recovered sample A3 | 60 µl | 60 µl | 16.61 | 31.0% |
| Original sample B | 40 µl | 40 µl | 15.92 | - |
| Recovered sample B | 400 µl | 400 µl | 21.6 | 29.2% |
| Original sample B | 40 µl | 250 µl | 19.40 | - |
| Recovered sample B | 1000 µl | 1000 µl | 23.55 | 28.2% |
In recovery sample A, the Ct value was 16.8±0.6, corresponding to an expected virus titer of ~6×106 EID50/ml (EID50 = Embryo Infectious Dose 50%). Such high virus concentration would satisfy the sample requirement for virus cultivation if the viruses were alive. To test the viability of the released viruses and the feasibility of virus culture after enrichment by the pSiNW microdevice, we inoculated the released viruses into embryonated chicken eggs for virus cultivation, and used HA test to measure the virus presence and concentration in harvested allantoic fluid (AF) after 72 hour incubation. The virus HA titer in the harvested AF sample was measured as 1:29 HA titer (Fig. 6C). Additionally, we also tested the AF sample in 2-fold serial dilutions by RT-qPCR assay. The measured Ct value of the AF sample at 1:210 dilution was 16.45, which is equivalent to virus concentration of ~4×106 EID50/ml. These findings demonstrated that the captured viruses remained viable during the entire capture and release processing in pSiNW-embedded microdevice, the viable viruses provide great potentials for subsequent virus propagation, pathogenicity and other characterization or vaccine development studies.
3. Conclusion
We developed a pSiNW forest embedded microfluidic POC device for label-free capture and release of viruses. We demonstrate that particles at sub-micrometer scale can be physically trapped inside the inter-wire spacing of pSiNW forest. The inter-wire spacing of the pSiNW is tunable during device fabrication. The microfluidic design with curved microchannels enhances Dean flow in the transverse plane of the sample flow. Approximately 50% of viruses can be physically captured in the pSiNW forest after 3 iterative cycles. The continuous flow and curved channel design enable the device to peak its virus capture efficiency at a relatively high sample flow rate of 8 µl/min for a single device of six parallel microfluidic channels. Viruses can be released through the degradation of the pSiNWs forest in 24 hours and remain viable. The release efficiency is ~60% and the overall recovery efficiency including virus capture and release is ~29%. Moreover, we also demonstrated the released H5N2 AIV could be lysed for RT-qPCR and cultured in embryonated chicken eggs. With this POC device, virus with specific size could be isolated from 100 µl in 30 minutes with triple passes and recovered by degrading pSiNWs in PBS for another 24 hours. This high sample processing capability, simple operation and facile integration with other virus analysis methods make this device suitable for real clinical and field applications.
4. Experimental Section
Design of pSiNWs forest-embedded microfluidic devices
The complete pSiNW microdevice has one inlet, one outlet and six parallel curved microfluidic channels. Each microfluidic channel is curved and 9.5 mm in length, 100 µm in width and 20–60 µm in height (Fig. 1B). In our device, the pSiNWs forest bearing inter-wire spacing was created on both sidewalls and bottom of the flow channels (Fig. 2C & 2D).
Fabrication of pSiNWs forest-embedded microfluidic devices
The fabrication process is illustrated in Fig. S1. Channels are etched on the silicon wafer by the standard photolithography and deep reactive ion etching (DRIE). Then, pSiNWs are etched on the bottom and sidewalls of the channel by two steps (Equation S1 & S2): SNP deposition and pSiNW etching. SNPs are deposited in the solution of 0.085% sliver nitrate and 9.8% HF. pSiNWs are etched in the etchant containing 0.35% hydrogen peroxide and 9.8% HF. Finally, after removing SNPs, the channels are covered with Polydimethylsiloxane (PDMS).
Simulation of flows in the channels
A FEM model was established to simulate the fluidic field inside the device by the software COMSOL Multiphysics. Laminar flow stationary study was used. In the inlet of the channel, the flow rate was 1.33 µl/min for a single channel. The results of velocity field in the transverse plane (Fig. S3–S5) were plotted to visualize the flow.
Measurement of virus capture efficiency using fluorescence nanobeads
Virus capture efficiency with different channel geometry was measured by comparing the fluorescence intensities of the nanobeads suspension before they were introduced into channels and after they were gathered in the outlet, respectively (Equation S3). The fluorescence intensities were measured by a microplate reader.
Measurement and optimization of capture and recovery efficiency for H5N2 AIV
The effect of flow rate on capture efficiency of H5N2 AIV was optimized first. During optimization, the capture efficiency was measured by the fluorescence intensity in the channels with immunostaining of viruses. To quantify capture and recovery efficiency of H5N2 AIV, the virus concentrations in the original suspension, the flow-through and recovery solution were measured using RT-qPCR. The cycle threshold (Ct) value of each sample was calculated to measure the capture (Equation S4) and recovery efficiency (Equation S5)
Propagation in embryonated chicken egg and hemagglutination (HA) assay
released H5N2 AIV suspension (200 µl) was inoculated in 10 day old special-pathogen free embryonated chicken egg for virus propagation. The inoculated eggs were placed inside an incubator for 72 hours and then were removed for virus harvest. The HA assay using 0.5% chicken RBCs[75] was conducted to detect the presence of H5N2 AIV and measure the virus titer. The propagated virus in AF suspension was made 2-fold serial dilutions for the HA test in 96-well microplate.
Detailed methods are in the supporting material.
Supplementary Material
Acknowledgments
The authors appreciate assistance and help from the Penn State Material Research Institute (MRI), the Penn State Huck Institutes of the Life Sciences and Mr. Gene Gerber of Biomedical Engineering department. We acknowledge the financial support from the National Institutes of Health under Award Number DP2CA174508.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Reference
- 1.Fauci AS, Morens DM. N. Engl. J. Med. 2012;366:454. doi: 10.1056/NEJMra1108296. [DOI] [PubMed] [Google Scholar]
- 2.Binder S, Levitt AM, Sacks JJ, Hughes JM. Science. 1999;284:1311. doi: 10.1126/science.284.5418.1311. [DOI] [PubMed] [Google Scholar]
- 3.Morens DM, Fauci AS. PLoS Pathog. 2013;9:e1003467. doi: 10.1371/journal.ppat.1003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lederberg J, Hamburg MA, Smolinski MS. Microbial Threats to Health:: Emergence, Detection, and Response. National Academies Press; 2003. [PubMed] [Google Scholar]
- 5.Organization, W.H. Situation summary. 2015:3.
- 6.Kanaji N, Bandoh S, Ishii T, Fujita J, Ishida T, Matsunaga T, Kubo A. Oncol. Rep. 2011;26:763. doi: 10.3892/or.2011.1357. [DOI] [PubMed] [Google Scholar]
- 7.Morens DM, Fauci AS. J. Infect. Dis. 2007;195:1018. doi: 10.1086/511989. [DOI] [PubMed] [Google Scholar]
- 8.Khan K, Arino J, Hu W, Raposo P, Sears J, Calderon F, Heidebrecht C, Macdonald M, Liauw J, Chan A, Gardam M. N. Engl. J. Med. 2009;361:212. doi: 10.1056/NEJMc0904559. [DOI] [PubMed] [Google Scholar]
- 9.Wilson ME. J. Appl. Microbiol. 2003;94:1. [Google Scholar]
- 10.Herring AJ, Inglis NF, Ojeh CK, Snodgrass DR, Menzies JD. J. Clin. Microbiol. 1982;16:473. doi: 10.1128/jcm.16.3.473-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y-F, Lien K-Y, Lei H-Y, Lee G-B. Biosens. Bioelectron. 2009;25:745. doi: 10.1016/j.bios.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leland DS, Ginocchio CC. Clin. Microbiol. Rev. 2007;20:49. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeh Y-T, Nisic M, Yu X, Xia Y, Zheng S-Y. Ann. Biomed. Eng. 2014;42:2333. doi: 10.1007/s10439-014-1044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y-G, Moon S, Kuritzkes DR, Demirci U. Biosens. Bioelectron. 2009;25:253. doi: 10.1016/j.bios.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tam PD, Van Hieu N, Chien ND, Le A-T, Anh Tuan M. J. Immunol. Methods. 2009;350:118. doi: 10.1016/j.jim.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Reimer CB, Baker RS, Newlin TE, Havens ML. Science. 1966;152:1379. doi: 10.1126/science.152.3727.1379. [DOI] [PubMed] [Google Scholar]
- 17.Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS, Gorcyca D, Chladek D. J. Vet. Diagn. Invest. 1992;4:117. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- 18.Benfield DA, Nelson E, Collins JE, Harris L, Goyal SM, Robison D, Christianson WT, Morrison RB, Gorcyca D, Chladek D. J. Vet. Diagn. Invest. 1992;4:127. doi: 10.1177/104063879200400202. [DOI] [PubMed] [Google Scholar]
- 19.Morgan H, Hughes MP, Green NG. Biophys. J. 1999;77:516. doi: 10.1016/S0006-3495(99)76908-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green NG, Morgan H, Milner JJ. J. Biochem. Biophys. Methods. 1997;35:89. doi: 10.1016/s0165-022x(97)00033-x. [DOI] [PubMed] [Google Scholar]
- 21.Klimpel GR. Medical Microbiology. 4th. Chapter 50. Galveston Texas: University of Texas Medical Branch at Galveston; 1996. [Google Scholar]
- 22.Brady-Estévez AS, Kang S, Elimelech M. Small. 2008;4:481. doi: 10.1002/smll.200700863. [DOI] [PubMed] [Google Scholar]
- 23.Yang SY, Ryu I, Kim HY, Kim JK, Jang SK, Russell TP. Adv. Mater. 2006;18:709. [Google Scholar]
- 24.Yeh Y-T, Tang Y, Sebastian A, Dasgupta A, Perea-Lopez N, Albert I, Lu H, Terrones M, Zheng S-Y. Sci. Adv. 2016;2:e1601026. doi: 10.1126/sciadv.1601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patolsky F, Zheng G, Lieber CM. Nat. Protoc. 2006;1:1711. doi: 10.1038/nprot.2006.227. [DOI] [PubMed] [Google Scholar]
- 26.Yeh YT, Xia Y, Yu X, Zheng SY. presented at the SENSORS, 2015 IEEE; Nov., 2015; Busan, South Korea. [Google Scholar]
- 27.Xia Y, Zheng SY. presented at 18th Int. Conf. on TRANSDUCERS; June, 2015; Anchorage, AK. [Google Scholar]
- 28.Chen GD, Fachin F, Fernandez-Suarez M, Wardle BL, Toner M. Small. 2011;7(8):1061–1067. doi: 10.1002/smll.201002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, Wang H, Jiao J, Chen K-J, Owens GE, Kamei K-i, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng H-R. Angew. Chem. 2009;121:9132. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Liu X, Su B, Li J, Jiang L, Han D, Wang S. Adv. Mater. 2011;23:4376. doi: 10.1002/adma.201102435. [DOI] [PubMed] [Google Scholar]
- 31.Wang S, Liu K, Liu J, Yu ZTF, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee Y-K, Chung LWK, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CKF, Tseng H-R. Angew. Chem. Int. Ed. 2011;50:3084. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, Liu X, Meng J, Zhang P, Yang G, Su B, Sun K, Chen L, Han D, Wang S, Jiang L. Adv. Mater. 2013;25:922. doi: 10.1002/adma.201203826. [DOI] [PubMed] [Google Scholar]
- 33.Zhang P, Chen L, Xu T, Liu H, Liu X, Meng J, Yang G, Jiang L, Wang S. Adv. Mater. 2013;25:3566. doi: 10.1002/adma.201300888. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Li Y, Sun K, Fan J, Zhang P, Meng J, Wang S, Jiang L. J. Am. Chem. Soc. 2013;135:7603. doi: 10.1021/ja401000m. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Wang S. Chem. Soc. Rev. 2014;43:2385. doi: 10.1039/c3cs60419e. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Lu Q, Liu H, Wang J, Zhang P, Liang H, Jiang L, Wang S. Adv. Mater. 2015;27:6848. doi: 10.1002/adma.201502615. [DOI] [PubMed] [Google Scholar]
- 37.Huang C, Yang G, Ha Q, Meng J, Wang S. Adv. Mater. 2015;27:310. doi: 10.1002/adma.201402213. [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Liu H, Zhang F, Li G, Wang S. Small. 2016;12:4697. doi: 10.1002/smll.201601275. [DOI] [PubMed] [Google Scholar]
- 39.Mota M, Teixeira JA, Yelshin A. Sep. Purif. Technol. 2002;27:137. [Google Scholar]
- 40.Hochbaum AI, Chen R, Delgado RD, Liang W, Garnett EC, Najarian M, Majumdar A, Yang P. Nature. 2008;451:163. doi: 10.1038/nature06381. [DOI] [PubMed] [Google Scholar]
- 41.Boukai AI, Bunimovich Y, Tahir-Kheli J, Yu J-K, Goddard Iii WA, Heath JR. Nature. 2008;451:168. doi: 10.1038/nature06458. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt V, Wittemann JV, Senz S, Gösele U. Adv. Mater. 2009;21:2681. doi: 10.1002/adma.200803754. [DOI] [PubMed] [Google Scholar]
- 43.Brönstrup G, Jahr N, Leiterer C, Csáki A, Fritzsche W, Christiansen S. ACS Nano. 2010;4:7113. doi: 10.1021/nn101076t. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell JI, Zhou N, Nam W, Traverso LM, Xu X. Sci. Rep. 2014;4:3908. doi: 10.1038/srep03908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu Y, Peng K-Q, Liu L, Qiao Z, Huang X, Wu X-L, Meng X-M, Lee S-T. Sci. Rep. 2014;4:3667. doi: 10.1038/srep03667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holmes JD, Johnston KP, Doty RC, Korgel BA. Science. 2000;287:1471. doi: 10.1126/science.287.5457.1471. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt V, Wittemann JV, Gösele U. Chem. Rev. 2010;110:361. doi: 10.1021/cr900141g. [DOI] [PubMed] [Google Scholar]
- 48.Cui Y, Lieber CM. Science. 2001;291:851. doi: 10.1126/science.291.5505.851. [DOI] [PubMed] [Google Scholar]
- 49.Colinge J-P, Lee C-W, Afzalian A, Akhavan ND, Yan R, Ferain I, Razavi P, O'Neill B, Blake A, White M, Kelleher A-M, McCarthy B, Murphy R. Nat. Nanotechnol. 2010;5:225. doi: 10.1038/nnano.2010.15. [DOI] [PubMed] [Google Scholar]
- 50.Liu D, Li L, Gao Y, Wang C, Jiang J, Xiong Y. Angew. Chem. Int. Ed. 2015;54:2980. doi: 10.1002/anie.201411200. [DOI] [PubMed] [Google Scholar]
- 51.Barrelet CJ, Greytak AB, Lieber CM. Nano Lett. 2004;4:1981. [Google Scholar]
- 52.Su Y, Liu C, Brittman S, Tang J, Fu A, Kornienko N, Kong Q, Yang P. Nat. Nanotechnol. 2016;11:609. doi: 10.1038/nnano.2016.30. [DOI] [PubMed] [Google Scholar]
- 53.Chan CK, Peng H, Liu G, McIlwrath K, Zhang XF, Huggins RA, Cui Y. Nat. Nanotechnol. 2008;3:31. doi: 10.1038/nnano.2007.411. [DOI] [PubMed] [Google Scholar]
- 54.Peng K-Q, Wang X, Li L, Hu Y, Lee S-T. Nano Today. 2013;8:75. [Google Scholar]
- 55.Peng F, Su Y, Wei X, Lu Y, Zhou Y, Zhong Y, Lee S-T, He Y. Angew. Chem. Int. Ed. 2013;52:1457. doi: 10.1002/anie.201206737. [DOI] [PubMed] [Google Scholar]
- 56.Chiappini C, De Rosa E, Martinez JO, Liu X, Steele J, Stevens MM, Tasciotti E. Nat. Mater. 2015;14:532. doi: 10.1038/nmat4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noor MO, Krull UJ. Anal. Chim. Acta. 2014;825:1. doi: 10.1016/j.aca.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 58.Zhang G-J, Zhang L, Huang MJ, Luo ZHH, Tay GKI, Lim E-JA, Kang TG, Chen Y. Sens. and Actuators B: Chem. 2010;146:138. [Google Scholar]
- 59.Chen K-I, Li B-R, Chen Y-T. Nano Today. 2011;6:131. [Google Scholar]
- 60.Anderson SHC, Elliott H, Wallis DJ, Canham LT, Powell JJ. Phys. Status Solidi A. 2003;197:331. [Google Scholar]
- 61.Chiappini C, Liu X, Fakhoury JR, Ferrari M. Adv. Funct. Mater. 2010;20:2231. doi: 10.1002/adfm.201000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di Carlo D, Irimia D, Tompkins RG, Toner M. Proc. Natl. Acad. Sci. 2007;104:18892. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS-W, Lim W-T, Han J, Bhagat AAS, Lim CT. Sci. Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, Martel JM, Kojic N, Smith K, Chen P-i, Yang J, Hwang H, Morgan B, Trautwein J, Barber TA, Stott SL, Maheswaran S, Kapur R, Haber DA, Toner M. Nat. Protoc. 2014;9:694. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.R A Lamb a, Choppin PW. Annu. Rev. Biochem. 1983;52:467. doi: 10.1146/annurev.bi.52.070183.002343. [DOI] [PubMed] [Google Scholar]
- 66.Di Carlo D. Lab Chip. 2009;9:3038. doi: 10.1039/b912547g. [DOI] [PubMed] [Google Scholar]
- 67.Wan Y, Kim Y-t, Li N, Cho SK, Bachoo R, Ellington AD, Iqbal SM. Cancer Res. 2010;70:9371. doi: 10.1158/0008-5472.CAN-10-0568. [DOI] [PubMed] [Google Scholar]
- 68.Lipowsky HH, Kovalcheck S, Zweifach BW. Circ. Res. 1978;43:738. doi: 10.1161/01.res.43.5.738. [DOI] [PubMed] [Google Scholar]
- 69.Abe T, Segawa Y, Watanabe H, Yotoriyama T, Kai S, Yasuda A, Shimizu N, Tojo N. Lab Chip. 2011;11:1166. doi: 10.1039/c0lc00519c. [DOI] [PubMed] [Google Scholar]
- 70.Reichmuth DS, Wang SK, Barrett LM, Throckmorton DJ, Einfeld W, Singh AK. Lab Chip. 2008;8:1319. doi: 10.1039/b801396a. [DOI] [PubMed] [Google Scholar]
- 71.Vella SJ, Beattie P, Cademartiri R, Laromaine A, Martinez AW, Phillips ST, Mirica KA, Whitesides GM. Anal. Chem. 2012;84:2883. doi: 10.1021/ac203434x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, Wang J, Moore H, Rouse R, Umviligihozo G, Karita E, Mwambarangwe L, Braunstein SL, van de Wijgert J, Sahabo R, Justman JE, El-Sadr W, Sia SK. Nat. Med. 2011;17:1015. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 73.Yang S-Y, Lien K-Y, Huang K-J, Lei H-Y, Lee G-B. Biosens. Bioelectron. 2008;24:855. doi: 10.1016/j.bios.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 74.Wang Z, Wu H-j, Fine D, Schmulen J, Hu Y, Godin B, Zhang JXJ, Liu X. Lab Chip. 2013;13:2879. doi: 10.1039/c3lc41343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hirst GK. J. Exp. Med. 1942;75:49. doi: 10.1084/jem.75.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.