Abstract
Following extracellular stress signals, all eukaryotic cells choose whether to elicit a pro-survival or pro-death response. The decision over which path to take is governed by the severity and duration of the damage. In response to mild stress, pro-survival programs are initiated (unfolded protein response, autophagy, mitophagy) whereas severe or chronic stress forces the cell to abandon these adaptive programs and shift towards regulated cell death to remove irreversibly damaged cells. Both pro-survival and pro-death programs involve regulated communication between the endoplasmic reticulum (ER) and mitochondria. In yeast, recent data suggest this inter-organelle contact is facilitated by the endoplasmic reticulum mitochondria encounter structure (ERMES). These membrane contacts are not only important for the exchange of cellular signals, but also play a role in mitochondrial tethering during mitophagy, mitochondrial fission and mitochondrial inheritance. This review focuses on recent findings in yeast that shed light on how ER-mitochondrial communication mediates critical cell fate decisions.
Keywords: Mitophagy, Signal Transduction, Programmed cell death, Mitochondria, ERMES, endoplasmic reticulum
Introduction
The mitochondria and ER are organelles that play critical roles in the decision to live or die following environmental stress. Adaptive pro-life stress responses include mounting the unfolded protein response and/or the mitophagy pathway that are mediated through the ER and mitochondria respectively. However, if the stress outweighs the capability of the adaptive response to restore homeostasis, then regulated cell death (also referred to as programmed cell death –PCD [1]) can be triggered [2] (Figure 1). In recent years, studies in the budding yeast Saccharomyces cerevisiae have revealed that physical communication between the mitochondria and ER at mitochondrial–ER junctions play a critical role in this pro-life/pro-death decision. In addition, this communication is required for other basic biological processes including lipid and calcium signaling, mitochondrial morphology, inheritance and mitophagy. Understanding the molecular details of the mitochondria-ER junction is very important as ER-mitochondrial communication plays a role in the etiology of many diseases including neurodegenerative disorders, cardiomyopathies, metabolic syndrome, cancer, obesity and aging [3-6]. Interestingly, different aspects of ER-mitochondria communication are affected in different diseases. For example, deficiencies in removal of defective mitochondria may contribute to the accumulation of protein aggregates, which are apparent in a number of neurodegenerative diseases [7]. Many cancers are defective triggering the intrinsic programed cell death pathway whose components converge at the ER-mitochondria interface [8]. Given the conservation of many ER-mitochondria communication pathways, yeast represents an excellent model to dissect the intricate molecular details governing ER-mitochondria communication. This bipartite review focuses on recent advances in our understanding of how the interaction between these organelles aids pro-life and pro-death decisions in this model system.
Figure 1.
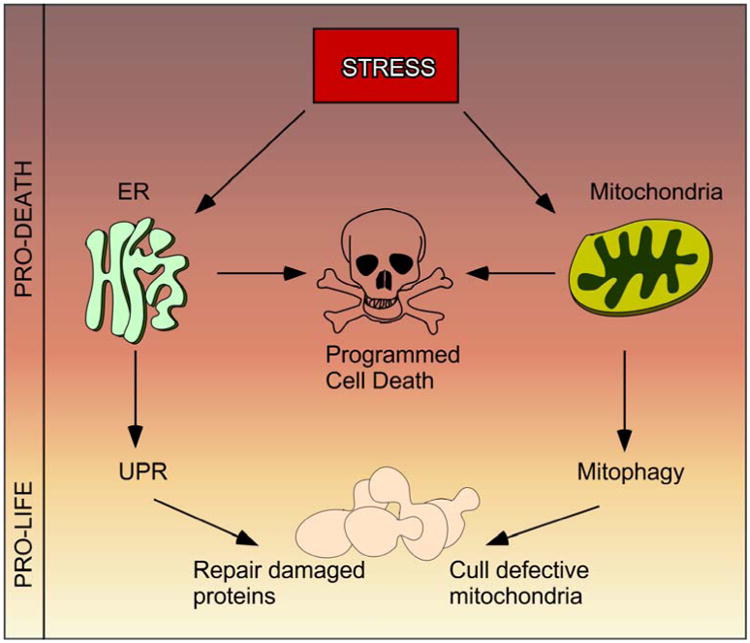
Following stress cells have to decide whether to elicit a pro-life or a pro-death response. The ER-Mitochondrial interface co-ordinates this decision.
PART 1. The role of ERMES in maintaining cellular homeostasis
ERMES provides contact sites between ER and mitochondria in yeast
Sites of close proximity between various organelles including the ER and mitochondrial membranes have been observed for many years [9]. Since then, it has become well established that membrane contact sites (MCSs) are a nexus for the exchange of lipids, small molecules and other signals crucial to cellular function and homeostasis [10-15]. In higher eukaryotes, regions of close interaction between ER and mitochondria are known as MAMs (for Mitochondrial Associated ER Membranes). Over 30 proteins have been identified at the ER-mitochondrial juxtaposition associated with MAMs (reviewed in [16]) which are involved a variety of processes including lipid metabolism, physical tethering, mitochondrial fission and autophagy. In S. cerevisiae, a multisubunit complex coined ERMES for ER–mitochondria encounter structure was discovered to be the primary complex that tethers the ER to the mitochondria in S. cerevisiae [17, 18]. The ERMES complex is composed of four core proteins called either MDM (for mitochondria distribution and morphology or MMM (for mitochondria morphology maintenance). Mdm10 and Mmm1 are anchored in the mitochondrial outer membrane (OMM) and ER membrane, respectively [17, 19] (Figure 2). The two other proteins, the cytosolic protein Mdm12 and the mitochondrial-associated protein Mdm34, form a bridge between Mmm1 and Mdm10 [20, 21]. Similar to MAM, ERMES has been implicated in numerous cellular processes including maintenance of mitochondrial morphology, mitochondrial protein import, phospholipid transport between the ER and mitochondria, mitochondrial attachment to the actin cytoskeleton, and mitochondrial division and inheritance of mitochondrial DNA [17, 22-27].
Figure 2.
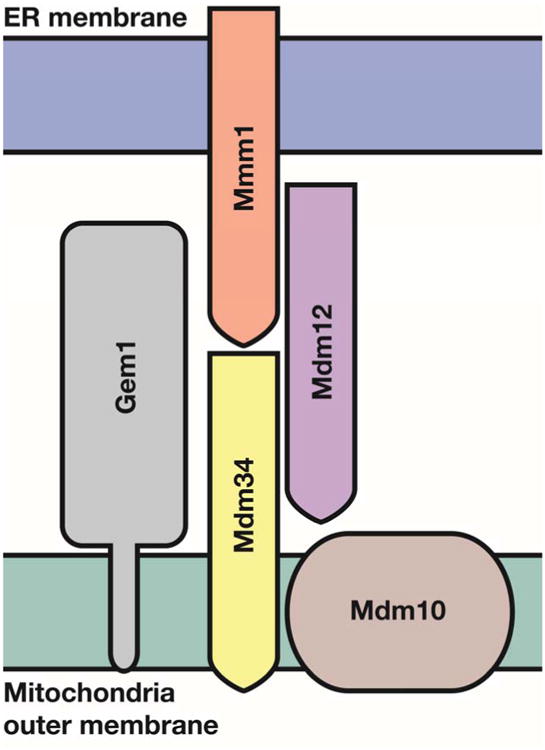
Topology of the ERMES complex at the ER-mitochondria contact site. Mdm34 and Mdm10 are embedded in the mitochondrial outer membrane, as is the regulatory GTPase Gem1. Mmm1 is an ER protein and is bridged to the mitochondrial components by the cytosolic Mdm12. Mmm1, Mdm12 and Mdm34 all contain SMP domains. Modified from [32].
Considering that the ERMES complex is implicated in so many processes, it is surprising that no clear ERMES homolog has been identified in mammals [28] despite being discovered in non-fungal linages [29, 30]. Thus, it has been proposed that ERMES may have been lost in metazoans through evolution [28]. In support of this theory, a recent study reported that the universally conserved vacuole protein Vsp13 suppresses all phenotypic consequences of ERMES deficiency [31]. Alternatively, others have suggested that mammalian ERMES homologues may be found by analyzing membrane-bound proteins that contain SMP (synaptotagmin-like, mitochondrial and lipid-binding proteins) domains [32]. This domain may be a signature motif for ERMES function as three of the four members of the ERMES complex contain this domain [32]. Alternatively, and considering the importance of the role it plays in budding yeast, it is also possible that functional homologs of ERMES exist that exhibit low sequence similarity to the yeast proteins. For example, human homologs of other essential yeast proteins (for example securin [33, 34]) have been identified by function and not sequence similarity. Thus, metazoan proteins providing ERMES function may still await discovery.
ERMES directs sites of mitochondrial fission
When visualized using fluorescent markers, ERMES appears as discrete puncta in regions of close contact between ER and mitochondria [19-21, 35]. Remarkably, in the absence of an ERMES complex member, the complex falls apart and the puncta disappear, indicating that all the members of the complex are required for the mitochondria-ER association. Loss of ERMES function results in mitochondrial aggregates indicating a defect in fission. Intriguingly, the ER morphology stays intact indicating that ERMES plays a distinct role in maintaining normal mitochondrial morphology [19, 21, 36]
The machinery regulating mitochondrial fission in yeast has been described in many excellent reviews [37-40]. In brief, fission is achieved after the conserved cytosolic GTPase protein Dnm1 docks onto the OMM protein Fis1 via two bridging proteins (Mdv1 and Caf4). Dnm1 then self-assembles into helical structures that wrap around mitochondria and coordinately divide the outer and inner membranes. Recently, work from Jodi Nunnari's group demonstrated that in yeast this mitochondrial constriction and division occurs at ER contact sites. This finding led to the notion that these sites create hotspots for Dnm1 assembly and eventual mitochondrial scission [41]. This process was coined ERMD for ER-associated Mitochondrial Division and is consistent with a model that the aberrant mitochondrial shape observed in ERMES mutants is caused by dysregulated fission [19, 42].
It has been proposed that a constriction of the mitochondria occurs prior to Dnm1 binding, due to the difference in diameter of the Dnm1 helix and the mitochondrion [41, 43]. However, it is not known whether ERMES facilitates mitochondrial constriction or plays another role in this process. In mammalian cells, mitochondrial division is dependent on an ER-localized actin-polymerizing formin protein called INF2 [44]. This has led to a model in which INF2-mediated actin polymerization drives the initial mitochondrial constriction directing the activity of the Dnm1 homologue Drp1 to complete the secondary constriction [44]. Myosin II, which binds INF2, is proposed to generate the force needed to drive constriction [45]. The role of actin polymerization in this process in yeast is not yet clear. The two yeast formins (Bni1 and Bnr1) have not been shown to localize to the mitochondria, although Bni1 is cytosolic and therefore could still induce actin filament growth at ER-mitochondria contact sites [46, 47]. However, the ERMES complex is implicated in mediating the attachment of mitochondria to the actin cytoskeleton in S. cerevisiae [20]. This interaction enables the mitochondria to move along actin cables in yeast, an event important for both morphological control and critical distribution of the organelle to emerging buds [19, 42].
ERMES role in other ER-mitochondrial linked processes
In addition to maintaining mitochondrial morphology, the ERMES complex is involved in many processes that maintain cellular homeostasis (see Table 1). The Miro GTPase Gem1, that is found in substoichiometric amounts in ERMES complexes has been linked to some of these roles. Gem1 is a ubiquitous dynamin-related MOM-anchored GTPase that has been proposed to be a regulatory subunit of the ERMES complex [48-50]. Importantly, Gem1 has been reported to facilitate proper mitochondrial distribution after cell division as it antagonize ERMES driven ER- mitochondrial contacts. The exact role Gem1 plays in regulating ERMES remains unsettled as Janet Shaw's group reported that the assembly and maintenance of ERMES complexes do not depend upon Gem1 [51]. Interestingly, Gem1 is the only member of ERMES that has a conserved mammalian homologue called Miro. Miro is thought to function as a calcium sensor connecting mitochondria to motor proteins facilitating cellular transport along the cytoskeleton [52]. Consistent with the proposed role for Gem1, dysfunctional Miro results in reduced mitochondrial mobility possibly due to enhanced mitochondria-ER contacts [27]. Thus the role of Miro and Gem1 may be similar in mammals and yeast but more work is necessary to clarify this activity.
Table 1.
Summary of the different roles ERMES members play to maintain cellular homeostasis.
| Proteins | Function | Reference |
|---|---|---|
| Mdm10 and Mdm12 | ERMES proteins control mitochondrial genome maintenance. | [18, 41] |
| Mmm1, Mdm10 and Mdm12 | Attach mitochondria to the actin cytoskeleton for polarized transport to the growing yeast bud. | [19, 21] |
| Mmm1 | Co-localizes with replicating mtDNA nucleoids | [117] |
| Mmm1 | Link mitochondria to the actin cytoskeleton of yeast and is required for mitochondrial inheritance. | [34] |
| Mdm10, Mmm1, Mdm12 | Mdm10, Mmm1 and Mdm12 function in the import and assembly of mitochondrial β-barrel proteins. Mdm10 is also a genuine subunit of the SAM complex, specifically required for the assembly of the TOM complex. | [118-120] |
| Mdm10, Mmm1, Mdm12 and Mdm34 | Linked to the transport of phosphatidylserine (PS) from the ER into mitochondria where it serves as a substrate for phosphatidylethanolamine (PE) synthesis. | [16, 121] |
| Mmm1, Mdm12 and Mdm34 | May be involved in lipid transport as have SMP (synaptotagmin-like, mitochondrial and lipid-binding proteins) domains implicated in binding hydrophobic ligands, including lipids. | [31, 79, 122, 123]. |
| Gem1 | Mitochondrial inheritance during cell division and regulation of lipid synthesis by ERMES. | [47, 50, 88, 124] |
Other contact sites between ER and mitochondria in yeast
Loss of ERMES alters mitochondrial membrane lipid composition and disrupts mitochondrial phospholipid metabolism, indicating that this complex is also important in lipid exchange [51]. However, cells lacking a functional ERMES complex are not without ER-derived lipids. Recent evidence has suggested that they received lipids via VCLAMP (vacuole and mitochondria patch). Consistent with this model, elimination of both vCLAMP and ERMES contact sites leads to significant defects in phospholipid transfer to mitochondria [53]. Recent studies have revealed that an additional protein complex called the EMC (ER membrane protein complex) can also facilitate ER-mitochondrial tethering, independent of the ERMES [54]. One possible explanation for why there are multiple tethering complexes is that each tether may facilitate a different type of communication between the ER and mitochondria. Interestingly, the EMC (but not ERMES) may mediate phospholipid trafficking between the between these two organelles. Consistent with another complex playing a role in phosphatidylserine (PS) exchange is the observation that ERMES mutants are viable [55] whereas PS transfer to mitochondria is an essential event as it is required for phosphatidylethanolamine (PE) synthesis [56]. Likewise, there is no significant decrease in ER to mitochondria PS transfer in cells lacking ERMES whereas there is in EMC mutant cells [26, 51]. Taken together, although precise molecular details still need to be resolved, the EMC is thought to have a predominant role in mediating phospholipid trafficking between the ER and mitochondria. In addition, it is not known if the relationship between ERMES and EMC is synergistic but it has been postulated that ERMES facilitates different ER-mitochondrial communications, for example the exchange of different lipids. Finally, it is also important to note that the EMC tether is conserved [30, 57] but whether the mammalian EMC homologue also facilitates ER–mitochondria tethering and PS transfer remains unknown.
Part 2. ERMES and the stress response
ERMES and mitophagy – pro-life response
Mitophagy is a highly conserved process in which damaged or unwanted mitochondria are culled from the cell using autophagic turnover [58, 59]. Like mammals, mitophagy can be executed by either ubiquitin or receptor mediated processes [9, 60-62]. The receptor mediated pathway is the best characterized and summarized in many excellent reviews [63, 64]. In short, receptor-dependent mitophagy is triggered when the exposed N-terminal domain of Atg32, a mitophagy-specific mitochondrial outer membrane protein, binds to two cytosolic proteins, Atg8 and Atg11 (Figure 3). Atg32 activation first triggers the Atg32-Atg11 interaction to recruit damaged mitochondria to the pre-autophagosomal structure/phagophore assembly site (PAS), where sequestering cytosolic vesicles are generated [46, 65-67]. Thereafter, Atg8 associates with Atg32 leading the core nonselective autophagic machinery to anchor the phagophore isolation membrane (IM), which in turn facilitates the formation of the autophagosome surrounding the mitochondria [47, 66, 67]. Autophagosomes carrying mitochondria eventually fuse with vacuoles to degrade their contents [68]. Interestingly, a dynamic actin cytoskeleton is required for mitophagy. Actin nucleation by the Arp2/3 complex facilitates the cycling of Atg9 between the mitochondria and the pre-autophagosome (PAS) while Atg9 co-localizes with Arp2 [65]. Atg11, which interacts with Atg9, is mis-localized when actin filament dynamics is disrupted [66].
Figure 3.
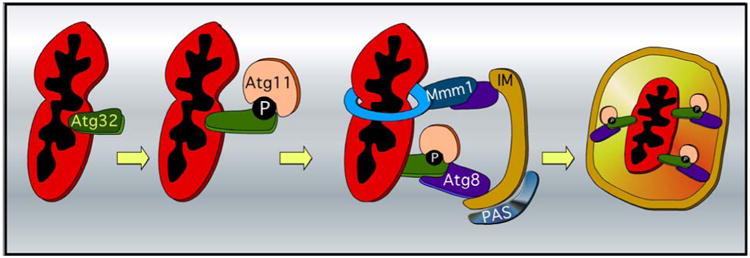
Molecular mechanisms of mitophagy in yeast. Mitophagy is induced upon CK2 mediated phosphorylation of Atg32. This event triggers the Atg32-Atg11 receptor-adaptor interaction and recruits the mitochondria to the pre-autophagosomal structure/phagophore assembly site (PAS). Subsequently the Atg8-Atg32 interaction anchors the mitochondria on the isolation membrane (IM) which also facilitates the formation of the autophagosome surrounding the mitochondria. Atg8 also interacts with the ERMES member Mmm1, thereby tethering the mitochondria to the ER. This interaction also supplies lipids from the endoplasmic reticulum (blue ring) for extension of the isolation membrane and thereby facilitates mitophagy.
In light of the fact that ERMES plays a role in ER tethering to the mitochondria, it is not surprising that ERMES plays a role in mediating mitophagy. By using an elegant experimental approach, Westerman and co-workers [69] revealed that Mmm1 and Atg8 interact and tether the mitochondrion destined for degradation to the ER and the isolation membrane of the growing phagophore (Figure 3). Consistent with this result, ERMES mutants have severe mitophagy defects [61] including accumulating immature autophagosomes. These results led the authors to hypothesize that these connections might provide an efficient supply of lipids from the ER to promote phagophore growth.
In mammalian cells, there is a growing body of evidence suggesting that mitochondrial fission is a pre-requisite for mitophagy [70-72]. In spatial terms alone, this makes sense as the size of the cargo has to be smaller than the autophagosome. In yeast, the role of mitochondrial fission in mitophagy is still controversial. Firstly, only deletion of the GTPase Dnm1 but not other fission factors (Fis1, Mdv1/Caf4) suppress mitophagy [73] suggesting that mitochondrial fission may be a pre-requisite for efficient mitophagy. In support of this model, Atg11 interacts with Dnm1 once the mitophagic pathway initiates and this association is required for efficient mitophagy [74]. On the other hand, a genetic screen performed by Okamoto et al. looking for proteins involved in mitophagy did not reveal Dnm1 or any of the other fission factors to be required [67]. Furthermore, Reichert's group has shown that mitochondrial fragmentation per se is not sufficient to trigger mitophagy [11]. In addition, this group has proposed that Dnm1 is not required for rapamycin-induced mitophagy [11]. Instead, these researchers propose that the activation of Whi2, a scaffold-like stress response protein, plays a significant role in mitophagy. Although its exact mechanism of action is not clear, Whi2 inhibits the Ras-cAMP-PKA pathway during the diauxic shift [75]. Also, Whi2 physically binds to the transcription factor Msn2 affecting both its transcriptional activity and nuclear localization [76]. Further studies will be required to determine whether fission is required in some, but not all types of mitophagy.
Signaling and mitophagy
Mitophagy can be induced by several methods. For example, mitophagy is observed in cells after treating with rapamycin, an inhibitor of the TOR pathway [10, 11], following nitrogen starvation [65], following the diauxic shift [67, 73] and upon mitochondrial dysfunction [32, 59]. It is well documented that under nitrogen starvation conditions, TORC1 activity is suppressed [43]. This results in rapid dephosphorylation of Atg13, an event necessary for binding to the Atg11 complex and the consequential phagophore formation and nonselective autophagy (Figure 4) [77]. Although phagophores are necessary for mitophagy, the relationship between TORC1 as well as other pro-life pathways (PKA and Sch9) and mitophagy has only recently emerged. Importantly, Rim15, a transcription factor that is negatively regulated by TORC1, PKA and Sch9, indirectly upregulates ATG gene expression including ATG32 following nitrogen starvation [78]. In addition, activated Rim15 also suppresses the Ume6–Sin3–Rpd3 transcriptional repressor complex following nitrogen starvation leading to elevated ATG32 transcription in both S. cerevisiae [79] and P. pastoris [80]. The combination of enhanced ATG32 expression and Atg32 protein phosphorylation is important as failure to do either reduces mitophagic efficiency.
Figure 4.
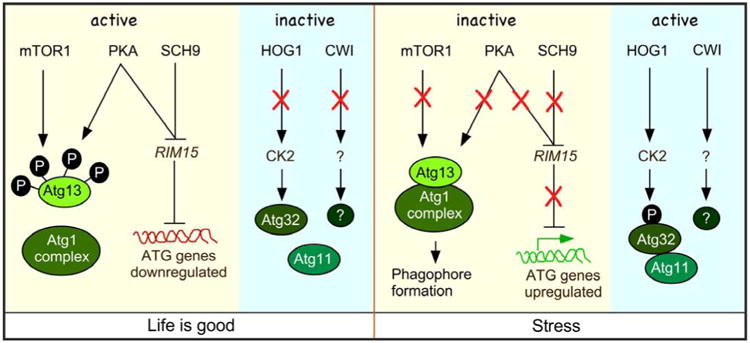
Autophagy induction in yeast. Under nutrient-rich condition, the activation of both the mTOR1 and PKA pathways results in Atg13 phosphorylation. This in turn inhibits the formation of the Atg1 complex and formation of the pre-autophagosomal structure (PAS). PKA and SCH9 pathways also inhibit the expression of the kinase Rim15. Rim15 activity is needed to upregulate genes needed for mitophagy. In addition under nutrient-rich conditions the HOG1 and CWI pathways are not activated. When mitophagy is induced (see text for details) the mTOR1, PKA and SCH9 pathways are inactivated. The resulting dephosphorylated Atg13 can consequently associated with the Atg1 complex. Also inactivation of PKA and SCH9 leads to Rim15 activation and upregulation of ATG specific genes. The HOG1 and CWI pathways are also activated which leads to activation of CK2 and an unknown kinase.
In mammals, nitrogen starvation is signaled by AMPK-dependent inhibition of Tor1 activity [81]. Recently, AMPK mediated inhibition of Tor1 was also identified in the fission yeast S. pombe [82]. However, in S. cerevisiae, how nitrogen starvation is sensed and the signal propagated is less well understood. Recently, it has been shown that the High Osmolarity Glycerol (HOG) and the Cell Wall Integrity (CWI) mitogen-activated protein kinase (MAPK) signal transduction pathways [83] are required for Atg32 phosphorylation (Figure 4) [84]. This modification enhances Atg32-Atg11 interaction leading to delivery of the targeted mitochondria to the phagophore assembly site [85]. However, the MAPK kinases from the HOG (Hog1) and CWI (Slt2) pathways do not directly phosphorylate Atg32. Rather, Hog1 activates casein kinase-2 (CK2) which directly modifies Atg32 [86]. This group demonstrated that although mitophagy is inhibited in cells deleted for both slt2Δ and hog1Δ mutant cells, Atg32 phosphorylation is prevented only in hog1Δ cells. This suggests that the Hog1, but not Slt2, signaling pathway is upstream of CK2 and regulates Atg32 phosphorylation. Taken together, these studies show that mitophagy regulation is complex and likely to require the coordination of different pathways. For example, recent work has shown that mitophagy induction is regulated in concert with phospholipid methylation [87]. In addition, genes involved in a broad range of cellular processes have been identified in genome wide screens for mitophagy mutants. Thus, further studies are needed to untangle the complexity of mitophagy regulation as it appears to be a well-integrated and fundamental process of cellular life.
ERMES and apoptosis – pro-death decision
It has been known for many years that both the ER and the mitochondria play dominant roles in the cellular response to stress. In short, following stress the ER-mitochondria interface coordinates the relevant response (see Figure 1), the outcome of which is dependent on the severity of the insult.
UPR and cell death
Perturbing ER function induces a pro-life response that activates genes that restore protein folding (coined the unfolded protein response or UPR) [88, 89]. Recently, it has been demonstrated that unfolded proteins can also be removed by autophagy [90]. However, in cases of chronic or unresolved ER stress, the UPR response declines and the cell moves from a pro-survival to a pro-apoptotic state by inducing several signal transduction events [8]. Thus, as the UPR induces not only survival but also cell death signals, understanding the nature of the switch between cellular outcomes is of great importance. The role ERMES plays in this switch is currently unknown in yeast. However, in mammals, it has recently been found that a protein called cell death–involved p53 target-1 (CDIP1) binds to Bap31 during ER stress and promotes apoptotic signaling from the ER to mitochondria [91] suggesting how ER stress signals are transmitted from the ER to mitochondria through MCSs.
Mitochondria Fission and cell death
In recent years, it has become increasingly clear that yeast are able to induce programmed cell death as a last resort to stress exposure [92]. Like all eukaryotes, the mitochondria play a critical role in this pro-death decision, which most resembles the mammalian intrinsic pathway. The proteins involved in mitochondrial dynamics have been shown to play critical roles in this process [93, 94]. Indeed, deleting genes encoding the mitochondrial fission machinery (Dnm1, Mdv1, Fis1) all result in cells being better able to survive various types of external stresses as well as expanding lifespans [95, 96]. As would be anticipated from this model, in higher eukaryotes, hyperfusion also represents a recognized strategy to allow survival during nutrient deprivation and cellular stress [97]. However, in yeast the role fusion plays in survival mechanisms remains to be elucidated [92]. This has led to the widely accepted notion that Dnm1 mediated stress-induced mitochondrial hyper-fission as possibly impaired fusion facilitates programed cell death.
In yeast the morphological change in mitochondria is accompanied by changes in mitochondrial outer membrane permeabilization (MOMP) resulting in cytochrome c release into the cytosol via mechanisms not yet fully understood [98]. Recent work in mammalian systems has shown that mitochondrial fragmentation per se is not a key factor in MOMP. Instead the mitochondrial constriction site marked by ERMD may play a critical role in Bax-dependent mitochondrial outer membrane permeabilization [41, 99]. A current model is that binding of Drp1, the mammalian homologue of Dnm1, which is massively recruited to the OMM following stress, and constriction of the OMM alters its topology such that it becomes amenable to Bax insertion [100]. This is supported by the observation that under apoptotic conditions, Drp1 is found in foci with Bax on mitochondria [101].
In yeast, many orthologues of metazoan apoptotic mediators (e.g., caspases, cytochrome c, EndoG) have been discovered suggesting that the core PCD machinery is highly conserved [102]. This includes the discovery of a yeast BH3-only protein reported to translocate to mitochondria inducing PCD following oxidative stress [103]. In addition, we have found that the nuclear protein cyclin C translocates to the cytoplasm following oxidative stress. In the cytoplasm, cyclin C associates with the fission machinery and is required for stress-induced mitochondrial hyper-fission, MOMP and cell death [104, 105] and (reviewed in [106]). This role of cyclin C is conserved in mammalian cells [107]. As cyclin C translocation to the mitochondria only occurs following stress, its presence at the OMM could potentially play a role in MOMP initiation and differentiate fission leading to PCD from those evoking mitophagy. However, relocating cyclin C to the mitochondrial in the absence of stress induces mitochondrial fragmentation but does not induce PCD [108]. Rather, these cells die faster after the addition of low concentrations of H2O2. These results suggest that relocating cyclin C to the mitochondria induces stress-induced fission and primes the cells to execute PCD following an additional stress signal. The molecular details of this unexpected role of a nuclear cyclin will be made clear with further research.
ERMES and cell death
Our recent data show that cells exhibiting constitutive mitochondrial fragmentation do not execute PCD [108]. This finding is consistent with the emerging idea that mitochondrial fragmentation itself does not commit a cell to MOMP and subsequent cell death. Rather, Drp1's role in regulating MOMP may be independent of its mitochondrial fission activity [109, 110]. Interestingly, the ER-mitochondrial contact sites create Drp1 hotspots (termed microdomians) that may direct Bax-dependent MOMP [37]. Consistent with this model, we observed an increase in the number of ER-mitochondrial contact sites in yeast following H2O2 stress (K. F. C. unpublished observations). In mammals Mff1, an OMM receptor for Drp1, localizes at these sites even in the absence of Drp1 [41]. As these ER mitochondrial sites are associated with lipid and Ca2+ exchange, it has been suggested that these contact sites may facilitate lipid effectors that promote Bax binding [101, 111]. Also, Stefan Grimm's group has shown that Fis1 conveys an apoptotic signal from the mitochondria to the ER by interacting with the ER protein Bap31 [112]. This interaction occurs at ER–mitochondria contact sites and results in the cleavage of Bap31 by caspase-8 to form p20Bap31, which is pro-apoptotic.
Concluding remarks
It is clear that ER-mitochondria connections play an important role in many diseases including cancer. Thus there is no doubt that the role ERMES, as well as other ER contact sites, play in regulating cell death is an area of research that will expand in the future. It is envisioned that the outcomes of such studies will improve our understanding of the molecular basis of related disorders associated with defective ER-mitochondria communication with the ultimate aim of developing better therapeutics.
Highlights.
Revised Manuscript “ER fatalities - the role of ER-mitochondrial contact sites in yeast life and death decisions”
For consideration in MAD special issue “Yeast on the corner of life and death decisions”
The revised manuscript has considerably changed. It now focuses on the role of the ERMES complex in maintaining cellular homeostasis (part 1) and then discusses what the roel this complex plays in cell death decisions (part 2).
Acknowledgments
We thank R. Strich for editorial help with this manuscript. We also thank Beth Cone and other members of KFC's early morning running group for help with the title. This work was supported by the National Institutes of Health, USA (GM R15-113196) awarded to K. F. C. D.G.J.S. is supported by an award from the National Institutes of Health, USA (GM RO1-113052) to R. Strich.
Footnotes
Conflict of interest: The authors declare no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Galluzzi L, et al. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ. 2015;22(1):58–73. doi: 10.1038/cdd.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14(1):20–8. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51(3):794–8. [PubMed] [Google Scholar]
- 5.Shutt TE, McBride HM. Staying cool in difficult times: mitochondrial dynamics, quality control and the stress response. Biochim Biophys Acta. 2013;1833(2):417–24. doi: 10.1016/j.bbamcr.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 6.Yoon YS, et al. Formation of elongated giant mitochondria in DFO-induced cellular senescence: involvement of enhanced fusion process through modulation of Fis1. J Cell Physiol. 2006;209(2):468–80. doi: 10.1002/jcp.20753. [DOI] [PubMed] [Google Scholar]
- 7.Takalo M, et al. Protein aggregation and degradation mechanisms in neurodegenerative diseases. Am J Neurodegener Dis. 2013;2(1):1–14. [PMC free article] [PubMed] [Google Scholar]
- 8.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327(5964):425–31. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copeland DE, Dalton AJ. An association between mitochondria and the endoplasmic reticulum in cells of the pseudobranch gland of a teleost. J Biophys Biochem Cytol. 1959;5(3):393–6. doi: 10.1083/jcb.5.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vance JE. Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem. 1990;265(13):7248–56. [PubMed] [Google Scholar]
- 11.Achleitner G, et al. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur J Biochem. 1999;264(2):545–53. doi: 10.1046/j.1432-1327.1999.00658.x. [DOI] [PubMed] [Google Scholar]
- 12.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456(7222):605–10. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 13.Baile MG, Lu YW, Claypool SM. The topology and regulation of cardiolipin biosynthesis and remodeling in yeast. Chem Phys Lipids. 2014;179:25–31. doi: 10.1016/j.chemphyslip.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010;11(10):739–50. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- 15.Kajiwara K, et al. Perturbation of sphingolipid metabolism induces endoplasmic reticulum stress-mediated mitochondrial apoptosis in budding yeast. Mol Microbiol. 2012;86(5):1246–61. doi: 10.1111/mmi.12056. [DOI] [PubMed] [Google Scholar]
- 16.Naon D, Scorrano L. At the right distance: ER-mitochondria juxtaposition in cell life and death. Biochim Biophys Acta. 2014;1843(10):2184–94. doi: 10.1016/j.bbamcr.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325(5939):477–81. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lang A, John Peter AT, Kornmann B. ER-mitochondria contact sites in yeast: beyond the myths of ERMES. Curr Opin Cell Biol. 2015;35:7–12. doi: 10.1016/j.ceb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126(6):1361–73. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boldogh IR, et al. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol Biol Cell. 2003;14(11):4618–27. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Youngman MJ, et al. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J Cell Biol. 2004;164(5):677–88. doi: 10.1083/jcb.200308012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boldogh I, et al. Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J Cell Biol. 1998;141(6):1371–81. doi: 10.1083/jcb.141.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Javaux EJ, Knoll AH, Walter MR. Morphological and ecological complexity in early eukaryotic ecosystems. Nature. 2001;412(6842):66–9. doi: 10.1038/35083562. [DOI] [PubMed] [Google Scholar]
- 24.Wideman JG, et al. Analysis of mutations in Neurospora crassa ERMES components reveals specific functions related to beta-barrel protein assembly and maintenance of mitochondrial morphology. PLoS One. 2013;8(8):e71837. doi: 10.1371/journal.pone.0071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahana S, et al. Functional dissection of IME1 transcription using quantitative promoter-reporter screening. Genetics. 2010;186(3):829–41. doi: 10.1534/genetics.110.122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–27. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murley A, et al. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helle SC, et al. Organization and function of membrane contact sites. Biochim Biophys Acta. 2013;1833(11):2526–41. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 29.Flinner N, et al. Mdm10 is an ancient eukaryotic porin co-occurring with the ERMES complex. Biochim Biophys Acta. 2013;1833(12):3314–25. doi: 10.1016/j.bbamcr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Wideman JG, et al. The ancient and widespread nature of the ER-mitochondria encounter structure. Mol Biol Evol. 2013;30(9):2044–9. doi: 10.1093/molbev/mst120. [DOI] [PubMed] [Google Scholar]
- 31.Lang AB, et al. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J Cell Biol. 2015;210(6):883–90. doi: 10.1083/jcb.201502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.AhYoung AP, et al. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc Natl Acad Sci U S A. 2015;112(25):E3179–88. doi: 10.1073/pnas.1422363112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen-Fix O, Koshland D. Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit. Genes Dev. 1999;13(15):1950–9. doi: 10.1101/gad.13.15.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou H, et al. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285(5426):418–22. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 35.Hobbs AE, et al. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J Cell Biol. 2001;152(2):401–10. doi: 10.1083/jcb.152.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgess SM, Delannoy M, Jensen RE. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994;126(6):1375–91. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoppins S, Nunnari J. Cell Biology. Mitochondrial dynamics and apoptosis--the ER connection. Science. 2012;337(6098):1052–4. doi: 10.1126/science.1224709. [DOI] [PubMed] [Google Scholar]
- 38.Voeltz GK, et al. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124(3):573–86. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 39.Westermann B. Bioenergetic role of mitochondrial fusion and fission. Biochim Biophys Acta. 2012;1817(10):1833–8. doi: 10.1016/j.bbabio.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 40.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11(12):872–84. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 41.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science. 2011;334(6054):358–62. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berger KH, Sogo LF, Yaffe MP. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J Cell Biol. 1997;136(3):545–53. doi: 10.1083/jcb.136.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nezich CL, Youle RJ. Make or break for mitochondria. Elife. 2013;2:e00804. doi: 10.7554/eLife.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339(6118):464–7. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen XJ. Mechanism of homologous recombination and implications for aging-related deletions in mitochondrial DNA. Microbiol Mol Biol Rev. 2013;77(3):476–96. doi: 10.1128/MMBR.00007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11(1):62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 47.Pon LA. Mitochondrial fission: rings around the organelle. Curr Biol. 2013;23(7):R279–81. doi: 10.1016/j.cub.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kornmann B, Osman C, Walter P. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc Natl Acad Sci U S A. 2011;108(34):14151–6. doi: 10.1073/pnas.1111314108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stroud DA, et al. Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J Mol Biol. 2011;413(4):743–50. doi: 10.1016/j.jmb.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 50.Klinge S, et al. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334(6058):941–8. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 51.Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5(12):931–42. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- 52.Oertle T, et al. A reticular rhapsody: phylogenic evolution and nomenclature of the RTN/Nogo gene family. FASEB J. 2003;17(10):1238–47. doi: 10.1096/fj.02-1166hyp. [DOI] [PubMed] [Google Scholar]
- 53.Elbaz-Alon Y, et al. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell. 2014;30(1):95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 54.Lahiri S, et al. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol. 2014;12(10):e1001969. doi: 10.1371/journal.pbio.1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kornmann B, Walter P. ERMES-mediated ER-mitochondria contacts: molecular hubs for the regulation of mitochondrial biology. J Cell Sci. 2010;123(Pt 9):1389–93. doi: 10.1242/jcs.058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gohil VM, Greenberg ML. Mitochondrial membrane biogenesis: phospholipids and proteins go hand in hand. J Cell Biol. 2009;184(4):469–72. doi: 10.1083/jcb.200901127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323(5922):1693–7. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kopec KO, Alva V, Lupas AN. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 2010;26(16):1927–31. doi: 10.1093/bioinformatics/btq326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shutt T, et al. The intracellular redox state is a core determinant of mitochondrial fusion. EMBO Rep. 2012;13(10):909–15. doi: 10.1038/embor.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Otsu K, Murakawa T, Yamaguchi O. BCL2L13 is a mammalian homolog of the yeast mitophagy receptor Atg32. Autophagy. 2015;11(10):1932–3. doi: 10.1080/15548627.2015.1084459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pereira C, et al. A yeast model of the Parkinson's disease-associated protein Parkin. Experimental Cell Research. 2015;333(1):73–79. doi: 10.1016/j.yexcr.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 63.May AI, Devenish RJ, Prescott M. The many faces of mitochondrial autophagy: making sense of contrasting observations in recent research. Int J Cell Biol. 2012;2012:431684. doi: 10.1155/2012/431684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rabouille C. Old dog, new tricks: Arf1 required for mitochondria homeostasis1. EMBO J. 2014;33(22):2604–5. doi: 10.15252/embj.201489899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Monastyrska I, et al. Arp2 links autophagic machinery with the actin cytoskeleton. Mol Biol Cell. 2008;19(5):1962–75. doi: 10.1091/mbc.E07-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He C, et al. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175(6):925–35. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17(1):87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 68.Fortsch J, et al. The myosin-related motor protein Myo2 is an essential mediator of bud-directed mitochondrial movement in yeast. J Cell Biol. 2011;194(3):473–88. doi: 10.1083/jcb.201012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klecker T, Bockler S, Westermann B. Making connections: interorganelle contacts orchestrate mitochondrial behavior. Trends Cell Biol. 2014;24(9):537–45. doi: 10.1016/j.tcb.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 70.Bagnat M, Simons K. Cell surface polarization during yeast mating. Proc Natl Acad Sci U S A. 2002;99(22):14183–8. doi: 10.1073/pnas.172517799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27(2):433–46. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bernstein C, Johns V. Sexual reproduction as a response to H2O2 damage in Schizosaccharomyces pombe. J Bacteriol. 1989;171(4):1893–7. doi: 10.1128/jb.171.4.1893-1897.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tatsuta T, Scharwey M, Langer T. Mitochondrial lipid trafficking. Trends Cell Biol. 2014;24(1):44–52. doi: 10.1016/j.tcb.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 74.Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9(12):971–80. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 75.Leadsham JE, Gourlay CW. cAMP/PKA signaling balances respiratory activity with mitochondria dependent apoptosis via transcriptional regulation. BMC Cell Biol. 2010;11:92. doi: 10.1186/1471-2121-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brina D, et al. Translational control by 80S formation and 60S availability: the central role of eIF6, a rate limiting factor in cell cycle progression and tumorigenesis. Cell Cycle. 2011;10(20):3441–6. doi: 10.4161/cc.10.20.17796. [DOI] [PubMed] [Google Scholar]
- 77.Fujioka Y, et al. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol. 2014;21(6):513–21. doi: 10.1038/nsmb.2822. [DOI] [PubMed] [Google Scholar]
- 78.Boldogh IR, et al. Arp2/3 complex and actin dynamics are required for actin-based mitochondrial motility in yeast. Proc Natl Acad Sci U S A. 2001;98(6):3162–7. doi: 10.1073/pnas.051494698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–77. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 80.Aihara M, et al. Tor and the Sin3-Rpd3 complex regulate expression of the mitophagy receptor protein Atg32 in yeast. J Cell Sci. 2014;127(Pt 14):3184–96. doi: 10.1242/jcs.153254. [DOI] [PubMed] [Google Scholar]
- 81.Alers S, et al. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32(1):2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davie E, Forte GM, Petersen J. Nitrogen regulates AMPK to control TORC1 signaling. Curr Biol. 2015;25(4):445–54. doi: 10.1016/j.cub.2014.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gustin MC, et al. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62(4):1264–300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mao K, et al. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol. 2011;193(4):755–67. doi: 10.1083/jcb.201102092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reinisch KM, De Camilli P. SMP-domain proteins at membrane contact sites: Structure and function. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbalip.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aoki Y, et al. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol Biol Cell. 2011;22(17):3206–17. doi: 10.1091/mbc.E11-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cronin SR, Rao R, Hampton RY. Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J Cell Biol. 2002;157(6):1017–28. doi: 10.1083/jcb.200203052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Conrad M, et al. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2014;38(2):254–99. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fok V, et al. Multiple domains of EBER 1, an Epstein-Barr virus noncoding RNA, recruit human ribosomal protein L22. RNA. 2006;12(5):872–82. doi: 10.1261/rna.2339606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fahl SP, et al. Rpl22 Loss Impairs the Development of B Lymphocytes by Activating a p53-Dependent Checkpoint. J Immunol. 2015;194(1):200–9. doi: 10.4049/jimmunol.1402242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Delaney JR, et al. Stress profiling of longevity mutants identifies Afg3 as a mitochondrial determinant of cytoplasmic mRNA translation and aging. Aging Cell. 2013;12(1):156–66. doi: 10.1111/acel.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Westermann B. Mitochondrial dynamics in model organisms: what yeasts, worms and flies have taught us about fusion and fission of mitochondria. Semin Cell Dev Biol. 2010;21(6):542–9. doi: 10.1016/j.semcdb.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 93.Braun RJ, Westermann B. Mitochondrial dynamics in yeast cell death and aging. Biochem Soc Trans. 2011;39(5):1520–6. doi: 10.1042/BST0391520. [DOI] [PubMed] [Google Scholar]
- 94.Guaragnella N, et al. The role of mitochondria in yeast programmed cell death. Front Oncol. 2012;2:70. doi: 10.3389/fonc.2012.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fannjiang Y, et al. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 2004;18(22):2785–97. doi: 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scheckhuber CQ, et al. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat Cell Biol. 2007;9(1):99–105. doi: 10.1038/ncb1524. [DOI] [PubMed] [Google Scholar]
- 97.Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505(7483):335–43. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Corte-Real M, Madeo F. Yeast programed cell death and aging. Front Oncol. 2013;3:283. doi: 10.3389/fonc.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337(6098):1062–5. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Montessuit S, et al. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 2010;142(6):889–901. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Karbowski M, et al. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol. 2002;159(6):931–8. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carmona-Gutierrez D, et al. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 2010;17(5):763–73. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- 103.Buttner S, et al. A yeast BH3-only protein mediates the mitochondrial pathway of apoptosis. EMBO J. 2011;30(14):2779–92. doi: 10.1038/emboj.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cooper KF, et al. Stress-induced nuclear-to-cytoplasmic translocation of cyclin C promotes mitochondrial fission in yeast. Dev Cell. 2014;28(2):161–73. doi: 10.1016/j.devcel.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cooper KF, et al. Oxidative-stress-induced nuclear to cytoplasmic relocalization is required for Not4-dependent cyclin C destruction. J Cell Sci. 2012;125(Pt 4):1015–26. doi: 10.1242/jcs.096479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Clark AD, Oldenbroek M, Boyer TG. Mediator kinase module and human tumorigenesis. Crit Rev Biochem Mol Biol. 2015:1–34. doi: 10.3109/10409238.2015.1064854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xilouri M, et al. Abberant alpha-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS One. 2009;4(5):e5515. doi: 10.1371/journal.pone.0005515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khakhina S, Cooper KF, Strich R. Med13p prevents mitochondrial fission and programmed cell death in yeast through nuclear retention of cyclin C. Mol Biol Cell. 2014;25(18):2807–16. doi: 10.1091/mbc.E14-05-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cassidy-Stone A, et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14(2):193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517(7534):302–10. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chi X, et al. Regulating cell death at, on, and in membranes. Biochim Biophys Acta. 2014;1843(9):2100–13. doi: 10.1016/j.bbamcr.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 112.Iwasawa R, et al. Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 2011;30(3):556–68. doi: 10.1038/emboj.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]


