Abstract
The development of synergistically pathogenic communities of Porphyromonas gingivalis and Streptococcus gordonii is controlled by a tyrosine phosphorylation dependent signaling pathway in P. gingivalis. The Ptk1 bacterial tyrosine (BY) kinase of P. gingivalis is required for maximal community development and for the production of extracellular polysaccharide. Herein we show that the consensus BY kinase Walker A and B domains, the RK cluster and the YC domain are necessary for autophosphorylation and for substrate phosphorylation. Mass spectrometry showed that 6 tyrosine residues in a 16 aa C-terminal region were phosphorylated in recombinant (r) Ptk1. Complementation of a ptk1 mutant with the wild type ptk1 allele in trans could restore community development between P. gingivalis and S. gordonii, and extracellular polysaccharide production by P. gingivalis. In contrast, complementation of Δptk1 with ptk1 containing a mutation in the Walker A domain failed to restore community development of extracellular polysaccharide production. rPtk1 was capable of phosphorylating the tyrosine phosphatase Ltp1 and the transcriptional regulator CdhR, both of which are involved in the development of P. gingivalis communities with S. gordonii.
Introduction
Bacterial tyrosine (BY) kinases, enzymes which are widely distributed in gram-positive and gram-negative bacteria, are structurally and functionally distinct from their eukaryotic cell tyrosine kinase counterparts (Grangeasse et al., 2012). The elemental catalytic structure of BY kinases is similar to P-loop kinases and comprises Walker A, Walker A′ and Walker B motifs which are responsible for nucleotide binding and hydrolysis. The catalytic domain transfers phosphate groups to a C-terminal (YC) domain with several closely spaced tyrosine residues which can subsequently phosphorylate client proteins (Grangeasse et al., 2007). Furthermore, an activator domain, which is typically transmembrane, enhances the ATP-binding affinity of the catalytic region (Mijakovic et al., 2016). In proteobacteria, the activator and catalytic domains are usually on one polypeptide chain, whereas in firmicutes the activator and catalytic domains often reside on two independent polypeptides. In the single polypeptide configuration, an additional arginine- and lysine- rich RK cluster contributes to catalytic activity (Bechet et al., 2010). The most extensively documented role of BY kinases is in the synthesis and export of extracellular polysaccharides, and consequently also in biofilm formation (Grangeasse et al., 2012; Whitmore & Lamont, 2012). BY kinases have also been shown to be involved in stress responses, antibiotic resistance lysogeny, and DNA metabolism; and phosphorylation can affect the location as well as the activity of substrate proteins (Grangeasse et al., 2012; Mijakovic et al., 2016; Whitmore & Lamont, 2012).
Periodontitis is a common chronic inflammatory disease of the gingival tissues that ensues from the action of a polymicrobial bacterial community (Hajishengallis & Lamont, 2012; 2014; 2016). Bacterial constituents of periodontally pathogenic communities often exhibit functional specialization and polymicrobial synergy. For example, the accessory pathogen Streptococcus gordonii can elevate the virulence of the keystone pathogen Porphyromonas gingivalis (Daep et al., 2011). S. gordonii and P. gingivalis are physically associated in vivo, and form heterotypic communities in vitro (Kuboniwa & Lamont, 2010; Wright et al., 2013). Community formation by P. gingivalis is controlled by a protein tyrosine (de)phosphorylation signaling pathway involving a BY kinase (Ptk1) and a phosphatase (Ltp1) and which converges on regulation of the Mfa1 fimbrial adhesin through the CdhR transcription factor (Chawla et al., 2010; Maeda et al., 2008; Wright et al., 2014). Similar to other bacteria, Ptk1 is also involved in the control of extracellular polysaccharide production (Wright et al., 2014). Ptk1 has an archetypal proteobacterial structure with Walker A, A′, B domains along with an RK region and a C-terminal Y cluster (Wright et al., 2014). While this general scheme is conserved among BY kinases, the specific functional roles of the domains can vary among individual enzymes. For example, autophosphorylation of PtkB, a BY kinase of Bacillus subtilis, can inhibit kinase activity (Elsholz et al., 2014), and PtkA is capable of substrate phosphorylation in the absence of a functional YC cluster (Mijakovic et al., 2003).
The functionality of the BY kinase domains in Ptk1 of P. gingivalis is yet to be addressed. Through the use of targeted mutations we defined the role of the Walker A and B domains, the RK domain and the YC cluster in autophosphorylation and substrate phosphorylation. The involvement of phosphotransfer in community formation between P. gingivalis and S. gordonii, and in extracellular polysaccharide production has also been established.
Materials and Methods
Bacteria and culture conditions
Wild type Porphyromonas gingivalis ATCC 33277 and isogenic mutants 33277 + pT-COW, Δptk1, CΔptk1 (Wright et al., 2014), and CMΔptk1 (this study), were cultured anaerobically in trypticase soy broth (TSB) supplemented with yeast extract (1 mg/ml), hemin (5 μg/ml) and menadione (1 μg/ml). When necessary, erythromycin at 10 μg ml−1, and tetracycline at 1 μg ml−1, were incorporated into the medium.
Construction of complemented strain CMΔptk1
To generate a Δptk1 strain complemented with ptk1 containing a K614M/S615C mutation in the Walker A domain (CMΔptk1), the promoter and coding region of ptk1 were amplified and cloned into pUC19. Site-specific mutations were introduced into ptk1 using the QuikChange Multi Site Directed Mutagenesis Kit (Agilent Technologies) with primers listed in Supplementary Table 1. Specifically, the Lys614 and Ser615 of the Walker A box were replaced with methionine and cysteine respectively, as described (Doublet et al., 1999). The construct was confirmed by sequencing and cloned into pT-COW. The resulting plasmid was electroporated into Δptk1 creating strain CMΔptk1. Transformants were selected with erythromycin and tetracycline, the presence of plasmid was confirmed by sequencing and the expression of mutated ptk1 was determined by RT-PCR.
Expression and purification of recombinant proteins
The C-terminal catalytic domain of Ptk1 (FPtk1, aa541–821), and the catalytically inactive Ltp1:C10S were expressed as His-tag fusion proteins and purified from BL21 Star as described previously (Wright et al., 2014). Site-specific mutations were introduced into FPtk1 using the QuikChange Multi Site Directed Mutagenesis Kit with plasmid pET200Fptk1 (Wright et al., 2014) as template and primers listed in Supplementary Table 1. The following mutations were created: K614M/S615C (Walker A domain), D717N (Walker B domain), E581A/E584A/R587A (RK cluster), Y782F/Y784F/Y786F/Y788F/Y790F (C-terminal tyrosine rich domain. The entire coding regions of epsD (PGN_0224), and cdhR (PGN_1373) were amplified from a P. gingivalis 33277 genomic template using primers listed in Supplementary Table 1 and cloned into pET200. Following confirmation by sequencing, derivatives of pET200 were transformed into BL21 Star (Invitrogen). Soluble protein was obtained using MagneHIS particles (GE). The purity of the resulting protein was determined by SDS-PAGE and Coomassie staining.
Auto- and substrate- phosphorylation
For autophosphorylation, recombinant FPtk1 (5 μg) was treated with 20 units of calf intestinal alkaline phosphatase (New England) for 2 h at 37°C. ATP (0.1 mmol/L) and phosphatase inhibitor cocktail 2 (Sigma-Aldrich) were added for an additional 15 min before reactions were stopped by boiling in an equal volume of 2× SDS-PAGE sample buffer. Following SDS-PAGE and electrotransfer to nitrocellulose, membranes were blocked with 10% BSA and probed with phosphotyrosine antibodies (Clone PY20; Sigma-Aldrich) at 1:1000 for 2 h. Membranes were washed three times with TBST (TBS and 0.1% Tween 20) before addition of a horseradish peroxidase-conjugated secondary antibody (1:1000) and incubation for 2 h at room temperature. Antibody binding was detected with Enhanced chemiluminescence (ECL) reagent and a ChemiDoc XRS imaging system (BioRad,).
For the substrate phosphorylation assay, FPtk1 (5 μg) was incubated with 10 μg of recombinant substrate. Five mM ATP and 0.5 μL of phosphatase inhibitor cocktail 2 were added and incubated at 37°C for 30 min. The reaction was stopped by boiling for 10 min in 2× SDS sample buffer. Samples were analyzed by Western immunoblotting as described above.
Mass spectrometry
To determine protein phosphorylation sites on FPtk1, protein samples were diluted into reducing SDS-PAGE buffer and 10 μg of each assay were loaded onto 4–12% Bis-Tris gels (Invitrogen, San Diego, CA). The samples were electrophoresed into the gel for 1 cm and the 1 cm gel lane excised for in-gel trypsin proteolysis as described previously (Caster et al., 2015). The concentrated and desalted peptides were re-suspended in 40 μL of 2% acetonitrile/0.1% formic acid, and filtered through a 0.2 μm regenerated cellulosic filter, prior to analysis by 1-dimensional (1D) reversed phase liquid chromatography (RP-LC) mass spectrometric analysis (MS). Tryptic peptides (0.5 μg) were separated using an EASY n-LC (Thermo) UHPLC system and a 360 μm OD × 100 μm ID fused silica tip packed with 10 cm of Jupiter 5 μm C18 300 Å material (Phenomenex, Torrance, CA). Following injection of the sample onto the column, separation was accomplished with a 75 min linear gradient from 2% acetonitrile to 40% acetonitrile in 0.1% formic acid.
The eluate was introduced into the LTQ-Orbitrap ELITE + ETD mass spectrometer using a Nanospray Flex source (ThermoElectron, Waltham, MA). A Data Dependent Neutral Loss MS3 method was created in Xcalibur v2.2. Scan event one of the method obtained an FTMS MS1 scan (normal mass range; 60,000 resolution, full scan type, positive polarity, profile data type) for the range 300–2000m/z. Scan event two obtained ITMS MS2 scans (CID activation type, normal mass range, rapid scan rate, centroid data type) on up to ten peaks that had a minimum signal threshold of 10,000 counts from scan event one. If a neutral loss of 24.5, 32.7, or 49 was observed within the top three peaks of the MS2 scan, the scan event was repeated using ETD fragmentation. The lock mass option was enabled (0% lock mass abundance) using the 371.101236m/z polysiloxane peak as an internal calibrant.
Proteome Discoverer v1.3.0.339 was used to analyze the data collected by the mass spectrometer. The database used in Mascot v2.1 and Sequest searches was the 12/9/2013 version of the P. gingivalis strain ATCC 33277 protein sequences from the UniProtKB. In order to estimate the false discovery rate, a decoy database was generated from this database with the program decoy.pl (from matrixscience.com). This decoy database of reverse sequences was appended to the original database. Proteome Discoverer was used for extraction of MS2 scan data from the Xcalibur RAW file and compile separate searches of CID and ETD MS2 scans in Mascot and Sequest into a single file (.msf extension). The resulting .msf files from Proteome Discoverer were loaded together into Scaffold v3.6.5. Scaffold was used to calculate the false discovery rate using the Peptide and Protein Prophet algorithms (Keller et al., 2002; Nesvizhskii et al., 2003). Proteins were accepted if they had at least two peptides with a Peptide Prophet confidence of at least 95% and a Protein Prophet confidence of at least 99%. Proteins were grouped to satisfy the parsimony principle.
Extracellular polysaccharide detection
Exopolysaccharide production was quantified with fluorescent lectins as described previously (Maeda et al., 2008). In brief, P. gingivalis cells were labeled with Syto-17 (Invitrogen) and deposited on glass coverslips. Polysaccharide was labeled with concanavalin A-FITC and wheat germ agglutinin-FITC (100 μg/mL) for 30 min at room temperature. After washing, images were examined on a Leica SP8 confocal microscope images using 488 nm (FITC) and 648 nm (Syto17) lasers. Fluorescent levels were quantified with Volocity software (Perkin Elmer).
Heterotypic P. gingivalis-S. gordonii communities
Communities of P. gingivalis and S. gordonii were generated as described previously (Kuboniwa et al., 2006). S. gordonii cells (2 × 108) were stained with hexidium iodide (15 μg/mL, Invitrogen) and deposited on glass coverslips for 16 h anaerobically at 37°C. P. gingivalis cells (2 × 107) were labeled with 5-(and-6)-carboxyfluorescein, succinimidyl ester (FITC, 4 μg/mL, Invitrogen) and reacted with S. gordonii for 18 h in prereduced PBS anaerobically at 37°C with rocking. After washing with PBS, coverslips were examined on a Leica SP8 confocal microscope using 488 nm (FITC) and 558 nm (hexidium iodide) lasers. XYZ stacks were digitally reconstructed using Volocity software (Perkin Elmer). Quantitation of the volume of P. gingivalis fluorescence was obtained using the Find Objects algorithm in the Volocity program. This process analyzed all P. gingivalis fluorescence in the 3D digitally recreated confocal images. To estimate microcolony formation, the Find Objects process was used with a threshold for 3D objects greater than 30 μm3.
Statistics
P-values were determined using two-tailed t-test. Data presented are representative of at least three independent experiments.
Results
Autophosphorylation and substrate phosphorylation
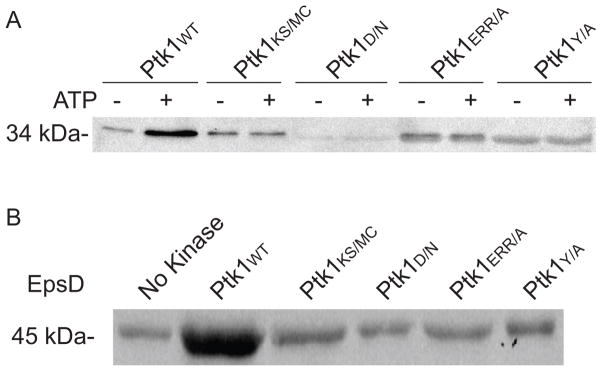
BY kinases undergo autophosphorylation and can phosphorylate a range of substrate proteins (Grangeasse et al., 2012). The Ptk1 BY kinase of P. gingivalis possesses the domain architecture typical of BY kinases; however, the functionality of these domains has not been investigated. We examined the role of Ptk1 domains in autophosphorylation and substrate phosphorylation through the generation of domain mutations as depicted in Fig 1. Full-length Ptk1 is toxic to E. coli (Wright et al., 2014), thus a recombinant Ptk1 fragment (FPtk1) was utilized as the parental protein. FPtk1, spanning amino acids 541–821, lacks the predicted N-terminal membrane domains but is capable of autophosphorylation and substrate phosphorylation (Wright et al., 2014). When FPtk1 and its derivatives were dephosphorylated with CIP and then reacted with ATP, we observed that mutation of the Walker A, Walker B, RK, and YC domains all prevented autophosphorylation of Ptk1 (Fig 2A), One documented substrate of Ptk1 is EpsD, a predicted UDP-acetyl-mannosamine dehydrogenase involved in carbohydrate production (Grangeasse et al., 2003). Thus we utilized PGN_0224 to test the concordance between autophosphorylation and substrate phosphorylation with Ptk1. Figure 2B shows that all of the domain mutants of Ptk1 were unable to phosphorylate EpsD, demonstrating that in the absence of autophosphorylation of Ptk1, phospho-transfer to a substrate does not occur.
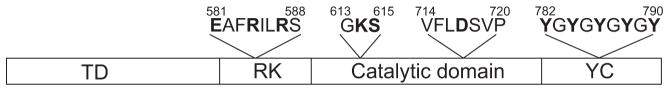
Figure 1.
Domain architecture of the Ptk1 tyrosine kinase in P. gingivalis. Amino acid residues that were mutated are indicated in bold. TD: transmembrane domain, RK: arginine rich domain, YC: tyrosine rich C-terminal cluster.
Figure 2.
Auto (A) and substrate (B) phosphorylation by Ptk1 and its phosphotransfer domain mutants. For autophosphorylation, recombinant FPtk1 (Ptk1:541–821) or FPtk1 with mutations in the Walker A (KS/MC), Walker B (D/N), RK (ERR/A) and YC (YA) domains were dephosphorylated with alkaline phosphatase and incubated with or without ATP in the presence of a phosphatase inhibitor. For substrate phosphorylation, Fptk1 and mutant derivatives were incubated with recombinant EpsD (PGN_0224) in the presence of ATP and a phosphatase inhibitor. The phosphorylation level of proteins was determined by Western blotting with phosphotyrosine antibodies.
C-terminal phosphorylation of Ptk1
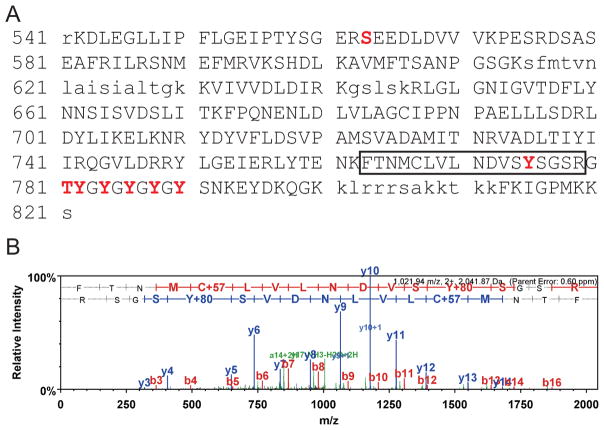
Recombinant FPtk1 purified from E. coli can both autophosphorylate and phosphorylate substrates. Using mass spectrometry we identified the amino acid residues in FPtk1 that were phosphorylated in the recombinant protein. Tyrosine residue 775 and the 5 tyrosines in the Y cluster spanning residues 782–790 were all found to be phosphorylated (Fig 3). Thus, Ptk1 would appear to possess a conventional YC cluster. Additionally, the finding that all of the tyrosines in the YC cluster were phosphorylated provides evidence that FPtk1 is fully active in the absence of the N-terminal region. The role of the phosphorylated tyrosine at 775 remains to be determined. A number of proteobacterial BY kinases possess an internal tyrosine near the Walker A domain that is susceptible to autophosphorylation (Grangeasse et al., 2007). However, Y775 of Ptk1 is closer to the YC domain than to the Walker A domain, and indeed phosphorylation at a homologous site is not apparent in other documented BY kinases. S563 and T781 are also phosphorylated in rFtpk1 (Fig 3). The functional role of serine and threonine phosphorylation is currently under investigation.
Figure 3.
Mass spectroscopy sequence coverage for FPtk1 (Ptk1:541–821). A. High confidence MS/MS data for 37 unique peptides comprising 87 exclusive unique and 394 total spectra were used to assign sequence coverage for 246 (blue upper case font) of 281 amino acids achieving 87.5% coverage. Non-detected residues are in lower case. MS/MS spectra for peptide in box is shown in (B). Phosphorylated residues are shown in red upper case, and bolded font. B. Example of MS/MS spectra used to assign site specific phosphorylation to MS/MS, in this case to tyrosine-775 (Y775) in the carbamidomethylated (c) tryptic peptide FTNMcLVLNDVSYSGSR.
Signaling substrates of Ptk1
Ptk1 is essential for heterotypic community formation between P. gingivalis and S. gordonii, and loss of Ptk1 causes an increase in the levels of CdhR, a transcriptional repressor of mfa1 which encodes a P. gingivalis fimbrial adhesin (Chawla et al., 2010). We sought to determine whether Ptk1 may also act directly on CdhR and hence we examined phosphorylation of CdhR by Ptk1. Fig 4 shows that rFPtk1 can phosphorylate rCdhR, indicating that Ptk1 can regulate the activity of CdhR through direct phosphorylation.
Figure 4.
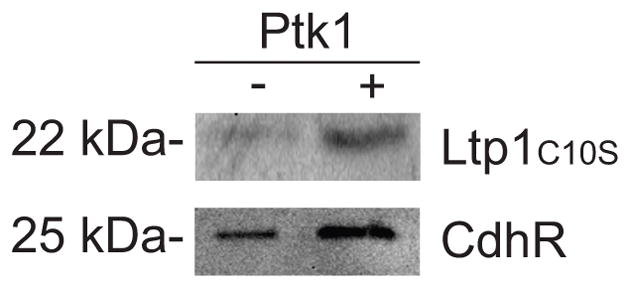
Phosphorylation of Ltp1:C10S and CdhR by FPtk1. Recombinant proteins were dephosphorylated with alkaline phosphatase and incubated individually with or without FPtk1 along with ATP and in the presence of a phosphatase inhibitor. Protein phosphorylation was determined by Western blotting with phosphotyrosine antibodies.
Following Ptk1-dependent initiation of P. gingivalis-S. gordonii communities, accumulation is regulated by the Ltp1 phosphatase which can dephosphorylate Ptk1 (Maeda et al., 2008). Currently, these enzymes are the only annotated tyrosine kinase and phosphatase in the P. gingivalis strain 33277 genome (Naito et al., 2008), and thus may comprise a cognate pair. Moreover, Ltp1 can dephosphorylate Ptk1 (Wright et al., 2014), concordant with the established activities of other cognate pairs (Mijakovic et al., 2016). However, the ability of the Ptk1 to phosphorylate its Ltp1 has not been documented. As shown in Fig. 4, interaction between FPtk1 and Ltp1 resulted in increased phosphorylation of Ltp1, suggesting that Ptk1 and Ltp1 can phosphorylate/dephosphorylate each other, which could comprise a mutual feedback mechanism to control the activity of tyrosine phosphorylation dependent networks.
Role of Ptk1 functional domains in community formation and extracellular polysaccharide production
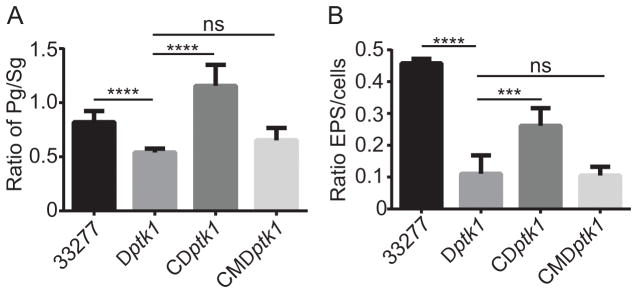
While the requirement for Ptk1 in P. gingivalis-S. gordonii community formation and in extracellular polysaccharide has been established (Wright et al., 2014), the involvement of phosphotransfer per se was not addressed. In this regard, some kinase homologs can act as a pseudo-kinase and provide scaffold or allosteric functions to regulate the organization or function of other proteins (Gee et al., 2012; Gruszczyk et al., 2013). To determine whether functional phosphotransfer is necessary for Ptk1-mediated control of community formation and EPS production, we complement the Δptk1 mutant strain with the ptk1 allele containing the K614M/S615C substitutions in the Walker A domain. As shown in Fig 5A, while complementation of Δptk1 with the wild type ptk1 allele restored levels of P. gingivalis accumulation with S. gordonii, complementation with the Walker A mutant allele did not restore community development. Similarly, complementation with the Walker A mutant failed to increase extracellular polysaccharide production by P. gingivalis (Fig 5B). These results confirm the necessity for phosphotransfer in two of the major phenotypic properties controlled by Ptk1.
Figure 5.
Phosphotransfer by Ptk1 is required for P. gingivalis community development with S. gordonii and for extracellular polysaccharide production. A. P. gingivalis 33277, Δptk1 (Dptk1), Δptk1 + pptk1 (CDptk1) and Δptk1 + pptk1 containing a K614M/S615C mutation in the Walker A domain (CMDptk1) were reacted with a substrate of S. gordonii for 18 h. Total biovolumes of P. gingivalis and S. gordonii in a 213 × 213 μm area were obtained with by confocal microscopy, analyzed by Volocity and data are expressed as ratio of P. gingivalis (Pg) to S. gordonii (Sg). B. P. gingivalis strains were stained for extracellular polysaccharide with FITC-labeled concanavalin A and wheat germ agglutinin. Bacterial cells were stained with Syto-17. Fluorescent images were collected by confocal microscopy and the ratio of lectin staining of EPS to whole cell staining determined using Volocity. Error bars are SD, n = 3. ***P < 0.001; ****P < 0.0001, ns P > 0.05.
Discussion
BY kinases, present in a large array of bacteria, possess a characteristic domain architecture. Walker A, A′ and B motifs constitute the ATP binding site and this catalytic domain transfers the gamma phosphate to the hydroxyl group of tyrosine residues in the YC domain (Mijakovic et al., 2016). This is thought to be a transphosphorylation event, and structural studies show that in the dephosphorylated state BY kinases form an ocatamer in which the YC cluster of one polypeptide resides in the catalytic site of an adjacent polypeptide (Olivares-Illana et al., 2008). Upon phosphorylation of the YC cluster, the ocatmer dissociates and the monomeric polypeptides can interact with substrate proteins (Olivares-Illana et al., 2008). Additionally an arginine and alanine rich RK cluster contributes to subunit interactions and nucleotide binding (Bechet et al., 2010). Within this overall format, different bacteria can utilize BY kinase domains in a distinct manner and catalytic activity is not always required for all of the functions contributed by BY kinases, which can also include scaffolding and allosteric functions (Mijakovic et al., 2016). The Ptk1 BY kinase of P. gingivalis possesses the Walker A and B, RK and YC domains, and in this study utilizing mutant recombinant proteins we show for the first time that all of these domains are required for autophosphorylation and phosphorylation of a substrate protein, UDP-acetyl-mannosamine dehydrogenase.
The typical YC cluster of BY kinases varies from 10–20 amino acids and contains 3–7 tyrosine residues (Grangeasse et al., 2012). Ptk1 possesses a 10 amino acid region spanning residues 781 to 790 that contains 5 tyrosines, and MS analysis of the C-terminal of rPtk1 showed all of these tyrosines were phosphorylated. The phosphorylation status of Ptk1 in P. gingivalis cells will vary with activation state; however, as the recombinant protein is functional it is likely that this represents the highly phosphorylated state. Indeed, it has been proposed that it is simplistic to consider BY kinases as ‘active’ or ‘inactive’, but rather activity varies between a state where the YC motif is highly phosphorylated to a state where YC phosphorylation is greatly reduced (Doublet et al., 2002; Nadler et al., 2012; Temel et al., 2013). Dephosphorylation of YC occurs through the action of the tyrosine phosphatase, in many cases a member of the low molecular weight protein-tyrosine phosphatase (LMW-PTP) family (Nadler et al., 2012; Temel et al., 2013). In Gram-negative organisms the genes encoding the kinase and phosphatase are usually located in the same operon (Grangeasse et al., 2007). However, in the current P. gingivalis annotation, the gene for only predicted tyrosine phosphatase (Ltp1) an LMW-PTP family member, is located remotely from ptk1 (Naito et al., 2008). Nonetheless Ltp1 can dephosphorylate Ptk1 and the enzymes have opposing activities in P. gingivalis (Wright et al., 2014). In this study we found that Ptk1 can phosphorylate Ltp1, and may thus impact the activity of the phosphatase. In Mycobacterium tuberculosis the PtkA tyrosine kinase can phosphorylate the tyrosine phosphatase PtpA and enhance activity (Zhou et al., 2015). Additionally, phosphorylation of eukaryotic LMW-PTPs can regulate activity (Rigacci et al., 1996; Schwarzer et al., 2006), and it has recently been shown that Etp, a LMW-PTP in E. coli for Etk, is phosphorylated on a D-loop tyrosine, although this reaction is mediated by an unidentified enzyme that is neither of the cognate tyrosine kinases (Etk and Wzc) (Nadler et al., 2012).
In periodontal disease P. gingivalis is keystone pathogen which can increase the nososymbiocity of the heterotyptic bacterial community (Hajishengallis & Lamont, 2016). P. gingivalis is synergistically pathogenic with S. gordonii, and development of dual species communities is regulated by the activity of Ptk1 and Ltp1 (Maeda et al., 2008; Wright et al., 2014). The results of the current study show that the phosphotransfer functionality of Ptk1 is necessary for the accretion of P. gingivalis on a S. gordonii substratum. A P. gingivalis Δptk1 mutant is deficient in community development with S. gordonii (Wright et al., 2014), and herein we show that complementation of this mutation in trans with the wild type ptk1 allele restores the community phenotype, whereas complementation with the ptk1 Walker A mutation retains the mutant phenotype. Mechanistically, the role of tyrosine kinase activity may be in the phosphorylation of CdhR, a transcriptional regulator of the mfa1 gene encoding the Mfa1 fimbrial subunit protein which mediates attachment of P. gingivalis to S. gordonii. The effect of CdhR tyrosine phosphorylation on activity is currently under investigation. BY kinases in general tend to be promiscuous in substrate recognition, and the most extensively documented role of the enzymes is in the synthesis and export of extracellular polysaccharides (Mijakovic et al., 2016). In the archetypal Wzy-dependent biosynthesis pathway in E. coli, the Wzc and Etk proteins are BY-kinases which are thought to function as co-polymerases controlling both the polymerization and translocation of polysaccharide (Cuthbertson et al., 2009; Whitfield, 2006). The precise nature of BY kinase involvement in the polysaccharide assembly machinery is still unknown, and they may also function as molecular scaffolds for activity of the flippase Wzx, the polymerase Wzy and the outer membrane translocon Wza (Bechet et al., 2010). In P. gingivalis the Wza homolog PGN_1523 is immediately upstream of ptk1 and both are co-transcribed in an operon with a gene encoding a putative capsular polysaccharide biosynthesis protein (Wright et al., 2014). P. gingivalis also possess a functional homolog of Wzz, (WzzP) (Shoji et al., 2013), and potential Wzy and Wzx proteins have also been identified (Paramonov et al., 2009; Shoji & Nakayama, 2016); however these are involved in the biosynthesis of O polysaccharide in LPS (Shoji & Nakayama, 2016). While the machinery for capsular and extracellular polysaccharide production and secretion remains to be unraveled, the results of the current study, combined with previous reports from our laboratory (Wright et al., 2014), establish the importance of tyrosine phosphorylation/dephosphorylation in EPS production by P. gingivalis.
Supplementary Material
Table S1. Primers used in this study
Acknowledgments
We thank the NIH/NIDCR for support through DE012505. The authors have no conflicts of interests to declare.
References
- Bechet E, Gruszczyk J, Terreux R, et al. Identification of structural and molecular determinants of the tyrosine-kinase Wzc and implications in capsular polysaccharide export. Mol Microbiol. 2010;77:1315–1325. doi: 10.1111/j.1365-2958.2010.07291.x. [DOI] [PubMed] [Google Scholar]
- Caster DJ, Korte EA, Merchant ML, et al. Autoantibodies targeting glomerular annexin A2 identify patients with proliferative lupus nephritis. Proteomics Clin Appl. 2015;9:1012–1020. doi: 10.1002/prca.201400175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Hirano T, Bainbridge BW, Demuth DR, Xie H, Lamont RJ. Community signalling between Streptococcus gordonii and Porphyromonas gingivalis is controlled by the transcriptional regulator CdhR. Mol Microbiol. 2010;78:1510–1522. doi: 10.1111/j.1365-2958.2010.07420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L, Mainprize IL, Naismith JH, Whitfield C. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol Mol Biol Rev. 2009;73:155–177. doi: 10.1128/MMBR.00024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daep CA, Novak EA, Lamont RJ, Demuth DR. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect Immun. 2011;79:67–74. doi: 10.1128/IAI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublet P, Grangeasse C, Obadia B, Vaganay E, Cozzone AJ. Structural organization of the protein-tyrosine autokinase Wzc within Escherichia coli cells. J Biol Chem. 2002;277:37339–37348. doi: 10.1074/jbc.M204465200. [DOI] [PubMed] [Google Scholar]
- Doublet P, Vincent C, Grangeasse C, Cozzone AJ, Duclos B. On the binding of ATP to the autophosphorylating protein, Ptk, of the bacterium Acinetobacter johnsonii. FEBS Lett. 1999;445:137–143. doi: 10.1016/s0014-5793(99)00111-8. [DOI] [PubMed] [Google Scholar]
- Elsholz AK, Wacker SA, Losick R. Self-regulation of exopolysaccharide production in Bacillus subtilis by a tyrosine kinase. Genes Dev. 2014;28:1710–1720. doi: 10.1101/gad.246397.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee CL, Papavinasasundaram KG, Blair SR, et al. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci Signal. 2012;5:ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeasse C, Cozzone AJ, Deutscher J, Mijakovic I. Tyrosine phosphorylation: an emerging regulatory device of bacterial physiology. Trends Biochem Sci. 2007;32:86–94. doi: 10.1016/j.tibs.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Grangeasse C, Nessler S, Mijakovic I. Bacterial tyrosine kinases: evolution, biological function and structural insights. Philos Trans R Soc Lond B Biol Sci. 2012;367:2640–2655. doi: 10.1098/rstb.2011.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeasse C, Obadia B, Mijakovic I, Deutscher J, Cozzone AJ, Doublet P. Autophosphorylation of the Escherichia coli protein kinase Wzc regulates tyrosine phosphorylation of Ugd, a UDP-glucose dehydrogenase. J Biol Chem. 2003;278:39323–39329. doi: 10.1074/jbc.M305134200. [DOI] [PubMed] [Google Scholar]
- Gruszczyk J, Olivares-Illana V, Nourikyan J, et al. Comparative analysis of the Tyr-kinases CapB1 and CapB2 fused to their cognate modulators CapA1 and CapA2 from Staphylococcus aureus. PLoS One. 2013;8:e75958. doi: 10.1371/journal.pone.0075958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur J Immunol. 2014;44:328–338. doi: 10.1002/eji.201344202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. Dancing with the Stars: How Choreographed Bacterial Interactions Dictate Nososymbiocity and Give Rise to Keystone Pathogens, Accessory Pathogens, and Pathobionts. Trends Microbiol. 2016 doi: 10.1016/j.tim.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kuboniwa M, Lamont RJ. Subgingival biofilm formation. Periodontol 2000. 2010;52:38–52. doi: 10.1111/j.1600-0757.2009.00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M, Tribble GD, James CE, et al. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol Microbiol. 2006;60:121–139. doi: 10.1111/j.1365-2958.2006.05099.x. [DOI] [PubMed] [Google Scholar]
- Maeda K, Tribble GD, Tucker CM, et al. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol Microbiol. 2008;69:1153–1164. doi: 10.1111/j.1365-2958.2008.06338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I, Grangeasse C, Turgay K. Exploring the diversity of protein modifications: special bacterial phosphorylation systems. FEMS Microbiol Rev. 2016;40:398–417. doi: 10.1093/femsre/fuw003. [DOI] [PubMed] [Google Scholar]
- Mijakovic I, Poncet S, Boel G, et al. Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J. 2003;22:4709–4718. doi: 10.1093/emboj/cdg458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler C, Koby S, Peleg A, et al. Cycling of Etk and Etp phosphorylation states is involved in formation of group 4 capsule by Escherichia coli. PLoS One. 2012;7:e37984. doi: 10.1371/journal.pone.0037984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito M, Hirakawa H, Yamashita A, et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 2008;15:215–225. doi: 10.1093/dnares/dsn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Olivares-Illana V, Meyer P, Bechet E, et al. Structural basis for the regulation mechanism of the tyrosine kinase CapB from Staphylococcus aureus. PLoS Biol. 2008;6:e143. doi: 10.1371/journal.pbio.0060143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramonov NA, Aduse-Opoku J, Hashim A, Rangarajan M, Curtis MA. Structural analysis of the core region of O-lipopolysaccharide of Porphyromonas gingivalis from mutants defective in O-antigen ligase and O-antigen polymerase. J Bacteriol. 2009;191:5272–5282. doi: 10.1128/JB.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigacci S, Degl’Innocenti D, Bucciantini M, Cirri P, Berti A, Ramponi G. pp60v-src phosphorylates and activates low molecular weight phosphotyrosine-protein phosphatase. J Biol Chem. 1996;271:1278–1281. doi: 10.1074/jbc.271.3.1278. [DOI] [PubMed] [Google Scholar]
- Schwarzer D, Zhang Z, Zheng W, Cole PA. Negative regulation of a protein tyrosine phosphatase by tyrosine phosphorylation. J Am Chem Soc. 2006;128:4192–4193. doi: 10.1021/ja0585174. [DOI] [PubMed] [Google Scholar]
- Shoji M, Nakayama K. Glycobiology of the oral pathogen Porphyromonas gingivalis and related species. Microb Pathog. 2016;94:35–41. doi: 10.1016/j.micpath.2015.09.012. [DOI] [PubMed] [Google Scholar]
- Shoji M, Yukitake H, Sato K, et al. Identification of an O-antigen chain length regulator, WzzP, in Porphyromonas gingivalis. Microbiologyopen. 2013;2:383–401. doi: 10.1002/mbo3.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temel DB, Dutta K, Alphonse S, Nourikyan J, Grangeasse C, Ghose R. Regulatory interactions between a bacterial tyrosine kinase and its cognate phosphatase. J Biol Chem. 2013;288:15212–15228. doi: 10.1074/jbc.M113.457804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- Whitmore SE, Lamont RJ. Tyrosine phosphorylation and bacterial virulence. Int J Oral Sci. 2012;4:1–6. doi: 10.1038/ijos.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CJ, Burns LH, Jack AA, et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CJ, Xue P, Hirano T, et al. Characterization of a bacterial tyrosine kinase in Porphyromonas gingivalis involved in polymicrobial synergy. Microbiologyopen. 2014 doi: 10.1002/mbo3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Li W, Wong D, Xie J, Av-Gay Y. Phosphorylation control of protein tyrosine phosphatase A activity in Mycobacterium tuberculosis. FEBS Lett. 2015;589:326–331. doi: 10.1016/j.febslet.2014.12.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used in this study