Abstract
Companion diagnostics rely on genomic testing of molecular alterations to enable effective cancer treatment. Here we report the clinical application and validation of the Oncomine Focus Assay (OFA), an integrated, commercially available next-generation sequencing (NGS) assay for the rapid and simultaneous detection of single nucleotide variants, short insertions and deletions, copy number variations, and gene rearrangements in 52 cancer genes with therapeutic relevance. Two independent patient cohorts were investigated to define the workflow, turnaround times, feasibility, and reliability of OFA targeted sequencing in clinical application and using archival material. Cohort I consisted of 59 diagnostic clinical samples from the daily routine submitted for molecular testing over a 4-month time period. Cohort II consisted of 39 archival melanoma samples that were up to 15 years old. Libraries were prepared from isolated nucleic acids and sequenced on the Ion Torrent PGM sequencer. Sequencing datasets were analyzed using the Ion Reporter software. Genomic alterations were identified and validated by orthogonal conventional assays including pyrosequencing and immunohistochemistry. Sequencing results of both cohorts, including archival formalin-fixed, paraffin-embedded material stored up to 15 years, were consistent with published variant frequencies. A concordance of 100% between established assays and OFA targeted NGS was observed. The OFA workflow enabled a turnaround of 3½ days. Taken together, OFA was found to be a convenient tool for fast, reliable, broadly applicable and cost-effective targeted NGS of tumor samples in routine diagnostics. Thus, OFA has strong potential to become an important asset for precision oncology.
Abbreviations: CLPv2, Ion AmpliSeq Colon and Lung Cancer Research Panel v2; CRC, colorectal cancer; DNA, deoxyribonucleic acid; FFPE, formalin-fixed,paraffin-embedded; H&E, hematoxylin and eosin; IHC, immunohistochemistry; NSCLC, non–small cell lung cancer; NSCLC (adeno), non–small cell lung cancer, adenocarcinoma; NSCLC (squamous), non–small cell lung cancer, squamous cell carcinoma; NSCLC (pleo), non–small cell lung cancer, pleomorphic carcinoma; NSCLC (NOS), non–small cell lung cancer, not otherwise specified; NGS, next generation sequencing; OFA, Oncomine Focus Assay; pT, tumor stage as determined by pathological assessment; RNA, ribonucleic acid; SSM, superficially spreading melanoma; SCNV, somatic copy number variation
Introduction
Next-generation sequencing (NGS) is an emerging technology for molecular diagnostics. It enables the parallel identification of multiple genomic variants even from small tissue samples [1], [2]. Scalable and cost-effective NGS solutions to reliably identify therapeutically relevant genomic driver alterations of tumors are the prerequisite for precision oncology. However, implementing multiplexed and comprehensive NGS assays into the clinical routine is challenging because data analysis and interpretation require specialty infrastructure and expertise [3]. In addition, most established routine tests cannot assess somatic copy number variations (SCNVs) and/or gene fusions, which presently guide treatment selection for several common cancers [4]. To make precision medicine approaches available for all cancer patients, there is a need for fast, reliable, and cost-effective NGS systems that can detect all classes of currently clinically relevant genomic targets from routine formalin-fixed,paraffin-embedded (FFPE) tissues [5], [6], [7]. To address these challenges, targeted NGS solutions have been developed to identify recurrently altered oncogenes as well as tumor suppressors, genes with frequent high-level amplifications or deletions, and driving gene fusions in a variety of cancers [8]. However, this emerging approach has so far not been sufficiently evaluated on routine diagnostic FFPE material in terms of feasibility, reliability, cost, and capacity [6], [9], [10].
The Oncomine Focus Assay (OFA, Thermo Fisher Scientific, San Francisco, CA) is a targeted, multibiomarker NGS assay that enables fast simultaneous detection of hundreds of variants across 52 genes relevant to solid tumors [8], [11]. These variants are treatable by on-market oncology drugs approved by the U.S. Food and Drug Administration as well as drugs that are part of the National Comprehensive Cancer Network guidelines or are currently listed in clinical trials [8], [11]. The assay analyzes clinically relevant gene alterations including single nucleotide variants, short insertions and deletions, SCNVs, and gene fusions from DNA and RNA in a single workflow. It enables the detection of tumor-specific genomic alterations using low-input FFPE samples such as needle biopsies and fine needle aspirates and is compatible with benchtop Ion Torrent sequencers. The power of the OFA technology for identification of genetic alterations is underlined by the present application in the nationwide NCI–Molecular Analysis for Therapy Choice Trial [12]. Here we evaluated the performance and applicability of this novel targeted NGS assay for transfer into daily diagnostic practice.
Materials and Methods
Study Cohorts and Patient Selection
This study was carried out in accordance with the guidelines of the Cantonal Ethics Committee Basel (KEK-EKBBno. 326-12, 2016-01134, and 2016-01499). Cohort I consisted of 59 diagnostic FFPE tissue samples from 51 consecutive patients that were analyzed in the diagnostic routing during a 4-month time period (March-June 2016). All patients had a clinical indication for molecular testing and were informed about the purpose of the molecular analysis by the treating physician. The demographic and histopathological features of these prospectively collected samples are listed in Table 1. Cohort II consisted of 39 archival FFPE tissue samples from 39 patients with cutaneous malignant melanoma. Tissue blocks were up to 15 years old. Median follow-up was 19.5 months (range 0-62 months) for cases with lymph node or distant metastasis and 27 months (range 12-144 months) for cases without metastases. Patients in this cohort were characterized and described in a previous study (cohort 1 with 57 patients in Garg et al.). [13] Cases with insufficient material remaining on the tissue block were excluded from molecular analysis (n = 18). The clinicopathological features of cohort II are provided in Table 2.
Table 1.
Demographic and Histopathological Features of the Prospective Clinical Samples (Cohort I)
| Characteristic | n = 51 | % |
|---|---|---|
| Gender | ||
| Male | 28 | 54.9 |
| Female | 23 | 45.1 |
| Age | ||
| Median | 67 | – |
| Range | 31-85 | – |
| Tumor entities (n = 51) | ||
| Colorectal cancer | 20 | 39.2 |
| Non–small cell lung cancer | 19 | 27.3 |
| Thyroid cancer | 3 | 5.9 |
| Melanoma | 2 | 3.9 |
| Pancreatic cancer | 2 | 3.9 |
| Breast cancer | 2 | 3.9 |
| GIST | 1 | 2.0 |
| Nonmelanoma skin cancer | 1 | 2.0 |
| Erdheim Chester disease | 1 | 2.0 |
| Non–small cell lung cancer (n = 19) | ||
| Adenocarcinoma | 15 | 78.9 |
| Squamous cell carcinoma | 1 | 5.3 |
| Pleomorphic carcinoma | 1 | 5.3 |
| Combined⁎ | 1 | 5.3 |
| NOS | 1 | 5.3 |
| Primary tumor/metastasis | ||
| Primary tumor only | 32 | 62.7 |
| Metastasis only | 13 | 25.5 |
| Matched primary and metastasis | 3 | 5.9 |
| Two different primary tumors† | 3 | 5.9 |
| Nucleic acid analysis | ||
| DNA and RNA | 39 | 76.5 |
| DNA only | 12 | 23.5 |
| Tumor cell content | ||
| 10%-30% | 7 | 13.7 |
| 31%-70% | 15 | 29.4 |
| >70% | 29 | 56.9 |
Combined small cell carcinoma with adenocarcinoma and sarcoma components.
Two different, synchronous or metachronous primary tumors or different manifestations of the disease (for Erdheim Chester disease case) were analyzed.
Table 2.
Demographic and Histopathological Features of 39 Archival Cutaneous Melanoma Samples (Cohort II)
| Characteristic | n = 39 | % |
|---|---|---|
| Gender | ||
| Male | 24 | 61.5 |
| Female | 15 | 38.5 |
| Age | ||
| Median | 70 | – |
| Range | 31-91 | – |
| Melanoma types | ||
| Superficial spreading melanoma | 24 | 61.5 |
| Nodular melanoma | 15 | 38.5 |
| Metastases⁎ | ||
| Melanoma with metastases | 12 | 30.8 |
| Melanoma without metastases | 27 | 69.2 |
| Tumor cell content | ||
| 10%-30% | 9 | 23.1 |
| 31%-70% | 7 | 17.9 |
| >70% | 23 | 59.0 |
The cohort consisted of cases with lymph node and/or distant metastases detected at the time of first diagnosis or during a median follow-up of 19.5 months (range 0-62 months) and of cases with no evidence of no lymph node and/or distant metastases at the time of first diagnosis or during a median follow-up of 27 months (range 12-144 months).
Genomic Profiling of Samples by Targeted NGS
All samples in this study were analyzed using the commercially available OFA platform. The genes targeted in this panel are carefully selected biomarkers derived from expertly curated cancer genomics data [8]. The assay analyzes a maximum of six parallel samples per run for DNA and RNA. It can be used on FFPE samples (10 ng DNA and 10 ng DNase-treated RNA per reaction) and is compatible with benchtop Ion Torrent sequencers (Ion Personal Genome Machine, Ion Proton System, Ion S5 System, Thermo Fisher Scientific).
The percentage of tumor cells relative to other cells (e.g., stromal, inflammatory, and preexisting epithelial cells) was estimated on one hematoxylin and eosin (H&E) stained tumor section by a board-certified molecular pathologist (K.D.M.), and an area with a minimum tumor cell content of >20% was designated for the analysis. FFPE tissue sections (4 μm) on positively coated slides were deparaffinized by a standard procedure (2 × 15 minutes in xylol, 5 minutes in absolute EtOH). The defined tumor area was transferred into an Eppendorf LoBind PCR tube (Eppendorf, Hamburg, Germany). For the direct DNA extraction, the dissected tissue was mixed with digestion buffer (50mM Tris–-HCl pH 8.5, 1 mM EDTA pH 8.0, 5% Tween 20, 6 mg/ml of proteinase K; Qiagen, Hilden, Germany) followed by thermomixer incubation (1 hour at 56°C and 5 minutes at 95°C). This crude extract was directly processed or frozen at −20°C for long-term storage. RNA was extracted from FFPE samples using the Recover All Total Nucleic Acid Isolation kit by the “RNA Only” protocol as described by the manufacturer (Ambion, Thermo Fisher Scientific). Nucleic acid concentrations were measured by the Qubit dsDNA HS and the Qubit RNA HS Assay kits, respectively, with the Qubit Fluorometer, and RNA was reverse transcribed to cDNA using the SuperScript VILO cDNA synthesis kit (all from Thermo Fisher Scientific). Libraries were prepared using the Ion PGM Select Library kit and were equalized to 100 pm concentration using the Ion PGM Select Library Equalizer kit. Templation and enrichment were performed using the Ion OneTouch Select Template Kit (all from Thermo Fisher Scientific) on Ion OneTouch 2 and Ion OneTouch ES instruments. The library was sequenced using the Ion PGM sequencer (Thermo Fisher Scientific). The run was considered successful and the sequencing quality adequate when the following quality metrics were met: 1) mapped reads ≥300,000; 2) average base coverage depth ≥1000; 3) amplicons having at least 500 reads: ≥90%; 4) no strand bias: ≥90%; 5) amplicons read end-to-end: ≥85%. Five thousand mapped reads were set as the limit for a single RNA library. Primary analysis was performed by Torrent Server™ (v 5.0) and further by Ion Reporter™ Server hosting informatic tools (Ion Reporter™ Software v5.0) for variant analysis, filtering, and annotations. Automatic workflow (Oncomine Focus v2.0, DNA and fusions/DNA/fusions, Single Sample) with preconfigured parameter settings (Oncomine Variants 5% CI SCNV ploidy ≥ gain of 2 over normal) was utilized.
The performance of the OFA panel was ascertained using a highly multiplexed test panel, the AcroMetrix Oncology Hotspot Control (Thermo Fisher Scientific) targeting various mutations in 25 of the genes represented in the OFA panel (AKT1, ALK, BRAF, CTNNB1, EGFR, ERBB2, ERBB4, FGFR2, FGFR3, GNA11, GNAQ, HRAS, IDH1, IDH2, JAK2, JAK3, KIT, KRAS, MAP2K1, MET, NRAS, PDGFRA, PIK3CA, RET, SMO), with a frequency near the limit of detection (5%-35%). The performance of the OFA RNA part was ascertained using the Lung Panel (ALK-RET-ROS1) FFPE RNA Reference Standard (Horizon Discovery, Cambridge, UK).
Confirmation of Genetic Alterations by Alternative Methods
A random subset of samples of the two cohorts analyzed by the OFA was selected for additional validation by alternative methods. For validation of common hotspot mutations, pyrosequencing was performed using the PyroMark Q24 system (Qiagen, Hilden, Germany) and the following commercially available kits according to the manufacturer's instructions: therascreen BRAF Pyro Kit (BRAF codons 600 and 464-469), therascreen EGFR Pyro Kit (EGFR codons 719, 768, 790, 858-861, exon 9 deletions), therascreen KRAS Pyro Kit (KRAS codons 12,13, and 61), therascreen NRAS Pyro Kit (NRAS codons 12,13, and 61), and therascreen RAS Extension Pyro Kit (KRAS codons 59, 61, 117, 146; NRAS codons 59, 61, 117, 146). Four samples of cohort I [n = 4, the first three cases of non–small cell lung cancer (NSCLC) and the first case of colorectal cancer (CRC) during the 4-month time period] were analyzed by the Ion AmpliSeqColon and Lung Cancer Research Panel v2 (CLPv2, Thermo Fisher Scientific), an alternative targeted NGS panel (Supplementary Table S1). For cases with ALK1, ROS1, or MET inter- or intragenic fusions detected in the RNA part of the assay (n = 7) and for cases with SCNVs/amplifications affecting the genes CCND1 (Cyclin D1; n = 1) or MYC (n = 2), results were confirmed by immunohistochemistry (IHC) (Table 3; Supplementary Table S2). For the two breast cancer cases analyzed, HER2 silver in situ hybridization (SISH; Ventana/Roche Diagnostics, Mannheim, Germany) and HER2 IHC were performed according to the manufacturer's instructions (Supplementary Table S2). Our laboratory, including the above mentioned standard assays (IHC, SISH, pyrosequencing), is certified by the Swiss Accreditation Service (STS 0599) according to the ISO/IEC 17025 und ISO 15189.
Table 3.
Validation of the OFA Results by Orthogonal Testing (Cohort I)
| Patient | Diagnosis | OFA Result | Validation Method |
|---|---|---|---|
| 1 | NSCLC (squamous) | BRAF p. G469V FGFR1 amplification |
Pyrosequencing, CLPv2 – |
| 2 | NSCLC (adeno) | KRAS p. G12C KRAS p. A59G |
CLPv2⁎ |
| 3 | CRC | KRAS p. G13C PIK3CA p. H1047R |
CLPv2⁎ |
| 4 | Melanoma | NRAS p. A146T | Pyrosequencing |
| 5 | NSCLC (pleo) | KRAS p. G12A | CLPv2 |
| 8 | NSCLC (adeno), primary NSCLC (sarcoma), primary NSCLC (sarcoma), metastasis SCLC, primary |
EGFR p. L858R EGFR p. L858R FGFR1 amplification MYC amplification EGFR p. L858R, FGFR1 amplification, MYC amplification – |
Pyrosequencing Pyrosequencing – – Pyrosequencing – – Pyrosequencing |
| 10 | NSCLC (adeno) | – | Pyrosequencing† |
| 11 | Breast cancer | PIK3CA p. N345K | HER2 IHC, HER2 SISH |
| 13 | Breast cancer | PIK3CA p. E545K | HER2 IHC, HER2 SISH |
| 17 | NSCLC (adeno) | BRAF p. V600E | Pyrosequencing |
| 19 | NSCLC (adeno) | KRAS p. G12D EGFR amplification EGFRvIII MET exon 14 mutation |
– – – MET IHC |
| 23 | CRC | KRAS p. Q61K CCND1 amplification MYC amplification |
– CCND1 IHC MYC IHC |
| 25 | NSCLC (adeno) 1 NSCLC (adeno) 2 |
CD74 – ROS1 fusion CD74 – ROS1 fusion |
ROS1 IHC ROS1 IHC |
| 27 | CRC | BRAF p. V600E MYC amplification |
– MYC IHC |
| 29 | CRC | PIK3CA p. R93Q MAP2K1 (MEK) p. K57T MET exon 14 mutation |
– – MET IHC |
| 30 | Pancreatic cancer | KRAS p. G12D MET exon 14 mutation |
– MET IHC |
| 32 | NSCLC (adeno) | EML4 – ALK fusion | ALK IHC |
| 35 | CRC | KRAS p. A146T | Pyrosequencing |
| 39 | Melanoma | BRAF amplification CDK4 amplification |
– CDK4 IHC |
| 40 | NSCLC (adeno) | CD74 – ROS1 fusion | ROS1 IHC |
| 43 | Thyroid carcinoma (anaplastic) | NRAS p. Q61K MET exon 14 mutation |
– MET IHC |
| 44 | Thyroid carcinoma (papillary), primary Thyroid carcinoma (papillary), metastasis |
BRAF p. V600E BRAF p. V600E |
Pyrosequencing Pyrosequencing |
| 45 | NSCLC (adeno) | EGFR amplification MET amplification |
– MET IHC |
| 48 | CRC | KRAS p. G12V | Pyrosequencing |
| 50 | NSCLC (NOS) | MET amplification | MET IHC |
| 51 | Erdheim Chester disease 1 Erdheim Chester disease 2 |
BRAF p. V600E BRAF p. V600E |
Pyrosequencing Pyrosequencing |
Abbreviations: NSCLC, non–small cell lung cancer; NSCLC (adeno), non–small cell lung cancer, adenocarcinoma; NSCLC (squamous), non–small cell lung cancer, squamous cell carcinoma; NSCLC (pleo), non–small cell lung cancer, pleomorphic carcinoma; NSCLC (NOS), non–small cell lung cancer, not otherwise specified.
CLPv2 detected an additional TP53 mutation.
No EGFR or KRAS mutation detected by pyrosequencing.
Results
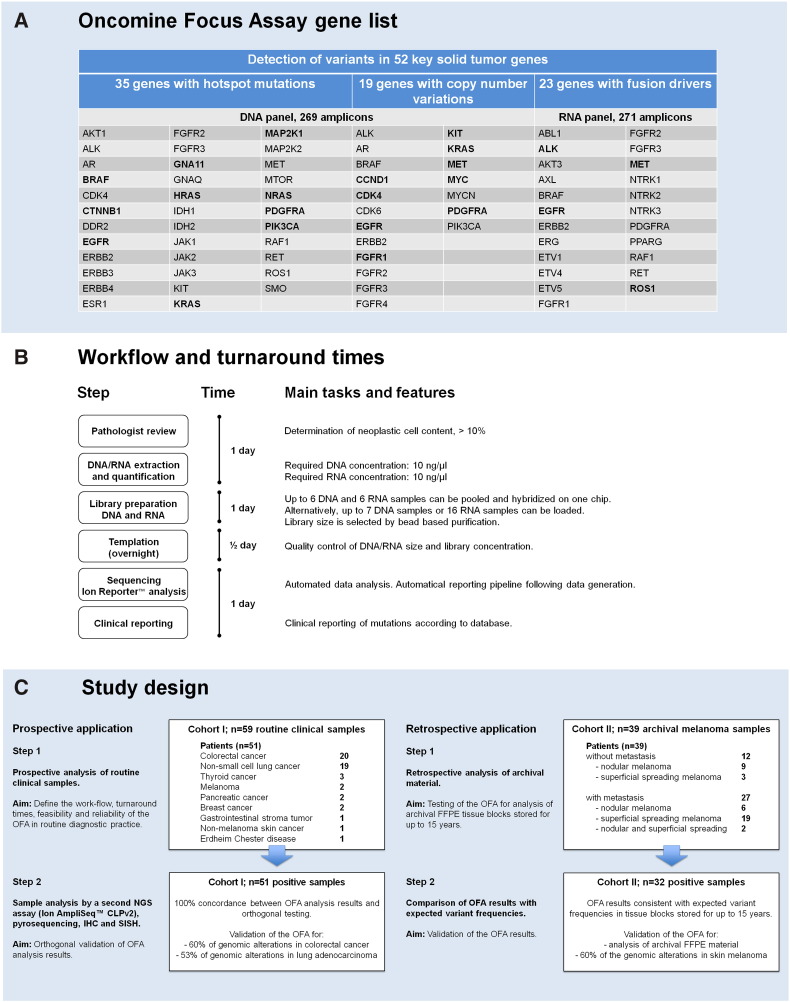
First, we evaluated the performance of the OFA in the prospective clinical setting of 59 diagnostic FFPE tissue samples from 51 consecutive patients of cohort I that were routinely submitted to our molecular pathology service during a 4-month time period (March-June 2016). The main entities that were analyzed during this time included CRC (20 patients, 39.2%) and NSCLC (19 patients, 37.3%). For the vast majority of patients, the treating physician was interested in tumor-specific genomic alterations with predictive value. In six cases, the treating physician requested information on clonal relationships between different primary tumors of the patient or between a primary tumor and metastases. Therefore, two different tissues were analyzed for five patients (two primary tumors or primary tumor and corresponding metastasis for patient nos. 16, 20, 25, 44, and 51;Supplementary Table S3), and four different tissues were analyzed for one patient (three different morphologic components of a combined small cell/non –small cell lung carcinoma and one corresponding metastasis in the small bowel, patient no. 8;Supplementary Table S3). All samples underwent successful targeted sequencing of the 52 OFA genes (Figure 1A). The OFA workflow enabled a fast turnaround of 3½ days (Figure 1B). Importantly, the nested design thereby enables simultaneous DNA and RNA analysis, with RNA extraction and analysis requiring an additional working time of approximately 2 hours.
Figure 1.
Molecular profiling with the OFA, a targeted multibiomarker NGS assay. (A) The OFA is a targeted NGS panel that includes 52 solid tumor genes associated with current oncology drugs and published evidence. It enables sequencing of 35 hotspot genes, 19 genes associated with copy number gain, and 23 fusion genes, all in a single workflow using the Ion PGM system. Genes printed in bold were detected in cases of the two cohorts studied here. (B) Workflow and turnaround time for molecular profiling of clinical FFPE samples using targeted NGS with the OFA. (C) Design of this study. The OFA was tested and validated on two different cohorts. Cohort I (left) consisted of 59 routine FFPE samples from 51 patients with various solid tumors that were routinely submitted to the molecular pathology service. Half of these samples were randomly selected for orthogonal validation by alternative tests. In addition, validation was performed by comparing the OFA results with expected mutation frequencies from the literature. Cohort II (right) was an archival cohort of 39 FFPE melanoma samples from 39 patients. Mutation frequencies were retrieved from the literature, and the samples of cohort II were analyzed by the OFA retrospectively. The OFA results of the melanomas in cohort II were consistent with the data that can be expected from large published cohorts of cutaneous melanomas.
In the OFA DNA analysis, 51 (86.4%) of the 59 prospectively collected samples from routine diagnostics showed at least 1 alteration, whereas only 8 samples (13.6%) did not show any alterations with this panel (Supplementary Table S3). Of the 51 samples with alterations detected by OFA, 34 (66.7%) had just 1 alteration, 11 (21.6%) had 2 alterations, 5 (9.8%) had 3 different alterations, and 1 sample (no. 19; 2%) had 4 different alterations. The most frequently mutated genes in these routine diagnostic cases across all tumor types included KRAS (20 patients, 39.2%), BRAF (7 patients, 13.7%), PIK3CA (5 patients, 9.8%), MET (4 patients, 7.8%), EGFR (3 patients, 5.9%), NRAS (3 patients, 5.9%), and CTNNB1 (3 patients, 5.9%). Analysis of copy number data revealed the most frequent copy number gains in MYC (4 patients, 7.8%), EGFR (2 patients, 3.9%), MET (2 patients, 3.9%), and FGFR1 (2 patients, 3.9%). One of the two melanomas included in this routine diagnostics cohort was found to have amplifications in the region of the BRAF (chromosome 7q34, copy number 18.25) and CDK4 (chromosome 12q14.1, copy number 48.65) gene (no. 39, 2%). Most copy number gains co-occurred with other relevant hotspot mutations, and only three patients (nos. 39, 45, 50; 5.9%) revealed isolated amplifications and no additional hotspot mutations.
For the two breast cancer cases included in this study (no. 11, 13), HER2 IHC showed a strong membranous positivity (score 3+) without evidence of ERBB2 (HER2) amplification by SISH. Molecular testing was performed due to these inconsistent results. In both cases, the OFA showed neither ERBB2 amplification nor ERBB2 mutation. Thus, the OFA supports the in situ hybridization results, considered the gold standard for analysis of HER2 in breast cancer.
In all five patients with two different samples analyzed (nos. 16, 20, 25, 44, 51;Supplementary Table S3), both samples (two primary tumors or one primary and one metastasis) showed exactly the same alterations. In one single patient with a combined smallcell lung carcinoma with adenocarcinoma and sarcoma components (no. 8), four different samples were analyzed. In this case, an activating EGFR mutation in exon 21 (p. L858R) was detected both in the adenocarcinoma and in the sarcoma components of the primary tumor as well as in a metastasis with sarcomatous differentiation. The sarcomatous primary tumor component and the sarcomatous metastasis additionally showed FGFR1 and MYC amplifications. No alterations were detected in the small cell component of the primary tumor by OFA.
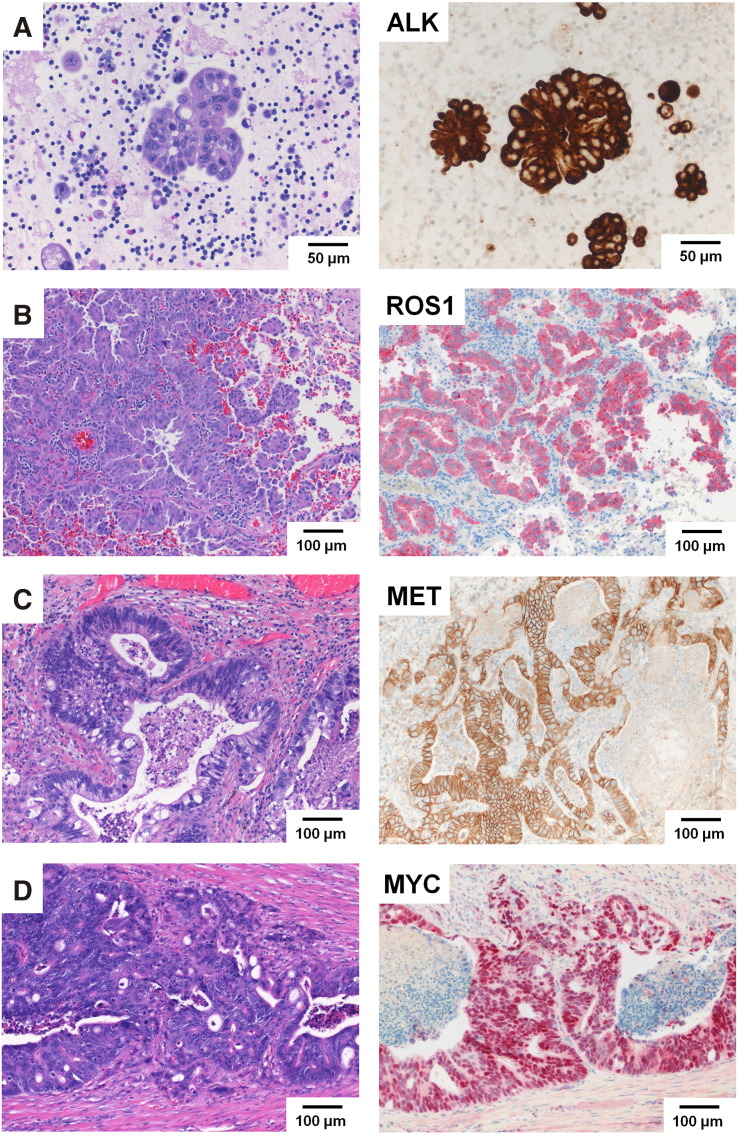
In the OFA RNA analysis, a total of eight driver fusions were identified in MET (four patients, 7.8%), ROS1 (two patients, 3.9%), ALK (one patient, 2%), and EGFR (EGFRvIII, one patient, 2%). The inter- or intragenic MET, ROS1, and ALK fusions in seven of these patients were confirmed by IHC demonstrating protein overexpression of MET, ROS1, and ALK, respectively (Figure 2, Table 3). All four patients with MET mutations detected in the OFA RNA panel showed a fusion of MET exon 13 with exon 15 (so-called MET “exon 14 skipping” mutations). This was a relatively frequent event in our routine diagnostic cohort and was detected in various unexpected entities (one adenocarcinoma of the lung, one CRC, one pancreatic cancer, and one anaplastic thyroid carcinoma; Figure 2). The MET exon 14 skipping mutations were not associated with MET genomic amplifications [14]. The intragenic MET mutations were not isolated events but co-occurred with hotspot mutations of KRAS, NRAS, PIK3CA, or MAP2K1 (nos. 19, 29, 30, 43 in Table 3 and Supplementary Table S3). Similarly, the EGFRvIII intragenic mutation (EGFR variant III) co-occurred in an NSCLC (adenocarcinoma) with a classic driver mutation in the KRAS gene (p. G12D), an EGFR amplification, and a MET exon 14 mutation (no. 19 in Table 3 and Supplementary Table S3) [15].
Figure 2.
Validation of gene fusions and SCNVs detected in routine FFPE samples. (A) EML4-ALK intrachromosomal fusion found in sample no. 32 (NSCLC, adenocarcinoma, H&E left). Confirmation of ALK overexpression due to this EML4-ALK gene fusion by IHC (right). (B) CD74-ROS1 fusion found in sample no. 25 (NSCLC, adenocarcinoma, H&E left). Confirmation of ROS1 overexpression due to this gene fusion between CD74 (Exon 6) and ROS1 (Exon 34) by IHC (right). (C) Intragenic MET fusion (Exon 13-Exon 15; MET “exon 14 skipping”) found in sample no. 29 (lung metastasis of CRC, H&E left). Confirmation of membranous MET overexpression due to this MET exon 14 mutation by IHC (right). (D) MYC amplification found in sample no. 27 (CRC, H&E left). The gene region 8q24.21 on chromosome 8 had a copy number of 25. Confirmation of MYC amplification by IHC (right).
Half of the above patients (n = 26) were picked at random for orthogonal validation of the alterations identified by OFA using a variety of other established analysis techniques (Table 3). Orthogonal testing included pyrosequencing and an alternate NGS assay (CLPv2) to confirm the presence or the absence of selected hotspot mutations. IHC and for some samples also SISH were used to validate the predicted copy number gains or gene fusions observed by OFA (Figure 2, Table 3). Overall, a concordance rate of 100% between the results of the established assays and the novel targeted NGS approach with OFA was observed. In one case of Erdheim Chester disease (no. 51), two different archival samples obtained at two different time points were retrospectively analyzed by OFA to confirm a BRAF p. V600E mutation previously detected by pyrosequencing. Indeed, OFA confirmed this result. In patient nos. 2 and 3 of cohort I, an additional TP53 mutation was detected by the CLPv2 (p. M246I in no. 2, p. R267W in no. 3). However, this is not considered as a discrepancy between the two assays because the TP53 gene is not a therapeutically relevant target and, as such, is not represented in the OFA panel.
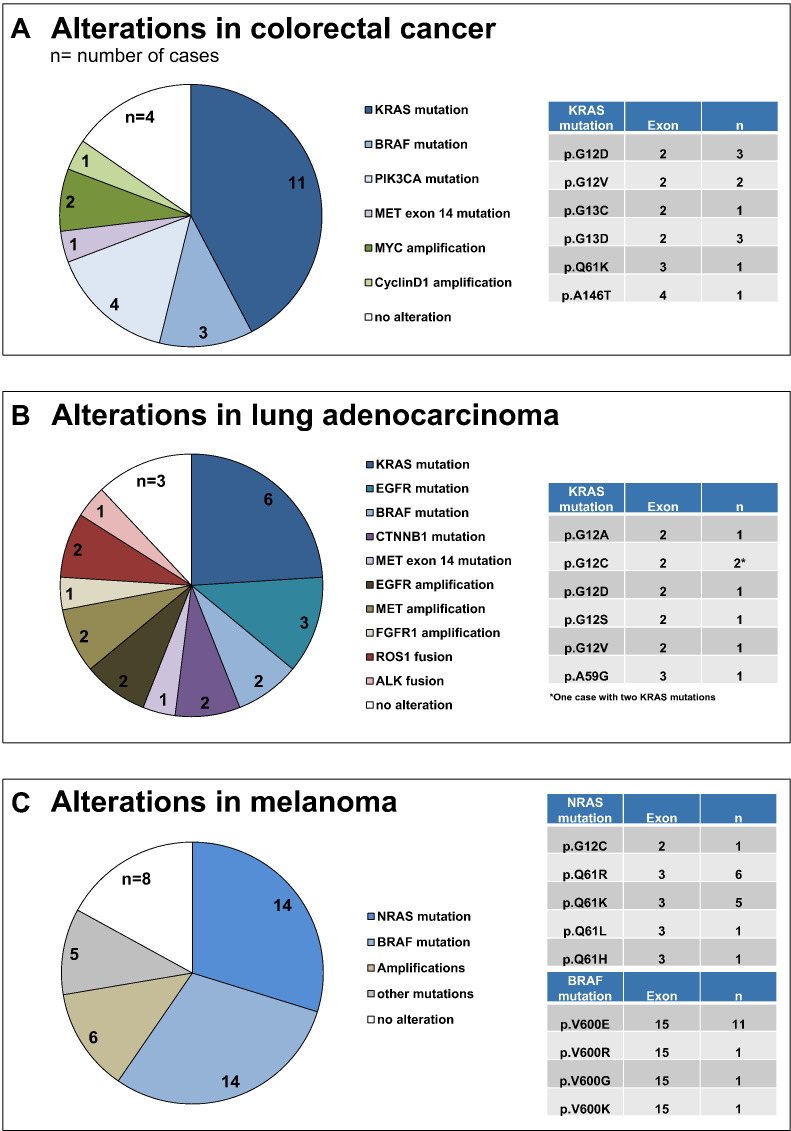
Comparison of the OFA results with the expected variant frequencies from the literature showed consistent results. Of the 20 CRC patients, 11 (55%) harbored a KRAS mutation and 3 (15%) a BRAF mutation. Of the 19 NSCLC patients, 6 (31.6%) had a KRAS mutation [1 of these 6 patients (no. 2) had two different KRAS mutations in codons 12 and 59;Supplementary Table S3], 2 (10.5%) an EGFR mutation, 2 (10.5%) a ROS1 rearrangement, and 1 (5.3%) an ALK rearrangement. This means that, for most known mutations, the OFA detection rate was in the range or above expected variant frequencies (Figure 3) [16], [17], [18]. The OFA found additional actionable alterations as a result of the broader target range (ability of OFA to detect SCNVs and driver gene fusions that cannot be detected by other commercial NGS assays) compared with traditional methods. In some cases, major driver mutations coexisted with other relevant variants, as in the CRC case nos. 3, 6, and 14 which showed KRAS mutations (p. G13C, p. G12D, p. G13D) as well as a mutation in PIK3CA (p. H1047R). PIK3CA mutations were not detected as an isolated event in our cohort [19].
Figure 3.
Genomic aberrations in different tumor types. (A) Actionable variants in 20 CRC patients (n = number of patients) (left). Distribution of KRAS mutations that were detected in 11 (55%) CRC patients (right). (B) Actionable variants detected in 19 NSCLC patients (left). Distribution of KRAS mutations that were detected in six (31.6%) NSCLC patients (right). One NSCLC patient harbored two different KRAS mutations (p. G12C and p. A59G). (C) Actionable variants detected in 39 melanoma patients (left). Distribution of BRAF (right, top panel) and NRAS (right, bottom panel) mutations identified in 14 (35.9%) melanomas.
Next, we applied the OFA to an independent, well-characterized cohort of 39 archival cutaneous malignant melanoma samples [13]. For these cases, DNA was isolated from archival FFPE blocks of tumor samples that were up to 15 years old. All samples underwent successful targeted sequencing of the 41 genes included in the OFA DNA panel. Eight samples (20.5%) showed no relevant genomic alteration detectable in the OFA DNA panel, 25 samples (64.1%) had 1 alteration, 4 samples (10.3%) had 2 alterations, 2 samples (5.1%) had 3 alterations, and 1 sample (2.6%) had 4 alterations. As expected from large cohort studies, the most frequently mutated genes were BRAF (14 samples, 35.9%) and NRAS (14 samples, 35.9%) (Figure 3, Supplementary Table S4) [20], [21]. Amplifications were rare events and were found in the regions of the CCND1, CDK4, KIT, KRAS, and PDGFRA genes (Figure 3, Supplementary Table S4) [20]. Altogether, amplifications were detected in 4 of the 39 patients (10.3% of patients). In two of these patients (no. 2, 24), copy number gains co-occurred with relevant driver hotspot mutations of the NRAS gene. Five melanoma samples were randomly selected for further validation by pyrosequencing which confirmed the OFA results.
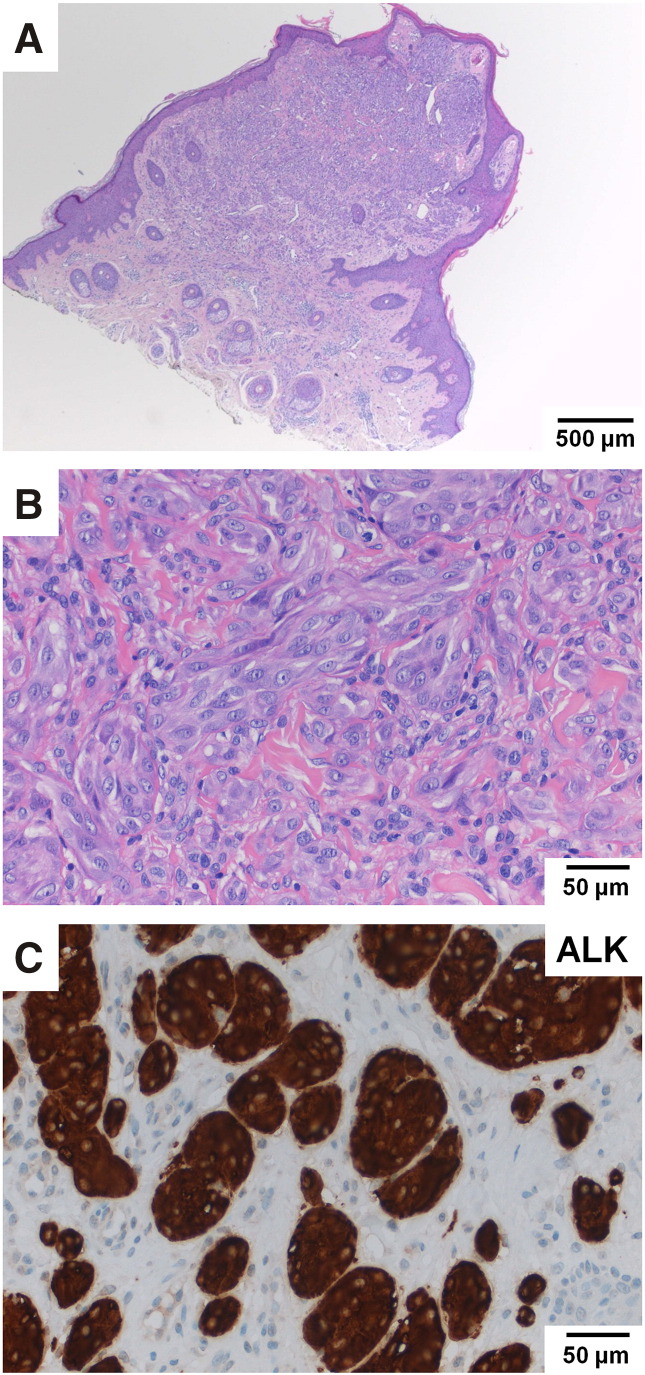
Kinase fusions that can be detected in the OFA RNA panel occur in a mutually exclusive pattern in cutaneous melanomas and are not detected in melanomas with the classic BRAF or NRAS driver mutations [21], [22]. Therefore, RNA analysis was performed only in the 10 melanoma samples without a detectable driver mutation in the OFA DNA panel (“wild-type subtype”) (Supplementary Table S4). Seven cases did not show any relevant alteration in the OFA RNA panel. One case (2.6% of all patients) showed an ALK rearrangement leading to a fusion between the TPM3 gene (exon 7) on chromosome 1q21 and the ALK gene (exon 20) on chromosome 2p23. Because of this genomic rearrangement, TPM3 exon 7 is fused in-frame with ALK exon 20, preserving the transforming tyrosine kinase domain of ALK. The expression of this chimeric transcript was confirmed by IHC showing a strong cytoplasmic ALK positivity of the archival melanoma (no. 5, Figure 4, Supplementary Table S4). For two cases (nos. 17, 21), it was not possible to isolate RNA because of insufficient material remaining in the tissue block.
Figure 4.
Detection and validation of an ALK translocation in one archival melanoma. (A) Relatively symmetrical, exophytic, predominantly intradermal melanocytic tumor with focal epidermal hyperplasia from the right earlobe of a 30-year-old man. (B) Plexiform growth pattern and intersecting fascicles of fusiform melanocytes. Proliferation of spindle and epithelioid melanocytes. (C) The neoplastic melanocytes are positive for ALK in IHC, with strong staining of the cytoplasm. The ALK expression confirms the presence of a TPM3-ALK fusion in this melanoma which was retrospectively reclassified as a melanoma with spitzoid features.
Discussion
Molecular profiling of cancer has become essential to predict therapeutic response to targeted therapies. NGS has enabled genomewide personalized oncology efforts for actionable variant identification and prioritization. Here, we conducted a comprehensive analysis and validation of a novel amplicon-based NGS solution for identification of relevant somatic alterations in solid tumors. First, we characterized the workflow, turnaround times, feasibility, and reliability in the analysis of routine clinical tissue samples. OFA performance was assessed using independent methods to confirm the detected alterations. Second, we tested the retrospective analysis of archival FFPE tissue samples in an independent cohort of well-characterized malignant melanoma cases. In both settings, the OFA was found to be a convenient tool for fast, reliable, easy-to-use, broadly applicable, and cost-effective targeted NGS analysis.
Multibiomarker NGS assays from numerous commercial and academic providers are rapidly reaching clinical application. Lower cost and wider availability of NGS now raise the debate over the merit of routine tumor genomic analysis. Although some genetic lesions are targeted by a new generation of cancer therapies and certain treatment regimens are coupled to single gene assays, we still do not know if the vast majority of information on other genomic alterations is worth the added cost and if assignment of patients to off-label treatment with a targeted agent might carry potentially serious side effects. We tested the OFA NGS solution which focuses on a carefully selected panel of genomic alterations with therapeutic relevance in comparison to other amplicon-based assays on the market (Supplementary Table S1) [8]. The OFA thereby promises to avoid time consuming and costly analyses of molecular changes without known predictive value. As defined molecular alterations are currently used to drive treatment selection, the OFA appears to be a highly suitable tool for molecular routine diagnostics [23], [24], [25], [26]. Further, the high sensitivity of the OFA allows to study cases with intratumoral genetic heterogeneity and potential low-level alterations, as is the case for acquired mutations conferring resistance to tyrosine kinase inhibitor therapies [27], [28]. In the present study, several actionable somatic alterations were detected in NSCLC samples with tumor purity as low as 10% to 20%.
Before NGS technologies can be applied in the daily routine, systematic studies are needed to ensure consistent and reliable assay performance. Comparison with current conventional tests with respect to turnaround time, sensitivity, specificity, mutation detection limits, costs, and feasibility is required. In the present study, we identify a 100% concordance between the OFA results and alternative methods including pyrosequencing and an alternate NGS assay (CLPv2) in two independent patient cohorts. Of the 35 hotspot genes included in the OFA panel, mutations in 10 different genes were detected in the 2 cohorts of this study (Figure 1A, printed in bold). Alterations were identified in 9/19 genes associated with copy number gains and 4/23 fusion genes (Figure 1A, printed in bold). Overall, genomic alterations including hotspot mutations, amplifications, and gene fusions in 18 (34.6%) of the 52 therapy relevant genes covered by the OFA were detected. Of these 18 alterations, we validated 11 (61.1%, 21.2% of all 52 cancer genes included in the OFA) with alternative methods (BRAF, EGFR, KRAS, NRAS hotspot mutations, CCND1, CDK4, MET, MYC amplifications, ALK, MET, ROS1 driver gene fusions). These alterations represent the most common changes observed in CRC, NSCLC, and malignant melanoma [16], [17], [18], [20]. KRAS/NRAS mutations occur in approximately 50% of CRCs, 32% of NSCLCs (adenocarcinoma), and 28% of cutaneous melanomas. BRAF mutations are found in 10% of CRCs, 7% of NSCLCs (adenocarcinoma), and 52% of cutaneous melanomas. EGFR mutations, and ALK and ROS1 gene fusions are identified in 11.3%, 1.3%, and 1.7% of NSCLCs (adenocarcinoma), respectively [16], [17], [18], [20]. Taken together, this study validated the OFA for detection of approximately 60% of genomic alterations occurring in CRCs, 53% of those occurring in NSCLCs (adenocarcinoma), and 70% of the alterations commonly found in cutaneous melanomas. The conformity of the data with published variant frequencies further supports that this novel targeted NGS approach is a reliable and reproducible method for detection of therapeutically relevant genomic alterations in these entities. Application of the OFA was also successfully tested in samples of breast, gastrointestinal, and thyroid cancer but needs to be independently confirmed due to the limited number of cases under study.
We illustrate that the OFA can reliably detect single genomic events in rare tumor entities, as illustrated by the detection of a rare TPM3-ALK translocation in one archival melanoma sample that was previously classified as an ulcerated cutaneous melanoma (nodular type). The patient was initially diagnosed with stage pT4b disease and skin metastases but was alive without evidence for disease recurrence 9 years after the initial diagnosis. ALK fusions are typically found in Spitz tumors of the skin, including Spitz naevi, atypical Spitz tumors, and spitzoid melanomas [21], [22], [29], [30]. In these cases, ALK fusions are associated with a good prognosis and might be targetable with tyrosine kinase inhibitors such as crizotinib [21], [22], [29], [30]. The OFA result therefore provided a retrospective explanation for the unusual clinical course of this melanoma patient. Further, the OFA analysis also offered valuable information for a potential systemic therapy in the case of future recurrence and prompted a reclassification of the tumor based on molecular changes. This case illustrates the importance of a correlation between genomic aberrations and histopathological evaluation. For example, a patient with an ALK fusion–positive spitzoid melanoma can be expected to have a favorable prognosis, but a tumor with the same morphological characteristics and a BRAF mutation should raise the suspicion of an aggressive clinical course and should result in timely clinical intervention [20], [21]. In these situations, integration of clinical, histopathological, and molecular data, ideally in the context of an interdisciplinary molecular tumor board, plays an essential role for diagnosis, prognostic assessment, and targeted treatment of tumors [31].
MET intragenic fusions with skipping of exon 14 were relatively frequently detected by the OFA in our routine diagnostic cohort (cohort I) [14], [32], [33], [34], [35], [36]. All four cases with MET exon 14 mutations were validated by IHC in this study and showed a strong and homogenous membranous MET overexpression in the tumor cells. The high frequency of these MET fusions could be due to a bias by the small number of cases included in this study and therefore requires further validation in larger cohorts. MET mutations were also unexpectedly detected in tumor entities such as pancreatic and thyroid cancer that are not routinely submitted for molecular analysis by NGS [33], [34], [36]. Thus, the high number of MET intragenic fusions reported here could also be due to the fact that molecular diagnostic approaches are not routinely applied on these rare and fatal malignancies with limited or no conventional therapeutic options. In addition, splicing of the MET gene is complex, variable, and poorly understood [14], [37]. Thus, we cannot exclude that the OFA might detect splicing variants of unknown significance. As it is presently unknown whether METmutations in these tumor entities are also responsive to MET inhibition, further studies are needed to investigate the therapeutic relevance of these findings.
The retrospective analysis of archival material and tissue bank samples is of central importance for patients with relapsed disease. However, the analysis of FFPE tissue can be challenging due to fixation-induced crosslinking and fragmentation of nucleic acids. [38], [39] Here, we successfully applied the OFA for analysis of archival FFPE tissue blocks of malignant melanoma aged up to 15 years. The frequency of detected genomic alterations in the 41 genes included in the OFA DNA panel was consistent with published data. This indicates that the OFA is a reliable and reproducible method for analysis of archival FFPE material. We successfully performed RNA extraction and analysis in 8 out of 10 cases where no relevant driver mutations were detected in the DNA panel. Importantly, RNA quality from FFPE material has been shown to be less affected than DNA by fixation-induced crosslinking and fragmentation, allowing the application of the OFA RNA panel for identification of relevant driver gene fusions in the retrospective setting [38], [39].
Eight samples of the routine diagnostics cohort (13.6%, cohort I) and seven melanoma samples (17.9%, cohort II) did not show any detectable alterations with the OFA. For these cases, we cannot exclude the possibility that they are “false negative” and may show genomic alterations with other NGS assays analyzing targets that are not covered by the OFA and thus cannot be detected with this assay. However, the vast majority of additional genetic alterations likely have no predictive value at the present time. The issue of “false-negative” results is exemplified by the four samples that underwent additional analysis by the CLPv2 assay. Additional TP53 mutations were identified in two of these four samples. However, we do not consider these findings as a relevant discrepancy or disadvantage of the OFA because the TP53 gene is not a therapy-relevant target and as such is not included in the OFA panel.
In the retrospective cohort, 18 cases were excluded because insufficient material remained on the tissue blocks for analysis. This high proportion is explained by the prior use of material in research studies and is likely not representative of the exclusion rate in a standard setting. This is underlined by successful retrieval of tissue material in all of the 59 cases of the routine diagnostics cohort. Nevertheless, the careful disposition of diagnostic material is highly important for personalized molecular diagnostics. In the present study, several samples with a low tumor cell content of only 10% to 30% were successfully analyzed using both the DNA and RNA panel, underlining previous data on the feasibility of targeted, high-depth NGS in low-input samples [10].
Turnaround time and costs are key issues for the transfer of NGS to routine molecular diagnostics. The direct material costs for analysis of one sample by the OFA is approximately $450 in our laboratory including chemicals. Depending on the type of sample analyzed, the cost–-benefit ratio appears to be generally more favorable than that of conventional assays. As an example, the direct material costs for comprehensive testing of one lung cancer sample for EGFR, BRAF, KRAS, and NRAS mutations using pyrosequencing and fluorescence in situ hybridization for ALK and ROS presently amount to approximately $1000 in our laboratory. Another benefit of the OFA is a fast turnaround time which saves resources and associated costs. The required time from specimen acquisition to clinical reporting is only 3½ days, which is faster than any other NGS assay currently available on the market.
In conclusion, our study validates the OFA in clinical routine diagnostics and proves the reliability of this targeted NGS system for identification of actionable genetic alterations in FFPE tumor samples. Compared with conventional gene-specific assays, the OFA showed significant advantages in terms of sensitivity, costs, turnaround time, and a broader range of detectable relevant alterations. We are confident that the OFA has strong potential for routine clinical tumor testing and will promote precision oncology efforts at an unforeseeable pace.
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. In particular, no funding from Thermo Fisher Scientific was received for this study.
The following are the supplementary data related to this article.
Comparison of Amplicon-Based Sequencing Assays for Hotspot Mutations
List of Antibodies Used for Validation of the OFA Results
OFA Analysis Results of 59 Routine Diagnostic Samples from 51 Patients (Cohort I)
Alterations Detected by the OFA in 39 Archival Melanoma Samples (Cohort II)
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2017.01.003.
References
- 1.Shyr D, Liu Q. Next generation sequencing in cancer research and clinical application. Biol Proced Online. 2013;15:4. doi: 10.1186/1480-9222-15-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tripathy D, Harnden K, Blackwell K, Robson M. Next generation sequencing and tumor mutation profiling: are we ready for routine use in the oncology clinic? BMC Med. 2014;12:140. doi: 10.1186/s12916-014-0140-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strom SP. Current practices and guidelines for clinical next-generation sequencing oncology testing. Cancer Biol Med. 2016;13:3–11. doi: 10.28092/j.issn.2095-3941.2016.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luthra R, Chen H, Roy-Chowdhuri S, Singh RR. Next-generation sequencing in clinical molecular diagnostics of cancer: advantages and challenges. Cancers. 2015;7:2023–2036. doi: 10.3390/cancers7040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, Kalyana-Sundaram S, Sam L, Balbin OA, Quist MJ. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoogstraat M, Hinrichs JW, Besselink NJ, Radersma-van Loon JH, de Voijs CM, Peeters T, Nijman IJ, de Weger RA, Voest EE, Willems SM. Simultaneous detection of clinically relevant mutations and amplifications for routine cancer pathology. J Mol Diagn. 2015;17:10–18. doi: 10.1016/j.jmoldx.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Grasso C, Butler T, Rhodes K, Quist M, Neff TL, Moore S, Tomlins SA, Reinig E, Beadling C, Andersen M. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J Mol Diagn. 2015;17:53–63. doi: 10.1016/j.jmoldx.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hovelson DH, McDaniel AS, Cani AK, Johnson B, Rhodes K, Williams PD, Bandla S, Bien G, Choppa P, Hyland F. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia. 2015;17:385–399. doi: 10.1016/j.neo.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh RR, Patel KP, Routbort MJ, Reddy NG, Barkoh BA, Handal B, Kanagal-Shamanna R, Greaves WO, Medeiros LJ, Aldape KD. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15:607–622. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Hadd AG, Houghton J, Choudhary A, Sah S, Chen L, Marko AC, Sanford T, Buddavarapu K, Krosting J, Garmire L. Targeted, high-depth, next-generation sequencing of cancer genes in formalin-fixed, paraffin-embedded and fine-needle aspiration tumor specimens. J Mol Diagn. 2013;15:234–247. doi: 10.1016/j.jmoldx.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conley BA, Doroshow JH. Molecular analysis for therapy choice: NCI MATCH. Semin Oncol. 2014;41:297–299. doi: 10.1053/j.seminoncol.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Garg K, Maurer M, Griss J, Bruggen MC, Wolf IH, Wagner C, Willi N, Mertz KD, Wagner SN. Tumor-associated B cells in cutaneous primary melanoma and improved clinical outcome. Hum Pathol. 2016;54:157–164. doi: 10.1016/j.humpath.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 14.Awad MM, Oxnard GR, Jackman DM, Savukoski DO, Hall D, Shivdasani P, Heng JC, Dahlberg SE, Janne PA, Verma S. MET exon 14 mutations in non–small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J Clin Oncol. 2016;34:721–730. doi: 10.1200/JCO.2015.63.4600. [DOI] [PubMed] [Google Scholar]
- 15.Padfield E, Ellis HP, Kurian KM. Current therapeutic advances targeting EGFR and EGFRvIII in glioblastoma. Front Oncol. 2015;5:1–8. doi: 10.3389/fonc.2015.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research N Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cathomas G. PIK3CA in colorectal cancer. Front Oncol. 2014;4:1–4. doi: 10.3389/fonc.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas N Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiesner T, Kutzner H, Cerroni L, Mihm MC, Jr., Busam KJ, Murali R. Genomic aberrations in spitzoid melanocytic tumours and their implications for diagnosis, prognosis and therapy. Pathology. 2016;48:113–131. doi: 10.1016/j.pathol.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiesner T, He J, Yelensky R, Esteve-Puig R, Botton T, Yeh I, Lipson D, Otto G, Brennan K, Murali R. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun. 2014;5:3116. doi: 10.1038/ncomms4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwaederle M, Daniels GA, Piccioni DE, Fanta PT, Schwab RB, Shimabukuro KA, Parker BA, Kurzrock R. On the road to precision cancer medicine: analysis of genomic biomarker actionability in 439 patients. Mol Cancer Ther. 2015;14:1488–1494. doi: 10.1158/1535-7163.MCT-14-1061. [DOI] [PubMed] [Google Scholar]
- 24.Schwaederle M, Kurzrock R. Actionability and precision oncology. Oncoscience. 2015;2:779–780. doi: 10.18632/oncoscience.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwaederle M, Parker BA, Schwab RB, Daniels GA, Piccioni DE, Kesari S, Helsten TL, Bazhenova LA, Romero J, Fanta PT. Precision oncology: the UC San Diego Moores Cancer Center PREDICT experience. Mol Cancer Ther. 2016;15:743–752. doi: 10.1158/1535-7163.MCT-15-0795. [DOI] [PubMed] [Google Scholar]
- 26.Schwaederle M, Zhao M, Lee JJ, Eggermont AM, Schilsky RL, Mendelsohn J, Lazar V, Kurzrock R. Impact of precision medicine in diverse cancers: a meta-analysis of phase ii clinical trials. J Clin Oncol. 2015;33:3817–3825. doi: 10.1200/JCO.2015.61.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart EL, Tan SZ, Liu G, Tsao MS. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations—a review. Transl Lung Cancer Res. 2015;4:67–81. doi: 10.3978/j.issn.2218-6751.2014.11.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sundaresan TK, Sequist LV, Heymach JV, Riely GJ, Janne PA, Koch WH, Sullivan JP, Fox DB, Maher R, Muzikansky A. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin Cancer Res. 2016;22:1103–1110. doi: 10.1158/1078-0432.CCR-15-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeh I, de la Fouchardiere A, Pissaloux D, Mully TW, Garrido MC, Vemula SS, Busam KJ, LeBoit PE, McCalmont TH, Bastian BC. Clinical, histopathologic, and genomic features of Spitz tumors with ALK fusions. Am J Surg Pathol. 2015;39:581–591. doi: 10.1097/PAS.0000000000000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Busam KJ, Kutzner H, Cerroni L, Wiesner T. Clinical and pathologic findings of Spitz nevi and atypical Spitz tumors with ALK fusions. Am J Surg Pathol. 2014;38:925–933. doi: 10.1097/PAS.0000000000000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwaederle M, Parker BA, Schwab RB, Fanta PT, Boles SG, Daniels GA, Bazhenova LA, Subramanian R, Coutinho AC, Ojeda-Fournier H. Molecular tumor board: the University of California–San Diego Moores Cancer Center experience. Oncologist. 2014;19:631–636. doi: 10.1634/theoncologist.2013-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Awad MM. Impaired c-Met receptor degradation mediated by MET Exon 14 mutations in non–small-cell lung cancer. J Clin Oncol. 2016;34:879–881. doi: 10.1200/JCO.2015.64.2777. [DOI] [PubMed] [Google Scholar]
- 33.Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, Akimov M, Bufill JA, Lee C, Jentz D. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5:850–859. doi: 10.1158/2159-8290.CD-15-0285. [DOI] [PubMed] [Google Scholar]
- 34.Lee J, Ou SH, Lee JM, Kim HC, Hong M, Kim SY, Jang J, Ahn S, Kang SY, Lee S. Gastrointestinal malignancies harbor actionable MET exon 14 deletions. Oncotarget. 2015;6:28211–28222. doi: 10.18632/oncotarget.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Jia Y, Stoopler MB, Shen Y, Cheng H, Chen J, Mansukhani M, Koul S, Halmos B, Borczuk AC. Next-generation sequencing of pulmonary sarcomatoid carcinoma reveals high frequency of actionable MET gene mutations. J Clin Oncol. 2016;34:794–802. doi: 10.1200/JCO.2015.62.0674. [DOI] [PubMed] [Google Scholar]
- 36.Wasenius VM, Hemmer S, Karjalainen-Lindsberg ML, Nupponen NN, Franssila K, Joensuu H. MET receptor tyrosine kinase sequence alterations in differentiated thyroid carcinoma. Am J Surg Pathol. 2005;29:544–549. doi: 10.1097/01.pas.0000156103.37756.e2. [DOI] [PubMed] [Google Scholar]
- 37.Paik PK, Drilon A, Fan PD, Yu H, Rekhtman N, Ginsberg MS, Borsu L, Schultz N, Berger MF, Rudin CM. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015;5:842–849. doi: 10.1158/2159-8290.CD-14-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludyga N, Grunwald B, Azimzadeh O, Englert S, Hofler H, Tapio S, Aubele M. Nucleic acids from long-term preserved FFPE tissues are suitable for downstream analyses. Virchows Arch. 2012;460:131–140. doi: 10.1007/s00428-011-1184-9. [DOI] [PubMed] [Google Scholar]
- 39.Hedegaard J, Thorsen K, Lund MK, Hein AM, Hamilton-Dutoit SJ, Vang S, Nordentoft I, Birkenkamp-Demtroder K, Kruhoffer M, Hager H. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One. 2014;9:1–16. doi: 10.1371/journal.pone.0098187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of Amplicon-Based Sequencing Assays for Hotspot Mutations
List of Antibodies Used for Validation of the OFA Results
OFA Analysis Results of 59 Routine Diagnostic Samples from 51 Patients (Cohort I)
Alterations Detected by the OFA in 39 Archival Melanoma Samples (Cohort II)