Abstract
Medulloblastoma is the most common malignant brain tumor in children. Therapeutic approaches to medulloblastoma (combination of surgery, radiotherapy, and chemotherapy) have led to significant improvements, but these are achieved at a high cost to quality of life. Alternative therapeutic approaches are needed. Genetic mutations leading to the activation of the Hedgehog pathway drive tumorigenesis in ~30% of medulloblastoma. In a yeast two-hybrid proteomic screen, we discovered a novel interaction between GLI1, a key transcription factor for the mediation of Hedgehog signals, and PIN1, a peptidylprolyl cis/trans isomerase that regulates the postphosphorylation fate of its targets. The GLI1/PIN1 interaction was validated by reciprocal pulldowns using epitope-tagged proteins in HEK293T cells as well as by co-immunoprecipiations of the endogenous proteins in a medulloblastoma cell line. Our results support a molecular model in which PIN1 promotes GLI1 protein abundance, thus contributing to the positive regulation of Hedgehog signals. Most importantly, in vivo functional analyses of Pin1 in the GFAP-tTA;TRE-SmoA1 mouse model of Hedgehog-driven medulloblastoma demonstrate that the loss of Pin1 impairs tumor development and dramatically increases survival. In summary, the discovery of the GLI1/PIN1 interaction uncovers PIN1 as a novel therapeutic target in Hedgehog-driven medulloblastoma tumorigenesis.
Abbreviations: AD, activation domain; CGNP, cerebellar granular neuron progenitor cells; CIP, calf intestinal alkaline phosphatase; DB, DNA-binding domain; EGCG, epigallocatechin gallate; FFPE, formalin-fixed, paraffin-embedded; H&E, hematoxylin/eosin staining; Hh, Hedgehog; IHC, immunohistochemistry; IP, immunoprecipitation; MRI, magnetic resonance imaging; PPIase, peptidylprolyl cis/trans isomerase; pSer/Thr-Pro, phosphorylated serine-proline or threonine-proline motifs; tTA, tetracycline-regulated transactivators; TRE, tetracycline responsive element; SAG, SMO agonist; SBP, streptavidin binding peptide tag; WB, Western blot; Y2H, yeast two-hybrid
Introduction
Medulloblastoma is the most common malignant brain tumor of childhood [1]. Genomics applied to medulloblastoma identified four medulloblastoma subgroups, each characterized by a distinct molecular and clinical profile (Wnt, Hh, groups 3 and 4) [1], [2]. Current therapeutic approaches to medulloblastoma are based on surgery, radiation, and nontargeted chemotherapy, and are indistinguishably applied to all medulloblastoma subgroups. These therapies have led to significant improvements, with a 70% 5-year survival rate, but these results are achieved at a high cost to quality of life, e.g., cognitive or hormonal deficiencies, resulting from the effects of nonspecific, antimitotic agents on the developing brains of young medulloblastoma patients [3], [4]. Alternative therapeutic approaches are needed, and molecular stratification of medulloblastoma patients has yet to be routinely implemented in the clinic.
Hedgehog (Hh) signaling is an essential contributor to tumorigenesis for ~30% of medulloblastoma patients [1], [2]. At the molecular level, canonical Hh signaling involves the binding of a secreted Hh ligand, e.g., Sonic Hedgehog, to a membrane-bound Patched receptor, e.g., PTCH1. In the absence of ligand binding, PTCH1 inhibits SMO, a membrane-bound G-protein–coupled receptor-like molecule and a positive regulator of Hh signals. In the absence of activated SMO, GLI transcription factors are sequestered in the cytoplasm in a complex containing the negative regulator SUFU (applicable to GLI1, 2 and 3) and/or present as processed repressor forms (applicable to GLI2 and 3, not GLI1). Upon ligand binding, inhibition of PTCH1 leads to the de-repression of SMO. Activated SMO orchestrates a signaling cascade that eventually results in the release and translocation of activated GLI transcription factors into the nucleus. GLI transcription factors positively regulate the expression of various context-specific Hh-signal effectors that govern cell fate, e.g., CCND1 and MYCN, as well as PTCH1 itself, thus forming a negative feedback loop [5], [6]. Genetic alterations observed in Hh-medulloblastoma patients include loss of function mutations in the genes of negative regulators of Hh, e.g., PTCH1 and SUFU, as well as gain-of-function mutations of SMO and gene amplifications of other positive regulators or downstream targets of Hh, e.g., GLI2 and MYCN[7].
Our objective is to discover Hh pathway protein interactors and signal modulators that are essential for maintaining oncogenic Hh signaling and tumorigenesis in medulloblastoma. Using a proteomic platform for systematic protein interaction mapping, we discovered a novel interaction between GLI1 and PIN1, a conserved enzyme that catalyzes the cis/trans isomerization of peptidyl-prolyl peptide bonds [8], [9], [10], [11]. The peptidylprolyl cis/trans isomerase (PPIase) activity of PIN1 is specifically aimed at phosphorylated Serine-Proline or Threonine-Proline motifs (pSer/Thr-Pro), thereby regulating the postphosphorylation conformation of its substrates in various physiological and pathophysiological conditions [8], [9], [10], [11], [12]. Although Pin1 KO mice display a range of cell-proliferative abnormalities, e.g., decreased body weight [13], they develop essentially normally [14]. PIN1 may be implicated in the amplification of oncogenic signals, as shown by its frequent overexpression in several human malignancies [15], [16], [17], including brain tumors [18]. However, there are no reports to date linking PIN1 to medulloblastoma tumorigenesis.
In light of the novel GLI1/PIN1 interaction and the previous reports that PIN1 interacts with other key positive regulators of Hh-medulloblastoma, e.g., CCND1 [13], NANOG [19], NOTCH1 [20] and PLK1 [21], we hypothesized that PIN1 promotes Hh-medulloblastoma tumorigenesis. In the present study, we investigated the loss of Pin1 in a mouse model of Hh-medulloblastoma. Our results demonstrate that loss of Pin1 suppresses tumorigenesis, thus identifying a novel therapeutic target in this disease context.
Materials and Methods
Reagents
The protein-encoding ORFs of GLI1 and PIN1 cloned as Gateway Entry (Thermo Fisher Scientific, Waltham, MA) clones were obtained from the Center for Cancer Systems Biology (CCSB, Dana-Farber Cancer Institute, Boston, MA) human ORFeome v8.1 collection or cloned by Gateway recombination cloning from cDNA plasmids as previously described [22]. The PIN1 mutant PIN1W34A was generated by site-directed mutagenesis from WT PIN1 Entry clone. The pcDNA3-HA-DEST and pDEST-GEX5X protein expression vectors were kindly provided by Dr. Siming Li (University of Michigan). The pBABE-SFB (S-FLAG-SBP triple tags) vector was provided by Dr. Jun. Huang (Zhejiang University, China). The yeast two-hybrid (Y2H) pDEST-DB and pDEST-AD vectors were generously provided by the CCSB. The Sonic hedgehog N-Terminus (Shh-N) plasmid was provided by Dr. Benjamin Allen (University of Michigan). The sh-PIN1 construct was obtained from Open Biosystems (Oligo ID#: V2LHS58415). The following primary antibodies were used: PIN1 (Santa Cruz Biotechnology, Santa Cruz, CA, sc15340 and sc46660), GLI1 (Cell Signaling, Danvers, MA, #2534, #2643; Novus, Littleton, CO, NB600-600), NeuN (Zymed, Thermo Fisher Scientific, Waltham, MA, #18-7373), Ki67 (Abcam, Cambridge, MA, ab16667), HA (Roche, Reinach, Switzerland, #12013819001), FLAG (Sigma, St. Louis, MO, A8592), and β-actin (Cell Signaling, #5125). Secondary antibodies were purchased from Cell Signaling (goat α-rabbit IgG, #7074 and horse α-mouse IgG, #7076). Juglone was purchased from Sigma (H47003). SAG was purchased from EMD Millipore, Billerica, MA (Cat: 566660). CRISPR/Cas9 guide sequences targeting the GLI1 and PIN1 genes were designed as previously described [23] and cloned into the lentiCRISPR CRISPR/Cas9 plasmid (Addgene, Cambridge, MA, 49535) using a previously described method [24].
Cell Lines and Cell Culture Conditions
MED-311FH is a low-passage, patient-derived cell line derived from a medulloblastoma tumor, which was recently generated by the Fred Hutchinson Cancer Research Center (FHCRC) Brain Tumor Resource Laboratory. MED-311FH was obtained by the Rual lab from FHCRC on 10/2015. Cell line authentication was performed by STR profiling. We note that MED-311FH had originally been classified as Hh-medulloblastoma by nanoString [25]; however, it was recently reclassified as an atypical medulloblastoma in genomewide 450k methylation analyses. Molecular studies were also performed in the following cell lines: 22Rv1 (human prostate carcinoma) and HEK293T (human embryonic kidney). 22Rv1 and HEK293T were obtained from ATCC, Manassas, VA prior to 2013. Cells were maintained in cell culture by following provider's instructions. Protein expression vectors were transfected into the cells using polyethylenimine, as described previously [26], or by lentiviral/retroviral transduction, using standard protocols. After transduction with the sh-PIN1 or sh-Scramble short hairpin RNA interference constructs, puromycin (2 μg/ml; Sigma, P8833) was included to ensure selection for presence of the plasmids. All cell lines have been tested for mycoplasma contamination.
Y2H
Y2H screens were performed as described previously [27].
Co-Immunoprecipitation of the Endogenous PIN1 and GLI1 Proteins
MED-311FH and 22Rv1 cells were grown in Shh-N conditioned medium for 48 hours. Cells were treated with 10 μM MG132 (Sigma, C2211) for 12 hours to block the 26S proteasome. For immunoprecipitation, cells were harvested and lysed with lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.5% NP-40, 2 mM EDTA, Complete protease inhibitor (Roche, Reinach, Switzerland). The supernatant fraction was recovered and immunoprecipitated with α-PIN1antibody (Epitomics, Abcam, Cambridge, MA, 2136–1, 1:50 or Santa Cruz Biotechnology, Santa Cruz, CA, sc15340, 1:30) and 30 μl Protein A/G-Sepharose (Sigma, P9424/P3296, 1:1). After three washes with lysis buffer, purified protein extracts were resuspended in 2× LDS sample buffer, separated on acrylamide gels, transferred to PVDF membranes, and immunoblot-analyzed with either α-GLI1 antibody (Cell Signaling, #2643) or α-PIN1 antibody (Santa Cruz Biotechnology, sc46660).
SBP-Tag Protein Pulldowns in HEK293T Cells
HEK293T cells transfected with protein expression vector or “empty” negative control vector were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Cells were harvested and lysed on ice for 30 minutes in a lysis buffer [50 mM Tris pH 8.0, 150 mM NaCl, 0.5% NP-40, 2 mM EDTA, Complete protease inhibitor (Roche, 05056489001)]. Cell lysates were cleared by centrifugation for 10 minutes at 16,000×g, and protein complexes were incubated with 30 μl of streptavidin agarose resin (Thermo Fisher Scientific, Waltham, MA, 20361) for 4 hours at 4°C. The beads were washed three times with 1 ml of lysis buffer. Then proteins were eluted by boiling for 5 minutes in 2× LDS sample buffer (Invitrogen, Thermo Fisher Scientific, Waltham, MA, NP0008). Purified protein extracts and input control lysate samples were then separated on acrylamide gels and transferred to PVDF membranes, and proteins were detected using standard immunoblotting techniques using the antibodies described above.
GST Pulldowns
For purification of recombinant PIN1 protein, WT or mutant PIN1 protein-encoding ORFs were Gateway-cloned into pDEST-GEX5X (an N-terminal GST-tagged protein, bacterial expression vector) and transformed into Escherichia coli BL21(DE3). Expression of the recombinant proteins was induced with 0.3 mM IPTG at 16°C. After overnight culture, cells were collected by centrifugation, resuspended in lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.05% NP-40, 1 mM PMSF, and Complete protease inhibitor), and sonicated. After centrifugation, the cleared supernatants were collected and incubated with 50 μl of washed glutathione sepharose (GE Healthcare, Livonia, MI, 17-0756-01) for 4 hours at 4°C. The “GST-alone” or the GST-PIN1 protein-bound glutathione beads were then washed three times with 1 ml of lysis buffer. For the GST pulldown assay, HA-GLI1 was transient transfected into HEK293T cells. Cells were harvested 48 hours after transfection and lysed with lysis buffer. The cleared cell lysates were incubated with the “GST-alone” or the GST-PIN1 protein-bound glutathione beads for 2 hours at 4°C. The beads were washed five times with lysis buffer and subjected to immunoblotting analysis.
Confocal Microscopic Analysis
HEK293T cells were transfected with plasmids expressing GFP-GLI1 and RFP-PIN1 for 24 hours in a glass-bottom culture dish coated with Poly-d-lysine (MatTek, Ashland, MA, P35GC-1.5-14-C). Fluorescence images were obtained using a Nikon A1 (Melville, NY) confocal microscope.
Luciferase Reporter Assay
Shh-Light2 cells (GRCF Biorepository & Cell Center, Johns Hopkins School of Medicine) were treated with Shh-N conditioned medium for 48 hours. Dual-luciferase reporter assays were performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to the manufacturer's instructions, and data were collected on a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA).
Gene Expression Analysis
Total RNA was extracted with Trizol reagent (Ambion, 15596018) using manufacturer's instructions and further purified with the RNeasy Mini Kit (Qiagen, 74106). Five micrograms of RNA was reverse-transcribed in cDNA using oligo(dT)18-primed reverse transcription and SuperScript III RT First-Strand kit (Invitrogen, 18080-051) as described by the manufacturer. The cDNA was analyzed via quantitative polymerase chain reaction (qPCR) analysis using the Power SYBR Green PCR master mix (Applied Biosystems, 4367662) and the CFX96 Touch Real-Time PCR Detection System (BioRad) according to manufacturer's recommendations. Data were normalized to the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Primers used in RT-qPCR experiments are listed in Supplementary Table 1.
[GFAP-tTA;TRE-SmoA1] Mouse Model
We studied medulloblastoma tumorigenesis in the absence or presence of Pin1 in the previously published bitransgenic [GFAP-tTA;TRE-SmoA1] model [28], where the expression of the tetracycline-regulated transactivators (tTA) is driven by a GFAP promoter and the expression of oncogenic SmoA1 is under the control of the tetracycline responsive element (TRE). The experimental breeders used in this study were a Pin1 KO mouse [14], a TRE-SmoA1 mouse [28], and a GFAP-tTA mouse [29], all of which were maintained on a C57/BL6 background for at least five generations prior to initiating experiments. TRE-SmoA1, [GFAP-tTA;TRE-SmoA1], and [GFAP-tTA;TRE-SmoA1;Pin1−/−] mouse littermates were generated by crossing [GFAP-tTA;Pin1−/+] mice with [TRE-SmoA1;Pin1−/+] mice. Maintenance of mouse colonies and experimental procedures were approved by the University of Michigan University Committee on the Use and Care of Animals.
Mouse Genotyping
Pups were genotyped between P14 and P21 by PCR analysis. The presence of the GFAP-tTA transgene was ascertained by individual PCR for the tetracycline transactivator (5′-ctcgcccagaagctaggtgt-3′ and 5′-ccatcgcgatgacttagt-3′). A TRE-SmoA1 genotype was assessed by PCR amplification of the SV40 poly-A tail (5′-ggaactgatgaatgggagca-3′ and 5′-gggaggtgtgggaggttt-3′). The Pin1 WT versus KO genotype was ascertained by PCR targeting the Pin1 gene locus (forward: 5′-gcacccgatcctgttctgcaaactcag-3′, WT reverse: 5′-catgagaagggattagaagcaagattcgactgg-3′, or mutant reverse: 5′-gccagaggccacttgtgtagcgc-3′).
Magnetic Resonance Imaging (MRI)
Medulloblastoma tumor growth was monitored in vivo, noninvasively, by MRI analysis at the University of Michigan Center for Molecular Imaging. Isoflurane-anesthetized animals were laid prone, head first in a 7.0-T Agilent (Santa Clara, CA) MR scanner with the body temperature maintained at 37°C using forced heated air. A quadrature volume radiofrequency coil was used to scan the head region of the mice. Axial and coronal T2-weighted images were acquired using a fast spin-echo sequence with the following parameters: repetition time/effective echo time, 4000/60 milliseconds; number of echoes, 8; field of view, 20 × 20 mm; matrix, 256 × 128; slice thickness, 0.5 mm; number of slices, 25.
Immunohistochemistry
Immunohistochemistry (IHC) was performed on 5-μm–thick formalin-fixed, paraffin-embedded (FFPE) tissue sections. Antigen retrieval was performed with Reveal Decloaker (Biocare Medical, Concord, CA, RV1000; for Ki67 IHC) or by heat-induced epitope retrieval in 10 mM sodium citrate pH 6 buffer (for NeuN IHC). IHC was performed using the DAKO Autostainer (Troy, MI). Sections were stained with the following primary antibodies for 1 hour at ambient temperature: a rabbit polyclonal antibody to Ki67 (Abcam, ab16667, 1:500-diluted) and a purified mouse monoclonal antibody to NeuN (EMD Millipore, Billerica, MA, MAB377, 1:200-diluted). After primary antibody incubation and washing, a polymer horseradish peroxidase secondary antibody [Biocare Medical, RMR622 (for Ki67 IHC) or Envision+, Agilent, Santa Clara, CA, K4000 (for NeuN IHC)] was applied. Chromogen diaminobenzidine was applied for visualization and slides counterstained with hematoxylin.
Results
GLI1 Interacts with PIN1
GLI1 is a key factor in the Hh pathway that contributes to the amplification of Hh signals. In mouse, Gli1 is not required for development or viability [30], [31]. However, loss of Gli1 impairs Hh-medulloblastoma tumorigenesis [32], [33], [34]. Using a proteomic approach, we obtained multiple, independent lines of evidence supporting a molecular interaction between GLI1 and PIN1. First, we discovered the GLI1/PIN1 interaction in a Y2H screen (Figure 1A). Second, the GLI1/PIN1 interaction was validated by reciprocal pulldowns using epitope-tagged proteins in HEK293T cells (Figure 1B). Third, we performed immunoprecipitations of endogenous PIN1 in MED-311FH cells, a low-passage cell line derived from a medulloblastoma patient, and in 22Rv1, a human prostate carcinoma cell line. In both immunoprecipitates, we observed that endogenous GLI1 co-purifies with endogenous PIN1 (Figure 1C and Supplementary Figure 1). Fourth, in a confocal microscopy analysis, we observed that GFP-GLI1 co-localizes with RFP-PIN1 in the nucleus of HEK293T cells (Supplementary Figure 2). Fifth, we observed that the WW domain of PIN1 is required for the interaction, as verified by the analysis of the PIN1W34A point mutant, which fails to co-purify with GLI1 (Supplementary Figure 3).
Figure 1.
GLI1 interacts with PIN1. (A) Detection of the GLI1/PIN1 interaction using the Y2H assay. In this Y2H experiment, PIN1 is fused to the GAL4 DNA-binding (DB) domain and GLI1 is fused to the GAL4 activation domain (AD). The six Y2H controls have been previously described [27]. The DB-PIN1 and AD-GLI1 fusion proteins interact with each other, leading to the activation of the HIS3 reporter gene and allowing yeast cells to grow on selective medium lacking histidine. Experiment was replicated three times. (B) HA-GLI1 co-purifies with SBP-FLAG-PIN1 (top panels), and HA-PIN1 co-purifies with SBP-FLAG-GLI1 (bottom panels). Immunoprecipitation (IP) of SBP-FLAG–tagged PIN1 (top panels) or SBP-FLAG–tagged GLI1 (bottom panels) in HEK293T cells in the presence of HA-tagged GLI1 or HA-tagged PIN1, respectively. WB: Western blot; SBP: Streptavidin binding peptide tag. Experiment was replicated three times. (C) Endogenous GLI1 co-purifies with endogenous PIN1 in MED-311FH cells, a low-passage, patient-derived cell line derived from a medulloblastoma tumor. The IP of endogenous PIN1 was performed using an α-PIN1 monoclonal antibody on a protein extract derived from Shh-N ligand-treated MED-311FH cells, followed by Western blot analyses using either α-PIN1 or α-GLI1 antibody. Experiment was replicated twice. WB: Western blot; IP: immunoprecipitation. (D) PIN1 interacts with GLI1 in a phosphorylation-dependent manner. GST pulldown with glutathione beads of bacteria-purified GST-tagged PIN1 in the presence of a protein extract derived from mammalian HEK293T cells transduced with HA-tagged GLI1 and in the presence or absence of calf intestinal alkaline phosphatase (CIP). Experiment was replicated twice. WB: Western blot. (E) PIN1 leads to increased GLI1 protein abundance in vitro. Analysis of GLI1 protein levels in the presence of an increasing level of WT PIN1 and mutant PIN1W34A in HEK293T cells. HEK293T cells were transfected with 300 ng of HA-GLI1 protein expression vector as well as 0, 50, 150, or 450 ng of the SBP-FLAG-PIN1 or the SBP-FLAG-PIN1W34A protein expression vector. Experiment was replicated twice. WB: Western blot; SBP: Streptavidin binding peptide tag.
PIN1 binds pSer/Thr-Pro motifs [8], [9], [10], [11]. Thus, a phosphorylation event likely precedes the binding of PIN1 to its target. Upon investigating the phosphorylation dependence of the GLI1/PIN1 interaction, we verified that the ability for PIN1 to co-purify with GLI1 is significantly decreased in the presence of phosphatase (Figure 1D). We also note that GLI1 protein levels are increased in the presence of PIN1, which we propose is due to protein stabilization (see input controls in Figure 1B and Supplementary Figure 3A). Similarly, we analyzed GLI1 protein levels in the presence of an increasing level of either wild-type (WT) PIN1 or mutant PIN1W34A in HEK293T cells. We observed that co-expression of WT PIN1, but not mutant PIN1W34A, results in a greater GLI1 protein abundance (Figure 1E). These observations indicate that PIN1 leads to increased GLI1 protein abundance in vitro. Further investigation of the protein kinase responsible for the phosphorylation event leading to the regulation of the GLI1/PIN1 interaction as well as the molecular mechanisms involved in the PIN1-dependent regulation of GLI1 protein levels is warranted.
PIN1 Inhibition Impairs Hh Signaling
As a GLI1 interactor and regulator of GLI1 protein levels, PIN1 may contribute to the GLI1-mediated modulation of Hh target genes. To test this hypothesis, we investigated the effect of depletion of PIN1 on Hh signaling using an NIH3T3 Shh-Light2-based GLI-dependent luciferase reporter assay [35]. We observed that the Shh-N–mediated activation of Hh signaling is impaired in NIH3T3 Shh-Light2 cells upon the CRISPR/Cas9-mediated depletion of either GLI1 or PIN1 (Figure 2A). Next, we analyzed by quantitative reverse transcriptase (RT)-PCR the levels of expression of Hh target genes upon treatment with SAG, a chlorobenzothiophene-containing compound which acts as an SMO agonist, and juglone, a PIN1 inhibitor [36], in the MED-311FH cell line. In the presence of SAG, the levels of mRNA expression of GLI1, PTCH1, PTCH2, and FOXA2 increase in MED-311FH (Figure 2B). Remarkably, we observed that the pharmacological inhibition of PIN1 by juglone in SAG-activated MED-311FH cells results in the strong reduction of the levels of expression of these Hh target genes (Figure 2B). Altogether, these results indicate that PIN1 is a positive regulator of Hh signaling.
Figure 2.
PIN1 promotes Hh signaling. (A) GLI-dependent luciferase reporter assay. Shh-Light2 is an NIH 3T3 cell line stably transfected with GLI-dependent Firefly luciferase and constitutive Renilla luciferase reporters, which can be used to study modulation of Hh signaling [35]. Shh-N–mediated activation of luciferase activities was measured upon CRISPR/Cas9-mediated depletion of either GLI1 or PIN1. Control: Shh-Light2 cells stably transfected with empty CRISPR/Cas9 vector. First, for each one of the 10 conditions tested, the Firefly luciferase activity signals were measured using the Dual-Luciferase Reporter Assay System (Promega) and were normalized by the Renilla luciferase signals. Second, the 10 signal ratios were normalized by the signal ratio obtained in the [control medium/control cell line] condition. Shown are means ±S.D. of three biological replicates. We fit analysis of variance (ANOVA) models to log-transformed data and tested whether the increase with Shh-N versus no treatment was smaller under Gli1 and Pin1 KD conditions than the same increase under control conditions. The difference of fold-changes was significant for all four KD conditions tested; [***] P < 3 × 10−5. Inset: Western blot analyses validate the CRISPR/Cas9-mediated depletion of PIN1 and GLI1. sg: CRISPR/Cas9 single guide RNA; WB: Western blot. (B) Expression analysis by quantitative RT-PCR of Hh target genes upon treatment with SAG (0.3 μM) and/or juglone (5 μM) in MED-311FH. Shown are means ± S.D. of two biological replicates measured twice each. Statistical significances of the difference between the “SAG” and “SAG + juglone” conditions were computed using one-way ANOVA models on log-transformed data for each gene; [*] P < .05, [**] P < .01.
Loss of Pin1 Suppresses Hh-Medulloblastoma Tumorigenesis In Vivo
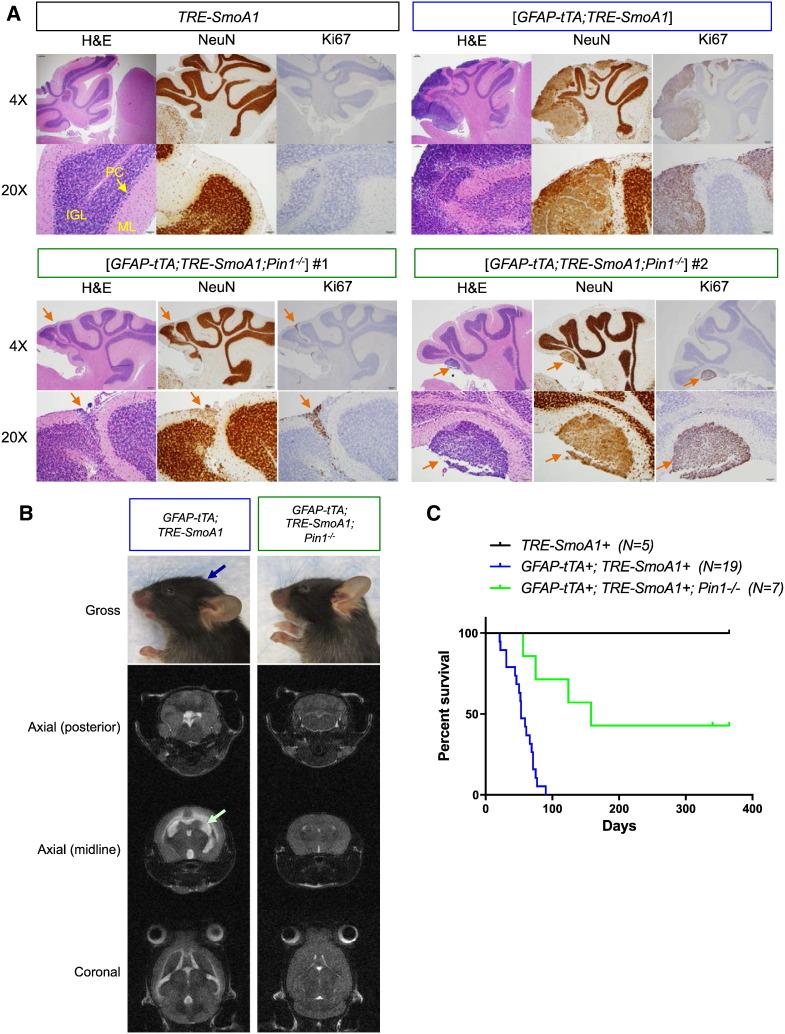
In addition to the novel GLI1/PIN1 interaction, PIN1 has been previously shown to interact with other key modulators of Hh-medulloblastoma tumorigenesis [13], [19], [20], [21]. We sought to investigate the loss of Pin1 in vivo using a genetically engineered mouse model of Hh-medulloblastoma tumorigenesis. SmoA1 is an oncogenic gain-of-function allele of Smo[35], a key positive regulator of Hh signals [5], [6]. The ND2 or GFAP promoter-driven expression of SmoA1 in mouse cerebellar granular neuron progenitor cells (CGNP), i.e., a “cell of origin” in Hh-medulloblastoma [37], results in CGNP hyperproliferation and medulloblastoma formation, as observed in the ND2:SmoA1[38] and the [GFAP-tTA;TRE-SmoA1] [28] mouse models of Hh-medulloblastoma. To assess in vivo the role of Pin1 in medulloblastoma, we studied the genetic loss of Pin1 in the bitransgenic [GFAP-tTA;TRE-SmoA1] model, where the expression of the tetracycline-regulated transactivators (tTA) is driven by a GFAP promoter and the expression of oncogenic SmoA1 is under the control of the tetracycline responsive element (TRE) [28]. Three groups of C57BL/6 mice were studied: 1) a TRE-SmoA1–negative control group (SmoA1 is not expressed in the absence of GFAP-tTA), 2) a positive control group of [GFAP-tTA;TRE-SmoA1] mice, which develops medulloblastoma; and 3) the [GFAP-tTA;TRE-SmoA1;Pin1−/−] test group (genotyping data are available in Supplementary Figure 4). Each group was assessed by histological examination of the cerebellum by hematoxylin/eosin (H&E) staining, Ki67 (marker of proliferation) and NeuN (marker of neuronal differentiation) IHC, MRI evaluation, as well as Kaplan-Meier survival analysis.
At postnatal day (P)21, TRE-SmoA1 mice exhibit normal cerebellar development with properly laminated molecular, Purkinje cell, and internal granular cell layers, as expected. Also expected at P21, the external granular cell layer has completely disappeared. Moreover, normal cerebellar neurons in the internal granular layer strongly stain for the neuronal marker NeuN and are in quiescent, nonproliferative stage, as demonstrated by the absence of staining for Ki-67 (Figure 3A). As previously described, [GFAP-tTA;TRE-SmoA1] mice develop medulloblastoma tumors detectable by histological analysis as early as P7, with 100% penetrance by P21 [28]. In P21 [GFAP-tTA;TRE-SmoA1] mice, large tumors can extend along the entire rostral-caudal length of the cerebellum; account for about a quarter of the total cerebellar volume; express NeuN, indicative of neuronal origin; and show marked proliferative activity (positive for Ki67) (Figure 3A). For comparison, two [GFAP-tTA;TRE-SmoA1;Pin1−/−] mice were euthanized at P21 for histological analysis. The cerebellums of both [GFAP-tTA;TRE-SmoA1;Pin1−/−] mice exhibit well-formed folia with normal lamination of the cerebellar cortex, and only minimal, microscopic evidence of early tumor development is observed (best appreciated by NeuN and Ki67 stainings in Figure 3A). The cerebellums of these two mice were entirely sectioned and analyzed; the microscopic tumors comprised <5% of the cerebellar volume.
Figure 3.
Loss of Pin1 impairs Hh-medulloblastoma tumorigenesis in vivo. (A) Histopathological examination of the cerebellums of one P21 TRE-SmoA1 mouse, one P21 [GFAP-tTA;TRE-SmoA1] mouse, and two P21 [GFAP-tTA;TRE-SmoA1;Pin1−/−] mice by H&E staining as well as by Ki67 (marker of proliferation) and NeuN (marker of neuronal differentiation) IHC on 5-μm–thick FFPE tissue sections (magnification: 4× and 20×). The different cell layers of the cerebellum, i.e., the molecular (ML), Purkinje cell (PC), and internal granular cell (IGL) layers, are labeled in the 20× TRE-SmoA1 control H&E picture. Note: the 20× pictures of the [GFAP-tTA;TRE-SmoA1;Pin1−/−] mice focus on the microscopic tumors (orange arrows), which are detected in <5% of the cerebellums, and thus are not representative of the whole [GFAP-tTA;TRE-SmoA1;Pin1−/−] cerebellums which, for the most part, appear normal. (B) MRI analysis of the cerebellums of P56 [GFAP-tTA;TRE-SmoA1;Pin1−/−] and [GFAP-tTA;TRE-SMOA1] littermates. For the [GFAP-tTA;TRE-SmoA1] mouse, 1) doming of the skull can be grossly appreciated (blue arrow); 2) the axial (posterior) MRI film shows an enlarged [GFAP-tTA;TRE-SmoA1] cerebellum; and 3) the axial (midline) and coronal films demonstrate dilatation of the ventricles and subarachnoid spaces (green arrow). In contrast, analysis of the [GFAP-tTA;TRE-SmoA1;Pin1−/−] mouse does not reveal any such signs of tumor burden. (c) Kaplan-Meier analysis of Pin1 in SmoA1-driven medulloblastoma. Three groups of C57BL/6 mice were assessed for survival: 1) TRE-SmoA1, 2) [GFAP-tTA;TRE-SmoA1], and 3) [GFAP-tTA;TRE-SmoA1;Pin1−/−]. [GFAP-tTA;TRE-SmoA1;Pin1−/−] mice survived significantly longer than [GFAP-tTA;TRE-SmoA1] mice [P = .0005, log rank test]. No randomization was used, and no blinding was done.
CNS presentation was also assessed by MRI for a P56 [GFAP-tTA;TRE-SmoA1;Pin1−/−] animal and its [GFAP-tTA;TRE-SmoA1] littermate. Whereas the pathophysiological effects can be appreciated in the [GFAP-tTA;TRE-SmoA1] mouse with the observation of dilated ventricles and subarachnoid spaces as well as significant doming of the skull, MRI analysis of the [GFAP-tTA;TRE-SmoA1;Pin1−/−] mouse brain revealed no such signs of tumor burden (Figure 3B). In vitro, PIN1 promotes Hh signaling (Figure 2). We analyzed the level of mRNA expression of Hh target genes in the cerebellums of [GFAP-tTA;TRE-Smoa1;Pin1−/−] mice and their [GFAP-tTA;TRE-Smoa1] littermates. As shown in Supplementary Figure 5, the level of expression of Gli1, Gli2, Ptch2, Hhip1, and Foxa2 is significantly reduced in the cerebellums of [GFAP-tTA;TRE-SmoA1;Pin1−/−] mice compared with the cerebellums of [GFAP-tTA;TRE-SmoA1] mice, in agreement with the in vitro data.
To assess the effect of loss of Pin1 on normal cerebellar development, we also performed the histological analysis of P6 and P10 cerebellums isolated from Pin1 KO mouse littermates. Cerebellums appear properly developed with normal foliation and normal lamination of the external granular cell, molecular, Purkinje cell, and internal granular cell layers. The external granular cell layer decreases in thickness as these cells begin to migrate through the molecular layer to produce the internal granular layer from P6 to P10, as expected (Supplementary Figure 6). Altogether, loss of Pin1 has no noticeable histological effect on normal cerebellar development yet inhibits Hh-medulloblastoma tumorigenesis, thus echoing the genetic studies of loss of Gli1 in physiological and pathophysiological contexts [30], [31], [34].
Finally, we performed a Kaplan-Meier survival analysis of the three aforementioned mouse groups. As previously described [28], [GFAP-tTA;TRE-SmoA1] animals succumb to medulloblastoma tumor within ~53 days. Remarkably, we observed that the median survival of the [GFAP-tTA;TRE-SmoA1;Pin1−/−] mice is about three times greater than for [GFAP-tTA;TRE-SmoA1] mice (158 versus 53 days, respectively; P = .0005; Figure 3C). Although four of the seven [GFAP-tTA;TRE-SmoA1;Pin1−/−] mice succumbed to medulloblastoma tumorigenesis at day 56, 75, 124, and 158, three of them survived beyond ~1 year. Moreover, the histological analysis of the cerebellums of two of these long-term survivors mice did not show evidence of tumor, either macroscopic or microscopic (>80% of each cerebellum was analyzed; Supplementary Figure 7). Thus, in agreement with our observations that PIN1 promotes GLI1 stability and Hh signaling, these in vivo analyses strongly support the hypothesis that Pin1 plays an essential role in Hh-medulloblastoma tumorigenesis.
Discussion
We discovered a novel interaction between GLI1 and PIN1, and our in vivo analyses of Pin1 in a genetically engineered Smo-induced mouse model of medulloblastoma tumorigenesis demonstrate that the loss of Pin1 impairs tumor development and increases survival considerably. We speculate that the pro-oncogenic role of Pin1 in Hh-medulloblastoma is not solely mediated via GLI1 but also via the regulation of other PIN1 targets [13], [19], [20], [21]. Beyond its impact on disease progression, the potential of PIN1 as a therapeutic target in Hh-medulloblastoma is also dependent on the side effect associated with the inhibition of this protein during normal physiological development. Although Pin1 KO mice display a range of cell-proliferative abnormalities, e.g., decreased body weight [13], they develop essentially normally [14]. The absence of any noticeable effect associated with the complete loss of Pin1 on cerebellar development strongly suggests that therapeutic benefit may be achieved with tolerable doses of PIN1 inhibitors.
We are currently investigating whether pharmacological inhibition of PIN1 by either juglone or the flavonoid epigallocatechin gallate improves survival in mouse models of Hh-medulloblastoma. Interestingly, we note that quercetin, another flavonoid which inhibits PIN1, was recently identified in a chemical screen as a putative radiosensitizer for human medulloblastoma cells [39]. If our hypothesis is validated, i.e., PIN1 inhibitors can improve survival, it will strongly justify testing the clinical relevance of PIN1 blockade in Hh-medulloblastoma patients, either alone or in combination treatment regimens, e.g., in combination with SMO antagonists, which are currently being tested in clinical trials [40]. As a modulator of GLI1, the downstream position of PIN1 in the Hh genetic pathway underscores its potential as a complement to SMO inhibition and may be specifically beneficial for overcoming resistance to SMO inhibitors [7], thereby paving the way toward a multitherapy approach to Hh-medulloblastoma. Beyond Hh-medulloblastoma, we speculate that PIN1 can also modulate Hh signals in other neoplastic conditions in which Hh is involved [6]. Based on our findings, studies aiming to test the requirement of Pin1 in mouse models of basal cell carcinoma [41], basaloid follicular hamartomas [42], and embryonal rhabdomyosarcoma [43] are highly justified and will help establish whether the role of Pin1 in Hh-driven tumorigenesis is either general or specific to medulloblastoma.
The following are the supplementary data related to this article.
Target Guide Sequences for CRISRR/Cas9 (Wang et al., 2014).
Supplementary figures
Author Contributions
J. F. R. conceived and directed the project. T. X. and K. H. designed and performed the Y2H experiments. T. X., H. Z., S. S. P., and J. M. O. designed and performed the molecular and cellular studies. H. Z., L. E. M., C. U., T. U., A. D., S. C. P., and J. F. R. designed and performed the in vivo mouse studies. H. Z., S. C. P., S. V., and M. S. performed and analyzed the IHC experiments. H. Z., S. C. P., J. F. R., and A. S. performed and analyzed the MRI experiments. R. K. performed statistical analyses. J. F. R. wrote the manuscript, with contributions from other coauthors.
Acknowledgements
We thank UM Comprehensive Cancer Center Tissue for technical support with the IHC experiments, specifically Dafydd Thomas and Tina Fields; UM Vector Core Facility for the lentiviral sh-RNA plasmids; UM Center for Molecular Imaging for technical support with the MRI experiments, specifically Amanda Fair; CCSB for sharing ORFeome and Y2H clones. This work was supported by Padnos Fund for Innovative Cancer Research of the UM Comprehensive Cancer Center awarded to J. F. R.; Bench to Bedside Translation Award awarded to J. F. R. by the Michigan Institute for Clinical and Health Research and the National Center for Advancing Translational Sciences of the National Institutes of Health (NIH) under Award Number UL1TR000433; M-Cubed Grant awarded to J. F. R. and S. C. P.; grant R01CA155360 awarded to J. M. O. by the National Cancer Institute of the NIH; Seattle Children's Neuro-oncology endowment awarded to J. M. O; UM Cancer Center Support Grant (P30CA046592); and funds from the UM Department of Pathology provided to J. F. R. The sponsors were not involved in any of the following: study design; collection, analysis, and interpretation of data; the writing of the report; and the decision to submit the article for publication.
Footnotes
Conflicts of Interest: The authors declare no conflict of interest.
Contributor Information
Sandra Camelo-Piragua, Email: sandraca@umich.edu.
Jean-François Rual, Email: jrual@umich.edu.
References
- 1.Gajjar AJ, Robinson GW. Medulloblastoma-translating discoveries from the bench to the bedside. Nat Rev Clin Oncol. 2014;11:714–722. doi: 10.1038/nrclinonc.2014.181. [DOI] [PubMed] [Google Scholar]
- 2.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mulhern RK, Merchant TE, Gajjar A, Reddick WE, Kun LE. Late neurocognitive sequelae in survivors of brain tumours in childhood. Lancet Oncol. 2004;5:399–408. doi: 10.1016/S1470-2045(04)01507-4. [DOI] [PubMed] [Google Scholar]
- 4.Laughton SJ, Merchant TE, Sklar CA, Kun LE, Fouladi M, Broniscer A, Morris EB, Sanders RP, Krasin MJ, Shelso J. Endocrine outcomes for children with embryonal brain tumors after risk-adapted craniospinal and conformal primary-site irradiation and high-dose chemotherapy with stem-cell rescue on the SJMB-96 trial. J Clin Oncol. 2008;26:1112–1118. doi: 10.1200/JCO.2008.13.5293. [DOI] [PubMed] [Google Scholar]
- 5.Ingham PW, AP McMahon. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 6.Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat Med. 2013;19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- 7.Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25:393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 9.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 10.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 11.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 12.Liou YC, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. 2011;36:501–514. doi: 10.1016/j.tibs.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liou YC, Ryo A, Huang HK, Lu PJ, Bronson R, Fujimori F, Uchida T, Hunter T, Lu KP. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc Natl Acad Sci U S A. 2002;99:1335–1340. doi: 10.1073/pnas.032404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimori F, Takahashi K, Uchida C, Uchida T. Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G(0) arrest. Biochem Biophys Res Commun. 1999;265:658–663. doi: 10.1006/bbrc.1999.1736. [DOI] [PubMed] [Google Scholar]
- 15.Ryo A, Nakamura M, Wulf G, Liou YC, Lu KP. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat Cell Biol. 2001;3:793–801. doi: 10.1038/ncb0901-793. [DOI] [PubMed] [Google Scholar]
- 16.Bao L, Kimzey A, Sauter G, Sowadski JM, Lu KP, Wang DG. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am J Pathol. 2004;164:1727–1737. doi: 10.1016/S0002-9440(10)63731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wulf G, Garg P, Liou YC, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J. 2004;23:3397–3407. doi: 10.1038/sj.emboj.7600323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atkinson GP, Nozell SE, Harrison DK, Stonecypher MS, Chen D, Benveniste EN. The prolyl isomerase Pin1 regulates the NF-kappaB signaling pathway and interleukin-8 expression in glioblastoma. Oncogene. 2009;28:3735–3745. doi: 10.1038/onc.2009.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moretto-Zita M, Jin H, Shen Z, Zhao T, Briggs SP, Xu Y. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc Natl Acad Sci U S A. 2010;107:13312–13317. doi: 10.1073/pnas.1005847107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rustighi A, Tiberi L, Soldano A, Napoli M, Nuciforo P, Rosato A, Kaplan F, Capobianco A, Pece S, Di Fiore PP. The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat Cell Biol. 2009;11:133–142. doi: 10.1038/ncb1822. [DOI] [PubMed] [Google Scholar]
- 21.Shen M, Stukenberg PT, Kirschner MW, Lu KP. The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. 1998;12:706–720. doi: 10.1101/gad.12.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Boehm JS, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, Green TM. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang L, Aiken C, McClelland R, Morrison LC, Tatari N, Remke M, Ramaswamy V, Issaivanan M, Ryken T, Del Bigio MR. Characterization of novel biomarkers in selecting for subtype specific medulloblastoma phenotypes. Oncotarget. 2015;6:38881–38900. doi: 10.18632/oncotarget.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longo PA, Kavran JM, Kim MS, Leahy DJ. Transient mammalian cell transfection with polyethylenimine (PEI) Methods Enzymol. 2013;529:227–240. doi: 10.1016/B978-0-12-418687-3.00018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dreze M, Monachello D, Lurin C, Cusick ME, Hill DE, Vidal M, Braun P. High-quality binary interactome mapping. Methods Enzymol. 2010;470:281–315. doi: 10.1016/S0076-6879(10)70012-4. [DOI] [PubMed] [Google Scholar]
- 28.Michael LE, Westerman BA, Ermilov AN, Wang A, Ferris J, Liu J, Blom M, Ellison DW, van Lohuizen M, Dlugosz AA. Bmi1 is required for Hedgehog pathway-driven medulloblastoma expansion. Neoplasia. 2008;10:1343–1349. doi: 10.1593/neo.81078. [1345p following 1349] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin W, Kemper A, McCarthy KD, Pytel P, Wang JP, Campbell IL, Utset MF, Popko B. Interferon-gamma induced medulloblastoma in the developing cerebellum. J Neurosci. 2004;24:10074–10083. doi: 10.1523/JNEUROSCI.2604-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593–1605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- 31.Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- 32.Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci U S A. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Infante P, Mori M, Alfonsi R, Ghirga F, Aiello F, Toscano S, Ingallina C, Siler M, Cucchi D, Po A. Gli1/DNA interaction is a druggable target for Hedgehog-dependent tumors. EMBO J. 2015;34:200–217. doi: 10.15252/embj.201489213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura H, Stephen D, Joyner A, Curran T. Gli1 is important for medulloblastoma formation in Ptc1+/- mice. Oncogene. 2005;24:4026–4036. doi: 10.1038/sj.onc.1208567. [DOI] [PubMed] [Google Scholar]
- 35.Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 36.Hennig L, Christner C, Kipping M, Schelbert B, Rucknagel KP, Grabley S, Kullertz G, Fischer G. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry. 1998;37:5953–5960. doi: 10.1021/bi973162p. [DOI] [PubMed] [Google Scholar]
- 37.Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, Schuller U, Machold R, Fishell G, Rowitch DH. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein ID, Beachy PA. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 39.Lagerweij T, Hiddingh L, Biesmans D, Crommentuijn MH, Cloos J, Li XN, Kogiso M, Tannous BA, Vandertop WP, Noske DP. A chemical screen for medulloblastoma identifies quercetin as a putative radiosensitizer. Oncotarget. 2016;7:35776–35788. doi: 10.18632/oncotarget.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson GW, Orr BA, Wu G, Gururangan S, Lin T, Qaddoumi I, Packer RJ, Goldman S, Prados MD, Desjardins A. Vismodegib Exerts Targeted Efficacy Against Recurrent Sonic Hedgehog-Subgroup Medulloblastoma: Results From Phase II Pediatric Brain Tumor Consortium Studies PBTC-025B and PBTC-032. J Clin Oncol. 2015;33:2646–2654. doi: 10.1200/JCO.2014.60.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grachtchouk M, Mo R, Yu S, Zhang X, Sasaki H, Hui CC, Dlugosz AA. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat Genet. 2000;24:216–217. doi: 10.1038/73417. [DOI] [PubMed] [Google Scholar]
- 42.Grachtchouk V, Grachtchouk M, Lowe L, Johnson T, Wei L, Wang A, de Sauvage F, Dlugosz AA. The magnitude of hedgehog signaling activity defines skin tumor phenotype. EMBO J. 2003;22:2741–2751. doi: 10.1093/emboj/cdg271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatley ME, Tang W, Garcia MR, Finkelstein D, Millay DP, Liu N, Graff J, Galindo RL, Olson EN. A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer Cell. 2012;22:536–546. doi: 10.1016/j.ccr.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Target Guide Sequences for CRISRR/Cas9 (Wang et al., 2014).
Supplementary figures