Abstract
Saccharomyces cerevisiae is the main microorganism responsible for the fermentation of wine. Nevertheless, in the last years wineries are facing new challenges due to current market demands and climate change effects on the wine quality. New yeast starters formed by non-conventional Saccharomyces species (such as S. uvarum or S. kudriavzevii) or their hybrids (S. cerevisiae x S. uvarum and S. cerevisiae x S. kudriavzevii) can contribute to solve some of these challenges. They exhibit good fermentative capabilities at low temperatures, producing wines with lower alcohol and higher glycerol amounts. However, S. cerevisiae can competitively displace other yeast species from wine fermentations, therefore the use of these new starters requires an analysis of their behavior during competition with S. cerevisiae during wine fermentation. In the present study we analyzed the survival capacity of non-cerevisiae strains in competition with S. cerevisiae during fermentation of synthetic wine must at different temperatures. First, we developed a new method, based on QPCR, to quantify the proportion of different Saccharomyces yeasts in mixed cultures. This method was used to assess the effect of competition on the growth fitness. In addition, fermentation kinetics parameters and final wine compositions were also analyzed. We observed that some cryotolerant Saccharomyces yeasts, particularly S. uvarum, seriously compromised S. cerevisiae fitness during competences at lower temperatures, which explains why S. uvarum can replace S. cerevisiae during wine fermentations in European regions with oceanic and continental climates. From an enological point of view, mixed co-cultures between S. cerevisiae and S. paradoxus or S. eubayanus, deteriorated fermentation parameters and the final product composition compared to single S. cerevisiae inoculation. However, in co-inoculated synthetic must in which S. kudriavzevii or S. uvarum coexisted with S. cerevisiae, there were fermentation performance improvements and the final wines contained less ethanol and higher amounts of glycerol. Finally, it is interesting to note that in co-inoculated fermentations, wine strains of S. cerevisiae and S. uvarum performed better than non-wine strains of the same species.
Keywords: Saccharomyces species, competition, wine fermentation, temperature, fitness, wine composition
Introduction
Wine is the product of complex interactions among yeast, bacteria and other fungi that begin in vineyards and continue with the fermentation process. Different yeast species are predominant on the surface of grape skins and in the winery environment (Sabate et al., 2002; Albergaria and Arneborg, 2016), and S. cerevisiae is recognized as being the main microorganism responsible for this process (Pretorius, 2000). However, other Saccharomyces species (Saccharomyces non-cerevisiae yeasts, SNC) may play an important role in wine fermentation under certain conditions. In this way S. uvarum is less frequent than S. cerevisiae in wines, but appears to be predominant in European wine regions with an oceanic climate where wine fermentations are performed at lower temperatures; e.g., the Basque Country, Spain (Rementeria, 2003), Alsace, France (Demuyter et al., 2004), Val de Loire, Sauternes, and Jurançon in France (Naumov et al., 2000), Valpolicella, Italy (Torriani et al., 1999), Tokaj in Hungary and Slovakia (Sipiczki et al., 2001; Naumov et al., 2002; Antunovics et al., 2005), and Yalta, the Ukraine (Naumov and Nikonenko, 1987). S. paradoxus is a natural species worldwide distributed with a fortuitous presence in vineyards and fermentation processes (Valero et al., 2007). However, some strains of this species have been described as predominant in Croatian vineyards (Redžepović et al., 2002), and exhibit interesting enological properties.
The fermentations conducted by natural interspecific Saccharomyces hybrids, such as S.cerevisiae × S.kudriavzevii and S.cerevisiae × S.uvarum, have also been described in European wine regions with oceanic and continental climates (northern Spain, Alsace, Germany, Switzerland, Austria, Croatia, Hungary, and Moldavia), close to the northern limit of grapevine distribution (Masneuf et al., 1998; González et al., 2006; Erny et al., 2012; Peris et al., 2012).
Despite these exceptions, presence of SNC in the final stages of the fermentation process is quite rare. This is because S. cerevisiae can competitively displace other yeast species from wine fermentations, both SNC (Arroyo-López et al., 2011; Williams et al., 2015) and non Saccharomyces yeasts (Holm Hansen et al., 2001; Pérez-Nevado et al., 2006). Different mechanisms have been proposed to explain the higher competing capability of S. cerevisiae compared to non Saccharomyces yeasts which, in most cases, are not mutually exclusive, but complementary.
The vigorous fermentative capacity of S. cerevisiae yeasts in both the presence (Crabtree effect) and absence of oxygen, has been recognized as the main strategy to outcompete other microbial species present in must. S. cerevisiae consumes sugar resources faster, and the ethanol and CO2 produced during fermentation can be harmful or less tolerated by their competitors. Once competitors are overcome, S. cerevisiae can then use accumulated ethanol as a substrate for aerobic respiration. This ecological strategy is called (ethanol) make-accumulate-consume (Thomson et al., 2005; Piškur et al., 2006), and provides a selective advantage to S. cerevisiae to outcompete other microorganisms. Different non Saccharomyces yeast, as well as bacteria, have also been proven to be very sensitive to the killer peptides or toxic compounds produced by S. cerevisiae (Pérez-Nevado et al., 2006; Albergaria et al., 2010; Branco et al., 2014; Wang et al., 2015, 2016), which may play a key role during competition. Finally, the higher S. cerevisiae cell density has also been postulated as being an important factor that contribute to the exclusion of non Saccharomyces yeasts (Holm Hansen et al., 2001; Nissen et al., 2003, 2004; Nissen and Arneborg, 2003; Arneborg et al., 2005).
Other Saccharomyces species share very similar physiological properties with S. cerevisiae and, hence, similar ecological strategies. However, wine S. cerevisiae yeasts show better adaptation to survive under the stressful environmental conditions occurring during alcohol fermentation; e.g., high concentrations of sugar or ethanol, low pH and nutritional depletion, which provides them with a competitive advantage (Albergaria and Arneborg, 2016).
Another important advantage of S. cerevisiae on SNC species is its efficient growth at a wide range of temperatures, especially at higher temperatures (32°C). This has also been considered an important trait that explains its dominance during wine fermentation (Salvadó et al., 2011a). Goddard (2008) also observed that S. cerevisiae is even able to significantly increase the environmental temperature during vigorous fermentation. Arroyo-López et al. (2011) also demonstrated that S. cerevisiae was able to outcompete S. kudriavzevii even at temperatures that are more suitable to the latter (Salvadó et al., 2011b). However, S. paradoxus has been shown to be present during grape fermentation processes when competing with S. cerevisiae at both 22 and 30°C (Williams et al., 2015). Therefore, very little is known about the behavior of other SNC in competition with S. cerevisiae in winemaking environments at low temperatures.
In the Twenty-First century, the wine industry must respond to the challenges posed by both new consumers' demands and changes in grape composition and properties due to climate change. Consumers demand products with lower alcohol content and fruitier aromas, which lead winemakers to lower fermentation temperatures, as far as 10–12°C, to preserve aroma compounds in wines. Climate change influences grape must characteristics (acidity, content in sugars or tannins, etc.), which has an impact on final product quality. Also due to climatic change there is a gap between the maturity according to sugar content and the maturity of the phenolic compounds of the grape. Therefore, sugar concentration in musts reaches higher levels, which leads to wines with higher ethanol content.
These facts strongly challenge the quality and acceptance of the final product which leads to the necessity of improvements in oenological practices, among which the development of new yeast starters adapted to the new imposed conditions are of chief importance. Previous studies have shown that unconventional SNC yeast species, such as S. kudriavzevii and S. uvarum, could be good candidates to achieve those goals. This is because they exhibit good fermentative capabilities at low temperatures (Salvadó et al., 2011b), produce wines with lower alcohol and higher glycerol amounts (Arroyo-López et al., 2010; Oliveira et al., 2014; Pérez-Torrado et al., 2016), and contribute with good aromatic profiles (Gonzalez et al., 2007; Lopandic et al., 2007; Díaz-Montaño et al., 2008; Gamero et al., 2011, 2013, 2014; Stribny et al., 2015). As well as S. uvarum and S. kudriavzevii, we also included S. paradoxus, the closest species to S. cerevisiae among those of the Saccharomyces genera, which has been already tested for its fermentative capacity as we mentioned above; and S. eubayanus, the cryotolerant and recently discovered parental of lager yeast, found in natural fermented beverages from indigenous South American communities (Rodríguez et al., 2014), which makes it a good candidate for screening new properties that might increase the diversity of current commercial wines. Yet despite their potential, these species may have difficulties in competing at the industry level with S. cerevisiae, which in most of the cases exhibits better ethanol resistance and the ability to ferment at higher temperatures.
In the present study, we analyzed the survival capacity of SNC in competing with S. cerevisiae during fermentation at different temperatures to identify those traits that influence their competitive capabilities and to evaluate their industrial potential. Whereas, genetic markers are the standard to differentiate Saccharomyces strains in a complex culture, a quantitative PCR (QPCR)-based approach was designed to avoid them and their possible effect on gene expression or relative fitness. This approach consists on a relative quantification of the proportion of cells based on the QPCR amplification of a gene with species-specific primers using total DNA isolated from a mix of two strains.
Materials and methods
Yeast strains
Seven different Saccharomyces strains were used in our experiments. We chose a commercial strain, T73 (Lalvin T73 from Lallemand Montreal, Canada), as our wine S. cerevisiae representative. We also included YPS128, a S. cerevisiae strain isolated from Pennsylvania woodlands; S. paradoxus strain 54, isolated from Croatian vineyards; two S. uvarum strains, BMV58, selected in our laboratory and commercialized for winemaking (VELLUTOBMV58™ from Lallemand), and CECT12600, isolated from a non-fermented beverage (mistela) in Alicante, Spain; S. eubayanus strain NPCC1292 is a natural isolate from North Patagonian Mudai, traditional fermentation made with Araucaria araucana seeds; and S. kudriavzevii strain CR85, a natural isolate from oak tree bark in Agudo, Ciudad Real, Spain.
Synthetic must fermentations
For all our experiments, fermentations were performed in 3x or 6x replicates in 250 mL flasks that contained 200 mL of synthetic must (SM), which is frequently used in microvinification experiments (Rossignol et al., 2003), with 100 g/L of glucose and 100 g/L of fructose.
To assess the relative growth of S. cerevisiae and other Saccharomyces species under winemaking conditions, we performed competition experiments in which we measured the relative amount of both strains in co-cultures. We included a S. cerevisiae strain, either T73 or YPS128, and a non cerevisiae one, in all these experiments and measured their relative abundance at different fermentation times. As controls, we monitored the growth of each strain in monocultures under the same conditions as the competitions experiments. Overnight precultures were grown in YPD medium at 25°C. Afterward must was inoculated with the corresponding yeast strain to reach an initial concentration of 106 cells/mL, and was incubated at a fixed temperature (8, 12, 20, or 25°C) with agitation at 100 rpm during fermentation.
Cell samples were collected at several time points during fermentation and kept at −20°C for the subsequent total DNA isolation, used for the QPCR analysis, as described below. Cell counting was carried out in a Neubauer chamber to determine cell density at every sampling point. Growth curves were obtained by considering cell density and the proportion of competing strains given by the QPCR data.
Müller valves were used to monitor fermentation stage through weight loss, until it reached a constant weight, when it was considered to be over. At this point, samples of supernatant were kept at −20°C for further analyses.
Primer design
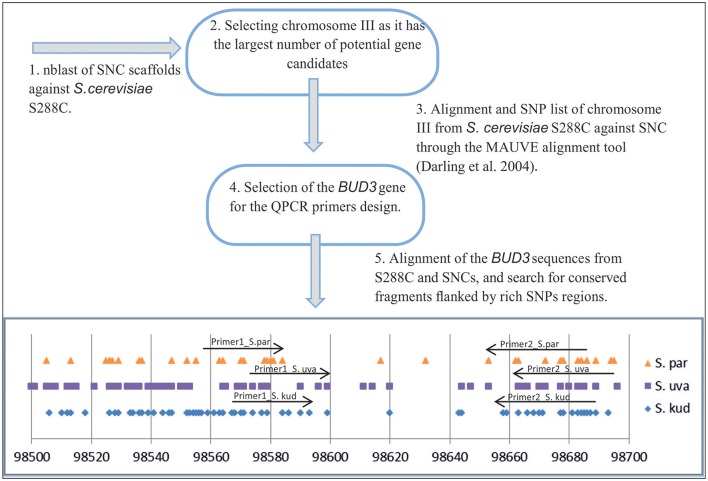
Alignments of homologous chromosomes from S. cerevisiae S288c, S. paradoxus, S. kudriavzevii, and S. uvarum were carried out by the Mauve alignment tool (Darling et al., 2004). Genomic sequences were downloaded from the Saccharomyces Sensu Stricto Resources website (Scannell et al., 2011) and Saccharomyces Genome Database (Engel, 2013). By way of example, a SNPs map of the gene BUD3 of S. paradoxus, S. kudriavzevii, and S. uvarum individually aligned against S. cerevisiae is shown in Figure 1. This highly conserved single copy gene was selected to look for strain-specific pairs of primers (Supplementary Table 1). All the resulting amplicons were approximately 100 bp in length and had a similar melting temperature when detected with our LightCycler 480 II instrument.
Figure 1.
Scheme used for QPCR primers design.
Specificity of the PCR assays
Total DNA samples were extracted from yeasts as described below. PCRs were carried out in a 20 μL final volume, including 1 μL of the DNA template, 0.25 μM of each primer, 200 μM of each dNTP, 2.5 mM of MgCl2, 10X buffer, and 0.75 U of Taq DNA polymerase (Takara, Bio, Shiga, Japan). For each case, total DNA from the competitor strain was used as a crossed amplification control.
The PCR program consisted of an initial denaturalization step at 94°C for 5 min, followed by 30 cycles of a denaturalization step at 94°C, an annealing step at either 53 or 54°C for 1 min, and an extension step at 72°C for 10 s, and a final extension step at 72°C for 5 min. PCR products were analyzed by electrophoresis on a 1.5% (w/v) agarose gel stained with RealSafe™ nucleic acid staining solution (20,000X) (Chembio Diagnosis Systems, Medford, NY, USA) in 1x TAE buffer, and were visualized under UV light. A 100-bp DNA ladder marker (Invitrogen™, Carlsbad, CA, USA) was used as the size standard.
DNA extraction and sample preparation
Total DNA samples from all the yeasts were extracted as described elsewhere (Querol et al., 1992). The concentration of the DNA samples was measured in a Nanodrop spectrophotometer ND-1000 (Nanodrop Technologies™, Wilmington, DE. USA) and adjusted to 20 ng/μL.
qPCR analysis
PCR amplification was performed in a 10 μL final volume that contained 2.5 μL of the DNA template, 1.5 μL MilliQ water, 0.2 μM of each primer, and 5 μL of LightCycler 480 SYBR Green I Master (Roche). Reactions were performed in 96-well plates in an LightCycler 480 (II) PCR amplification and detection instrument with an initial denaturalization step at 95°C for 5 min, followed by 45 cycles of 95°C for 10 s, either 53 or 54°C for 10 s and 72°C for 4 s. The CT values were calculated automatically by this instrument.
Plates were always divided into two symmetric halves. In each one, a different reaction mix was used where the pair of primers was specific for one of the two strains. For each half, an internal standard curve was included, made of six serial dilutions of the mixed total DNA from both competing strains in 1:1 proportions, the total DNA from the strain amplified in this half as a positive control, the total DNA from the other strain in competition as a control for cross amplification, and a negative control with PCR grade water instead of the template DNA. Three to six biological replicates were used.
The relative concentration of both strains in each biological replicate was given by the ratio of the means of the technical replicates concentrations calculated by the LightCycler 480 instrument software 1.5 (Roche Diagnosis, Darmstadt, Germany).
Method sensitivity
For every competition experiment, the following test was performed to assess the reliability of our method. The mix of cells of the corresponding strains was prepared from overnight GPY precultures in known proportions (10:90, 30:70, 50:50, 70:30, and 90:10). The QPCR analysis was carried out using total DNA extraction samples from the mixes of cells. The relative concentration of both strains in each sample was given by the ratio of the means of the concentrations of the replicates given by the LightCycler 480 instrument software 1.5 (Roche Diagnosis, Darmstadt, Germany). Three biological replicates were included.
Linear model adjustments were made for the cell proportions estimated with each used pair of primers against the theoretical values, and for all the collected data as a whole. The function lm() from R was used for this purpose.
Relative intrinsic growth rate determination
The intrinsic growth rate (r) can be calculated as in a previous work of Williams et al. (2015). Here the same method was followed with some modifications:
| (1) |
where Nt is cell density at a given time point, N0 corresponds to the initial cell density, and t is the time (in hours) when both strains reached their highest cell density in both competition and monoculture.
The effect that competition has on the involved strains can be assessed as the difference in their intrinsic growth rate in single culture and in competition (Δ = rsingle − rcompetition). For the sake of better clarifying the results, the relative intrinsic growth rate (RΔr = Δr/rsingle) was determined.
Growth kinetics parameters
On day 1, the precultures of all the used strains were grown o/n at 25°C in GPY medium. On day 2, cells were harvested by centrifugation, washed, suspended in dH2O and diluted to an OD600 of 2.7. Next 10 μL from each dilution were pipetted into one well of a 96-well plate, previously filled with 260 μL of SM (10 replicates). Four wells were filled with only sterile SM as a blank for the OD600 measurements. Four plates were set, one for each assayed temperature: 8, 12, 20, and 25°C.
OD600 was monitored in a SPECTROstar Omega instrument (BMG Labtech, Offenburg, Germany). Frequency of measurements varied according to temperature in order to obtain sufficient data points for a statistically significant adjustment to the reparametrized Gompertz equation proposed by Zwietering et al. (1990), which takes this expression:
| (2) |
where y = ln (ODt/OD0), OD0 is the initial OD and ODt is the OD at time t, D is the asymptotic maximum, the equivalent to ln (ODmax/OD0), μmax is the maximum specific growth rate (h−1) and λ is the lag phase period (h). An adjustment was made using a nonlinear regression procedure of minimizing the sum of the squares of the difference between the experimental data and the fitted model. This was done using version 7.0 of the Statistica software (Stat-Soft, Inc., Tulsa, OK, USA).
Strains were tested for the significant differences among them with an ANOVA using the one-way ANOVA module of the Statistica 7.0 software. Growth parameters μmax and λ were introduced as dependent variables. Means were grouped using the Tukey HSD test (α = 0.05).
Correlation of relative intrinsic growth rate and growth kinetics parameters
Linear regression models (y = Ax1 + B and y = Ax2 + B) were constructed, where y = RΔr for the non cerevisiae strain, x1 = (μmaxcompetitor − μmaxS. cerevisiae)/μmaxcompetitor (RΔμ) and x2 = (λS. cerevisiae − λcompetitor)/λcompetitor (RΔλ). This was done using the R function lm (R Core Team, 2015).
HPLC analysis
Residual sugars (glucose and fructose), glycerol, ethanol and acetic acid from the fermentation end point samples were determined by HPLC (Thermo Fisher Scientific, Waltham, MA. USA) using a refraction index detector and a HyperREZTM XP Carbohydrate H+ 8 μm column (Thermo Fisher Scientific) equipped with a HyperREZTM XP Carbohydrate Guard (Thermo Fisher Scientific). Samples were diluted 3-fold, filtered through a 0.22-μm nylon filter (Symta, Madrid, Spain) and injected in duplicate. The analysis conditions were: eluent, 1.5 mM of H2SO4; 0.6 ml min-1 flux and a 50°C oven temperature.
Statistical analysis of the fermentation kinetics and HPLC results
The recorded mass loss of the fermentation flasks correlates with sugar consumption, which was taken into consideration to fit our curve to Gompertz equation (Zwietering et al., 1990) and obtain fermentation parameters m (maximum sugar consumption rate, g L−1 h−1), l (lag phase period, h) and t90 (time taken to consume 90% of sugars, h) as in Pérez-Través et al. (2014).
Fermentations were tested for the significant differences among them with an ANOVA using the one-way ANOVA module of the Statistica 7.0 software. The concentrations of glucose, fructose, glycerol, ethanol and acetic acid obtained by HPLC, and the parameters m, l, and t90 were introduced as the dependent variables. Means were grouped using the Tukey HSD test (α = 0.05). The analysis was performed for each temperature condition used.
Results
Specificity and sensitivity of the qPCR assay
Six pairs of primers were designed, one for each strain, except for the S. uvarum strains, which share primers. To check for specificity, primers were tested by conventional PCR amplification. Bands of the desired size were observed in all cases. Absence of bands from the PCR reactions of the total DNA isolated from the competitor strain confirmed strain specificity.
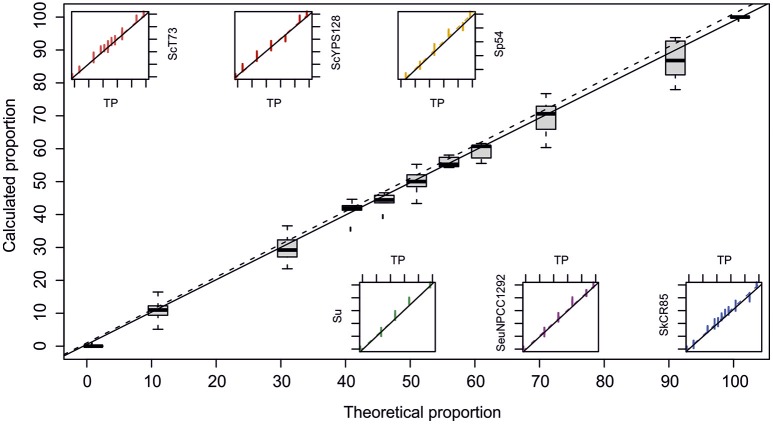
To assess the technique's sensitivity for the relative quantification of different yeast strains in co-culture, mixes of cells in known proportions were prepared for each assayed competition; i.e., our reference S. cerevisiae winery strain T73 against S. kudriavzevii strain CR85, S. uvarum strain BMV58, S. uvarum strain CECT12600, S. paradoxus strain 54 or S. eubayanus strain NPCC1292, and the wild S. cerevisiae strain YPS128 against S. kudriavzevii strain CR85. The obtained QPCR results about the theoretical proportions can be seen in Figure 2. Data were fitted to linear regression models and coefficients that came very close to the normal for all cases were obtained (Table 1). These results were statistically significant according to the Fisher test (Table 1). Thus, the method is suitable for the quantification of the different Saccharomyces strains mixed in a culture.
Figure 2.
Calculated relative quantification by QPCR against theoretical values. Boxplot shows the summary of all the data, while small graphics show the dispersion for each specific pair of primers. Data sets were adjusted to a linear model. Dotted lines represent normal distribution and full lines denote adjustments.
Table 1.
Linear model adjustment results for the calculated relative QPCR quantification (y) against the theoretical values (X).
| Pair of primers | A | B | R2 | p-value |
|---|---|---|---|---|
| S. cerevisiae T73 | 1.0089 | −0.5715 | 0.9924 | <2.2 × 10-16 |
| S. paradoxus 54 | 1.0074 | 1.2536 | 0.9829 | 1.473 × 10-15 |
| S. eubayanus NPCC1292 | 0.9834 | 1.4564 | 0.9905 | <2.2 × 10-16 |
| S. uvarum BMV58/CECT12600 | 0.9640 | 1.2897 | 0.9924 | 3.655 × 10-15 |
| S. cerevisiaeYPS128 | 1.0214 | −1.6341 | 0.9867 | <2.2 × 10-16 |
| S. kudriavzevii CR85 | 1.0182 | 0.5339 | 0.9905 | <2.2 × 10-16 |
| All | 10060 | 0.043 | 0.9889 | <2.2 × 10-16 |
A, is the regression coefficient and B, is the error term. p-values are obtained by the Fisher test.
Yeast competitions
To assess the effect of competition at low temperature on the intrinsic growth rate (r) of the SNC species, we performed a series of fermentations conducted by yeast strains S. paradoxus 54, S. uvarum BMV58, S. uvarum CECT12600, S. kudriavzevii CR85 and S. eubayanus NPCC1292 in competition with wine S. cerevisiae strain T73. We also tested the behavior of wild S. cerevisiae strain YPS128 in competition with S. kudriavzevii CR85. These competition experiments were performed in batch fermentations of SM at 8, 12, and 20°C. Fermentations at a moderate temperature condition (25°C) were also performed as a control of S. cerevisiae's imposition on cryotolerant yeasts. Monoculture fermentations, inoculated with the same strains, were performed as controls under the same conditions.
Figure 3 shows the percentages of the strains under competition when fermentation reached the stationary growth phase. These results offer an overview of the output of competitions during fermentation at different temperatures. We can see that wine S. cerevisiae T73 was able to exclude all the other Saccharomyces strains during fermentation at 25°C. At 20°C, T73 also outcompeted all the strains, except for wine S. uvarum BMV58, which was present in similar percentages. However, at low temperature, 12°C, T73 co-existed with S. eubayanus, S. kudriavzevii and S. paradoxus, but was displaced by both the S. uvarum strains.
Figure 3.
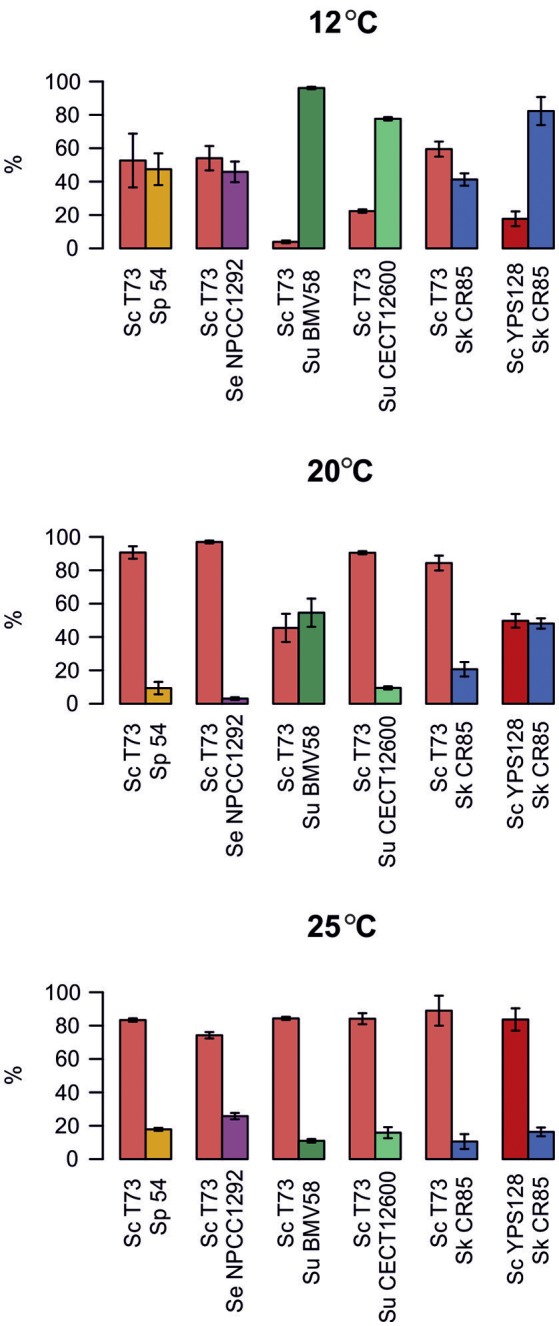
Presence of both strains from each competition when their highest cell densities were reached. Values are the mean of three replicates. Error bars represent SD.
In the competitions between wild strains S. cerevisiae YPS128 and S. kudriavzevii CR85, YPS128 clearly outcompeted CR85 at 25°C, they co-existed at 20°C, but CR85 certainly dominated at low temperatures.
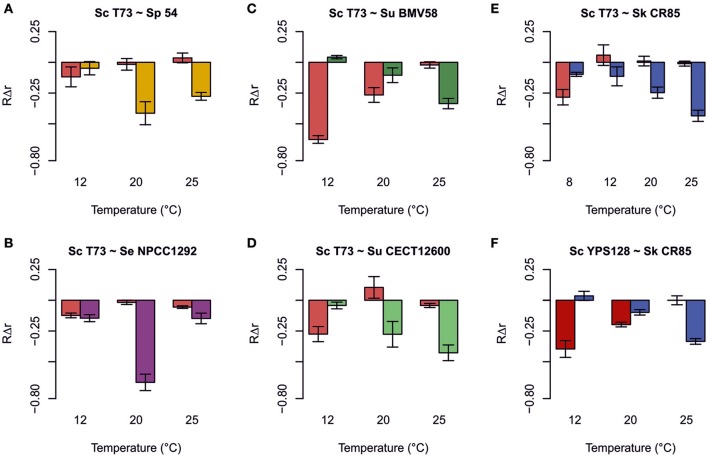
Despite these results being quite explicative about domination during competition, it is interesting to obtain a quantitative measurement of the effect that presence of a particular yeast can have on its competitor's growth. The most suitable indicator of these effects is the relative intrinsic growth rates (RΔr) based on the difference of growth rates when the strain is grown in a mixed culture and in a single culture (Figure 4). It is important to note that there was no significant positive effect on the growth of any strain as a result of the presence of a competitor. For S. cerevisiae, these effects were negative at low temperatures, but null or insignificant at high temperatures (Figure 4). For S. uvarum and S. kudriavzevii, a trend in the opposite direction was noted as the effect was less negative (or insignificant) at low temperatures, and the negative effect increased with temperature (Figures 4C–F). Finally for S. paradoxus and S. eubayanus, the strongest negative effect occurred at medium temperature (20°C) (Figures 4A,B).
Figure 4.
Relative intrinsic growth rate (RΔr = (rcompetition − rsingle)/rsingle) caused by the effect of competitions between Saccharomyces cerevisiae T73 and S. paradoxus 54 (A), S. cerevisiae T73 and S. eubayanus NPCC1292 (B), S. cerevisiae T73 and S. uvarum BMV58 (C), S. cerevisiae T73 and S. uvarum CECT12600 (D), S. cerevisiae T73 and S. kudriavzevii CR85 (E), and S. cerevisiae YPS128 and S. kudriavzevii CR85 (F). Values are the means of triplicate experiments. Error bars represent SD.
The comparison made between the performances of strains of the same species, but with different origins, showed that S. cerevisiae T73 and S. uvarum BMV58 wine strains were considerably less affected by their competitors than the strains with other origins, such as S. cerevisiae YPS128 and S. uvarum CECT12600 (Figures 4C–F).
Prevalence during fermentation seemed to be clearly related to temperature adaptation. The correlations of growth parameters maximum growth rate and lag phase duration (Table 2) with the relative increment in the intrinsic growth rate were calculated. Positive correlations with R2 ~ 0.4 were obtained for both parameters.
Table 2.
Growth parameters obtained by the Gompertz equation proposed by Zwietering et al. (1990) for the different strains in SM at different temperatures.
| 8°C | 12°C | 20°C | 25°C | |||||
|---|---|---|---|---|---|---|---|---|
| Strain | μmax (h−1) | λ (h) | μmax (h−1) | λ (h) | μmax (h−1) | λ (h) | μmax (h−1) | λ (h) |
| S. cerevisiae T73 | 0.0083±0.0002c | 247.77±9.15e | 0.0217±0.0005b | 82.5652±0.87c | 0.0602±0.0020a | 18.6771±0.43a | 0.1740±0.0060a | 9.3204±0.62a |
| S. paradoxus 54 | 0.0080±0.0003b, c | 260.69±9.10f | 0.0231±0.0011c | 104.5469±2.38a | 0.0504±0.0034c | 25.2771±1.01c | 0.1387±0.0021c | 12.7527±1.47d |
| S. eubayanus NPCC1292 | 0.0098±0.0012d | 143.58±2.47a | 0.0216±0.0006b | 63.4613±2.10e | 0.0471±0.0016d | 26.5521±0.44d | 0.1184±0.0022d | 10.3316±0.18a, b |
| S. uvarum BMV58 | 0.0181±0.0009f | 147.09±4.09a | 0.0320±0.0008e | 66.1784±1.07d | 0.0609±0.0024a | 27.7608±0.39e | 0.1668±0.0214a, b | 12.1358±1.04c, d |
| S. uvarum CECT12600 | 0.0153±0.0005e | 160.02±1.81b | 0.0301±0.0006d | 71.2131±1.95b | 0.0516±0.0009c | 26.2352±0.52d | 0.1539±0.0049b | 11.4711±0.30b, c |
| S. cerevisiae YPS128 | 0.0067±0.0003a | 220.15±7.35d | 0.0194±0.0003a | 83.1130±2.23c | 0.0554±0.0013b | 23.3442±0.49b | 0.1560±0.0040b | 9.5580±0.69a |
| S. kudriavzevii CR85 | 0.0074±0.0002a, b | 178.10±4.65c | 0.0199±0.0005a | 65.0655±2.06d, e | 0.0437±0.0020e | 25.8793±0.95c, d | 0.1023±0.0071e | 14.5462±0.55e |
μmax, is maximum growth rate and λ, is lag phase duration. Values are given as mean ± standard deviation. The values followed by different superindexes in the same column are significantly different according to the Tukey HSD test (α = 0.05, n = 10).
Competitions between S. cerevisiae T73 and S. paradoxus 54
When competing with S. paradoxus strain 54, T73 achieved slightly lower intrinsic growth rate at 12°C compared to a single fermentation. However, at 20 and 25°C, its growth fitness is maintained (Figure 4A). Strain 54 performed normally at low temperature, but was clearly affected at 20 and 25°C (Figure 4A), and was almost totally excluded from fermentation (Figure 3). Although both species were phylogenetically closely related, the wine S. cerevisiae strain seemed superior in this competition. Furthermore, it is interesting to note that at all tested temperatures, the dominant strain T73 had a shorter lag phase (λ, Table 2).
Competitions between S. cerevisiae T73 and S. eubayanus NPCC1292
In competition both strains maintained their capability to grow in co-cultures at 12 and 25°C, according to the slight drop in their intrinsic growth rate parameter compared to single cultures. Strikingly at intermediate temperatures, NPCC1292 was clearly outcompeted by T73 (Figure 4B), when its lag phase became noticeably longer (Table 2). Although the intrinsic growth rate of NPCC1292 was only slightly affected at 25°C, this strain was present at a low percentage during fermentation (Figure 3). This can be explained by a low cell density during not only competition, but also during single culture fermentation (data not shown).
Competitions between S. cerevisiae T73 and S. uvarum strains
Here we assessed the competitive adaptation capacity of a wine and a non fermentative S. uvarum. Wine S. uvarum strain BMV58 competed better at low temperatures (12 and 20°C), and severely affected T73 growth. This effect reverted as temperature rose. We can see that T73 shows a clear advantage at 25°C (Figure 4C).
To test whether the same trend could be observed with a non wine strain, we performed the same experiment using strain S. uvarum CECT12600. The behavior of the differential intrinsic growth rates was similar, but in this case S. uvarum CECT12600 obtained lower values and had a less intense effect on T73 (Figure 4D) than BMV58, which showed better competitive fitness in fermentative environments.
Finally, it is important to remark that S. cerevisiae T73 had a shorter lag phase (λ) than S. uvarum BMV58 during the competitions at 20 and 25°C (Table 2), but at similar maximum growth rates (μmax), and BMV58 was able to co-exist with T73 during the competition at 20°C, but not at 25°C (Figure 4). At 12°C, both S. uvarum strains had a shorter lag phase and higher maximum growth rates than S. cerevisiae (Table 2), and were dominant during fermentation (Figure 3).
Competitions between S. kudriavzevii CR85 and S. cerevisiae strains
Wine strain S. cerevisiae T73 is not affected by most temperature conditions when competing with S. kudriavzevii. However, at 8°C, a clear negative effect on the relative intrinsic growth rate (r) on T73 can be observed. S. kudriavzevii CR85 was always affected by presence of T73, although its impact was softer at 8°C and S. kudriavzevii became more competitive (Figure 4E).
To test if T73 resistance during competition, even at a very low temperature, was to some extent dependent on its better adaptation to fermentation environments, the wild S. cerevisiae strain YPS128, isolated from an oak bark, was used in the competitions assays with S. kudriavzevii. Figure 4F shows noticeable differences in the competition at 12°C, where CR85 clearly outcompetes YPS128, and it exhibited an intrinsic growth rate that was markedly affected. In the competitions at 20°C, in which S. kudriavzevii predominated (Figure 3), YPS128 underwent a greater negative effect (Figure 4). Contrarily at 25°C, CR85 was clear at a disadvantage (Figure 4F). Therefore, the fermentative origin of the S. cerevisiae yeasts seems to correlate with better performance in fermentation.
The growth parameters from Table 2 could explain most of these results. At 8°C, when S. Kudriavzevii outcompeted S. cerevisiae T73, the winner (Figure 3) had a higher maximum growth rate and a shorter lag phase. At 12°C, S. kudriavzevii presented a shorter lag phase than both the S. cerevisiae strains, and a higher maximum growth rate than the wild S. cerevisiae strain, which was clearly affected under these conditions (Figure 4F). S. cerevisiae wine strain T73 had a clearly higher μ at 12°C, which could be the reason why T73 became dominant as fermentation continued (Figure 3). At 20°C, both the S. cerevisiae strains already exhibited better growth capabilities in synthetic must (Table 2), but there were clear differences in their performance during the competition against S. kudriavzevii as the wine strain was a much better competitor than the wild strain (Figures 3, 4E,F).
Correlation between growth parameters and competitive advantage
We assessed whether there was any correlation between the fact of having better growth parameters in single culture and the imposition during our competition experiments (Table 2). Linear correlations between RΔr and RΔμ or RΔλ were obtained, with R2-values of 0.40 and 0.42, respectively. Significance was tested by the Fisher's test, and the resulting p-values were 0.005158 and 0.003681, respectively.
A graphical summary of the three parameters used in the analysis for each competition is depicted in Figure 5. In most cases, low RΔr values corresponded to low RΔμ and RΔλ. This indicates that a more affected strain during co-fermentations exhibits worse growth parameters in single culture than its competitor (Figure 4, Table 2).
Figure 5.
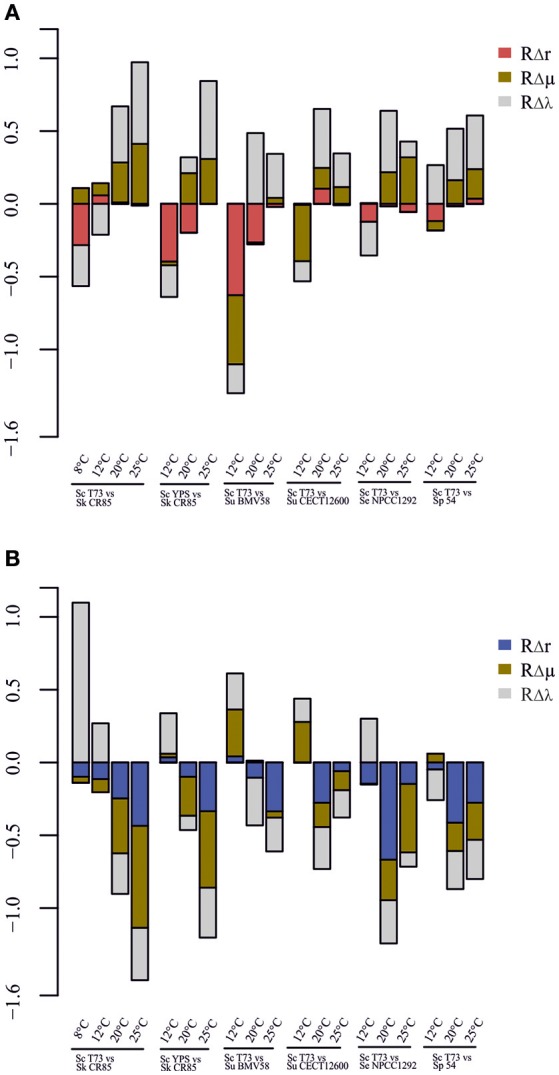
Comparative of performance in competition and growth kinetics parameters in single culture for S. cerevisiae (A) and competitor strains (B).
There are some exceptions however, as already mentioned above, such as the competition between T73 and BMV58 at 20°C, at which S. uvarum had a similar RΔμ and a notably worse RΔλ value, but competition had a remarkably negative effect on S. cerevisiae. Interestingly, Figure 5 shows comparatively slight RΔμ or RΔλ differences together with extreme RΔr values (T73-NPCC1292, 20°C), and vice versa (T73-NPCC1292, 25°C). This indicates that co-culture fermentations may be influenced by other competitive growth strategies.
Influence of competition on fermentation parameters
Yeast characterization as wine fermenters must include aspects like the ability to consume all the sugars present in must at a suitable pace, or the capability to produce a wine with high quality standards according to consumer demands. Table 3 includes different fermentation kinetic parameters: maximum sugar consumption rate (m) and fermentation lag phase (l), inferred from mass loss during fermentation, as well as the time taken to consume 90% of the initial sugar content (t90). Final product composition is also a key factor. Thus, we measured glucose, fructose, glycerol and ethanol concentrations at the end of fermentation, that is, when no mass loss was observed (Table 4). This data set is useful to determine the mixed yeast cultures that could potentially improve some wine characteristics.
Table 3.
Kinetics parameters of fermentations m is the maximum sugar consumption rate, l is the fermentation lag phase duration, and t90 is the time employed to consume 90% of the sugars present in the initial must.
| 12°C | 20°C | 25°C | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Fermentation | m (g L−1 h−1) | l (h) | t90 (h) | m (g L−1 h−1) | l (h) | t90 (h) | m (g L−1 h−1) | l (h) | t90 (h) |
| T73 | 0.31 ± 0.01a, b | 91.71 ± 1.10a, b, c | 472.92 ± 5.36a, b, c | 0.90 ± 0.06d, e | 26.70 ± 1.24a, b | 171.24 ± 10.73a, b | 1.14 ± 0.17b, c, d | 6.42 ± 1.69b, c | 149.22 ± 17.43b, c |
| 54 | 0.28±0.01a, b | 131.64±2.41e | 589.40±7.35c, d | 0.82±0.05b, c, d, e | 33.96±3.05c, d | 181.52±8.54a, b | 1.51±0.33d, e | 9.37±2.47b, c, d | 122.13±12.43a, b |
| T73-54 | 0.31±0.00a | 97.06±3.10b, c, d | 583.58±8.43c, d | 0.86±0.16b, c, d, e | 31.38±4.25b, c | 247.05±28.78c | 0.87±0.01a, b | 12.71±0.51d, e, f | 171.69±10.93c |
| NPCC1292 | 0.21±0.00a | 76.27±6.64a, b | Na | 0.59±0.01a, b | 30.14±0.89a, b, c | Na | 0.65±0.08a | 23.46±1.59h | Na |
| T73-NPCC1292 | 0.27±0.01a | 79.47±5.87a, b | 656.23±103.49d | 1.01±0.08e | 27.87±0.99a, b, c | 198.77±6.62b | 0.91±0.10a, b | 17.23±1.67g | 162.83±14.64c |
| BMV58 | 0.30±0.01a, b | 85.12±1.58a, b, c | 505.21±8.14b, c | 0.71±0.04a, b, c, d | 85.72±1.50e | 260.97±9.17c | 0.89±0.02a, b, c | 2.09±0.72a | 179.49±4.74c |
| T73-BMV58 | 0.49±0.01c, d | 71.20±2.32a | 320.76±6.27a | 0.63±0.03a, b, c | 84.49±1.58e | 283.37±17.12c, d | 1.47±0.14d, e | 15.86±0.46f, g | 104.81±7.54a |
| CECT12600 | 0.50±0.01d | 92.59±3.77a, b, c, d | 383.64±26.29a, b | 1.00±0.15e | 31.43±2.40b, c | 172.47±23.94a, b | 1.68±0.03e | 21.79±0.16h | 100.45±2.40a |
| T73-CECT12600 | 0.38±0.01b, c | 80.79±6.18a, b | 421.02±18.23a, b | 1.00±0.06e | 23.18±2.15a | 146.04±7.17a | 1.72±0.23e | 15.71±0.63e, f, g | 99.89±17.82a |
| YPS128 | 0.27±0.01a, b | 97.84±9.08b, c, d | 727.23±5.36d | 0.88±0.03c, d, e | 31.39±0.62b, c | 172.66±4.49a, b | 1.37±0.03c, d, e | 11.98±0.40d, e | 116.24±2.03a, b |
| YPS128-CR85 | 0.31±0.01a, b | 96.23±4.05b, c, d | 489.54±6.61b, c | 0.88±0.04c, d, e | 27.71±1.88a, b, c | 321.89±0.50d | 1.43±0.07d, e | 10.37±0.77c, d | 111.70±8.21a, b |
| CR85 | 0.32±0.01a, b | 116.35±2.49d, e | 502.88±7.59b, c | 1.02±0.00e | 38.50±1.16d | 156.49±0.87a, b | 0.87±0.07a, b | 15.68±0.62e, f, g | 184.99±13.71b |
| T73-CR85 | 0.52±0.14d | 103.32±19.91c, d | 382.17±26.05a, b | 0.54±0.07a | 24.15±1.86a | 259.85±19.33c | 1.65±0.11e | 6.17±0.78b | 98.44±9.57a |
Parameters are obtained through an adjustment to Gompertz (Zwietering et al., 1990). Values given as mean±standard deviation of three biological replicates. An ANOVA analysis was carried out. The values followed by different superindexes in the same column are significantly different according to the Tukey HSD test (α = 0.05).
Table 4.
Chemical composition of the fermented SM obtained through HPLC.
| 12°C | 20°C | 25°C | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fermentation | glucose | fructose | glycerol | ethanol | glucose | fructose | glycerol | ethanol | glucose | fructose | glycerol | ethanol |
| T73 | 0.10 ± 0.04a, b | 6.64 ± 1.04a, b, c, d | 5.09 ± 0.03a | 12.49 ± 0.28c, d | 0.02 ± 0.01a | 4.16 ± 0.60a, b, c | 5.53 ± 0.24a, b, c | 12.47 ± 0.48d, e | 0 ± 0a, b | 0 ± 0a | 5.78 ± 0.08a, b | 12.67 ± 0.10c, d, e |
| 54 | 0 ± 0a, b | 2.90 ± 0.33a, b | 5.03 ± 0.04a | 11.83 ± 0.10b, c, d | 0.02 ± 0.02a | 4.06 ± 2.11a, b, c | 6.08 ± 0.29b, c, d | 11.21 ± 0.34b, c, d | 0.21 ± 0.02a, b | 8.91 ± 0.31c | 7.57 ± 0.25e | 10.76 ± 0.04a, b |
| T73-54 | 0.51 ± 0.10a, b | 14.88 ± 0.70e | 5.06 ± 0.12a | 12.09 ± 0.28b, c, d | 0 ± 0a | 4.30 ± 0.41a, b, c | 5.95 ± 0.37b, c, d | 13.19 ± 0.44e, f | 0.28 ± 0.06a, b | 8.5 ± 1.94b, c | 5.92 ± 0.12a, b, c | 12.25 ± 0.40b, c, d, e |
| NPCC1292 | 6.66 ± 0.80c | 41.05 ± 0.50f | 7.68 ± 0.40d | 9.69 ± 0.54a | 2.25 ± 0.24b | 24.54 ± 2.45e | 6.66 ± 0.08d, e | 9.89 ± 0.01a, b | 2.73 ± 1.20c | 31.47 ± 5.03e | 7.46 ± 0.16e | 10.14 ± 0.23a |
| T73-NPCC1292 | 0.69 ± 0.58b | 13.45 ± 5.03d, e | 5.95 ± 0.64a, b | 11.40 ± 0.70b, c, d | 0.35 ± 0.34a | 9.31 ± 3.09c, d | 6.76 ± 0.27d, e | 13.97 ± 0.58f | 0 ± 0a | 0.56 ± 0.80a | 6.14 ± 0.09a, b, c | 12.70 ± 0.18e |
| BMV58 | 0 ± 0a, b | 0.70 ± 0.62a | 5.15 ± 0.17a | 11.71 ± 0.46b, c, d | 0 ± 0a | 2.93 ± 2.55a, b | 4.72 ± 0.14a | 9.51 ± 0.16a | 0 ± 0a, b | 4.15 ± 1.36a, b, c | 7.13 ± 0.20d, e | 12.27 ± 0.08b, c, d, e |
| T73-BMV58 | 0 ± 0a, b | 4.91 ± 1.34a, b | 5.85 ± 0.05a, b | 11.83 ± 0.16b, c, d | 0 ± 0a | 0 ± 0a | 5.50 ± 0.01a, b | 10.97 ± 0.74a, b, c, d | 0 ± 0a | 3.59 ± 1.45a | 5.95 ± 0.14a, b | 12.64 ± 0.09c, e |
| CECT12600 | 0.12 ± 0.12a, b | 9.80 ± 2.94b, c, d, e | 6.05 ± 0.08a, b, c | 11.26 ± 0.38a, b, c, d | 0 ± 0a | 0 ± 0a | 5.85 ± 0.08b, c, d | 12.27 ± 0.07d, e | 0 ± 0a, b | 3.11 ± 0.53a | 6.31 ± 0.31b, c, d | 11.49 ± 0.45a, b, c, d, e |
| T73-CECT12600 | 0 ± 0a | 3.92 ± 1.46a, b | 6.00 ± 0.42a, b | 12.44 ± 0.70c | 0.59 ± 0.23a | 12.76 ± 2.45d | 5.31 ± 0.01a, b | 11.51 ± 0.57c, d | 0 ± 0a, b | 1.13 ± 1.13a | 5.46 ± 0.36a, b | 11.11 ± 0.24a, b, d |
| YPS128 | 0.26 ± 0.09a, b | 8.35 ± 1.08a, b, c, d, e | 6.88 ± 0.57b, c, d | 10.87 ± 0.31b, c, d | 0.26 ± 0.07a | 8.35 ± 1.08b, c, d | 6.88 ± 0.57d, e | 12.03 ± 0.10c, d, e | 0.01 ± 0.01a, b | 3.52 ± 0.36a | 6.06 ± 0.15a, b, c | 10.78 ± 0.94a, b |
| YPS128-CR85 | 0 ± 0a, b | 3.13 ± 0.28a, b | 7.07 ± 0.07b, c, d | 11.76 ± 0.74b, c, d | 0.56 ± 0.36a | 11.31 ± 4.53d | 6.63 ± 0.29c, d, e | 11.78 ± 0.24c, d, e | 0.14 ± 0.11ab | 4.31 ± 1.87a, b, c | 6.17 ± 0.90a, b, c | 10.35 ± 0.77a |
| CR85 | 0.0 ± 0a, b | 4.89 ± 2.81c, d, e | 7.34 ± 0.23c, d | 10.87 ± 0.46a, b, d | 0.01 ± 0.01a | 4.13 ± 0.31a, b, c | 7.61 ± 0.94e | 10.54 ± 1.25a, b, c | 0.85 ± 0.14b | 16.35 ± 0.90d | 6.93 ± 0.51c, d, e | 10.46 ± 0.46a |
| T73-CR85 | 0.12 ± 0.17a, b | 6.42 ± 5.37a, b, c | 7.24 ± 1.32c, d | 10.36 ± 1.41a, b | 0.05 ± 0.75a | 2.73 ± 3.11a, b | 5.22 ± 0.51a, b | 11.44 ± 0.26c, d | 0.03 ± 0.04a, b | 3.95 ± 1.28a, b | 5.35 ± 0.24a | 11.17 ± 0.72a, b, c, d |
Glucose, fructose and glycerol are given in g L−1, and ethanol in %. Values are given as mean ± standard deviation of three biological replicates and two HPLC detection runs. An ANOVA analysis was carried out. The values followed by different superindexes in the same column are significantly different according to the Tukey HSD test (α = 0.05).
Reference wine strain S. cerevisiae T73 is characterized by the production of relatively low glycerol values (5–6 g L−1) and high ethanol content (>12 %), as observed in Table 4. It also accomplishes quite a high sugar consumption rate at 20°C (Table 3) and 25°C (Table 3), but a low one at 12°C (Table 3), which is consistent with the temperature adaptation of S. cerevisiae to grow at higher temperatures than cryotolerant species S. uvarum and S. kudriavzevii. In most cases, and according to the ANOVA analysis, its l and t90 belong to the group of the shortest times (Table 3).
Interestingly, some co-cultures improved these fermentation parameters; e.g., T73 with either S. kudriavzevii CR85 or S. uvarum BMV58 at 12°C increased the m, and reduced t90 (Table 3). At 25°C, the combinations of T73 with S. kudriavzevii and S. uvarum once again seemed to improve the fermentation kinetics. A reduction of t90 for the three co-cultures (T73-CR85, T73-BMV58 and T73-CECT12600) was also observed at 25°C (Table 3). These fermentation parameters improved compared to their respective single culture fermentations (Table 3), which is indicative of synergic interactions.
Unlike the 12and 25°C conditions, practically no fermentation parameters or compounds improved at 20°C (Tables 3, 4). The competitions against S. paradoxus seemed disadvantageous at 20°C and 25°C, which also occurred when competing with CR85 at 20°C and with NPCC1292 at 25°C (Table 3).
Despite their diverse origins, all the strains were able to complete their fermentations at 25°C except S. eubayanus NPCC1292 (Table 4). By the end of the fermentations conducted by this strain, the final product contained large amounts of glucose, and especially fructose. At low and medium temperatures (12 and 20°C) some strains also left a considerable amount of sugars, such as S. cerevisiae YPS128 or competences NPCC1292-T73, CECT12600-T73, 54-T73 and CR85-YPS128 (Table 4). Interestingly, most of them were able to ferment all the sugars when cultured alone (Table 4), so this could result in an antagonist effect for these pairs of strains. Moreover, some other parameters also reflected worse performances during co-fermentations, specifically t90 of T73-NPCC1292 at 12°C, or T73-54 and T73-CECT12600 at 20°C (Table 3).
As previously mentioned, a more profitable interaction is observed for CR85-T73 at low temperatures. At 12°C both strains co-existed during fermentation in similar proportions (Figure 3), which led to a final product with a lower ethanol concentration and a higher glycerol content than those obtained for the fermentations conducted by T73 alone (Table 4). Ethanol concentrations also lowered during co-inoculated fermentations at 20and at 25°C, but the conservative ANOVA test did not support the significance of these differences (Table 4). With the co-cultures of T73 with S. uvarum, no significant improvements in the final product composition were observed, although mean glycerol values and ethanol concentrations showed a positive tendency compared to the single S. cerevisiae fermentations at 12 and 20°C (Table 4).
Discussion
Accurate quantification of different Saccharomyces yeasts in co-cultures
Natural auxotrophic or drug-resistant mutants and strains genetically modified with reporter genes have been used to monitor yeast competences in co-cultures or during fermentation, which involves demanding tasks, such as mutant selection or construction, CFU enumeration in selective media, or flow cytometry (Arroyo-López et al., 2011; García-Ríos et al., 2014). Our results indicate that a more straightforward QPCR-based method, which does not require previous cell type separation, is suitable for the relative quantification of yeasts (Figure 2). In fact quantification by QPCR of different organisms in wine, including Saccharomyces yeasts, has already been applied (Neeley et al., 2005; Andorrà et al., 2010; Vendrame et al., 2014). However, to our knowledge, this technique has never been used to date to differentiate Saccharomyces yeasts during competition in the same environment. This novel approach can be applicable to broaden our knowledge about the ecology of the Saccharomyces yeast when competing for the same niche.
Temperature adaptation affects domination in a fermentative environment
Cryotolerant species S. kudriavzevii and S. uvarum have been used in this work given the trend to perform fermentation at lower temperatures in the wine industry to preserve the aroma fraction (Torija et al., 2001; Beltran et al., 2002; Gamero et al., 2013; Şener and Yildirim, 2013). Our results clearly show a longer prevalence of these species in fermentations at low temperatures. However, at 25°C, S. cerevisiae outgrow them. It can also be drawn from our data that this effect is modulated by the adaptation of strains to different habitats, where wine strains are always more competitive no matter what the temperature. This was observed not only for S. cerevisiae, but also for S. uvarum. Therefore, adaptation to high sugar environments could be another trait that influences fermentation domination as indicated by Barrajón et al. (2011).
Salvadó et al. (2011b) analyzed the thermotolerance of different Saccharomyces species using their growth kinetics parameters as measurable indicators. Growth ability under settled conditions could be considered as a suitable predictor for the imposition of one strain on another in competition. However, previous works (García-Ríos et al., 2014), as well as ours, have revealed that domination of environments is a more complex trait on Saccharomyces yeasts. Figure 5 shows that in most cases a higher μmax and a lower λ correlate to greater invulnerability in co-fermentation than the competitor strain. Nevertheless, we observed that the intensity of the effect is widely variable, and in some cases we found that the contrary happens; i.e., S. eubayanus NPCC1292 in the competition against S. cerevisiae T73 at 12°C, whose growth was affected despite having a shorter lag phase. S. cerevisiae YPS128, a strain isolated from oak trees, performed worse than expected against S. kudriavzevii CR85 at 20°C. Something similar occurred with the competition between S. cerevisiae T73 and S. uvarum BMV58 for both wine strains at 20°C: T73 had a noticeably shorter λ and a similar μmax, but was clearly wakened by BMV58. Thus, it is conceivable that an interaction among yeasts or their side products takes place as part of the competition mechanism. Whether this means that the presence of toxic compounds targets some specific Saccharomyces yeasts, the inhibitory physical contact among them, or some other strategy, is something that needs to be looked at in the future.
Coinoculation of Saccharomyces yeasts can be potentially beneficial for fermentations and final product composition
One of the main goals when studying alternative organisms for their use in food fermentation is achieving new characteristics of interest, that these organisms give rise to. With wine, numerous studies have been carried out with non Saccharomyces yeasts, and most have focuses on improving or enriching of aroma profiles, whereas others have focused more on controlling the final product concentration of specific compounds, such as ethanol or acetic acid (Andorrà et al., 2012; Rantsiou et al., 2012; Medina et al., 2013; Contreras et al., 2014; Izquierdo Cañas et al., 2014; Zara et al., 2014; Canonico et al., 2015, 2016; Rodrigues et al., 2016). However, fewer studies have been published about fermentation characterization by combining Saccharomyces strains or using uncommon Saccharomyces species (Cheraiti et al., 2005; Howell et al., 2006; King et al., 2010; Arroyo-López et al., 2011; Barrajón et al., 2011; Saberi et al., 2012; Williams et al., 2015; Gustafsson et al., 2016). Just as some of these investigations have suggested, our results showed that the final product composition of co-fermented musts cannot always be predicted from those of mono-fermentations. We observed a range of different scenarios: synergic or antagonist effects, as well as simply additive, depending on the strains and the assayed conditions. Nevertheless, we found some promising combinations of a wine S. cerevisiae strain with a SNC one; e.g., the remarkable case of the co-inoculation of S. cerevisiae T73 and S. kudriavzevii CR85 at low temperatures, which improved the efficiency of the process as regards single inoculations by increasing the maximum sugar consumption rate, and which also yielded a final product that contained less ethanol and more glycerol.
From the kinetics point of view, the co-fermentations of our wine S. cerevisiae strain with S. uvarum also revealed a positive effect, which was more visible at 12 and 25°C. This kind of synergic effect has been observed in previous works, where the addition of fructophilic yeast S. bombicola (similarly to S. uvarum) led to faster fructose and glucose consumption (Milanovic et al., 2012). In our case it was also noteworthy that the viability of T73 during competition against wine strain BMV58 was negatively influenced. However, against non winery strain CECT12600, it did not diminish, which means that winery strains could be more capable of sensing other yeasts in fermentative media and over-activate sugar consumption to take advantage of them. This hypothesis would also be supported by our results about the maximum sugar consumption rate (m) of the co-fermentations carried out by our reference wine strains S. cerevisiae T73 with S. uvarum or S. kudriavzevii. At any rate it is remarkable that reduced t90 values occurred during the fermentations run at 25°C for the cases of T73-CR85, T73-BMV58 and T73-CECT12600, when SNC strains were present in small proportions. This could be indicating that S. cerevisiae T73 responded to interactions during competences by increasing its metabolism.
To summarize, we have confirmed the great capacity of S. cerevisiae to dominate fermentative environments at traditional process temperatures (Holm Hansen et al., 2001; Pérez-Nevado et al., 2006; Arroyo-López et al., 2011; Williams et al., 2015). However, some cryotolerant Saccharomyces yeasts, particularly S. uvarum, can seriously compromise S. cerevisiae fitness during competences at lower temperatures, which explains why S. uvarum can replace S. cerevisiae during wine fermentations in European regions with oceanic and continental climates (Naumov et al., 2000; Sipiczki et al., 2001; Naumov et al., 2002; Redžepović et al., 2002; Rementeria, 2003; Demuyter et al., 2004). From a biotechnological point of view, the application of cryotolerant Saccharomyces species as starters for wine fermentation at low temperature could avoid its colonization by undesirable microorganisms that has been reported by other authors (Ciani and Comitini, 2006).
Our results also suggest that adaptation to winemaking establishes noticeable differences in the performance of Saccharomyces yeasts when competing during wine fermentation. Thus, profounder research on Saccharomyces yeasts' physical and biochemical interactions is necessary to optimize the composition of such starter cultures, which would make them even more interesting for industrial purposes. As a hint, at low temperatures we obtained improvements in the final composition of important compounds, such as higher glycerol contents and a lower ethanol yield, as well as the better fermentation performance of some yeast combinations, especially those of the S. cerevisiae with cryotolerant SNC species.
Author contributions
JA, EB, and AQ conceived and designed the experiments. JA and ML performed the experiments. JA, EB, and AQ analyzed the data and wrote the paper.
Funding
JA was supported by a FPI grant from the Ministerio de Economía y Competitividad (ref. BES-2013-066434). This work was supported by grants AGL2012-39937-C02-01 and AGL2015-67504-C3-1-R from the Spanish Government and FEDER to AQ, AGL2012-39937-C02-02 and AGL2015-67504-C3-3-R from the Spanish Government and FEDER to EB, and PROMETEO (project PROMETEOII/2014/042) from Generalitat Valenciana to AQ. We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00150/full#supplementary-material
References
- Albergaria H., Arneborg N. (2016). Dominance of Saccharomyces cerevisiae in alcoholic fermentation processes: role of physiological fitness and microbial interactions. Appl. Microbiol. Biotechnol. 100, 2035–2046. 10.1007/s00253-015-7255-0 [DOI] [PubMed] [Google Scholar]
- Albergaria H., Francisco D., Gori K., Arneborg N., Gírio F. (2010). Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl. Microbiol. Biotechnol. 86, 965–972. 10.1007/s00253-009-2409-6 [DOI] [PubMed] [Google Scholar]
- Andorrà I., Berradre M., Mas A., Esteve-Zarzoso B., Guillamón J. M. (2012). Effect of mixed culture fermentations on yeast populations and aroma profile. LWT-Food Sci. Technol. 49, 8–13. 10.1016/j.lwt.2012.04.008 [DOI] [Google Scholar]
- Andorrà I., Esteve-Zarzoso B., Guillamón J. M., Mas A. (2010). Determination of viable wine yeast using DNA binding dyes and quantitative PCR. Int. J. Food Microbiol. 144, 257–262. 10.1016/j.ijfoodmicro.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Antunovics Z., Irinyi L., Sipiczki M. (2005). Combined application of methods to taxonomic identification of Saccharomyces strains in fermenting botrytized grape must. J. Appl. Microbiol. 98, 971–979. 10.1111/j.1365-2672.2005.02543.x [DOI] [PubMed] [Google Scholar]
- Arneborg N., Siegumfeldt H., Andersen G. H., Nissen P., Daria V. R., Rodrigo P. J., et al. (2005). Interactive optical trapping shows that confinement is a determinant of growth in a mixed yeast culture. FEMS Microbiol. Lett. 245, 155–159. 10.1016/j.femsle.2005.03.008 [DOI] [PubMed] [Google Scholar]
- Arroyo-López F. N., Pérez-Torrado R., Querol A., Barrio E. (2010). Modulation of the glycerol and ethanol syntheses in the yeast Saccharomyces kudriavzevii differs from that exhibited by Saccharomyces cerevisiae and their hybrid. Food Microbiol. 27, 628–637. 10.1016/j.fm.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Arroyo-López F. N., Pérez-Través L., Querol A., Barrio E. (2011). Exclusion of Saccharomyces kudriavzevii from a wine model system mediated by Saccharomyces cerevisiae. Yeast 28, 423–435. 10.1002/yea.1848 [DOI] [PubMed] [Google Scholar]
- Barrajón N., Arévalo-Villena M., Úbeda J., Briones A. (2011). Enological properties in wild and commercial Saccharomyces cerevisiae yeasts: relationship with competition during alcoholic fermentation. World J. Microbiol. Biotechnol. 27, 2703–2710. 10.1007/s11274-011-0744-0 [DOI] [Google Scholar]
- Beltran G., Torija M. J., Novo M., Ferrer N., Poblet M., Guillamón J. M., et al. (2002). Analysis of yeast populations during alcoholic fermentation: a six year follow-up study. Syst. Appl. Microbiol. 25, 287–293. 10.1078/0723-2020-00097 [DOI] [PubMed] [Google Scholar]
- Branco P., Francisco D., Chambon C., Hébraud M., Arneborg N., Almeida M. G., et al. (2014). Identification of novel GAPDH-derived antimicrobial peptides secreted by Saccharomyces cerevisiae and involved in wine microbial interactions. Appl. Microbiol. Biotechnol. 98, 843–853. 10.1007/s00253-013-5411-y [DOI] [PubMed] [Google Scholar]
- Canonico L., Agarbati A., Comitini F., Ciani M. (2016). Torulaspora delbrueckii in the brewing process: a new approach to enhance bioflavour and to reduce ethanol content. Food Microbiol. 56, 45–51. 10.1016/j.fm.2015.12.005 [DOI] [PubMed] [Google Scholar]
- Canonico L., Comitini F., Ciani M. (2015). Influence of vintage and selected starter on Torulaspora delbrueckii/Saccharomyces cerevisiae sequential fermentation. Eur. Food Res. Technol. 241, 827–833. 10.1007/s00217-015-2507-x [DOI] [Google Scholar]
- Cheraiti N., Cheraiti N., Salmon J., Guezenec S., Salmon J. (2005). Redox interactions between. Appl. Environ. Microbiol. 71, 255–260. 10.1128/AEM.71.1.255-260.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani M., Comitini F. (2006). Influence of temperature and oxygen concentration on the fermentation behaviour of Candida stellata in mixed fermentation with Saccharomyces Cerevisiae. World J. Microbiol. Biotechnol. 22, 619–623. 10.1007/s11274-005-9080-6 [DOI] [Google Scholar]
- Contreras A., Hidalgo C., Henschke P. A., Chambers P. J., Curtin C., Varela C. (2014). Evaluation of non-Saccharomyces yeasts for the reduction of alcohol content in wine. Appl. Environ. Microbiol. 80, 1670–1678. 10.1128/AEM.03780-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. C., Mau B., Blattner F. R., Perna N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuyter C., Lollier M., Legras J.-L., Le Jeune C. (2004). Predominance of Saccharomyces uvarum during spontaneous alcoholic fermentation, for three consecutive years, in an Alsatian winery. J. Appl. Microbiol. 97, 1140–1148. 10.1111/j.1365-2672.2004.02394.x [DOI] [PubMed] [Google Scholar]
- Díaz-Montaño D. M., Délia M. L., Estarrón-Espinosa M., Strehaiano P. (2008). Fermentative capability and aroma compound production by yeast strains isolated from Agave tequilana Weber juice. Enzyme Microb. Technol. 42, 608–616. 10.1016/j.enzmictec.2007.12.007 [DOI] [Google Scholar]
- Engel S. (2013). The Reference Genome Sequence of Saccharomyces cerevisiae: Then and Now. Available online at: (Accessed January 28, 2016).
- Erny C., Raoult P., Alais A., Butterlin G., Delobel P., Matei-Radoi F., et al. (2012). Ecological success of a group of Saccharomyces cerevisiae/Saccharomyces kudriavzevii hybrids in the Northern European wine-making environment. Appl. Environ. Microbiol. 78, 3256–3265. 10.1128/AEM.06752-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamero A., Belloch C., Ibáñez C., Querol A. (2014). Molecular analysis of the genes involved in aroma synthesis in the species S. cerevisiae, S. kudriavzevii and S. bayanus var. uvarum in winemaking conditions. PLoS ONE 9:e97626. 10.1371/journal.pone.0097626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamero A., Manzanares P., Querol A., Belloch C. (2011). Monoterpene alcohols release and bioconversion by saccharomyces species and hybrids. Int. J. Food Microbiol. 145, 92–97. 10.1016/j.ijfoodmicro.2010.11.034 [DOI] [PubMed] [Google Scholar]
- Gamero A., Tronchoni J., Querol A., Belloch C. (2013). Production of aroma compounds by cryotolerant Saccharomyces species and hybrids at low and moderate fermentation temperatures. J. Appl. Microbiol. 114, 1405–1414. 10.1111/jam.12126 [DOI] [PubMed] [Google Scholar]
- García-Ríos E., Gutiérrez A., Salvadó Z., Arroyo-López F. N., Guillamon J. M. (2014). The fitness advantage of commercial wine yeasts in relation to the nitrogen concentration, temperature, and ethanol content under microvinification conditions. Appl. Environ. Microbiol. 80, 704–713. 10.1128/AEM.03405-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard M. R. (2008). Quantifying the complexities of Saccharomyces cerevisiae's ecosystem engineering via fermentation. Ecology 89, 2077–2082. 10.1890/07-2060.1 [DOI] [PubMed] [Google Scholar]
- González S. S., Barrio E., Gafner J., Querol A. (2006). Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res. 6, 1221–1234. 10.1111/j.1567-1364.2006.00126.x [DOI] [PubMed] [Google Scholar]
- Gonzalez S. S., Gallo L., Climent M. A., Barrio E., Querol A. (2007). Ecological characterization of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. Int. J. Food Microbiol. 116, 11–18. 10.1016/j.ijfoodmicro.2006.10.047 [DOI] [PubMed] [Google Scholar]
- Gustafsson F., Jiranek V., Neuner M., Scholl C., Morgan S., Durall D. (2016). The Interaction of two Saccharomyces cerevisiae strains affects fermentation-derived compounds in wine. Fermentation 2:9 10.3390/fermentation2020009 [DOI] [Google Scholar]
- Holm Hansen E., Nissen P., Sommer P., Nielsen J. C., Arneborg N. (2001). The effect of oxygen on the survival of non-saccharomyces yeasts during mixed culture fermentations of grape juice with Saccharomyces cerevisiae. J. Appl. Microbiol. 91, 541–547. 10.1046/j.1365-2672.2001.01426.x [DOI] [PubMed] [Google Scholar]
- Howell K. S., Cozzolino D., Bartowsky E. J., Fleet G. H., Henschke P. A. (2006). Metabolic profiling as a tool for revealing saccharomyces interactions during wine fermentation. FEMS Yeast Res. 6, 91–101. 10.1111/j.1567-1364.2005.00010.x [DOI] [PubMed] [Google Scholar]
- Izquierdo Cañas P. M., García-Romero E., Heras Manso J. M., Fernández-González M. (2014). Influence of sequential inoculation of Wickerhamomyces anomalus and Saccharomyces cerevisiae in the quality of red wines. Eur. Food Res. Technol. 239, 279–286. 10.1007/s00217-014-2220-1 [DOI] [Google Scholar]
- King E. S., Kievit R. L., Curtin C., Swiegers J. H., Pretorius I. S., Bastian S. E. P., et al. (2010). The effect of multiple yeasts co-inoculations on sauvignon Blanc wine aroma composition, sensory properties and consumer preference. Food Chem. 122, 618–626. 10.1016/j.foodchem.2010.03.021 [DOI] [Google Scholar]
- Lopandic K., Gangl H., Wallner E., Tscheik G., Leitner G., Querol A., et al. (2007). Genetically different wine yeasts isolated from Austrian vine-growing regions influence wine aroma differently and contain putative hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii. FEMS Yeast Res. 7, 953–965. 10.1111/j.1567-1364.2007.00240.x [DOI] [PubMed] [Google Scholar]
- Masneuf I., Hansen J., Groth C., Piskur J., Dubourdieu D. (1998). New hybrids between saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl. Envir. Microbiol. 64, 3887–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina K., Boido E., Fariña L., Gioia O., Gomez M. E., Barquet M., et al. (2013). Increased flavour diversity of Chardonnay wines by spontaneous fermentation and co-fermentation with Hanseniaspora vineae. Food Chem. 141, 2513–2521. 10.1016/j.foodchem.2013.04.056 [DOI] [PubMed] [Google Scholar]
- Milanovic V., Ciani M., Oro L., Comitini F. (2012). Starmerella bombicola influences the metabolism of Saccharomyces cerevisiae at pyruvate decarboxylase and alcohol dehydrogenase level during mixed wine fermentation. Microb. Cell Fact. 11:18. 10.1186/1475-2859-11-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov G. I., Naumova E. S., Antunovics Z., Sipiczki M. (2002). Saccharomyces bayanus var. uvarum in tokaj wine-making of slovakia and hungary. Appl. Microbiol. Biotechnol. 59, 727–730. 10.1007/s00253-002-1077-6 [DOI] [PubMed] [Google Scholar]
- Naumov G. I., Nikonenko T. A. (1987). Genomic divergence in cultivated and wild strains of the yeast, saccharomyces sensu stricto: four twin species. Dokl. Biol. Sci. 294, 330–332. [Google Scholar]
- Naumov G., Masneuf I. S., Naumova E., Aigle M., Dubourdieu D. (2000). Association of Saccharomyces bayanus var. uvarum with some french wines: genetic analysis of yeast populations. Res. Microbiol. 151, 683–691. 10.1016/S0923-2508(00)90131-1 [DOI] [PubMed] [Google Scholar]
- Neeley E. T., Phister T. G., Mills D. A. (2005). Differential real-time pcr assay for enumeration of lactic acid bacteria in wine differential real-time pcr assay for enumeration of lactic acid bacteria in wine. Appl. Environ. Microbiol. 71, 8954–8957. 10.1128/AEM.71.12.8954-8957.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P., Arneborg N. (2003). Characterization of early deaths of non-saccharomyces yeasts in mixed cultures with Saccharomyces cerevisiae. Arch. Microbiol. 180, 257–263. 10.1007/s00203-003-0585-9 [DOI] [PubMed] [Google Scholar]
- Nissen P., Neilsen D., Arneborg N. (2004). The relative glucose uptake abilities of non-saccharomyces yeasts play a role in their coexistence with Saccharomyces cerevisiae in mixed cultures. Appl. Microbiol. Biotechnol. 64, 543–550. 10.1007/s00253-003-1487-0 [DOI] [PubMed] [Google Scholar]
- Nissen P., Nielsen D., Arneborg N. (2003). Viable Saccharomyces cerevisiae cells at high concentrations cause early growth arrest of non-saccharomyces yeasts in mixed cultures by a cell - cell contact-mediated mechanism. Yeast 20, 331–341. 10.1002/yea.965 [DOI] [PubMed] [Google Scholar]
- Oliveira B. M., Barrio E., Querol A., Pérez-Torrado R. (2014). Enhanced enzymatic activity of glycerol-3-phosphate dehydrogenase from the cryophilic Saccharomyces kudriavzevii. PLoS ONE 9:e87290. 10.1371/journal.pone.0087290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Nevado F., Albergaria H., Hogg T., Girio F. (2006). Cellular death of two non-Saccharomyces wine-related yeasts during mixed fermentations with Saccharomyces cerevisiae. Int. J. Food Microbiol. 108, 336–345. 10.1016/j.ijfoodmicro.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Pérez-Torrado R., Oliveira B. M., Zemancíková J., Sychrová H., Querol A. (2016). Alternative glycerol balance strategies among Saccharomyces species in response to winemaking stress. Front. Microbiol. 7:435. 10.3389/fmicb.2016.00435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Través L., Lopes C. A., Barrio E., Querol A. (2014). stabilization process in saccharomyces intra and interspecific hybrids in fermentative conditions. Int. Microbiol. 17, 213–224. 10.2436/20.1501.01.224 [DOI] [PubMed] [Google Scholar]
- Peris D., Lopes C. A., Belloch C., Querol A., Barrio E. (2012). Comparative genomics among Saccharomyces cerevisiae × Saccharomyces kudriavzevii natural hybrid strains isolated from wine and beer reveals different origins. BMC Genomics 13:407. 10.1186/1471-2164-13-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piškur J., Rozpedowska E., Polakova S., Merico A., Compagno C. (2006). How did saccharomyces evolve to become a good brewer? Trends Genet. 22, 183–186. 10.1016/j.tig.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Pretorius I. S. (2000). Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16, 675–729. [DOI] [PubMed] [Google Scholar]
- Querol A., Barrio E., Huerta T., Ramón D. (1992). Molecular monitoring of wine fermentations conducted by active dry yeast strains. Appl. Environ. Microbiol. 58, 2948–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantsiou K., Dolci P., Giacosa S., Torchio F., Tofalo R., Torriani S., et al. (2012). Candida zemplinina can reduce acetic acid produced by Saccharomyces cerevisiae in sweet wine fermentations. Appl. Environ. Microbiol. 78, 1987–1994. 10.1128/AEM.06768-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2015). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; Available online at: https://www.R-project.org/ [Google Scholar]
- Redžepović S., Orlić S., Sikora S., Majdak a., Pretorius I. S. (2002). Identification and characterization of Saccharomyces cerevisiae and Saccharomyces paradoxus strains isolated from Croatian vineyards. Lett. Appl. Microbiol. 35, 305–310. 10.1046/j.1472-765X.2002.01181.x [DOI] [PubMed] [Google Scholar]
- Rementeria A. (2003). Yeast associated with spontaneous fermentations of white wines from the “Txakoli de Bizkaia” region (Basque Country, North Spain). Int. J. Food Microbiol. 86, 201–207. 10.1016/S0168-1605(03)00289-7 [DOI] [PubMed] [Google Scholar]
- Rodrigues A. J., Raimbourg T., Gonzalez R., Morales P. (2016). Environmental factors influencing the efficacy of different yeast strains for alcohol level reduction in wine by respiration. LWT-Food Sci. Technol. 65, 1038–1043. 10.1016/j.lwt.2015.09.046 [DOI] [Google Scholar]
- Rodríguez M. E., Pérez-Través L., Sangorrín M. P., Barrio E., Lopes C. A. (2014). Saccharomyces eubayanus and Saccharomyces uvarum associated with the fermentation of araucaria araucana seeds in Patagonia. FEMS Yeast Res. 14, 948–965. 10.1111/1567-1364.12183 [DOI] [PubMed] [Google Scholar]
- Rossignol T., Dulau L., Julien A., Blondin B. (2003). Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 20, 1369–1385. 10.1002/yea.1046 [DOI] [PubMed] [Google Scholar]
- Sabate J., Cano J., Esteve-Zarzoso B., Guillamón J. M. (2002). Isolation and identification of yeasts associated with vineyard and winery by RFLP analysis of ribosomal genes and mitochondrial DNA. Microbiol. Res. 157, 267–274. 10.1078/0944-5013-00163 [DOI] [PubMed] [Google Scholar]
- Saberi S., Cliff M. A., van Vuuren H. J. J. (2012). Impact of mixed S. cerevisiae strains on the production of volatiles and estimated sensory profiles of chardonnay wines. Food Res. Int. 48, 725–735. 10.1016/j.foodres.2012.06.012 [DOI] [Google Scholar]
- Salvadó Z., Arroyo-López F. N., Barrio E., Querol A., Guillamón J. M. (2011a). Quantifying the individual effects of ethanol and temperature on the fitness advantage of Saccharomyces cerevisiae. Food Microbiol. 28, 1155–1161. 10.1016/j.fm.2011.03.008 [DOI] [PubMed] [Google Scholar]
- Salvadó Z., Arroyo-López F. N., Guillamón J. M., Salazar G., Querol A., Barrio E. (2011b). Temperature adaptation markedly determines evolution within the genus Saccharomyces. Appl. Environ. Microbiol. 77, 2292–2302. 10.1128/AEM.01861-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D. R., Zill O. A., Rokas A., Payen C., Dunham M. J., Eisen M. B., et al. (2011). The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the saccharomyces sensu stricto genus. G3 (Bethesda). 1, 11–25. 10.1534/g3.111.000273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Şener H., Yildirim H. K. (2013). Influence of different maceration time and temperatures on total phenols, colour and sensory properties of cabernet sauvignon wines. Food Sci. Technol. Int. 19, 523–533. 10.1177/1082013212462229 [DOI] [PubMed] [Google Scholar]
- Sipiczki M., Romano P., Lipani G., Miklos I., Antunovics Z. (2001). Analysis of yeasts derived from natural fermentation in a tokaj winery. Antonie van Leeuwenhoek 79, 97–105. 10.1023/A:1010249408975 [DOI] [PubMed] [Google Scholar]
- Stribny J., Gamero A., Pérez-Torrado R., Querol A. (2015). Saccharomyces kudriavzevii and Saccharomyces uvarum differ from Saccharomyces cerevisiae during the production of aroma-active higher alcohols and acetate esters using their amino acidic precursors. Int. J. Food Microbiol. 205, 41–46. 10.1016/j.ijfoodmicro.2015.04.003 [DOI] [PubMed] [Google Scholar]
- Thomson J. M., Gaucher E. A., Burgan M. F., De Kee D. W., Li T., Aris J. P., et al. (2005). Resurrecting ancestral alcohol dehydrogenases from yeast. Nat. Genet. 37, 630–635. 10.1038/ng1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torija M. J., Rozès N., Poblet M., Guillamón J. M., Mas A. (2001). Yeast population dynamics in spontaneous fermentations: comparison between two different wine-producing areas over a period of three years. Antonie van Leeuwenhoek 79, 345–352. 10.1023/A:1012027718701 [DOI] [PubMed] [Google Scholar]
- Torriani S., Zapparoli G., Suzzi G. (1999). Genetic and phenotypic diversity of saccharomyces sensu stricto strains isolated from amarone wine. diversity of saccharomyces strains from amarone wine. Antonie Van Leeuwenhoek 75, 207–215. 10.1023/A:1001773916407 [DOI] [PubMed] [Google Scholar]
- Valero E., Cambon B., Schuller D., Casal M., Dequin S. (2007). Biodiversity of saccharomyces yeast strains from grape berries of wine-producing areas using starter commercial yeasts. FEMS Yeast Res. 7, 317–329. 10.1111/j.1567-1364.2006.00161.x [DOI] [PubMed] [Google Scholar]
- Vendrame M., Manzano M., Comi G., Bertrand J., Iacumin L. (2014). Use of propidium monoazide for the enumeration of viable Brettanomyces bruxellensis in wine and beer by quantitative PCR. Food Microbiol. 42, 196–204. 10.1016/j.fm.2014.03.010 [DOI] [PubMed] [Google Scholar]
- Wang C., Mas A., Esteve-Zarzoso B. (2015). Interaction between hanseniaspora uvarum and Saccharomyces cerevisiae during alcoholic fermentation. Int. J. Food Microbiol. 206, 67–74. 10.1016/j.ijfoodmicro.2015.04.022 [DOI] [PubMed] [Google Scholar]
- Wang C., Mas A., Esteve-Zarzoso B. (2016). The interaction between Saccharomyces cerevisiae and non-saccharomyces yeast during alcoholic fermentation is species and strain specific. Front. Microbiol. 7:502. 10.3389/fmicb.2016.00502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. M., Liu P., Fay J. C. (2015). Evolution of ecological dominance of yeast species in high-sugar environments. Evolution 69, 2079–2093. 10.1111/evo.12707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zara G., Mannazzu I., Del Caro A., Budroni M., Pinna M. B., Murru M., et al. (2014). Wine quality improvement through the combined utilisation of yeast hulls and candida zemplinina/Saccharomyces cerevisiae mixed starter cultures. Aust. J. Grape Wine Res. 20, 199–207. 10.1111/ajgw.12078 [DOI] [Google Scholar]
- Zwietering M. H., Jongenburger I., Rombouts F. M., van 't Riet K. (1990). Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56, 1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.