Abstract
Short oxygen–halogen interactions have been known in organic chemistry since the 1950s and recently have been exploited in the design of supramolecular assemblies. The present survey of protein and nucleic acid structures reveals similar halogen bonds as potentially stabilizing inter- and intramolecular interactions that can affect ligand binding and molecular folding. A halogen bond in biomolecules can be defined as a short C X···O
X···O Y interaction (C
Y interaction (C X is a carbon-bonded chlorine, bromine, or iodine, and O
X is a carbon-bonded chlorine, bromine, or iodine, and O Y is a carbonyl, hydroxyl, charged carboxylate, or phosphate group), where the X···O distance is less than or equal to the sums of the respective van der Waals radii (3.27 Å for Cl···O, 3.37Å for Br···O, and 3.50 Å for I···O) and can conform to the geometry seen in small molecules, with the C
Y is a carbonyl, hydroxyl, charged carboxylate, or phosphate group), where the X···O distance is less than or equal to the sums of the respective van der Waals radii (3.27 Å for Cl···O, 3.37Å for Br···O, and 3.50 Å for I···O) and can conform to the geometry seen in small molecules, with the C X···O angle ≈165° (consistent with a strong directional polarization of the halogen) and the X···O
X···O angle ≈165° (consistent with a strong directional polarization of the halogen) and the X···O Y angle ≈120°. Alternative geometries can be imposed by the more complex environment found in biomolecules, depending on which of the two types of donor systems are involved in the interaction: (i) the lone pair electrons of oxygen (and, to a lesser extent, nitrogen and sulfur) atoms or (ii) the delocalized π -electrons of peptide bonds or carboxylate or amide groups. Thus, the specific geometry and diversity of the interacting partners of halogen bonds offer new and versatile tools for the design of ligands as drugs and materials in nanotechnology.
Y angle ≈120°. Alternative geometries can be imposed by the more complex environment found in biomolecules, depending on which of the two types of donor systems are involved in the interaction: (i) the lone pair electrons of oxygen (and, to a lesser extent, nitrogen and sulfur) atoms or (ii) the delocalized π -electrons of peptide bonds or carboxylate or amide groups. Thus, the specific geometry and diversity of the interacting partners of halogen bonds offer new and versatile tools for the design of ligands as drugs and materials in nanotechnology.
Keywords: molecular folding, molecular recognition, molecular design
Two recent biomolecular single-crystal structures, a four-stranded DNA Holliday junction (1) and an ultrahigh-resolution structure (0.66 Å) of the enzyme aldose reductase complex with a halogenated inhibitor (2), revealed unusually short Br···O contacts [≈3.0 Å, or ≈12% shorter than the sum of their van der Waals radii (RvdW)]. The atypical contact in the enzyme complex was attributed to an electrostatic interaction between the polarized bromine and the lone pair electrons of the oxygen atom of a neighboring threonine side chain (3). Short halogen–oxygen interactions are not in themselves new: The chemist Odd Hassel (4) had earlier described Br···O distances as short as 2.7 Å (≈20% shorter than RvdW) in crystals of Br2 with various organic compounds.
These short contacts, originally called charge-transfer bonds, were attributed to the transfer of negative charge from an oxygen, nitrogen, or sulfur (a Lewis base) to a polarizable halogen (a Lewis acid) (5, 6). They are now referred to as halogen bonds (Fig. 1) by analogy to classical hydrogen bonds with which they share numerous properties (6) and are currently being exploited to control the crystallization of organic compounds in the design of new materials (7) as well as in supramolecular chemistry (6). Extensive surveys of structures in the Cambridge Structural Database (8–10) coupled with ab initio calculations (10) have characterized the geometry of halogen bonds in small molecules and show that the interaction is primarily electrostatic, with contributions from polarization, dispersion, and charge transfer. The stabilizing potential of halogen bonds is estimated to range from about half to slightly greater than that of an average hydrogen bond in directing the self-assembly of organic crystals (11, 12).
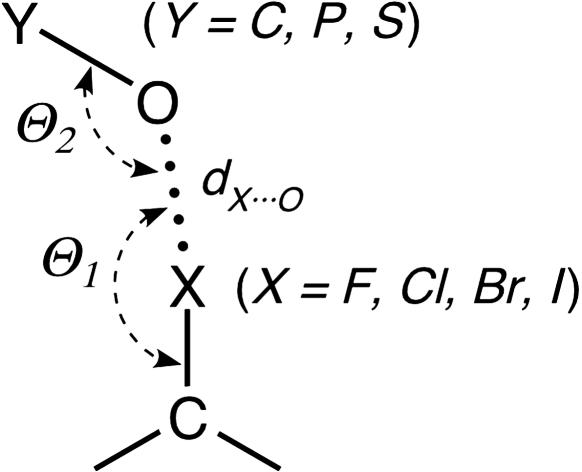
Fig. 1.
Schematic of short halogen (X) interactions to various oxygen-containing functional groups (where O Y can be a carbonyl, hydroxyl, or carboxylate when Y is a carbon; a phosphate when Y is a phosphorus; or a sulfate when Y is a sulfur). The geometry of the interaction is defined by the normalized RX···O distance [RX···O = dX···O/RvdW(X···O)], the Θ1 angle of the oxygen relative to the C
Y can be a carbonyl, hydroxyl, or carboxylate when Y is a carbon; a phosphate when Y is a phosphorus; or a sulfate when Y is a sulfur). The geometry of the interaction is defined by the normalized RX···O distance [RX···O = dX···O/RvdW(X···O)], the Θ1 angle of the oxygen relative to the C X bond, and the Θ2 angle of the halogen relative to the O
X bond, and the Θ2 angle of the halogen relative to the O Y bond.
Y bond.
Similar short halogen–oxygen contacts have rarely been described in biological systems, presumably because of the scarcity of available crystal structures of halogenated biomolecules. As such, their structural and functional roles have been largely ignored in biology. Halogens, however, do play important roles in natural systems. Thyroid hormones represent a class of naturally iodinated molecules for which halogen bonds appear to play a role in their recognition, as evident by the short I···O contacts between tetraiodothyroxine and its transport protein transthyretin (13). In addition, >3,500 halogen-containing metabolites, including the important antibiotics chloramphenicol, 7-chlorotetracyclin, and vancomycin (14), are currently known. Moreover, direct halogenation of proteins and nucleic acids can result from oxidative halogenation by a number of peroxidases involved in inflammatory responses. For example, levels of chlorotyrosines have been correlated with chronic respiratory disease in infants (15), whereas bromotyrosines are associated with allergen-induced asthma (16). In nucleic acids, DNA bases are oxidatively brominated by eosinophil peroxidase (17) and, specifically for brominated cytosines, have been suggested to induce a conformation that may be susceptible to spontaneous transition mutations (18).
Our interest in halogen-induced conformational effects initiated with the observation that the brominated DNA sequence d(CCAGTACbr5UGG) (br5U, 5-bromouridine) adopts a four-stranded Holliday junction with an associated short Br···O distance (≈3.0 Å). In contrast, standard B-DNA duplexes were seen in the closely related d(CCAGTACTGG) (1) and the nonbrominated d(CCAGTACUGG) sequences (unpublished data), where T (a methylated uridine) and U replace br5U (shown in bold italics). These results could not be explained by the known molecular interactions because of our own ignorance concerning halogen bonds, as is probably true for much of the biochemistry community.
In the current study, quantum mechanical calculations were used to generate electrostatic potential maps to compare the polarizability of halogen atoms within the context of biological molecules, and a data set of protein and nucleic acid structures with short halogen–oxygen distances was assembled and surveyed to characterize the prevalence and geometry of halogen bonds in biological systems.
Methods
Molecular models of halogenated methane and the two nucleobases, uridine and cytosine, were constructed, and their geometries were optimized at the 3-21G(*) level (Fig. 2). Electrostatic potential surfaces were generated by mapping the 3-21G(*) electrostatic potentials onto surfaces of molecular electron density (0.002 electron per Å3) by using the program spartan (Wavefunction, Irvine, CA). Note that the 3-21G(*) basis set was used because the 6-31G(*) basis set and those above are incomplete for atoms below the second row of the periodic table (i.e., bromine and iodine). The limitations related to the use of electrostatic potential surfaces are discussed at length by Mecozzi et al. (19), who demonstrated their usefulness in providing qualitative guidelines for evaluating the ability of aromatic compounds to form cation–π interactions.
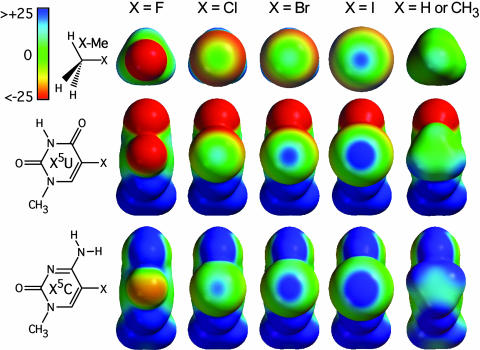
Fig. 2.
Ab initio electrostatic potential surfaces of halogenated model compounds. Halogenated methane (X–Me, Top), uridine nucleobase (X5U, Middle), and cytosine nucleobase (X5C, Bottom) are shown looking into the halogen atoms to compare the induced negative (red), neutral (green), and positive (blue) electrostatic potentials around the halogen surfaces. The potential energies are presented only in the –25 to +25 kcal/mol range to emphasize the variation in electrostatic potential associated with the halogen atoms (note that some regions of electrostatic potential, especially those associated with heteroatoms, may lie beyond this ±25 kcal/mol range). The compounds are ordered (from left to right) from least to most polarizable (F < Cl < Br < I), with the last column showing, for comparison, the potential surface of methane, methylated uridine, and methylated cytosine.
The Protein Data Bank (PDB) (20) (July 2004 release) contains 226 entries of single-crystal structures (at resolutions of 3.0 Å or better) of proteins showing C X bonds (where X is fluorine, chlorine, bromine, or iodine; 475 C
X bonds (where X is fluorine, chlorine, bromine, or iodine; 475 C Cl, 281 C
Cl, 281 C Br, and 208 C
Br, and 208 C I bonds for a total of 964 C
I bonds for a total of 964 C X) and 87 entries of halogenated nucleic acids (2 C
X) and 87 entries of halogenated nucleic acids (2 C Cl, 232 C
Cl, 232 C Br, and 87 C
Br, and 87 C I bonds for a total of 321 C
I bonds for a total of 321 C X). From these structures, we culled those with X···O distances that are shorter than their respective RvdW (with rF = 1.47 Å, rCl = 1.75 Å, rBr = 1.85 Å, rI = 1.98 Å, and rO = 1.52 Å, leading to RvdW(F···O) = 2.99 Å, RvdW(Cl···O) = 3.27 Å, RvdW(Br···O) = 3.37 Å, and RvdW(I···O) = 3.50 Å) to define a distinct data set of potential halogen bonds (21). The C
X). From these structures, we culled those with X···O distances that are shorter than their respective RvdW (with rF = 1.47 Å, rCl = 1.75 Å, rBr = 1.85 Å, rI = 1.98 Å, and rO = 1.52 Å, leading to RvdW(F···O) = 2.99 Å, RvdW(Cl···O) = 3.27 Å, RvdW(Br···O) = 3.37 Å, and RvdW(I···O) = 3.50 Å) to define a distinct data set of potential halogen bonds (21). The C X groups belong to halogenated nucleotides or drugs and the oxygen atoms belong to nucleic acids, proteins, or ligands bound to nucleic acids or proteins (Fig. 1). All short contacts within the crystallographic asymmetric unit and any contacts between crystallographically related molecules were considered. To limit the number of contacts to those that are most likely to form halogen bonds, the data set was limited to structures with Θ1 ≥ 120°, thereby eliminating (i) stereochemically defined intramolecular interactions, such as the intrabase O4···X contacts in halogenated nucleotides and (ii) short C
X groups belong to halogenated nucleotides or drugs and the oxygen atoms belong to nucleic acids, proteins, or ligands bound to nucleic acids or proteins (Fig. 1). All short contacts within the crystallographic asymmetric unit and any contacts between crystallographically related molecules were considered. To limit the number of contacts to those that are most likely to form halogen bonds, the data set was limited to structures with Θ1 ≥ 120°, thereby eliminating (i) stereochemically defined intramolecular interactions, such as the intrabase O4···X contacts in halogenated nucleotides and (ii) short C X···O contacts that deviate significantly from linearity. (The small number of contacts observed with Θ1 < 120° does not allow a meaningful statistical evaluation of such interactions in relation to known halogen bond geometries.) The PDB ID codes and geometric characteristics of the structures included in this survey are tabulated in Table 1, which is published as supporting information on the PNAS web site. Normalized RX···O distances [RX···O = dX···O/RvdW(X···O)] will be used to provide a common reference frame for comparing X···O contacts.
X···O contacts that deviate significantly from linearity. (The small number of contacts observed with Θ1 < 120° does not allow a meaningful statistical evaluation of such interactions in relation to known halogen bond geometries.) The PDB ID codes and geometric characteristics of the structures included in this survey are tabulated in Table 1, which is published as supporting information on the PNAS web site. Normalized RX···O distances [RX···O = dX···O/RvdW(X···O)] will be used to provide a common reference frame for comparing X···O contacts.
To study the potential effects of resolution and other refinement parameters on the geometry of X···O contacts, we re-refined the structure of d(CCAGTACbr5UGG) (1) by using the programs cns (22) and refmac5 (23) with the diffraction data limited from 1.9 to 2.8 Å (in 0.1-Å increments), the van der Waals constraints on the bromine or the phosphate oxygens removed, the bond lengths and distances for these atoms to their respective bases relaxed or modified, or the Br···O distances set between 2.99 and 3.02 Å (for data at 1.9-Å resolution) in the refinement libraries. For each refinement, the structure was first subjected to one round of simulated annealing at 1,500 K to effectively “erase” any memory of the structure at higher resolution and then refined with maximum likelihood target. This resulted in bromine to phosphate oxygen distances of 2.95–3.03 Å (average distance ± SD, 2.98 ± 0.02 Å). The only significant extension of the Br···O distance (to 3.14 Å) was observed when the weights for bond lengths and angles were reduced to values of 0.02 in refmac5, which resulted in a structure with highly distorted geometries for all bond lengths and angles. The various refined models yielded an average Θ1 of 169.3° (1.2° SD) and an average Θ2 of 121.3° (1.4° SD). Thus, we concluded that the halogen–oxygen distances and angles in this and other well refined crystal structures are relatively robust (varying by at most 0.07 Å in distance), leaving us confident that the geometry of a halogen bond can be accurately determined in a properly refined crystal structure even at relatively low resolution.
Results
To study potential halogen bonding interactions in biological molecules, we first characterized the electrostatic properties of halogens within the context of functional groups relevant to protein and nucleic acid structures. A survey of single-crystal structures of protein and nucleic acids in the PDB then allowed us to compare and contrast halogen bonds in biological systems with previous descriptions from small molecule structures in the Cambridge Structural Database (8–10). With the growing use of halogens, particularly bromine and iodine, to help phase crystallographic data and the increasing number of halogenated inhibitors developed by medicinal chemists, the number of single-crystal structures available is now sufficiently large enough to allow a meaningful survey of interactions in just biological macromolecules.
Electrostatic Potentials. The electrostatic potentials were calculated for halogenated methane (X Me, where X is fluorine, chlorine, bromine, or iodine) to model halogenated aliphatic compounds and for the halogenated uridine [5-halouridine (X5U)] and cytosine [5-halocytosine (X5C)] nucleobases to model halogenated aromatic groups and the effects of neighboring substituents on polarization (Fig. 2). A clear trend of increasing electropositive potential along the C
Me, where X is fluorine, chlorine, bromine, or iodine) to model halogenated aliphatic compounds and for the halogenated uridine [5-halouridine (X5U)] and cytosine [5-halocytosine (X5C)] nucleobases to model halogenated aromatic groups and the effects of neighboring substituents on polarization (Fig. 2). A clear trend of increasing electropositive potential along the C X bond is seen when going from fluorine to iodine within each group of halogenated compounds (X
X bond is seen when going from fluorine to iodine within each group of halogenated compounds (X Me, X5U, and X5C), consistent with the increasing polarizability of the halogen atoms along this series. Fluorine atoms remain entirely electronegative, whereas each of the other three halogen atoms shows the emergence of an electropositive crown (Fig. 2, blue) along the C–X axis, which is surrounded by an electroneutral ring (Fig. 2, green) and, farther out, an electronegative belt (Fig. 2, red). The size of the electropositive crown increases with the radius or polarizability of the halogen, suggesting that iodines would form the strongest X···O halogen bonds, whereas fluorines are more likely to serve as hydrogen bond acceptors in F···H–O-type interactions. The magnitude of these polarization effects is modulated by the chemical context of the halogen atom. The size and intensity of the electropositive crown increases in the series X
Me, X5U, and X5C), consistent with the increasing polarizability of the halogen atoms along this series. Fluorine atoms remain entirely electronegative, whereas each of the other three halogen atoms shows the emergence of an electropositive crown (Fig. 2, blue) along the C–X axis, which is surrounded by an electroneutral ring (Fig. 2, green) and, farther out, an electronegative belt (Fig. 2, red). The size of the electropositive crown increases with the radius or polarizability of the halogen, suggesting that iodines would form the strongest X···O halogen bonds, whereas fluorines are more likely to serve as hydrogen bond acceptors in F···H–O-type interactions. The magnitude of these polarization effects is modulated by the chemical context of the halogen atom. The size and intensity of the electropositive crown increases in the series X Me < X5U < X5C. Thus, halogen bonds associated with aromatic compounds are expected to be stronger than those formed with analogous aliphatic molecules. Furthermore, an amino group positioned ortho to the C
Me < X5U < X5C. Thus, halogen bonds associated with aromatic compounds are expected to be stronger than those formed with analogous aliphatic molecules. Furthermore, an amino group positioned ortho to the C X bond (as in X5C) has a significantly stronger polarizing effect on the halogen atom than a carbonyl group (as in X5U). The electron withdrawing properties of amino groups in aromatic systems is further supported by the observation that a methyl substituent in place of the halogen is also more electropositive on a cytosine than on a uridine base (Fig. 2, X = H or CH3).
X bond (as in X5C) has a significantly stronger polarizing effect on the halogen atom than a carbonyl group (as in X5U). The electron withdrawing properties of amino groups in aromatic systems is further supported by the observation that a methyl substituent in place of the halogen is also more electropositive on a cytosine than on a uridine base (Fig. 2, X = H or CH3).
Database Survey of Short Halogen–Oxygen Interactions. The data set that was assembled for this study contains 66 different protein and six different nucleic acid structures (resolution, 0.66–3.0 Å; average, ≈2.1 Å) with halogen–oxygen distances (RX···O) shorter than their respective RvdW (the criteria used to develop this data set are described in Methods), leading to a total of 113 distinct X···O interactions (Table 1). Although a small set of short F···O contacts was observed, they will not be discussed further, given the generally highly electronegative character of fluorines revealed by the quantum mechanical calculations. Our data set primarily includes short Cl···O (27% of contacts), Br···O (34%), and I···O (39%) interactions with carbonyl (O C), hydroxyl (O
C), hydroxyl (O H), or negatively charged acid (–O
H), or negatively charged acid (–O C/P/S) groups. This data set is dominated by X···O
C/P/S) groups. This data set is dominated by X···O C interactions (78 short contacts to backbone O
C interactions (78 short contacts to backbone O C groups in proteins, 1 to an asparagine side chain, and 2 to O
C groups in proteins, 1 to an asparagine side chain, and 2 to O C groups in nucleic acids), whereas interactions involving hydroxyl (17 with serine, threonine, or tyrosine side chains in proteins and 1 with an hydroxyl group in RNA) and negatively charged acid groups (six with asparagine or glutamic acid side chains in proteins, seven with anionic phosphate oxygens in RNA and DNA, and one with a sulfate anion) are less numerous. These proportions mirror the overall occurrence of the halogenated structures in the PDB. As a consequence of the small number of interactions involving hydroxyl or negatively charged acid groups, no attempts will be made to analyze these interactions separately
C groups in nucleic acids), whereas interactions involving hydroxyl (17 with serine, threonine, or tyrosine side chains in proteins and 1 with an hydroxyl group in RNA) and negatively charged acid groups (six with asparagine or glutamic acid side chains in proteins, seven with anionic phosphate oxygens in RNA and DNA, and one with a sulfate anion) are less numerous. These proportions mirror the overall occurrence of the halogenated structures in the PDB. As a consequence of the small number of interactions involving hydroxyl or negatively charged acid groups, no attempts will be made to analyze these interactions separately
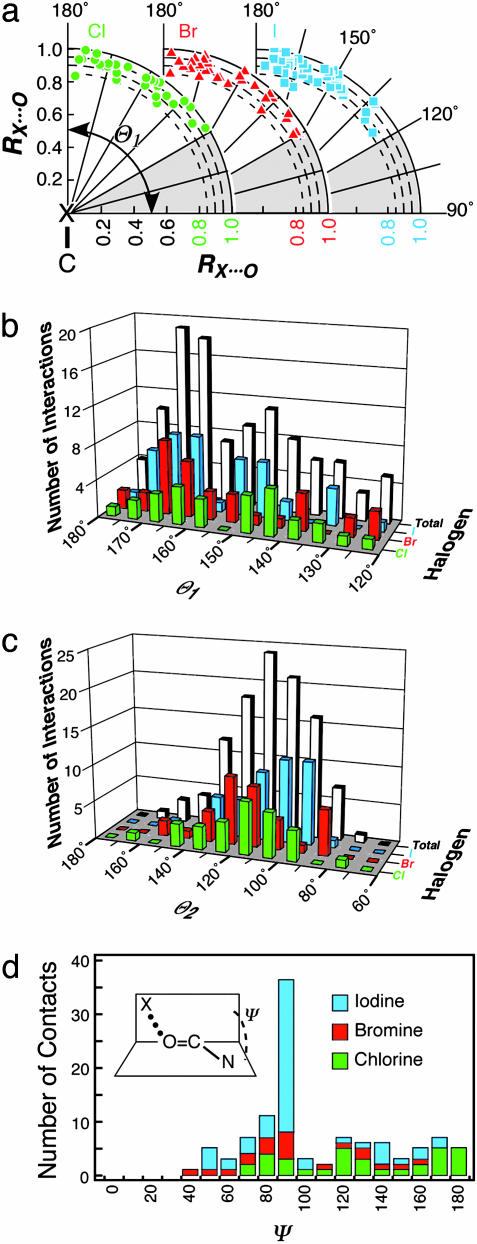
With the X···O distances (RX···O) normalized relative to their respective van der Waals radii (see Methods), we can analyze and compare the distance–angle relationships for all types of halogen bonds as polar scatter plots around any halogen or oxygen in a common reference frame (Fig. 3a). In this set, the shortest RX···O distances found for chlorine, bromine, and iodine were 2.72, 2.87, and 3.06 Å, respectively, whereas the corresponding average RX···O distances were 3.06, 3.15, and 3.24 Å, respectively. There was no significant correlation of distance to the interactions with any particular type of oxygen or to either angle, except that the shortest interactions tended to fall between 160–180° for Θ1 and ≈90° for Θ2.
Fig. 3.
Polar scatter plot and histogram distributions for halogen bonds. (a) Polar scatter plot relative to Θ1 and the normalized halogen (X) to oxygen distances (RX···O, where RX···O ≤ RvdW) are plotted for X& Cl (green circles), X
Cl (green circles), X Br (red triangles), and X
Br (red triangles), and X I (cyan squares). Both the x and y axes of the plot represent RX···O, with the y axis aligned along the C
I (cyan squares). Both the x and y axes of the plot represent RX···O, with the y axis aligned along the C X bond (180°) and the x axis perpendicular to the C
X bond (180°) and the x axis perpendicular to the C X bond (90°). The shaded region from 90° to 120° indicates the Θ1 angles that were excluded from our data set. (b) Histogram distribution of Θ1 angles. The number of short X···O interactions to chlorine (green), bromine (red), and iodine (cyan) halogen atoms, and their sum (gray) are counted and placed into 5° bins of Θ1 angle and plotted as a 3D histogram. (c) Histogram distribution of Θ2 angles. This plot is similar to b, except the interactions are placed into 10° bins of Θ2. (d) Histogram distribution of the dihedral angle Ψ calculated for short halogen bonds involving the O
X bond (90°). The shaded region from 90° to 120° indicates the Θ1 angles that were excluded from our data set. (b) Histogram distribution of Θ1 angles. The number of short X···O interactions to chlorine (green), bromine (red), and iodine (cyan) halogen atoms, and their sum (gray) are counted and placed into 5° bins of Θ1 angle and plotted as a 3D histogram. (c) Histogram distribution of Θ2 angles. This plot is similar to b, except the interactions are placed into 10° bins of Θ2. (d) Histogram distribution of the dihedral angle Ψ calculated for short halogen bonds involving the O C group of the peptide backbone.
C group of the peptide backbone.
The probability distributions of Θ1 angles (in 5° bins) for each of the (Cl, Br, I)···O subpopulations and the cumulative distribution for all X···O contacts found in the data set are shown in Fig. 3b. Interestingly, the total Θ1 angle distribution is clearly bimodal, with a first maximum of ≈160–170°, a second, broader maximum of ≈145–150°, and a minimum separation of ≈155°. The first peak, associated with a nearly linear C X···O interaction geometry, is consistent with results from previous Cambridge Structural Database surveys (8–10) and the alignment of the positive electrostatic potential peaks along the C
X···O interaction geometry, is consistent with results from previous Cambridge Structural Database surveys (8–10) and the alignment of the positive electrostatic potential peaks along the C X bond (Fig. 2).
X bond (Fig. 2).
The second, broader distribution maximum at 145–150° does not fit this general model. Interestingly, however, the highest-resolution structure of this data set (PDB ID code 1US0; 0.66 Å) shows a dBr···O distance of 2.97 Å and a Θ1 angle of 153° (2). Quantum mechanical calculations conducted at the density functional theory level (3) suggest that an additional polarization of the bromine atom induced by the contacting oxygen leads to a deformation of the electron density of the halogen that, consequently, loses its cylindrical symmetry. Thus, in complex environments, like those encountered in biomolecular systems, some deviation from linearity for halogen bonds can be expected. We note that this effect is directional and, therefore, could account for the distinct peak ≈145–150° for the Θ1 (Fig. 3b). Furthermore, as the electropositive potential along the C X bond increases, the interactions appear to become, on the average, slightly more linear: 〈Θ1〉 ≈ 151°, 154°, and 157° for chlorine, bromine, and iodine, respectively. The interactions with various types of oxygens are distributed across the entire range of angles for each halogen, indicating that such interactions cannot be distinguished within the current data set.
X bond increases, the interactions appear to become, on the average, slightly more linear: 〈Θ1〉 ≈ 151°, 154°, and 157° for chlorine, bromine, and iodine, respectively. The interactions with various types of oxygens are distributed across the entire range of angles for each halogen, indicating that such interactions cannot be distinguished within the current data set.
The average Θ2 angular value of ≈113° (Fig. 3c) would suggest an efficient approach of the lone pair electrons of the oxygen atom toward the electropositive crown of the halogen, which was verified by visual inspection for the halogen bonds involving hydroxyl groups. However, in all cases in which the halogen interacts with a carboxyl or an amide group and in most cases involving carbonyl groups in proteins (Table 1), it is often the π system of the O=C bond, rather than the lone pairs of the oxygen atom, that serves as the electron donor. A more detailed analysis of halogen to peptide bond interactions (Fig. 3d) shows a clear clustering of X···O C
C N dihedral angles ≈90° associated with interactions that involve primarily the π system of the O
N dihedral angles ≈90° associated with interactions that involve primarily the π system of the O C groups and a much broader distribution associated with interactions involving the in-plane lone pair electrons of the oxygen (the skewed distribution reflects the asymmetry of the peptide backbone). The halogen bonds to carbonyl π systems is rather surprising and largely undocumented (10), with the exception of one study describing the interaction of dihalogens with related thio- and selenocarbonyl compounds (9). One would be tempted to attribute the involvement of the carbonyl π system to the unavailability of the lone pair electrons of the C
C groups and a much broader distribution associated with interactions involving the in-plane lone pair electrons of the oxygen (the skewed distribution reflects the asymmetry of the peptide backbone). The halogen bonds to carbonyl π systems is rather surprising and largely undocumented (10), with the exception of one study describing the interaction of dihalogens with related thio- and selenocarbonyl compounds (9). One would be tempted to attribute the involvement of the carbonyl π system to the unavailability of the lone pair electrons of the C O groups, which are often involved in regular hydrogen bonds with the N
O groups, which are often involved in regular hydrogen bonds with the N H group of a neighboring peptide backbone chain. However, this pattern is not seen systematically across the entire data set and, therefore, a thermodynamic preference for the π system cannot be excluded.
H group of a neighboring peptide backbone chain. However, this pattern is not seen systematically across the entire data set and, therefore, a thermodynamic preference for the π system cannot be excluded.
The oxygen atoms of water molecules (OW) belong to another potential category of electron donor atoms that can participate in halogen bonds. Indeed, the present survey reveals the occurrence of a nontrivial number of short C X···OW contacts (26, 86, and 32 contacts to chlorine, bromine, and iodine atoms, respectively). However, because solvent atoms are not always precisely positioned and hydrogen atoms of water molecules are not observed crystallographically, it is more difficult to define short C
X···OW contacts (26, 86, and 32 contacts to chlorine, bromine, and iodine atoms, respectively). However, because solvent atoms are not always precisely positioned and hydrogen atoms of water molecules are not observed crystallographically, it is more difficult to define short C X···OW contacts as halogen bonds, although some of them certainly share several of their characteristics. Yet, the formation of C
X···OW contacts as halogen bonds, although some of them certainly share several of their characteristics. Yet, the formation of C X···H
X···H OW interactions appears less probable in the light of the present data.
OW interactions appears less probable in the light of the present data.
There are several reports (9, 10) that nitrogen and sulfur atoms could also be involved in halogen bonds (note that rN = 1.55 Å and rS = 1.80 Å; thus, the corresponding RvdW are increased by 0.03 and 0.28 Å, respectively). Short X···N contacts, although significantly less frequent than X···O contacts, were observed in two structures: PDB ID code 1J51 (Cl···N of tryptophan, dCl···N = 2.95 Å) and PDB ID code 1O5M (Br···N of histidine, dBr···N = 3.18 Å). In addition, short Cl···N contacts (3.03, 2.94, and 3.25 Å) to asparagine and glutamine side chains were observed in three structures (PDB ID codes 1R6N, 3PCH, and 7STD, respectively), implying the existence of unusual Cl···H N interactions. However, because the assignment of the oxygen and nitrogen atoms of the amide side chains in asparagine and glutamine is often ambiguous (at least in medium-resolution structures), reversing these atoms would lead to more typical Cl···O halogen bonds rather than to less probable Cl···H
N interactions. However, because the assignment of the oxygen and nitrogen atoms of the amide side chains in asparagine and glutamine is often ambiguous (at least in medium-resolution structures), reversing these atoms would lead to more typical Cl···O halogen bonds rather than to less probable Cl···H N bonds.
N bonds.
Given the small proportion of sulfur atoms in amino acids, it was surprising to observe several short X···S contacts. In sulfur-containing proteins, short Cl···S (methionine) contacts of 3.24 and 3.49 Å (in PDB ID codes 1GSF and 1PY2), and short I···S (cysteine–cysteine) as well as I···S(methionine) contacts of 3.66 and 3.61 Å (in PDB ID codes 1GJD and 1HK1, respectively) were observed. This finding suggests that sulfur can replace oxygen as the donor in the interaction.
Discussion
The present survey of short halogen–oxygen (C X···O) interactions in proteins and nucleic acids shows that the halogen bond geometries in biological systems conform generally to those seen in small molecules (8–10), with the interaction defined (i) by distances that are significantly shorter than the RvdW of the interacting atoms (as short as 80% of RvdW) and (ii) as directional relative to the C
X···O) interactions in proteins and nucleic acids shows that the halogen bond geometries in biological systems conform generally to those seen in small molecules (8–10), with the interaction defined (i) by distances that are significantly shorter than the RvdW of the interacting atoms (as short as 80% of RvdW) and (ii) as directional relative to the C X bond. There are, however, some significant deviations that can be ascribed uniquely to biomolecules. The nonlinear Θ1 angles (Fig. 3b) can be attributed to additional polarization of the halogen atom, suggested by quantum mechanical calculations (3) to be an environmentally induced distortion of the cylindrical shape of the electron distribution which, in turn, modifies the line of approach of the lone pair electrons of the interacting oxygen atom. The nearly perpendicular approach of the halogen toward certain O
X bond. There are, however, some significant deviations that can be ascribed uniquely to biomolecules. The nonlinear Θ1 angles (Fig. 3b) can be attributed to additional polarization of the halogen atom, suggested by quantum mechanical calculations (3) to be an environmentally induced distortion of the cylindrical shape of the electron distribution which, in turn, modifies the line of approach of the lone pair electrons of the interacting oxygen atom. The nearly perpendicular approach of the halogen toward certain O C oxygens in a separate group of halogen bonds can be attributed to the involvement of the π electrons of the peptide bonds. Perpendicular interactions to the peptide backbone are often ignored in molecular interactions (24, 25); however, the prevalence of π -system donors seen in this survey shows them to be very important in halogen bonding.
C oxygens in a separate group of halogen bonds can be attributed to the involvement of the π electrons of the peptide bonds. Perpendicular interactions to the peptide backbone are often ignored in molecular interactions (24, 25); however, the prevalence of π -system donors seen in this survey shows them to be very important in halogen bonding.
These deviations from the ideal halogen bond geometries, as defined in small molecules, reflect the more complex environments found in biomolecular systems as compared with simple organic crystals. Analogous effects are seen with hydrogen bonds in biomolecules. A recent study of the geometries of classic (N/O H···O) and weak (C
H···O) and weak (C H···O) interactions in a set of 28 high-resolution crystal structures of protein–ligand complexes concluded that the generally restrictive geometric criteria used to define hydrogen bonds in small-molecule crystal structures may need to be relaxed for macromolecules (26). Indeed, Hassel (4) had noted that stable halogen complexes resulting from charge-transfer interactions do not always result in bond distances that are significantly shorter than expected for van der Waals contacts. As with classical hydrogen bonds, the strength of which can vary from 40 to 0.25 kcal/mol (1 kcal = 4.18 kJ) depending on their environment (26), there may be a continuous scale for halogen bond interactions that extends from strong to weak, with the strength of the interaction modulated by their surrounding. Although halogen bonds are generally referred to as weak interactions, it has been shown that, in some instances, these interactions prevail over classical hydrogen bonds in the formation of supramolecular motifs (6). We thus looked more closely at the structures in which halogen bonds occur to better understand how they contribute to ligand binding and macromolecular folding.
H···O) interactions in a set of 28 high-resolution crystal structures of protein–ligand complexes concluded that the generally restrictive geometric criteria used to define hydrogen bonds in small-molecule crystal structures may need to be relaxed for macromolecules (26). Indeed, Hassel (4) had noted that stable halogen complexes resulting from charge-transfer interactions do not always result in bond distances that are significantly shorter than expected for van der Waals contacts. As with classical hydrogen bonds, the strength of which can vary from 40 to 0.25 kcal/mol (1 kcal = 4.18 kJ) depending on their environment (26), there may be a continuous scale for halogen bond interactions that extends from strong to weak, with the strength of the interaction modulated by their surrounding. Although halogen bonds are generally referred to as weak interactions, it has been shown that, in some instances, these interactions prevail over classical hydrogen bonds in the formation of supramolecular motifs (6). We thus looked more closely at the structures in which halogen bonds occur to better understand how they contribute to ligand binding and macromolecular folding.
The Thyroid Hormonal System. One of the best-characterized biological systems that naturally exploit halogenation are the thyroid hormones. Indeed, a very large number of short I···O contacts from thyroxine (PDB ID codes 1ETA, 1ETB, 1ICT, 1IE4, 1HK1–5, and 2ROX; see Table 1) and thyroxine derivatives (PDB ID codes 1THA, 1KED, 1KGI, 1NQ0, 1NQ2, and 1F86) to their associated proteins were identified in the current data set. This large number of contacts supports an earlier suggestion (8) that short I···O interactions (seen in the current survey to primarily involve the π system of the carbonyl groups along the polypeptide backbone; see Fig. 3d) play essential roles in the recognition of these hormones by their cognate proteins. In this particular case, as in many others, it is stunning to see how nature has exploited all possible intermolecular interactions, even the most “exotic” ones, to design very specific and efficient recognition systems.
Implications for Drug Design. In all other interactions involving halogenated ligands, halogen bonds seem to play a role in binding and recognition similar to that in the thyroid-related hormones. In the aldose reductase–inhibitor complex, a short Br···O contact was suggested to account for the >1,000–fold selectivity against the binding of the inhibitor to the related aldehyde reductase protein (2). The diversity of short X···O interactions that are involved in protein/ligand recognition is illustrated by the four short Br···O contacts between the inhibitor 4,5,6,7,-tetrabromobenzotriazole and the ATP binding site of phospho-CDK2-cyclin A (27). In this system, two C Br···O
Br···O C contacts involve the lone pair electrons of oxygen atoms, one involves the π electrons of a peptide O
C contacts involve the lone pair electrons of oxygen atoms, one involves the π electrons of a peptide O C group, and one is a short C
C group, and one is a short C Br···OW contact to solvent (Fig. 4a). In this case, the halogenated inhibitor very efficiently displaces the charged ATP ligand, mainly through halogen bonding interactions, stressing the potential role of halogen bonds in the design of new drugs and inhibitors. Indeed, the understanding that halogen atoms can be involved in electrostatic-type interactions that are strong enough to compete with standard hydrogen bonds (6), in addition to their better documented abilities to serve as electron withdrawing substituents or their supposed “hydrophilic” properties, would contribute to the design of ligands by providing a framework for the use of this interatomic interaction.
Br···OW contact to solvent (Fig. 4a). In this case, the halogenated inhibitor very efficiently displaces the charged ATP ligand, mainly through halogen bonding interactions, stressing the potential role of halogen bonds in the design of new drugs and inhibitors. Indeed, the understanding that halogen atoms can be involved in electrostatic-type interactions that are strong enough to compete with standard hydrogen bonds (6), in addition to their better documented abilities to serve as electron withdrawing substituents or their supposed “hydrophilic” properties, would contribute to the design of ligands by providing a framework for the use of this interatomic interaction.
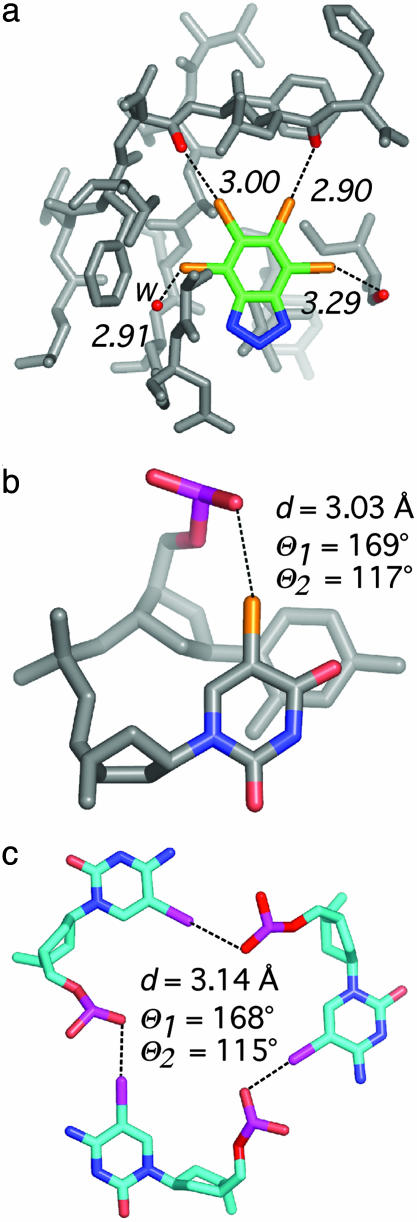
Fig. 4.
Examples of short X···O contacts in a ligand–protein complex and nucleic acids. (a) The 2.2-Å structure (PDB ID code 1P5E) of phospho-CDK2/cyclin A in complex with the inhibitor 4,5,6,7-tetrabromobenzotriazole (27). The inhibitor is shown with three bromine halogen bonds to peptide carbonyl oxygens of the protein. Two interactions (middle) involve the lone pairs of the oxygen atom and one (right) involves the π system of the C O group. In addition, one halogen bond to a water molecule (w) is seen (left). (b) Intramolecular halogen bond identified as stabilizing a DNA junction (PDB ID code 1P54) in the 1.9-Å structure of d(CCAGTACbr5UGG) (1). (c) View of the packing interactions involving three short I···O contacts in a unique six-stranded DNA structure (PDB ID code 1UE2; 1.4 Å) of the sequence d(Gi5CGAAAGCT) (i5C, 5-iodocytosine) (28).
O group. In addition, one halogen bond to a water molecule (w) is seen (left). (b) Intramolecular halogen bond identified as stabilizing a DNA junction (PDB ID code 1P54) in the 1.9-Å structure of d(CCAGTACbr5UGG) (1). (c) View of the packing interactions involving three short I···O contacts in a unique six-stranded DNA structure (PDB ID code 1UE2; 1.4 Å) of the sequence d(Gi5CGAAAGCT) (i5C, 5-iodocytosine) (28).
Effect on Conformational Equilibria. Halogens are now routinely introduced into macromolecules to help phase crystallographic data, with the assumption that these are conformationally neutral substitutions. However, there is good evidence that halogenation can induce significant conformational perturbations, particularly in nucleic acids. One clear example is found in the structure of d(CCAGTACbr5UGG) (1), which shows a very typical geometry for a Br···O P halogen bond (Fig. 4b). We were initially very surprised to find that this br5U sequence crystallized as a complex four-stranded junction, whereas the nonbrominated thymine (1) or uridine (unpublished results) containing sequences crystallized as resolved B-DNA duplexes. We now understand from the current study that this unique intramolecular Br···O halogen bond is responsible for the shift in equilibrium to favor the four-stranded junction and, consequently, is the primary stabilizing interaction within this complex.
P halogen bond (Fig. 4b). We were initially very surprised to find that this br5U sequence crystallized as a complex four-stranded junction, whereas the nonbrominated thymine (1) or uridine (unpublished results) containing sequences crystallized as resolved B-DNA duplexes. We now understand from the current study that this unique intramolecular Br···O halogen bond is responsible for the shift in equilibrium to favor the four-stranded junction and, consequently, is the primary stabilizing interaction within this complex.
An analogous situation is seen in the structure of d(Gi5CGAAAGCT) (i5C, 5-iodocytosine) (28), where short I···O P crystal contacts may contribute to the assembly of a unique six-stranded complex versus the parallel duplexes adopted by the noniodinated sequence (29). Chemists have already demonstrated the great potential of employing these very specific interactions in the design of new supramolecular motifs (6); therefore, it is not so surprising to observe that halogens can, through the formation of halogen bonds, alter conformational equilibrium in biomolecular systems. An important lesson from the current study is that one must be cautious when introducing halogen atoms into a biomolecular system, because this may sometimes lead to unexpected conformational effects. On the other hand, we suggest that halogen bonds, along with other more conventional interactions, can be introduced at specific positions of nucleic acids to help drive conformational equilibria in the course of designing new materials for nanotechnology (30).
P crystal contacts may contribute to the assembly of a unique six-stranded complex versus the parallel duplexes adopted by the noniodinated sequence (29). Chemists have already demonstrated the great potential of employing these very specific interactions in the design of new supramolecular motifs (6); therefore, it is not so surprising to observe that halogens can, through the formation of halogen bonds, alter conformational equilibrium in biomolecular systems. An important lesson from the current study is that one must be cautious when introducing halogen atoms into a biomolecular system, because this may sometimes lead to unexpected conformational effects. On the other hand, we suggest that halogen bonds, along with other more conventional interactions, can be introduced at specific positions of nucleic acids to help drive conformational equilibria in the course of designing new materials for nanotechnology (30).
Conclusions
This survey of halogen bonds in biological molecules clearly demonstrates the potential significance of this interaction in ligand binding and recognition, as well as in molecular folding. The rediscovery of this and other “nonstandard” types of interactions in proteins and nucleic acids will ultimately expand the variety of tools for molecular design using biological molecules. Molecular dynamics simulations, which are becoming more routinely used to explore the conformational dynamics of protein/drug and nucleic acid systems, rely on empirical force-fields (31) that generally do not explicitly take into account polarization effects. Thus, it should be recognized that, given the specific properties of halogen bonds described in this and previous studies (8–10), current molecular dynamics simulation protocols and force-fields should be used with great caution when applied to halogenated compounds.
Supplementary Material
Acknowledgments
We thank Dr. P. A. Karplus for his inquisitiveness, which helped to initiate this study. This work was supported by National Institutes of Health Grant R1GM62957A, National Science Foundation Grant MCB0090615, National Institutes of Environmental Health Sciences Grant ES00210, a Fulbright grant (to P.S.H.), and a grant from the Institut Universitaire de France (to E.W.).
Abbreviations: br5U, 5-bromouridine; X5U, 5-halouridine; X5C, 5-halocytosine; RvdW, sum of van der Waals radii; PDB, Protein Data Bank; OW, oxygen atom of a water molecule.
References
- 1.Hays, F. A., Vargason, J. M. & Ho, P. S. (2003) Biochemistry 42, 9586–9597. [DOI] [PubMed] [Google Scholar]
- 2.Howard, E. I., Sanishvili, R., Cachau, R. E., Mitschler, A., Chevrier, B., Barth, P., Lamour, V., Van Zandt, M., Sibley, E., Bon, C., et al. (2004) Proteins 55, 792–804. [DOI] [PubMed] [Google Scholar]
- 3.Muzet, N., Guillot, B., Jelsch, C., Howard, E. & Lecomte, C. (2003) Proc. Natl. Acad. Sci. USA 100, 8742–8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassel, O. (1972) in Nobel Lectures, Chemistry 1963–1970 (Elsevier, Amsterdam).
- 5.Foster, R. (1969) Organic Charge-Tranfer Complexes (Academic, London).
- 6.Metrangolo, P. & Resnati, G. (2001) Chem. Eur. J. 7, 2511–2519. [DOI] [PubMed] [Google Scholar]
- 7.Brisdon, A. (2002) Annu. Rep. Prog. Chem. 98, 107–114. [Google Scholar]
- 8.Cody, V. & Murray-Rust, P. (1984) J. Mol. Struct. 112, 189–199. [Google Scholar]
- 9.Ouvrard, C., Le Questel, J. Y., Berthelot, M. & Laurence, C. (2003) Acta Crystallogr. B 59, 512–526. [DOI] [PubMed] [Google Scholar]
- 10.Lommerse, J. P. M., Stone, A. J., Taylor, R. & Allen, F. H. (1996) J. Am. Chem. Soc. 118, 3108–3116. [Google Scholar]
- 11.Corradi, E., Meille, S. V., Messina, M. T., Metrangolo, P. & Resnati, G. (2000) Angew. Chem. Int. Ed. 112, 1852–1856. [DOI] [PubMed] [Google Scholar]
- 12.Lommerse, J. P. M., Price, S. L. & Taylor, R. (1997) J. Comput. Chem. 18, 757–774. [Google Scholar]
- 13.Wojtczak, A., Cody, V., Luft, J. R. & Pangborn, W. (2001) Acta Crystallogr. D 57, 1061–1070. [DOI] [PubMed] [Google Scholar]
- 14.van Pee, K. H. & Unversucht, S. (2003) Chemosphere 52, 299–312. [DOI] [PubMed] [Google Scholar]
- 15.Buss, I. H., Senthilmohan, R., Darlow, B. A., Mogridge, N., Kettle, A. J. & Winterbourn, C. C. (2003) Pediatr. Res. 53, 455–462. [DOI] [PubMed] [Google Scholar]
- 16.Wu, W., Samoszuk, M. K., Comhair, S. A., Thomassen, M. J., Farver, C. F., Dweik, R. A., Kavuru, M. S., Erzurum, S. C. & Hazen, S. L. (2000) J. Clin. Invest. 105, 1455–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen, Z., Mitra, S. N., Wu, W., Chen, Y., Yang, Y., Qin, J. & Hazen, S. L. (2001) Biochemistry 40, 2041–2051. [DOI] [PubMed] [Google Scholar]
- 18.Vargason, J. M., Eichman, B. F. & Ho, P. S. (2000) Nat. Struct. Biol. 7, 758–761. [DOI] [PubMed] [Google Scholar]
- 19.Mecozzi, S., West, A. P., Jr, & Dougherty, D. A. (1996) Proc. Natl. Acad. Sci. USA 93, 10566–10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berman, H. M., Battistuz, T., Bhat, T. N., Bluhm, W. F., Bourne, P. E., Burkhardt, K., Feng, Z., Gilliland, G. L., Iype, L., Jain, S., et al. (2002) Acta Crystallogr. D 58, 899–907. [DOI] [PubMed] [Google Scholar]
- 21.Bondi, A. (1964) J. Chem. Phys. 68, 441–451. [Google Scholar]
- 22.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 23.Collaborative Computational Project Number 4. (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 24.Karplus, P. A. (1996) Protein Sci. 5, 1406–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen, F. H., Baalham, C. A., Lommerse, J. P. M. & Raithby, P. R. (1998) Acta Crystallogr. B 54, 320–329. [Google Scholar]
- 26.Sarkhel, S. & Desiraju, G. R. (2004) Proteins 54, 247–259. [DOI] [PubMed] [Google Scholar]
- 27.De Moliner, E., Brown, N. R. & Johnson, L. N. (2003) Eur. J. Biochem. 270, 3174–3181. [DOI] [PubMed] [Google Scholar]
- 28.Sunami, T., Kondo, J., Hirao, I., Watanabe, K., Miura, K. I. & Takenaka, A. (2004) Acta Crystallogr. D 60, 90–96. [DOI] [PubMed] [Google Scholar]
- 29.Sunami, T., Kondo, J., Kobuna, T., Hirao, I., Watanabe, K., Miura, K. & Takenaka, A. (2002) Nucleic Acids Res. 30, 5253–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seeman, N. C. (2004) Sci. Am. 290, 71–75. [DOI] [PubMed] [Google Scholar]
- 31.Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. (2004) J. Comput. Chem. 25, 1157–1174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.