Abstract
Behcet disease (BD) is a chronic systemic vasculitis and considered as an autoimmune disease. Although rare, BD can be fatal due to ruptured vascular aneurysms or severe neurological complications. To date, no known biomarker has been reported for this disease, making it difficult to diagnosis in the clinics. To undertake this challenge, we employed the HuProt arrays, each comprised of ∼20,000 unique human proteins, to identify BD-specific autoantibodies using a Two-Phase strategy established previously. In Phase I, we profiled the autoimmunity on the HuProt arrays with 75 serum samples collected from 40 BD patients, 15 diagnosed autoimmune patients who suffer from Takayasu arteritis (TA; n = 5)), ANCA associated vasculitis (AAV; n = 5), and Sjogren's syndrome (SS; n = 5), and 20 healthy subjects, and identified 20 candidate autoantigens that were significantly associated with BD. To validate these candidates, in Phase II we constructed a focused array with these 20 candidate BD-associated antigens, and use it to profile a much larger cohort, comprised of serum samples collected from 130 BD patients, 103 autoimmune patients (i.e. 40TA, 40 AAV and 23 SS), and 110 healthy controls. This allowed us to validate CTDP1 (RNA polymerase II subunit A C-terminal domain phosphatase)as a BD-specific autoantigen. The association of anti-CTDP1 with BD patients was further validated using the traditional Western blotting analysis. In conclusion, anti-CTDP1 antibody serves a novel autoantibody for Behcet disease and is expected to help more accurate clinical diagnosis.
Behcet disease (BD)1 is a chronic systemic vasculitis affecting all sizes and all types of vessels. The presenting symptoms are mucocutaneous manifestations, including recurrent oralaphthae, genital ulcers and skin lesion. Arthritis, gastrointestine, and genitourinary can also be manifestations, while uveitis, major vascular and central nervous system involvement are the most serious (1, 2). Although BD occurs around the world, the highest prevalence is along the ancient Silk Road, including the Middle East, Mediterranean region, and Asia (3). People of different ages may be affected; however, increased incidence is observed around 30 years old. A continuous study during a period of two decades showed that the disease burden, including eye damage, is usually confined to the early stage. It is also worth noting that both the major vessel disease and neurologic involvement, mainly leading to mortality, have their onset early and late after the disease onset (4). Therefore, early diagnosis of BD is of great importance to therapy and prognosis.
The etiology of BD remains unknown. Previous studies showed that several genetic factors might contribute to BD. One of them, HLA-B51, was identified as a major susceptibility factor, whose subtypes were associated with the world distribution of BD (5–7). Immune-mediated mechanisms also play a vital role in the pathogenesis of the disease. It is generally believed that generation of autoantibodies is critical for many autoimmune diseases. However, unlike many autoimmune diseases, BD cannot be definitively diagnosed using those commonly known autoantigens, such as antinuclear antibody (ANA), antiphospholipid antibody (APLA), and antineutrophil cytoplasmic antibodies (ANCA), which are commonly found in many autoimmune diseases. Though the presence of anti-endothelial cell autoantibody (AECA) can partially account for the vascular damage of BD (8–10), AECA is observed at a low penetration rate of 13.1%, 26%, and 47.5% in Turkish, Spanish, and Chinese BD patients respectively, and its presence in other systemic vasculitis, such as systemic lupus erythematosus (SLE) and systemic sclerosis (SSc), is rather high, making accurate and timely diagnosis very challenging (11–13). Until now, diagnosis of BD is mainly dependent on clinical criteria. Although there is not a lack of effort to discover ideal BD biomarkers, including a-tropomyosin, selenium binding protein, haptoglobin, amyloidA, cofilin-1 mitochondrial carrier homolog 1and kinectin, via proteomic or express cloning approaches, the impact has been limited (14–19).
During the past decade, functional protein microarrays, in particular the HuProt arrays, comprised of ∼20,000 individually purified human proteins, have become a powerful tool for identifying novel biomarkers in many autoimmune diseases, such as rheumatoid arthritis (RA), primary biliary cirrhosis (PBC), SSc, and type 1 diabetes, to name a few (20–23), because it allows simultaneous profiling autoimmunity against nearly the entire human proteome with minimum consumption of the clinical samples (24). To take advantage of this unbiased proteomics tool, we applied a previously reported two-stage strategy to identify BD-specific biomarkers. In Phase I, the HuProt arrays were employed to identify candidate biomarkers using a relatively small cohort, to identify candidate autoantigens. In Phase II, a focused array, comprised of these candidate autoantigens, was employed to validate these autoantigens via screening against a much larger cohort (Fig. 1). On the basis of statistic analysis, CTDP1 was confirmed as a novel BD autoantigen and was further validated by Western blotting analysis.
Fig. 1.
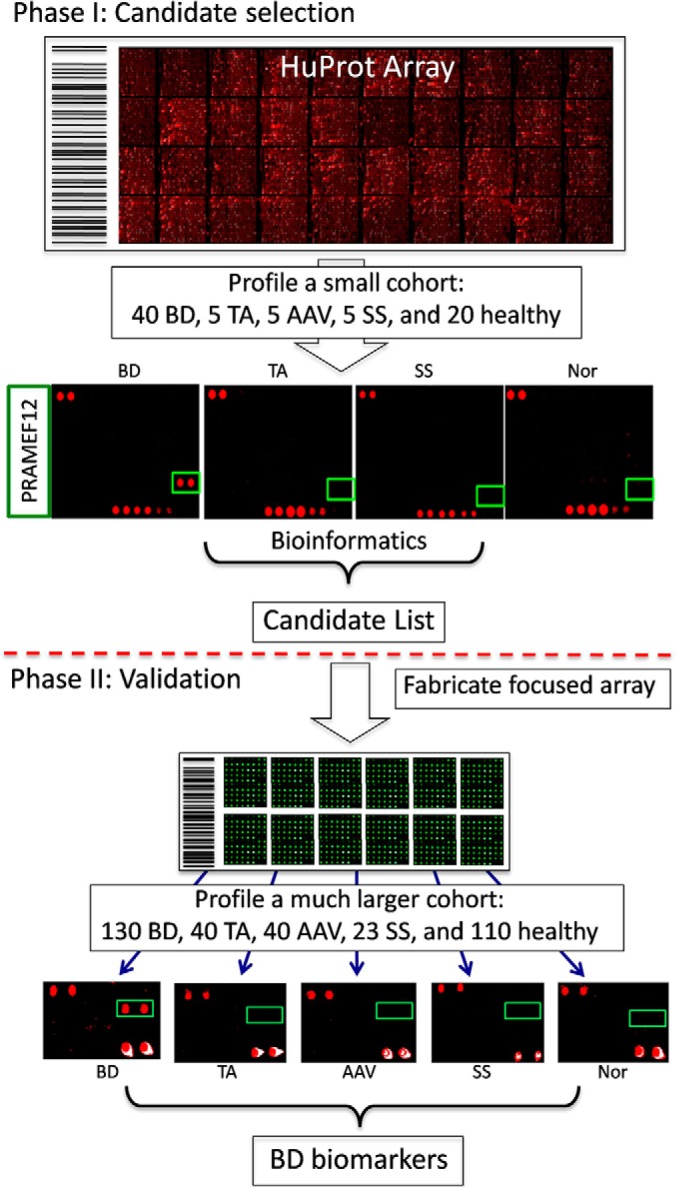
Two-Phase strategy to identify new BD biomarkers.
EXPERIMENTAL PROCEDURES
Serum Samples
All serum samples involved in this study were collected at Peking Union Medical College Hospital during a period from February 2012 to January 2014. This effort involved collecting serum samples from170 BD patients (37.1 ± 11.6 years of age; 79 females), the clinical symptoms information of the BD patient is shown in Table I, a disease control group of 118 patients diagnosed for TA (n = 45; 31.2 ± 10.2 years of age; 38 females), AAV (n = 45; 53.3 ± 15.9 years of age; 21females), and SS patients (n = 28; 50.8 ± 15.2 years of age; 28 females), and a healthy control group (n = 130; 37.6 ± 9.09 years of age; 41 females). All BD patients fulfilled the International Study Group (ISG) 1989 criteria of BD diagnoses (25) and were excluded from other autoimmune diseases, such as systemic lupus erythematosus, inflammatory bowel disease, RA, diabetes, and ankylosing spondylitis (AS). Disease controls were diagnosed according to the corresponding general criteria (25–27). Serum samples were obtained by separation from peripheral blood and stored at −80 °C until use. This study was approved by the Ethics Committee of Peking Union Medical College Hospital.
Table I. The Clinical symptoms information of the BD patients.
| Diagnostic symptom | Positive number/ total(positive rate) | Other symptom | Positive number/ total(positive rate) |
|---|---|---|---|
| Oral ulcers | 110/113(97.3%) | Untreated new-onset BD | 59/113(52.2%) |
| Genital ulcers | 81/113(71.7%) | Arthritis | 22/107(20.6%) |
| Eye lesions | 28/113(24.78%) | Epididymitis | 8/105(7.6%) |
| Skin lesions | 68/113(60.18%) | Gastrointestinal lesions | 23/109(21.1%) |
| Positive pathergy test | 31/73(42.47%) | Central nervous symptoms | 15/112(13.4%) |
| Vascular lesions | 21/105(20.0%) | ||
| Cardiac lesions | 7/101(6.9%) | ||
| Respiratory lesions | 2/104(1.9%) |
Construction and Quality Control Test of HuProt Arrays
The HuProt arrays, comprised of∼20,000 unique full-length proteins were constructed in Dr. Zhu's laboratory at Johns Hopkins University School of Medicine. Briefly, human ORFs cloned into a yeast expression vector (pEGH-A) were induced to produce N-terminally tagged GST fusions in Saccharomyces cerevisiae under control of the galactose-inducible GAL1 promoter. Using a high-throughput protein purification protocol, these GST fusions were purified in a 96-well format. Together with negative and positive control probes, all purified human proteins were printed in duplicate onto a single glass slide. The quality of the HuProt arrays was monitored by anti-GST probing, followed by the Cy5-labeled secondary antibodies (Fig. 2). The HuProt arrays were stored at-80 °C until use.
Fig. 2.
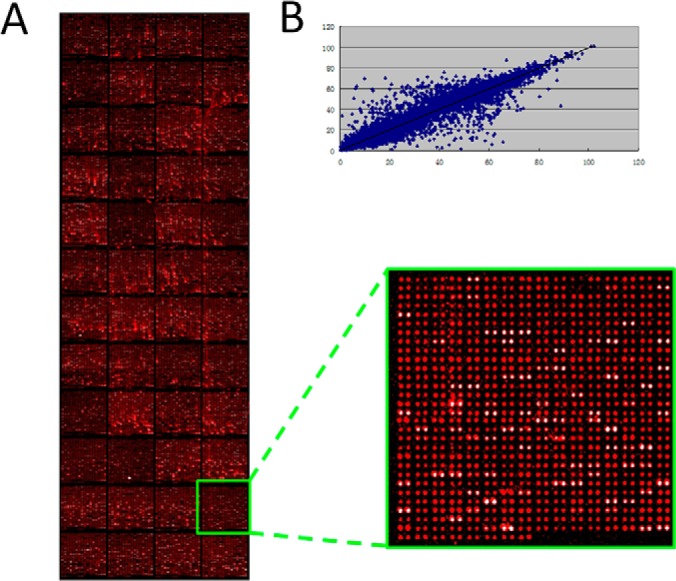
Quality control of HuProt arrays. A, The left image was the full view of a representative HuProt array detected with anti-GST signal. Statistical analysis shows 93.2% human proteins were detectable. There are 48 blocks on each microarray, and a magnified image of one of the 48 blocks on the HuProt array is shown on the right. B, The correlation coefficient between duplicate spots for each protein was determined as 0.978, suggesting a high reproducibility (Y = 0.99X + 0.53 R2 = 0.96).
Serum Profiling with HuProt Arrays
First, a small cohort of serum samples collected from 40 BD patients,15 autoimmune disease patients (i.e. 5 TA, 5 AAV, and 5 SS), and 20 healthy controls was probed individually to 75 HuProt arrays, the characteristics of study participants involved in this study is shown in Table II, as the first phage autoantibodies profiling assay (Fig. 1). HuProtarrays were taken out from −80 °C and warmed up at room temperature for half an hour and then incubated in a blocking buffer (3% BSA in PBS buffer with 0.1% Tween 20) at 37 °C for 1 h. Then a serum sample diluted1:1000 fold into 150 μl blocking buffer was added and incubated under coverslip (LifterSlip, Erie Scientific Company, Portsmouth, NH) at 37 °C for 1h. After 3 × 10 min washes with PBST, the microarray was incubated with 150 μl of 1:1000 diluted Alexa 647 conjugated goat anti-human IgG (the Jackson Laboratory, Bar Harbor, ME) at 37 °C for 1h in dark. Finally, after 3 × 10 min PBST washes the microarray was rinsed with double-distilled H2O and dried. The microarray was scanned with the GenePix 4000B Microarray Scanner (Molecular Devices, Sunnyvale, CA) and analyzed using GenePix Pro 6.0 software (Molecular Devices).
Table II. Characteristics of Study Participants in the study.
| Disease | No. | Gender(M/F) | Age |
|---|---|---|---|
| Phase I | |||
| BD | 40 | 16/14 | 40.0 ± 12.2 |
| Autoimmune disease (5TA,5AAV,5SS) | 15 | 5/10 | 45.0 ± 9.7 |
| Healthy controls | 20 | 11/9 | 41.5 ± 11.9 |
| Phase II | |||
| BD | 130 | 65/65 | 36.2 ± 11.3 |
| Autoimmune disease | 118 | 26/77 | 44.2 ± 18.0 |
| TA | 40 | 6/34 | 29.4 ± 8.8 |
| AAV | 40 | 20/20 | 54.9 ± 16.0 |
| SS | 23 | 0/23 | 51.2 ± 16.2 |
| Healthy controls | 110 | 78/32 | 36.9 ± 8.3 |
Construction of BD Focused Microarray and Serum Assay
Together with negative and positive controls, 20 candidate autoantigens potentially associated with BD as determined using the HuProt array data were purified and printed in duplicate in 12 identical sub-arrays on a single OPEpoxy SlideTM to construct the BD focused microarrays. A 12-hole rubber gasket divided each microarray to 12 individual chambers for 12 individual serum assays at same time. These low-density focused BD arrays were stored at −80 °C until use.
To validate these candidate autoantigens in Phase II, a larger cohort of serum samples collected from 130 BD patients, 103 autoimmune disease controls (DC) (i.e. 40TA, 40 AAV and 23 SS), and110 healthy controls were subjected to the focused BD array profiling. The protocol for the serum assay on BD focused microarray was basically identical to that of the HuProt array, except that 50 μl of 1/1000-diluted human serum and 50μL1/1000-diluted goat anti-human IgG were sequentially incubated in each chamber. After carefully removing the rubber gaskets, microarrays were washed and scanned.
Microarray Data Analysis
For HuProt array assays, the median foreground and background intensity for each spot on the arrays were acquired with GenePix Pro 6.0 software. The ratio of foreground to background signals for each spot was considered as the spot's signal value, and then the mean signal value of each duplicate pair was taken as the protein's signal value. We set the signal value cutoff at 2 to identify the positives. Differential proteins were identified using Gene Pattern platform (28). t test was chosen to assess the differential significance for each protein between different groups (BD) based on signal value. BD-specific candidate autoantigens were selected using p value <0.05 and fold change >1.3. To ensure appropriate sensitivity, only the antigens that showed a >20% positive rate (proportion of samples that are identified as positives in the BD group) in all the BD patients were considered as candidate autoantigens that were potentially associated with BD.
For the focused microarrays fabricated with those BD candidate autoantigens, the signal value for each protein was defined by ratio (dividing foreground intensity by the background intensity). Through comparing BD and control groups using t test, the proteins with p value<0.05 were considered as statistically significant BD-associated autoantigens.
Western Blotting Analysis
CTDP1 were expressed and purified from yeast as GST fusions. After electrophoresis on a 12% SDS-PAGE gel, CTDP1 protein was transferred to PVDF membrane, blocked with 5% nonfat milk, and incubated with 1:200 sera, followed by horseradish peroxidase conjugated anti-human IgG. The immuno reactive bands were visualized by ImageQuant (GE Healthcare).
RESULTS
Identification of Candidate Autoantigens Associated with BD Using HuProt Arrays
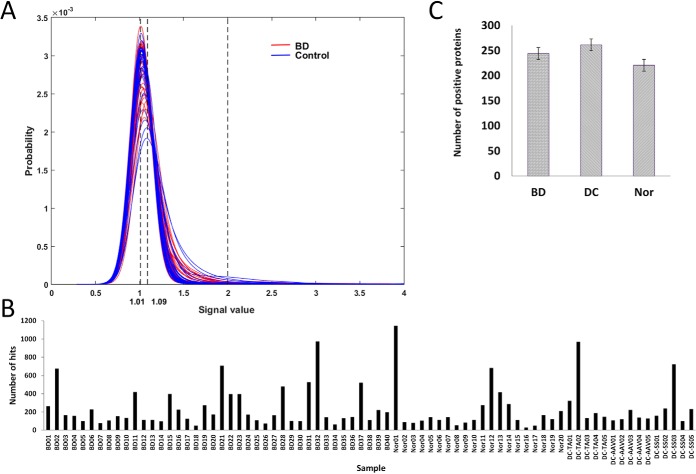
As described previously, in Phase I of the Two Phase strategy, we employed the HuProt arrays to profile a relatively small cohort. We assembled a cohort, comprised of serum samples collected from 40 BD patients, 15 autoimmune disease controls and 20 healthy controls (see Materials and Methods for more details). Each of the serum samples was individually incubated on the HuProt arrays, followed by incubation with a fluorescently labeled secondary anti-human IgG antibody to identify immune-reactive autoantigens. Using a relatively stringent cutoff value, human protein positively reacted to each serum sample were identified. However, number of identified positive hits identified by each serum sample varied dramatically, ranging from ∼100 to more than 1000 proteins, regardless of the origin of the samples (Fig. 3A). Comparison the number of hit number by each serum sample did not identify any significant difference between the BD patients and healthy control, or between autoimmune disease controls and healthy controls (Fig. 3B).
Fig. 3.
Comparison of positive hit distributions. A, Histogram analysis of foreground-to-background ratios of all the protein spots obtained with the 75 samples on the HuProt arrays. The major peaks of all the samples are located between 1.01 and 1.09, indicating that the majority of proteins are not recognized by the serum samples. B, The number of positive proteins identified in each serum profiling reaction varies dramatically from sample to sample. C, The number of positive proteins reacted with serum IgG in BD patients, autoimmune disease control group, and healthy controls were 244.05 ± 206.05, 261.45 ± 249.2, and 261.45 ± 249.2, respectively, without any significant difference.
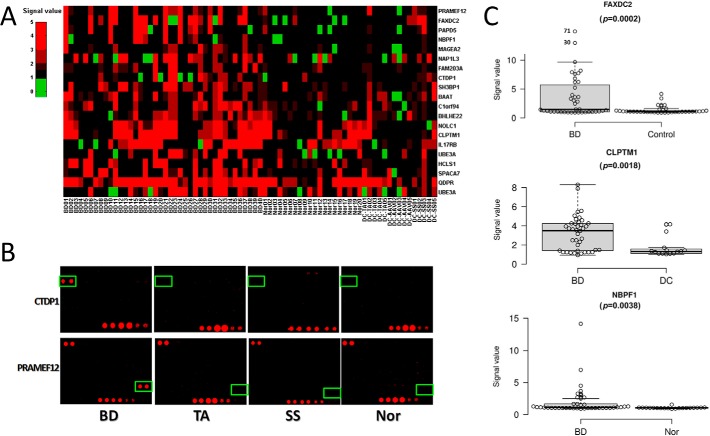
To identify potential biomarkers for BD diagnosis, data obtained on the HuProt arrays were first processed using Significant Analysis of Microarray (SAM) algorithm. BD-specific candidate autoantigens were identified between 3 groups, BD/Control, BD/Nor and BD/DC based on the p value <0.05 and fold change >1.3. We also set an arbitrary penetration rate at >20% among the BD patient samples, in order to improve the stringency. A total of 20 candidate BD-associated autoantigens were therefore identified (Table III). Using a one-dimensional clustering analysis, the autoimmunity of these 20 proteins can be visualized (Fig. 4A). For example, PRAMEF12 showed a sensitivity and specificity of 32.5% (i.e. 13positives identified among 40 BD samples) and 91.4% (i.e. 32negatives among 20 healthy controls and 15 disease controls). As illustrated in Fig. 4B, two representative proteins, namely CTDP1 and PRAMEF12, were specifically recognized by a BD serum sample, but are less likely to be recognized by serum samples either collected from the disease or healthy controls. CTDP1 is a phosphatase that dephosphorylates the C-terminal domain of RNA polymerase II, but little is known about the function of PRAMEF12. None of these proteins are known to be associated with any autoimmune diseases. Furthermore, box plot analysis also demonstrated a significant segregation between BD and control groups by three other proteins, namely FAXDC2, CLPTM1, and NBPF1 (Fig. 4C). Therefore, it was likely that some of these 20 potential biomarkers should be validated in Phase II.
Table III. Candidate proteins identified by Huprot array using 75 serum samples in Phase I.
| Name | Protein ID | Positive rate in BD | Positive rate in Nor | Positive rate in DC | BD VS Control (p value, fold change) | BD VS Nor (p value, fold change) | BD VS DC (p value, fold change) |
|---|---|---|---|---|---|---|---|
| FAXDC2 | NM_032385.1 | 45.00% | 5.00% | 26.70% | 0.0002, 3.99 | 0.0002, 4.79 | 0.0276, 3.26 |
| NBPF1 | BC034418.1 | 22.50% | 0.00% | 0.00% | 0.001, 1.82 | 0.0018, 1.87 | 0.0354, 1.76 |
| PAPD5 | NM_001040285 | 27.50% | 5.00% | 13.30% | 0.0012, 2.06 | 0.0014, 2.27 | — |
| HCLS1 | NM_005335.3 | 40.00% | 20.00% | 6.70% | 0.0034, 2.35 | 0.0396, 2.18 | 0.0152, 2.62 |
| C1orf94 | NM_032884.2 | 42.50% | 10.00% | 13.30% | 0.0044, 1.47 | 0.0072, 1.48 | — |
| MAGEA2 | BC013098 | 20.00% | 0.00% | 0.00% | 0.0066, 1.61 | 0.0288, 1.6 | 0.055, 1.62 |
| NOLC1 | BC001883.1 | 57.50% | 35.00% | 20.00% | 0.0068, 1.45 | — | 0.0066, 1.72 |
| NAP1L3 | BC034954.2 | 42.50% | 15.00% | 26.70% | 0.007, 1.76 | 0.0226, 1.84 | — |
| UBE3A | NM_000462.2 | 40.00% | 35.00% | 40.00% | 0.0088, 3.47 | — | 0.0314, 4.67 |
| SH3BP1 | BC008282 | 42.50% | 5.00% | 13.30% | 0.009, 1.38 | 0.0222, 1.4 | — |
| CLPTM1 | BC004865.2 | 65.00% | 50.00% | 20.00% | 0.0102, 1.47 | — | 0.0038, 1.79 |
| UBE3A | BC002582.2 | 27.50% | 15.00% | 13.30% | 0.0106, 2.38 | — | — |
| SPACA7 | BC016750.2 | 40.00% | 10.00% | 20.00% | 0.0122, 2.27 | 0.0056, 2.74 | — |
| BAAT | BC009567 | 30.00% | 5.00% | 33.30% | 0.0176, 1.35 | 0.0096, 1.46 | — |
| QDPR | BC000576.2 | 87.50% | 65.00% | 66.70% | 0.0182, 1.7 | — | 0.0098, 2.18 |
| FAM203A | NM_016458.2 | 25.00% | 5.00% | 0.00% | 0.019, 1.53 | — | — |
| PRAMEF12 | NM_001080830 | 32.50% | 5.00% | 13.30% | 0.0196, 3.1 | 0.0058, 3.51 | — |
| IL17RB | NM_018725.3 | 50.00% | 50.00% | 20.00% | 0.0208, 1.66 | — | 0.008, 2.28 |
| CTDP1 | NM_004715.4 | 22.50% | 5.00% | 6.70% | 0.0258, 1.58 | 0.0378, 1.65 | — |
| BHLHE22 | NM_152414 | 40.00% | 10.00% | 13.30% | — | 0.049, 1.33 | — |
Fig. 4.
20 candidate autoantigens identified in Phase I. A, Heat map of the signals obtained from the 20 candidate autoantigens in the HuProt array experiments. B, Two candidate autoantigens, namely CTDP1 and PRAMEF12, were found specifically identified by serums from the BD patients, but not from TA, SS and normal (Nor) groups on HuProt microarrays. C, Boxplot analysis of three other candidate autoantigens.
Validation of BD-associated Autoantigens with BD Focused Microarrays
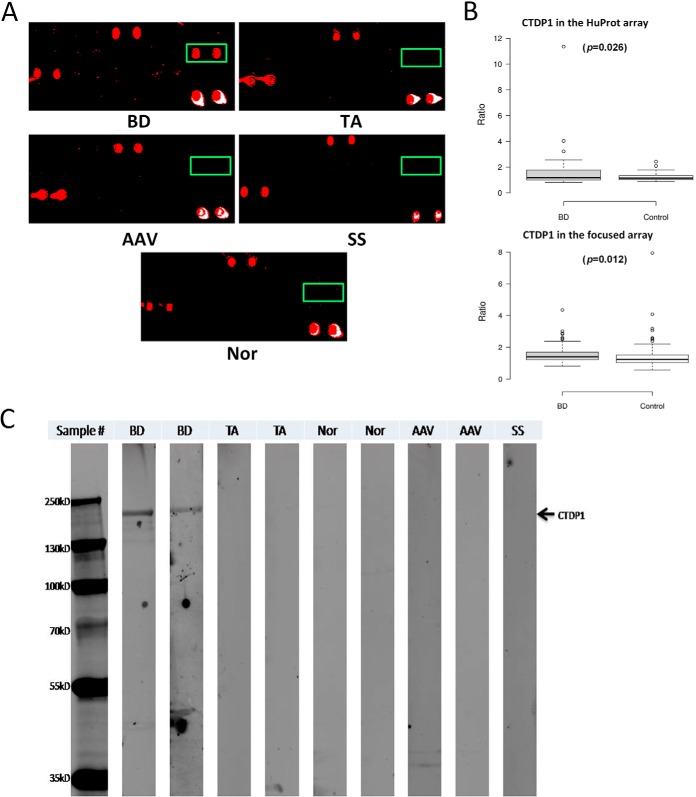
To validate the results obtained from HuProt arrays analysis in Phase II, we cherry-picked 20 yeast strains that each carries one of the 20 human ORFs on an expression vector, pEGH-A, from our human ORF collection. To purify these proteins, the 20 yeast strains were grown in 16 ml of culture, induced by galactose, and purified as N-terminal GST fusion proteins using a standard protocol described previously (29). After examination of the quantity and quality of the purified proteins using Coomassie stain and immunoblot analysis (data not shown), these purified proteins were spotted robotically onto glass sides to form the BD focused arrays. To save the cost, each glass slide contained 12 identical subarrays in a 2 × 6 format. Next, a much larger cohort, comprised of serum samples collected from 130 BD patients, 103 patients diagnosed of other types of autoimmune diseases, and 110 healthy subjects, was assembled and each serum sample was individually incubated on a subarray of the BD focused microarrays. The same assay conditions were applied to these focused arrays and the binding signals acquired, as described above. Using the same data analysis procedure, we validated anti-CTDP1 antibodies as the most significant biomarker that distinguishes BD patients from other autoimmune patients, as well as from healthy subjects (Fig. 5A). In this large cohort of 343 serum samples, anti-CTDP1 antibodies showed 23.86% sensitivity among the BD patients, which is significantly higher than that among disease control group (i.e. 4.9%; X2 = 15.87; p = 6.79 × 10–5), and than that among the healthy controls (i.e. 6.36%; X2 = 13.67; p = 0.000218). Overall, anti-CTDP1 antibodies showed a high specificity of 94.36% (= 201/213): 98 and 103 negatives in the 103 disease and 110 healthy controls, respectively. Moreover, the results obtained from the BD focused arrays agreed very well with that obtained from the HuProt arrays, as illustrated in boxplot analysis (Fig. 5B).
Fig. 5.
CTDP1 validated as a new BD biomarker in Phase II. A, CTDP1 (boxed in green) showed good sensitivity and high specificity in Phase II validation using the BD focused arrays. B, Boxplot analysis of CTDP1 indicates a similar behavior in HuProt and focused microarray assays. C, Western blotting analysis confirmed that CTDP1 can be specifically identified by BD serum samples but not by TA, AAV, or healthy (Nor) controls.
Detect Autoantibodies to CTDP1 by Western Blotting
To test the potential of transforming anti-CTDP1 antibodies as a more clinically friend biomarker, we purified anti-CTDP1 antibodies proteins and tested it using the traditional Western blotting analysis using serum samples collected from two anti-CTDP1 positive BD patients, as well as samples collected from patients diagnosed for other autoimmune disease, including TA and AAV. Samples from healthy two subjects were also tested as negative controls. As expected, anti-CTDP1 antibodies could only be detected with the two BD serum samples, but not by any other samples (Fig. 5C). Therefore, we believe that anti-CTDP1 antibodies test can be potentially converted to ELISA-based assays in the future.
DISCUSSION
BD is a rare immune-mediated small-vessel systemic vasculitis and is considered as an autoimmune disease in the clinics. Although a handful of recent studies showed that antiendothelial cell antibody (AECA) was found in serum samples of BD patients, AECA has a very low specificity in the clinics and the association of AECA with BD pathogenesis has not been firmly established. Therefore, it is in urgent need to employ an unbiased, high-throughput method to identify novel autoantibodies in BD patients. Protein microarrays, especially human proteome microarrays, are emerging as a highly effective approach for profiling new autoantibodies in numerous autoimmune diseases, as well as cancers (21, 30). Here, we employed the HuProt arrays, each comprised of ∼20,000 human recombinant proteins, to identify BD associated autoantibodies. Using a previously established Two-Phase strategy, we rapidly identified 20 potential BD-associated autoantigens and validated anti-CTDP1 antibody as BD-associated biomarker using a much larger cohort.
Although the pathology of BD is of great difference from the other autoimmune diseases, and let alone healthy persons, the number of autoantibody hits identified in each category is not significantly different from each other. However, the identity of the identified autoantigens varied dramatically from BD patients to the other groups. Thus, serum profiling assays performed on the HuProt arrays provide a proteome-wide fingerprints for each sample, a resolution that surpasses the traditional cell-based staining methods. Among the 20 candidate autoantigens of BD identified in the screening phase with HuProt arrays, some of which are play important role in transcription catalyzed by RNA polymerase I, such as NOLC1 and PAPD5; some of which are positive regulation of cell proliferation, such as PRAMEF12, CLPTM1, HCLS1, BHLHE22 and SH3BP1. It is very interesting SH3BP1 is act as a modulator of glutaredoxin biological activity to regulate actin cytoskeleton organization and blood vessel endothelial cell migration (31). The anti-SH3BP1 antibody in BD may affect the activity of SH3BP1 to have an important influence of actin cytoskeleton organization and blood vessel endothelial cell migration, which play important role in the pathogenesis of BD. It is also interesting to note that 11 of the 20 candidate autoantigens on BD focused arrays showed a similar sensitivity in Phase II between the BD and other autoimmune disease control cohorts, but a higher sensitivity than normal controls, suggesting that these 11 antibodies might play a similar role in the development of different autoimmune diseases. Function of these autoantibodies would be worthy further analysis.
CTDP1, also known as Fcp1, is a TFIIF-associating phosphatase, which encoding gene locates in the 14 exon of the 18q23. It is ubiquitously expressed in tissues (3) and locates in nucleus and cytoskeleton in relation to cell mitosis (32). CTDP1 is known to dephosphorylate the C-terminal domain (CTD) of RNA polymerase II subunit A and promote gene expression cycle. RNAP II plays key role in messenger RNA production, during which CTD undergoes a cycle of phosphorylation and dephosphorylation. Before the preinitiation complex formation and transcription, CTD is dephosphorylated; during transcription elongation it is phosphorylated (33). CTDP1, classic phosphatase for heptapeptide repeat of CTD (34–36), is necessary for the initiation of another mRNA synthesis cycle. CTDP1 also regulates other substrates. It inactivates crucial mitotic substrates (e.g. USP44, CDC20 and WEE1) to dephosphorylate M-phase-promoting factor (MPF)/CDK1, which results in promoting mitosis exit (32). In addition, CTDP1 regulates transcription elongation and stimulates the rate of elongation by RNAP II (37). Taken together, anti-CTDP1 autoantibodies may neutralize the CTDP1 role of dephosphorylation including RNAP resumption and mitosis exit, and transcription enlongation, inhibiting the production of mRNA and daughter cells. This may influence tissues which metabolism quickly, such as skin and mucosa, partly explaining the recurrent ulcer of BD.
CTDP1 partial deficiency has been reported in congenital cataracts facial dysmorphism neuropathy syndrome (CCFDN), which is an autosomal recessive developmental disease (38). As the name suggested, a key factor is nervous system involvement manifested with motor neuropathy leading to disability. Moderate nonprogressive cognitive deficit and pyramidal signs are also associated neurological features. Nerve biopsy shows demyelinating pathogenesis (39). Interestingly, Neuro- Behcet Disease, accounted for 5–10% of BD patients, can also involve the nervous system, primary characterized as subacute brainstem syndrome and hemiparesis. However, unlike CCFDN mainly injure periphery system, neuro-Behcet Disease mainly damages parenchymal of the central nervous system. The major histopathologic phenomenon are vasculitic manifestation and low-grade chronic nonspecific inflammation (40). The role of CTDP1in Behcet Disease, especially in neuro-Behcet Disease, should be further investigated.
Autoimmune diseases, including Behcet's disease, SS, AAV, and TA, are highly heterogeneous. In order to save research costs, we adapted two-phase strategy combines a proteome-wide screen for novel autoantigens followed by a stringent validation step using additional large cohorts to ensure the success of identification of useful autoantigens for a particular disease. In the screening phase, only a very small number of samples (i.e. 5TA,5AAV, and 5SS) were used as the disease control groups and therefore, the sensitivity and specificity of a candidate biomarker may be different from the real situation, presumably due to over fitting. As such, the additional large cohorts(including 130 BD, 118 autoimmune disease controls, and 110 healthy controls) used for the Phase II validation is extremely important to ensure the success of identification of true autoantigens for a particular disease. Therefore, it is understandable that the final biomarker of anti-CTDP1 antibody is not on the top list in the screening phase.
In previous studies, potential candidate biomarkers, such as anti-alpha-tropomyosin, anti-S-antigen, anti-alpha-enolase and anti-selenium binding protein, were reported for BD diagnosis. Although these proteins showed decent anti-GST signals (a proxy of purified protein amount) on the HuProt arrays, we did not observe any significant signal intensity in most of the serum profiling assays with BD-positive sera. One plausible explanation of this discrepancy is that these autoantigens can only be recognized by BD-positive sera in their denatured forms. Because most of the proteins on the HuProt arrays are in their native conformation, they failed to be recognized by the BD patient sera.
In summary, our study demonstrated that anti-CTDP1 is a BD-associated autoantibody, which might be very useful in clinical diagnosis of BD in the future.
Footnotes
Author contributions: C.H., J.P., G.S., F.Z., J.Q., H.Z., and Y.L. designed research; C.H., G.S., X.W., Z.W., S.C., and W.M. performed research; F.Z., H.Z., and Y.L. contributed new reagents or analytic tools; C.H., J.P., G.S., J.Q., H.Z., and Y.L. analyzed data; C.H., J.P., G.S., X.W., and H.Z. wrote the paper.
* This work was supported by the National Natural Science Foundation of China Grants No. 81373188, 81172857 (to YZ. L.), 81302610 to (to ChJ. H.), the Chinese National High Technology Research and Development Program, Ministry of Science and Technology Grants No. 2011AA02A113, the National Science Technology Pillar Program in the 12nd Five-year Plan No. 2014BAI07B00, the capital health research and development of special grants No. 2014-1-4011 (to YZ. L.). Youth Research Foundation of Peking Union Medical College No. 3332015091 (to ChJ. H.).
Authors' emails: Chao-Jun Hu: huchaojun818@qq.com, Jian-Bo Pan: jpan16@jhmi.edu, Guang Song: sguang1@jhmi.edu, Xiao-Ting Wen: xiaoting_wen@163.com, Zi-Yan Wu: wuziyanalice@163.com, Si Chen: chensi888.hi@163.com, Wen-Xiu Mo: mowenxiu8996@163.com, Feng-Chun Zhang: zhangfccra@aliyun.com, Jiang Qian: Jiang.Qian@jhmi.edu., Heng Zhu: hzhu4@jhmi.edu., Yong-Zhe Li: yongzhelipumch@163.com.
1 The abbreviations used are:
- BD
- Behcet disease
- TA
- Takayasu arteritis
- AAV
- ANCA associated vasculitis
- SS
- Sjogren's syndrome
- CTDP1
- RNA polymerase II subunit A C-terminal domain phosphatase
- ANA
- antinuclear antibody
- APLA
- anti-phospholipid antibody
- ANCA
- anti-neutrophil cytoplasmic antibodies
- AECA
- anti-endothelial cell autoantibody
- SLE
- systemic lupus erythematosus
- SSc
- systemic sclerosis
- RA
- rheumatoid arthritis
- PBC
- primary biliary cirrhosis
- DC
- disease controls.
REFERENCES
- 1. Yazici Y., Yurdakul S., and Yazici H. (2010) Behcet's syndrome. Curr. Rheumatol. Rep. 12, 429–435 [DOI] [PubMed] [Google Scholar]
- 2. Saadoun D., and Wechsler B. (2012) Behcet's disease. Orphanet J. Rare Dis. 7, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Archambault J., Pan G., Dahmus G. K., Cartier M., Marshall N., Zhang S., Dahmus M. E., and Greenblatt J. (1998) FCP1, the RAP74-interacting subunit of a human protein phosphatase that dephosphorylates the carboxyl-terminal domain of RNA polymerase IIO. J. Biol. Chem. 273, 27593–27601 [DOI] [PubMed] [Google Scholar]
- 4. Kural-Seyahi E., Fresko I., Seyahi N., Ozyazgan Y., Mat C., Hamuryudan V., Yurdakul S., and Yazici H. (2003) The long-term mortality and morbidity of Behcet syndrome: a 2-decade outcome survey of 387 patients followed at a dedicated center. Medicine 82, 60–76 [DOI] [PubMed] [Google Scholar]
- 5. Piga M., and Mathieu A. (2011) Genetic susceptibility to Behcet's disease: role of genes belonging to the MHC region. Rheumatology 50, 299–310 [DOI] [PubMed] [Google Scholar]
- 6. Verity D. H., Marr J. E., Ohno S., Wallace G. R., and Stanford M. R. (1999) Behcet's disease, the Silk Road and HLA-B51: historical and geographical perspectives. Tissue Antigens 54, 213–220 [DOI] [PubMed] [Google Scholar]
- 7. Gul A. (2014) Genetics of Behcet's disease: lessons learned from genomewide association studies. Curr. Opin. Rheumatol. 26, 56–63 [DOI] [PubMed] [Google Scholar]
- 8. Dinc A., Takafuta T., Jiang D., Melikoglu M., Saruhan-Direskeneli G., and Shapiro S. S. (2003) Anti-endothelial cell antibodies in Behcet's disease. Clin. Exp. Rheumatol. 21, S27–30 [PubMed] [Google Scholar]
- 9. Cervera R., Navarro M., Lopez-Soto A., Cid M. C., Font J., Esparza J., Reverter J. C., Monteagudo J., Ingelmo M., and Urbano-Marquez A. (1994) Antibodies to endothelial cells in Behcet's disease: cell-binding heterogeneity and association with clinical activity. Ann. Rheum. Dis. 53, 265–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zheng W. J., Zhao Y., Tang F. L., and Dong Y. (2005) [A study of antiendothelial cell antibodies in Behcet's disease]. Zhonghua nei ke za zhi 44, 910–913 [PubMed] [Google Scholar]
- 11. Praprotnik S., Blank M., Meroni P. L., Rozman B., Eldor A., and Shoenfeld Y. (2001) Classification of anti-endothelial cell antibodies into antibodies against microvascular and macrovascular endothelial cells: the pathogenic and diagnostic implications. Arthritis Rheum. 44, 1484–1494 [DOI] [PubMed] [Google Scholar]
- 12. Song J., Park Y. B., Lee W. K., Lee K. H., and Lee S. K. (2000) Clinical associations of anti-endothelial cell antibodies in patients with systemic lupus erythematosus. Rheumatol. Int. 20, 1–7 [DOI] [PubMed] [Google Scholar]
- 13. Mihai C., and Tervaert J. W. (2010) Anti-endothelial cell antibodies in systemic sclerosis. Ann. Rheum. Dis. 69, 319–324 [DOI] [PubMed] [Google Scholar]
- 14. Mor F., Weinberger A., and Cohen I. R. (2002) Identification of alpha-tropomyosin as a target self-antigen in Behcet's syndrome. Eur. J. Immunol. 32, 356–365 [DOI] [PubMed] [Google Scholar]
- 15. Okunuki Y., Usui Y., Takeuchi M., Kezuka T., Hattori T., Masuko K., Nakamura H., Yudoh K., Usui M., Nishioka K., and Kato T. (2007) Proteomic surveillance of autoimmunity in Behcet's disease with uveitis: selenium binding protein is a novel autoantigen in Behcet's disease. Exp. Eye Res. 84, 823–831 [DOI] [PubMed] [Google Scholar]
- 16. Mao L., Dong H., Yang P., Zhou H., Huang X., Lin X., and Kijlstra A. (2008) MALDI-TOF/TOF-MS reveals elevated serum haptoglobin and amyloid A in Behcet's disease. J. Proteome Res. 7, 4500–4507 [DOI] [PubMed] [Google Scholar]
- 17. Ooka S., Nakano H., Matsuda T., Okamoto K., Suematsu N., Kurokawa M. S., Ohtani-Kaneko R., Masuko K., Ozaki S., and Kato T. (2010) Proteomic surveillance of autoantigens in patients with Behcet's disease by a proteomic approach. Microbiol. Immunol. 54, 354–361 [DOI] [PubMed] [Google Scholar]
- 18. Vural B., Sehitoglu E., Cavus F., Yalcinkaya N., Haytural H., Kucukerden M., Ulusoy C., Ugurel E., Turan S., Bulut L., Turkoglu R., Shugaiv E., Kurtuncu M., Atakan S., Gure A. O., Gul A., Eraksoy M., Akman-Demir G., and Tuzun E. (2013) Mitochondrial carrier homolog 1 (Mtch1) antibodies in neuro-Behcet's disease. J. Neuroimmunol. 263, 139–144 [DOI] [PubMed] [Google Scholar]
- 19. Lu Y., Ye P., Chen S. L., Tan E. M., and Chan E. K. (2005) Identification of kinectin as a novel Behcet's disease autoantigen. Arthritis Res. Therapy 7, R1133–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Auger I., Balandraud N., Rak J., Lambert N., Martin M., and Roudier J. (2009) New autoantigens in rheumatoid arthritis (RA): screening 8268 protein arrays with sera from patients with RA. Ann. Rheum. Dis. 68, 591–594 [DOI] [PubMed] [Google Scholar]
- 21. Hu C. J., Song G., Huang W., Liu G. Z., Deng C. W., Zeng H. P., Wang L., Zhang F. C., Zhang X., Jeong J. S., Blackshaw S., Jiang L. Z., Zhu H., Wu L., and Li Y. Z. (2012) Identification of new autoantigens for primary biliary cirrhosis using human proteome microarrays. Mol. Cell. Proteomics 11, 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ayoglu B., Haggmark A., Khademi M., Olsson T., Uhlen M., Schwenk J. M., and Nilsson P. (2013) Autoantibody profiling in multiple sclerosis using arrays of human protein fragments. Mol. Cell. Proteomics 12, 2657–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koo B. K., Chae S., Kim K. M., Kang M. J., Kim E. G., Kwak S. H., Jung H. S., Cho Y. M., Choi S. H., Park Y. J., Shin C. H., Jang H. C., Shin C. S., Hwang D., Yi E. C., and Park K. S. (2014) Identification of novel autoantibodies in type 1 diabetic patients using a high-density protein microarray. Diabetes 63, 3022–3032 [DOI] [PubMed] [Google Scholar]
- 24. Zhu H., and Snyder M. (2003) Protein chip technology. Curr. Opin. Chem. Biol. 7, 55–63 [DOI] [PubMed] [Google Scholar]
- 25. International Study Group for Behcet's Disease (1990) Criteria for diagnosis of Behcet's disease. Lancet 335, 1078–1080 [PubMed] [Google Scholar]
- 26. Jennette J. C., Falk R. J., Andrassy K., Bacon P. A., Churg J., Gross W. L., Hagen E. C., Hoffman G. S., Hunder G. G., Kallenberg C. G., and et al. (1994) Nomenclature of systemic vasculitides. Proposal of an international consensus conference. Arthritis Rheum. 37, 187–192 [DOI] [PubMed] [Google Scholar]
- 27. Arend W. P., Michel B. A., Bloch D. A., Hunder G. G., Calabrese L. H., Edworthy S. M., Fauci A. S., Leavitt R. Y., Lie J. T., Lightfoot RW Jr and et al. (1990) The American College of Rheumatology 1990 criteria for the classification of Takayasu arteritis. Arthritis Rheum. 33, 1129–1134 [DOI] [PubMed] [Google Scholar]
- 28. Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., and Mesirov J. P. (2006) GenePattern 2.0. Nat. Genet. 38, 500–501 [DOI] [PubMed] [Google Scholar]
- 29. Jeong J. S., Jiang L., Albino E., Marrero J., Rho H. S., Hu J., Hu S., Vera C., Bayron-Poueymiroy D., Rivera-Pacheco Z. A., Ramos L., Torres-Castro C., Qian J., Bonaventura J., Boeke J. D., Yap W. Y., Pino I., Eichinger D. J., Zhu H., and Blackshaw S. (2012) Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol. Cell. Proteomics 11, O111.016253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anderson K. S., Sibani S., Wallstrom G., Qiu J., Mendoza E. A., Raphael J., Hainsworth E., Montor W. R., Wong J., Park J. G., Lokko N., Logvinenko T., Ramachandran N., Godwin A. K., Marks J., Engstrom P., and Labaer J. (2011) Protein microarray signature of autoantibody biomarkers for the early detection of breast cancer. J. Proteome Res. 10, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tata A., Stoppel D. C., Hong S., Ben-Zvi A., Xie T., and Gu C. (2014) An image-based RNAi screen identifies SH3BP1 as a key effector of Semaphorin 3E-PlexinD1 signaling. J. Cell Biol. 205, 573–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Visconti R., Palazzo L., Della Monica R., and Grieco D. (2012) Fcp1-dependent dephosphorylation is required for M-phase-promoting factor inactivation at mitosis exit. Nat. Commun. 3, 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meinhart A., Kamenski T., Hoeppner S., Baumli S., and Cramer P. (2005) A structural perspective of CTD function. Genes Develop. 19, 1401–1415 [DOI] [PubMed] [Google Scholar]
- 34. Archambault J., Chambers R. S., Kobor M. S., Ho Y., Cartier M., Bolotin D., Andrews B., Kane C. M., and Greenblatt J. (1997) An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 94, 14300–14305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kobor M. S., Simon L. D., Omichinski J., Zhong G., Archambault J., and Greenblatt J. (2000) A motif shared by TFIIF and TFIIB mediates their interaction with the RNA polymerase II carboxy-terminal domain phosphatase Fcp1p in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 7438–7449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Licciardo P., Ruggiero L., Lania L., and Majello B. (2001) Transcription activation by targeted recruitment of the RNA polymerase II CTD phosphatase FCP1. Nucleic Acids Res. 29, 3539–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mandal S. S., Cho H., Kim S., Cabane K., and Reinberg D. (2002) FCP1, a Phosphatase specific for the heptapeptide repeat of the largest subunit of RNA polymerase II, stimulates transcription elongation. Mol. Cell. Biol. 22, 7543–7552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Varon R., Gooding R., Steglich C., Marns L., Tang H., Angelicheva D., Yong K. K., Ambrugger P., Reinhold A., Morar B., Baas F., Kwa M., Tournev I., Guerguelcheva V., Kremensky I., Lochmuller H., Mullner-Eidenbock A., Merlini L., Neumann L., Burger J., Walter M., Swoboda K., Thomas P. K., von Moers A., Risch N., and Kalaydjieva L. (2003) Partial deficiency of the C-terminal-domain phosphatase of RNA polymerase II is associated with congenital cataracts facial dysmorphism neuropathy syndrome. Nat. Gen. 35, 185–189 [DOI] [PubMed] [Google Scholar]
- 39. Tournev I., Kalaydjieva L., Youl B., Ishpekova B., Guergueltcheva V., Kamenov O., Katzarova M., Kamenov Z., Raicheva-Terzieva M., King R. H., Romanski K., Petkov R., Schmarov A., Dimitrova G., Popova N., Uzunova M., Milanov S., Petrova J., Petkov Y., Kolarov G., Aneva L., Radeva O., and Thomas P. K. (1999) Congenital cataracts facial dysmorphism neuropathy syndrome, a novel complex genetic disease in Balkan Gypsies: clinical and electrophysiological observations. Ann. Neurol. 45, 742–750 [PubMed] [Google Scholar]
- 40. Saip S., Akman-Demir G., and Siva A. (2014) Neuro-Behcet syndrome. Handbook of Clin. Neurol. 121, 1703–1723 [DOI] [PubMed] [Google Scholar]