Abstract
Background
The recent investigations have rendered microRNAs (miRs) as a novel biomarker in cancer research. In fact, alteration in miR expression may be associated with tumor suppression, tumorigenesis, metastasis, and poor prognosis in human breast cancer (BC).
Objectives
The aim of this clinical experimental study was to measure the miR-328 expression level in breast cancer tissues, at first. Then, we tried to find out any possible correlation between miR-328 and prognostic and predictive biomarkers in BC. Both of these two objectives were investigated for the first time; and we did not find any similar survey measuring the expression level of miR-328 in both tumor and non-tumor breast tissues. This research was conducted in Iran (Ahvaz, Khuzestan), between December 2013 and April 2014. Furthermore, we did not find any previous document investigating the correlation between miR-328 expression level and prognostic factors in BC. Due to the lack of similar studies intending to measure the expression level of miR-328 in tumor and adjacent non-tumor tissues, we decided to carry out a pilot study.
Methods
We measured the expression level of miR-328 by Poly (A) real-time PCR based on SYBR Green-I in 28 fresh samples of BC tissues and 28 samples of normal adjacent tissues, including invasive ductal carcinoma (IDC), invasive lobular carcinoma (ILC), and ductal carcinoma in situ (DCIS). We tried to attribute the results to clinicopathologic features such as status of estrogen and progesterone receptors (ER/PR), HER2/neu (HER2), P53 and also Ki67 labeling (Ki67-LI).
Results
The results showed that the miR-328 median level of expression was 0.88 (2-ΔΔCt) (25th-75th percentile, 0.07 - 2.34). It means that the expression level increased in tumor tissues compared to normal adjacent tissues (NATs). However, a statistically significant correlation between the miR-328 median expression level and prognostic factors, including pathologic diagnosis, age, and also the status of ER, PR, HER2, and Ki67-LI was not observed (P > 0.05).
Conclusions
Therefore, it might be possible to consider miR-328 as an oncogene; but not necessarily an oncomiR, in human BC.
Keywords: Breast Carcinoma, Microrna-328, oncomir, Biomarkers, SYBR Green I Real-Time PCR
1. Background
BC is the most common cancer amongst women worldwide and it is also the leading cause of cancer-related mortality. Invasive breast tumors are commonly classified into IDC and ILC types (accounting for 70% - 75% and 10% - 14% of all invasive tumors, respectively), based on the assumption that ductal carcinomas originate from ducts and lobular carcinomas from lobules. However, it is now clear that both IDC and ILC originate from terminal duct lobular unit. Hence, this classification seems not to be more useful (1).
Age is a common risk factor in BC, as many other cancers. In fact, an accepted coding pattern is extensively postulated for the age-specific incidence rates (2). Contrarily, the curve that demonstrates the BC age-specific incidence rate in female shows a particular fluctuation, which is expected to happen near the age of 50, named as Clemmesen’s Hook (3). This undulation is thought to be associated with menopause (4).
Presumably, an important factor in designing a successful treatment is to determine the extent of tumor at time of diagnosis precisely. The most common cancer staging system used among clinicians is the TNM system adopted by the American joint committee on cancer (AJCC) and the international union for cancer control (UICC). In this system, the extent of the primary tumor (T), regional lymph nodes (N), and distant metastases (M) are coded; and a ‘‘stage grouping’’ based on T, N, and M is recommended (5). In this study, the 7th edition of the AJCC Cancer Staging Manual is used.
Obviously, to ensure effective therapy for BC, a precise and comprehensive knowledge of breast carcinoma is necessary. Extensive investigations have demonstrated that the expression of some genes such as ER, PR, and HER2 have deterministic roles in the determination of the prognosis of BC (6).
As reported by the college of American pathologists, both ER and PR status establish a first rank of prognostic factors in BC; so that low-grade tumors are positive for both ER and PR. Otherwise, high-grade tumors are usually ER and PR negative (7).
It has been shown that overexpression of HER2 occurs in 10% - 34% of invasive BCs (8). Obviously, the amplification of HER2 gene has been associated with increased cell proliferation, cell motility, tumor invasiveness, progressive local and distant metastases, accelerated angiogenesis, and reduced apoptosis in BC (6).
Another important character of BC is its high mitotic rate, which may be estimated by the measurement of Ki67, a protein which is expressed by proliferating cells. High Ki67 index is associated with poor prognosis in BC (9). The Ki67-LI with a cut-off < 20 % in ER-positive HER2-negative cancers is recommended in the St Gallen consensus (2013) to be used as a predictive factor (10). Moreover, Bustreo S et al. (11) showed that the > 20 % Ki67 cut-off is the best qualified value to accentuate high-risk patients in luminal BCs, and suggested to be incorporated with other prognostic factors. However, the existing biomarkers seem to be unreliable for precise prognosis and prediction in BC patients. Therefore, a significant effort would have been required to develop novel markers capable to predict the outcome of BC or even to strengthen the basis of our knowledge to be applied in efficient treatment strategies (12).
P53 is the most common mutated gene in various types of cancers, so that, it is postulated that the variation rate of the gene could be as high as 50% of human malignancies. However, the mutation of this gene can be seen in up to 80% of samples in some cancers such as triple-negative breast cancer, lung cancers, and high-grade serous ovarian tumors. P53 together with some genes is thought to be involved in different biological activities, including apoptosis, cell cycle arrest, senescence, metabolism, and autophagy (12).
MiRs are known as small noncoding RNA molecules, ranging in length from 17 to 25 nucleotides (miRBase; http://microrna.sanger.ac.uk/), which recognize their complementary target sites in the 3’-untranslated region (3’UTR) of mRNAs, resulting in a downregulation of target proteins through the degradation of this mRNA or through translational inhibition (13). MiRs are described to be abnormally expressed in cancer, with two distinct roles of either tumor suppressor or as oncogenes (oncomiRs). However, it depends on which genes or pathways are involved. MiRs are well known to be involved in almost all complex cellular processes, from the cell cycle to apoptosis to migration and invasion (14). The new concepts argue that miRs are abnormally expressed in cancers, let them intervene in a wide series of mechanisms such as tumor suppression, or even conversely, oncogenicity (oncomiRs). It now appears that about 60% of human genes are targets of miRs (15). New investigations have shown that one type of miR may regulate the expression of up to hundreds of target genes (16).
The importance of miRs in cancer is further authenticated by the fact that more than half of the human miR genes are located in cancer-associated genomic regions or at fragile sites (17, 18). Iorio et al. (19) reported the deregulation of miR expression in human BC, implying a possible role of miRs as prosperous biomarkers in BC diagnosis and prognosis.
MiR-328 gene is mapped to chromosome 16q22.1 based on an alignment of the mature miR-328 sequence (CUGGCCCUCUCUGCCCUUCCGU) with the genomic sequence (GRCh37) (20).
It seems that miR-328 downregulates the expression of breast cancer resistance protein (BCRP/ABCG2) in female BC cells. BCRP is known as an ATP-binding cassette (ABC) transporter protein, containing 655 amino acids with a single ATP-binding domain and six transmembrane domains (8). It has been elucidated that overexpression of BCRP can promote a multidrug chemotherapeutic resistance to agents such as topoisomerase I inhibitor topotecan and the antifolate agent methotrexate (21). A previous study carried out by Nogochi et al. (22) demonstrated a resistance as high as, 24-fold to SN-38, 10-fold to mitoxantrone, and 10-fold to topotecan in human myelogenous leukemia k562 cell line transduced by BCRP (k562/BCRP). However, there is not any document evaluating whether BCRP can affect radiation resistance in human BC.
Regarding the potential role of miRs in cancers, and lack of sufficient evidence of the importance of miR-328 in human BC, we decided to measure the miR-328 expression level in tumor samples and NATs, at first. Then, we tried to find out the likely relationship between miR-328 expression level and the status of prognostic and predictive biomarkers in invasive breast carcinomas, including ER/PR status, Ki67-LI, and HER2 status.
2. Objectives
We tried to perform more extensive investigations to elucidate miR-328 regulatory role in BC. These findings can give us novel insights into new strategies to reduce radiation resistance, which is now an obstacle in cancer treatment, using miR-328 either as a biomarker or a target molecule.
3. Methods
3.1. Patients and Sample Collection
In this clinical experimental study, all tissue samples were collected and used in accordance with the ethical rules confirmed by the ethics committee of Ahvaz Jundishapur University of Medical Sciences (Code no: U-92134 date: September 25, 2013). Ethical issues (including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors. Written informed consent was also obtained from all patients.
Due to the lack of such studies on BC samples, we decided to perform a pilot research on those 28 BC confirmed patients who had all of our inclusion criteria (between December 2013 and April 2014). Moreover, most studies measuring miR expression level have used formalin-fixed paraffin-embedded (FFPE)-paired normal and tumor tissue samples; whereas our study was performed on fresh frozen tumor samples (23-27), that their RNA yield was expected to be higher (28). Clearly, obtaining and preparing fresh frozen samples are much more difficult and time- consuming. Furthermore, a few studies carried out on BC tissues have chosen sample sizes as small as our study, possibly regarding them as pilot surveys (29, 30).
The exclusion criteria were previous breast surgery, radiotherapy and/ or chemotherapy. Out of 60 patients referred to surgery departments, 28 patients with histologically confirmed primary BC detected by two highly experienced independent pathologists, in a private pathology department (Pars Pathology Lab.) were selected to analyze the miR-328 expression level by real-time PCR. Fresh samples of human BC and paired normal adjacent tissues (NATs, > 2 cm from cancer tissue) -to minimize the effect of confounding variables- were obtained from almost all private hospitals (Ahvaz, Iran) between December 2013 and April 2014. Patients were mainly referred from nearly all cities of Khuzestan province. All of these patients were admitted to surgery departments.
The samples were divided into two groups. One part including resected paired samples (BC and NAT) was immersed in 1.8 mL cryotubes filled with RNAaseKiller Solution (5 PRIME), and stored at -80°C liquid nitrogen immediately before use. Cryotubes (SPL, South Korea) were labeled with the date of surgery and the sample code.
To obtain appropriate RNA yield, we followed the Approved Guideline MM13-A: (31) (Clinical and Laboratory Standards Institute, USA, 2005), carefully. Since tissues like breast ducts and lobules include a considerable content of fat and they supposed to have densities as same as water density (1 g.cm-3), a piece of tissue with 7 mm in diameter could provide us a suitable mass of breast tissue (about 50 g) and made us able to extract at least 10 µg of RNA, which is sufficient to make cDNA and lunch subsequent steps. The samples were quickly transported to the laboratory and stored in a -80°C freezer (WiseCryo, South Korea). An individual sheet was designed to register all patients’ clinical and pathological data including the sample code, the patient’s name, age, and the date of sampling, the name of the hospital, the surgeon’s name, and clinical and histopathological factors consisting of ER/ PR status, HER2, P53, and Ki67-LI. The median delay time to transport the samples into liquid nitrogen tank was about nine minutes (3 L MVE, USA). The second group of samples was fixed using 10% buffered formaldehyde for desired pathologic and immunohistochemical studies. Histopathological diagnosis of BC was done by a private laboratory of pathology (Pars Lab.), according to the criteria of the 7th edition of breast cancer staging of the American joint committee on cancer (32). The definite diagnosis and histological grade of all samples were confirmed by pathologists.
3.2. MicroRNA Extraction and Quantitative PCR
Tissue samples were thoroughly homogenized by a disruptor- homogenizer (WiseTis- HG15D, South Korea). Then, the products of extracted miR-328 were stored in a -80°C freezer.
In this study, we have adopted poly (A) real-time RT-PCR method to evaluate miR-328 expression to ensure whether miR-328 expression was correlated with clinical and pathological features, which is known as SYBR-Green I.
Moreover, to reduce possible errors induced by gene expression variations among different individuals, we used matched healthy control samples and applied 2-ΔΔCt equation to represent the level of miR-328 expression in tumor tissues in comparison with controls. As it was presumed, the results confirmed that miR-328 expression was up regulated in BC compared to NATs.
The master mix miScript SYBR Green PCR (Qiagen) kit was used to determine the expression of U6 small nuclear (sn) RNA (as an internal control) and miR-328 genes. All the primers for miR-328, U6 snRNA, and universal for the SYBR green miR assays were purchased from Qiagen. To minimize undesirable variations in PCR data, either tumor or non-tumor samples were evaluated on the same runs. The miR-328 was extracted using miRNeasy Mini Kit (Qiagen), according to its protocol.
QPCR reactions were performed with the following primers: for miR-328, 5’-GCTGGCCCTCTCTGCCC-3’ (forward) and 5’-CGTCAGATGTCCGAGTAGAGG-3’ (reverse); and for U6 snRNA, 5’-CTCGCTTCGGCAGCACA-3’ (forward) and 5’-AACGCTTCACGAATTTGCGT-3’ (reverse) (33).
The cDNA was synthesized using miScript II RT kit and HiSpec buffer (Qiagen) (Cat No. /ID: 218161), according to its protocol, and kept in a -20°C freezer.
Real- time PCR was performed on ABI- Step One real-time PCR system (USA). The reaction solutions were incubated at 95°C for 10 minutes, then 40 cycles at 95°C for 15 seconds and finally at 60°C for 1 minute. The cycle number at which the fluorescence level will pass a presumed threshold is described as threshold cycle (Ct). To analysis the miR-328 expression, we used 2-ΔΔCt formula, where ΔΔCt = (CtmiR-328 - CtU6 snRNA) tumor-(CtmiR-328 - CtU6 snRNA) mean normal (34).
To ensure the accuracy of results, all real-time PCR experiments were duplicated and the results were reviewed by two trained experts. The kappa coefficient was estimated at 100%, as well. Moreover, we developed the amplification, melting curves to affirm that this is specific and sensitive enough technique to detect miR-328.
Finally, considering the present canons, we classified breast tumors on the molecular basis as follows: The status of ER, PR, HER2, and P53 (considering positivity or negativity, for each factor), and the cut-off of 20% in Ki67- LI. Then, we tried to find out any possible correlation between these factors and the miR-328 expression level (2-ΔΔCt) in each sample.
3.3. Statistical Analysis
Normality of gathered data was not confirmed in Kolmogorov-Smirnov test (P < 0.05). Non-parametric tests, including Mann-Whitney and Kruskal-Wallis tests were used to determine the difference between the grades of breast tumor samples and miR-328 expression fold change. All tests were performed as two-tailed using SPSS 22 and a P < 0.05 was considered to be statistically significant.
4. Results
The well-sharply defined melting-curves of miR-328 with a narrow peak represent that pure, homogeneous PCR products were produced (Figure 1).
Figure 1. The Melting Curves of miR-328 are Presented as Sharply Defined Melting Curves With Narrow Peaks, Indicating Pure, Homogeneous PCR Products.
The expression level of miR-328 is considerably up-regulated in BC samples. The calculated Ct of miR-328 in tumor tissues (median expression level = 33.03; range = 30.39 - 35.47) was lower than that of miR-328 in NATs (median expression level = 33.52; range = 30.28 - 35.07), indicating that the expression level of miR-328 in tumor samples was higher than the controls (P = 0.87, Mann-Whitney Test).
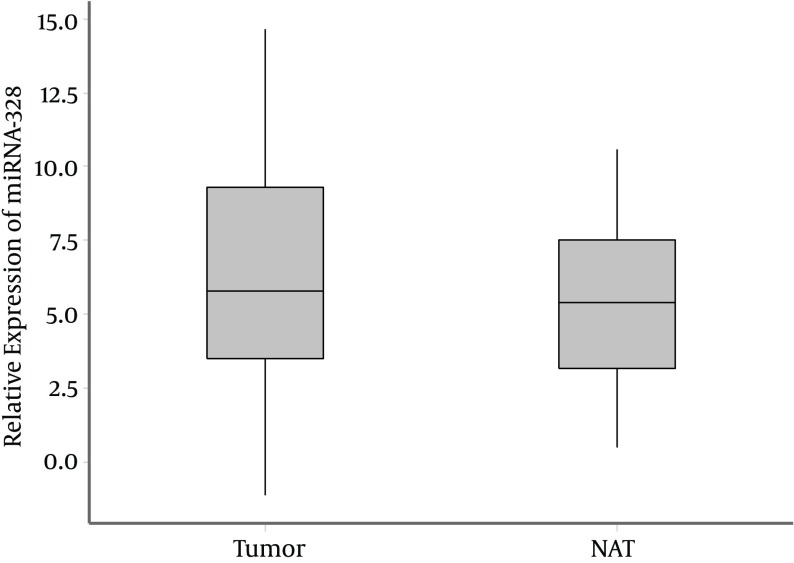
The median of relative expression of miR-328 (ΔCttumor = CtmiR-328-tumor-CtU6-tumor) was 5.80 (range = 3.49 - 9.30) in tumor samples. While, the median of relative expression of miR-328 in NATs (ΔCtcontrol = CtmiR-328-control-CtU6-control) was 5.40; range = 3.18 - 7.54 (P = 0.36; Mann-Whitney Test) (Figure 2).
Figure 2. The Median of Relative Expression Levels of miR-328 in Tumor Tissues of BC and NATs.
(Box-plot diagrams with median, 1st quartile, 3rd quartile).
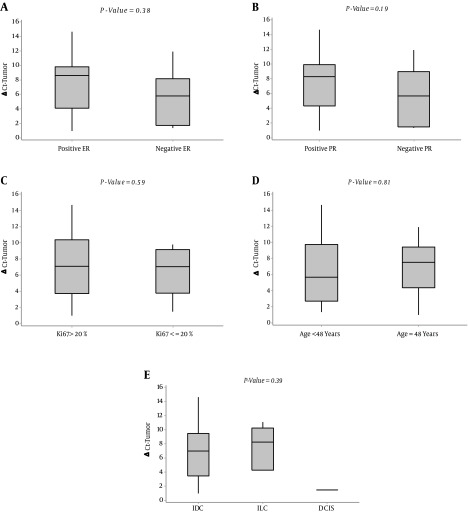
Therefore, despite the slight difference in the miR-328 expression between normal and tumor tissues, the miR-328 expression level (2-∆∆Ct) exhibited positive values in all 28 patients. The median of relative expression of miR-328 (2-∆∆Ct) was 0.88 (25th-75th percentile, 0.07 - 2.34). In our study, the relation of the expression level of miR-328 in tumor tissue (ΔCttumor) with prognostic factors was evaluated. The results showed that there is a statistically significant relationship between miR-328 expression level and HER2 status in tumor tissues (P = 0.03, Mann-Whitney Test). In this case, the median of relative expression level of miR-328 in HER2 -positive tumor samples (ΔCttumor) was 5.27 (first and third quartiles of 1.56 - 7.12), which is considerably lower than that of HER2-negatives, which was 9.08 (first and third quartiles of 4.90- 10.33) (Figure 3).
Figure 3. Association of miR-328 Expression Level in Tumor Samples With (A) ER Status, (B) PR Status, (C) Ki6, (D) Age, and (E) Pathologic Diagnosis.
(Box-plot diagrams with median, 1st quartile, 3rd quartile).
Nevertheless, we did not find any significant association between the expression of miR-328 in tumor tissues (ΔCttumor) and other prognostic factors, including ER, PR, Ki67, pathologic diagnosis, and age (Figure 3).
Furthermore, no statistically significant relationship was observed between the miR-328 expression level (2-∆∆Ct) and age, type of pathologic diagnosis, status of ER/PR and HER2, and Ki67-LI (Table 1).
Table 1. Association Between the Expression of miR-328 and Predictive/Prognostic Factors in BC Tissues.
| Variable | Frequency (%) | Median Relative Expression | P Valuea |
|---|---|---|---|
| Age, y | 0.81b | ||
| < 48 | 9 (32.1) | 0.96 | |
| ≥ 48 | 19 (67.9) | 0.42 | |
| Total | 28 (100) | - | |
| Pathologic diagnosis | 0.65c | ||
| IDC | 22 (78.6) | 0.91 | |
| ILC | 5 (17.9) | 0.42 | |
| DCIS | 1 (3.6) | 0.08 | |
| Total | 28 (100) | - | |
| ER status | 0.15b | ||
| Positive | 18 (64.3) | 1.08 | |
| Negative | 10 (35.7) | 0.22 | |
| Total | 28 (100) | - | |
| PR status | 0.31b | ||
| Positive | 17 (60.7) | 0.96 | |
| Negative | 11 (39.3) | 0.32 | |
| Total | 28 (100) | - | |
| HER2 status | 0.98b | ||
| Positive | 12 (42.9) | 0.91 | |
| Negative | 16 (57.1) | 0.49 | |
| Total | 28 (100) | - | |
| Ki67-LI | 0.48b | ||
| > 20% | 16 (57.1) | 0.49 | |
| ≤ 20% | 12 (42.9) | 0.91 | |
| Total | 28 (100) | - |
aP < 0.05 is significant.
bMann-Whitney Test.
cKruskal-Wallis Test.
5. Discussion
The first aim of this study was to show any possible alteration in the miR-328 expression in tumor tissues of breast carcinoma. Based on the obtained results, the miR-328 expression level was up regulated compared to NATs in all 28 samples. In other words, given that the miR-328 expression level gave a positive value, it may be considered as an oncomiR in BC.
We intended to elucidate the probable association between miR-328 expression level (applying the 2-∆∆Ct eq.) and prognostic factors in human BC, as well. Except for HER2 status, the other prognostic factors, including type of pathologic diagnosis, age, ER/PR, Ki67-LI, and P53 did not show any significant correlation with miR-328 expression level in all 28 tumor tissue samples. However, this result might be due to the lack of sufficient tumor samples, which restricted our study to obtain a statistical significance.
Interestingly, the median of relative expression level of miR-328 (ΔCttumor) in HER2 -positive tumor samples was considerably lower than that of HER2-negative samples.
In an extensive literature review on the alteration of miR-328 expression in tumors, we found only one study conducted by Arora et al., (2011) (35) that applied SYBR-Green I-based real- time RT-PCR technique in 13 non-small cell lung cancer (NCLC) patients, indicating that miR-328 expression level was significantly higher in patients with brain metastasis than those without brain metastasis.
Furthermore, the correlation between miR-328 expression level and predictive and prognostic factors such as, ER, PR, HER2, Ki67-LI, and P53 in human invasive breast carcinoma has not been evaluated yet. Our study may be considered as the first attempt to measure the miR-328 expression level, using SYBR-Green I- based poly (A) real- time RT-PCR technique in human BC.
5.1. Strengths
As mentioned above, no study has been still conducted to measure the miR-328 expression level and its relation to the predictive and prognostic factors such as, ER, PR, HER2, and Ki67 in human invasive breast carcinoma. Our study is the first one carried out to measure the expression level of miR-328 in human breast cancer tissues compared to NAT, using SYBR-Green I-based poly (A) real- time RT-PCR technique that tried to elucidate any possible relationship between the miR-328 expression and predictive and prognostic factors.
5.2. Restrictions
We suffered from lack of sufficient samples, which may be due to the fact that mastectomy in new cases of breast carcinoma is rarely performed at educational hospitals of Ahvaz. This limitation forced us to search the private hospitals, while we had not any authority over them to ask their cooperation. This basic constraint led to loss of a considerable number of samples.
Similar to the majority of malignancies, development, growth, and invasive behavior of breast carcinoma are due to multiple genetic interactions, with mechanisms which are still not clearly understood. It is supposed that one miR could be able to down regulate multiple target genes simultaneously. Thus, they may play an interesting role as powerful regulators of tumor-related genes in cancers. Recognition of genes targeted by these miRs is important to find out their mechanism of action in tumor pathogenesis.
5.3. Suggestions
We suggest more extensive studies on larger samples to ensure the oncogenicity of miR-328 in human breast carcinoma and elucidate the possible relation between miR-328 and predictive/prognostic factors in BC. It would be a stimulus to pursue our research in order to find probable solutions to make interventions in the miR-328 expression pathway by affecting tumor response through radiation therapy.
Acknowledgments
The authors would like to thank all patients for their participation, as well as Dr. Manouchehr Makvandi, Dr. Faramarz Pazyar, and Dr. Amir Ahmad Salmasi for their kind assistance.
Footnotes
Authors’ Contribution:Alihossein Saberi, Amir Danyaei and Mohammad Javad Tahmasbi Birgani conceived and designed the project, Amir Danyaei and Niloofar Neisi collected the data, Maryam Dastoorpoor analyzed and interpreted the data, Amir Danyaei and Maryam Dastoorpoor wrote the manuscript.
Funding/Support:This study was funded and supported by Ahvaz Jundishapur University of Medical Sciences, Grant No: U-92134.
References
- 1.Sainsbury JR, Anderson TJ, Morgan DA. ABC of breast diseases: breast cancer. BMJ. 2000;321(7263):745–50. doi: 10.1136/bmj.321.7263.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armitage P, Doll R. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br J Cancer. 2004;91(12):1983–9. doi: 10.1038/sj.bjc.6602297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemmesen J. Carcinoma of the breast; results from statistical research. Br J Radiol. 1948;21(252):583–90. doi: 10.1259/0007-1285-21-252-583. [DOI] [PubMed] [Google Scholar]
- 4.Gleason MX, Mdzinarishvili T, Sherman S. Breast cancer incidence in black and white women stratified by estrogen and progesterone receptor statuses. PLoS One. 2012;7(11):49359. doi: 10.1371/journal.pone.0049359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–4. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 6.Lowery AJ, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11(3):27. doi: 10.1186/bcr2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez-Aya LF, Chavez-Macgregor M, Lei X, Meric-Bernstam F, Buchholz TA, Hsu L, et al. Nodal status and clinical outcomes in a large cohort of patients with triple-negative breast cancer. J Clin Oncol. 2011;29(19):2628–34. doi: 10.1200/JCO.2010.32.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen JD, Schinkel AH. Multidrug resistance and pharmacological protection mediated by the breast cancer resistance protein (BCRP/ABCG2). Mol Cancer Ther. 2002;1(6):427–34. [PubMed] [Google Scholar]
- 9.Sahin S, Isik Gonul I, Cakir A, Seckin S, Uluoglu O. Clinicopathological Significance of the Proliferation Markers Ki67, RacGAP1, and Topoisomerase 2 Alpha in Breast Cancer. Int J Surg Pathol. 2016;24(7):607–13. doi: 10.1177/1066896916653211. [DOI] [PubMed] [Google Scholar]
- 10.Focke CM, van Diest PJ, Decker T. St Gallen 2015 subtyping of luminal breast cancers: impact of different Ki67-based proliferation assessment methods. Breast Cancer Res Treat. 2016;159(2):257–63. doi: 10.1007/s10549-016-3950-5. [DOI] [PubMed] [Google Scholar]
- 11.Bustreo S, Osella-Abate S, Cassoni P, Donadio M, Airoldi M, Pedani F, et al. Optimal Ki67 cut-off for luminal breast cancer prognostic evaluation: a large case series study with a long-term follow-up. Breast Cancer Res Treat. 2016;157(2):363–71. doi: 10.1007/s10549-016-3817-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duffy MJ, Synnott NC, McGowan PM, Crown J, O'Connor D, Gallagher WM. p53 as a target for the treatment of cancer. Cancer Treat Rev. 2014;40(10):1153–60. doi: 10.1016/j.ctrv.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 14.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: Current status and prospective. Int J Cancer. 2007;120(5):953–60. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 17.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 18.Manikandan J, Aarthi JJ, Kumar SD, Pushparaj PN. Oncomirs: the potential role of non-coding microRNAs in understanding cancer. Bioinformation. 2008;2(8):330–4. doi: 10.6026/97320630002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J, et al. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010;16(5):991–1006. doi: 10.1261/rna.1947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poy MN, Spranger M, Stoffel M. microRNAs and the regulation of glucose and lipid metabolism. Diabetes Obes Metab. 2007;9 Suppl 2:67–73. doi: 10.1111/j.1463-1326.2007.00775.x. [DOI] [PubMed] [Google Scholar]
- 21.Nakanishi T, Ross DD. Breast cancer resistance protein (BCRP/ABCG2): its role in multidrug resistance and regulation of its gene expression. Chin J Cancer. 2012;31(2):73–99. doi: 10.5732/cjc.011.10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noguchi K, Katayama K, Mitsuhashi J, Sugimoto Y. Functions of the breast cancer resistance protein (BCRP/ABCG2) in chemotherapy. Adv Drug Deliv Rev. 2009;61(1):26–33. doi: 10.1016/j.addr.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Gomes BC, Martins M, Lopes P, Morujao I, Oliveira M, Araujo A, et al. Prognostic value of microRNA-203a expression in breast cancer. Oncol Rep. 2016;36(3):1748–56. doi: 10.3892/or.2016.4913. [DOI] [PubMed] [Google Scholar]
- 24.Petrovic N, Davidovic R, Jovanovic-Cupic S, Krajnovic M, Lukic S, Petrovic M, et al. Changes in miR-221/222 Levels in Invasive and In Situ Carcinomas of the Breast: Differences in Association with Estrogen Receptor and TIMP3 Expression Levels. Mol Diagn Ther. 2016 doi: 10.1007/s40291-016-0230-3. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Guda C. Integrative exploration of genomic profiles for triple negative breast cancer identifies potential drug targets. Medicine (Baltimore). 2016;95(30):4321. doi: 10.1097/MD.0000000000004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei YF, Lei Y, Liu XQ. MiR-29a promotes cell proliferation and EMT in breast cancer by targeting ten eleven translocation 1. Biochim Biophys Acta. 2016;1862(11):2177–85. doi: 10.1016/j.bbadis.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 27.Cao ZG, Li JJ, Yao L, Huang YN, Liu YR, Hu X, et al. High expression of microRNA-454 is associated with poor prognosis in triple-negative breast cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luder Ripoli F, Mohr A, Conradine Hammer S, Willenbrock S, Hewicker-Trautwein M, Hennecke S, et al. A Comparison of Fresh Frozen vs. Formalin-Fixed, Paraffin-Embedded Specimens of Canine Mammary Tumors via Branched-DNA Assay. Int J Mol Sci. 2016;17(5) doi: 10.3390/ijms17050724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mollainezhad H, Eskandari N, Pourazar A, Salehi M, Andalib A. Expression of microRNA-370 in human breast cancer compare with normal samples. Adv Biomed Res. 2016;5:129. doi: 10.4103/2277-9175.186987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdel-Hamid NR, Mohammed EA, Abbas AH, Badr FM. MicroRNA-21 Expression in Primary Breast Cancer Tissue Among Egyptian Female Patients and its Correlation with Chromosome 17 Aneusomy. Mol Diagn Ther. 2015;19(6):365–73. doi: 10.1007/s40291-015-0161-4. [DOI] [PubMed] [Google Scholar]
- 31.Rainen L, Arbique JC, Asthana D, Earley MC, Geiszler RL, Krieg-Schneider F. Collection, transport, preparation, and storage of specimens for molecular methods: approved guideline. NCCLS; 2005. [Google Scholar]
- 32.Edge SBBD, Carducci M, Compton CC, Fritz AG, Greene FL. AJCC cancer staging manual. 7 ed. New York: Springer-Verlag; 2009. [Google Scholar]
- 33.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283(2):1026–33. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 34.Wang G, Wang L, Sun S, Wu J, Wang Q. Quantitative measurement of serum microRNA-21 expression in relation to breast cancer metastasis in Chinese females. Ann Lab Med. 2015;35(2):226–32. doi: 10.3343/alm.2015.35.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arora S, Ranade AR, Tran NL, Nasser S, Sridhar S, Korn RL, et al. MicroRNA-328 is associated with (non-small) cell lung cancer (NSCLC) brain metastasis and mediates NSCLC migration. Int J Cancer. 2011;129(11):2621–31. doi: 10.1002/ijc.25939. [DOI] [PMC free article] [PubMed] [Google Scholar]