Abstract
Whole-genome and exome sequence data can be cost-effectively generated for the detection of rare-variant (RV) associations in families. Causal variants that aggregate in families usually have larger effect sizes than those found in sporadic cases, so family-based designs can be a more powerful approach than population-based designs. Moreover, some family-based designs are robust to confounding due to population admixture or substructure. We developed a RV extension of the generalized disequilibrium test (GDT) to analyze sequence data obtained from nuclear and extended families. The GDT utilizes genotype differences of all discordant relative pairs to assess associations within a family, and the RV extension combines the single-variant GDT statistic over a genomic region of interest. The RV-GDT has increased power by efficiently incorporating information beyond first-degree relatives and allows for the inclusion of covariates. Using simulated genetic data, we demonstrated that the RV-GDT method has well-controlled type I error rates, even when applied to admixed populations and populations with substructure. It is more powerful than existing family-based RV association methods, particularly for the analysis of extended pedigrees and pedigrees with missing data. We analyzed whole-genome sequence data from families affected by Alzheimer disease to illustrate the application of the RV-GDT. Given the capability of the RV-GDT to adequately control for population admixture or substructure and analyze pedigrees with missing genotype data and its superior power over other family-based methods, it is an effective tool for elucidating the involvement of RVs in the etiology of complex traits.
Keywords: family-based association analysis, rare variants, whole-genome sequence data, exome sequence data, missing data, extended and nuclear pedigrees, Alzheimer disease, generalized disequilibrium test, pedigree disequilibrium test, TNK1
Introduction
The inability of common variants identified by genome-wide association studies (GWASs) to explain much of the heritability of most complex diseases and the advances of next-generation sequencing (NGS) technologies have led to an increased interest in investigating the etiology of complex disease due to rare variants.1, 2 Most NGS association studies use a population-based design, for which a large number of rare-variant association methods have been developed. There is also great interest in performing NGS association studies with the use of family data given that causal variants that aggregate in families usually have larger effect sizes than those found in sporadic cases. Family-based studies can therefore be more powerful than population-based studies given an equivalent number of cases.3 Study designs with familial cases might be preferred over case-control design when families with multiple affected individuals are available for study, especially for complex diseases for which loci with large effects have not been detected.4 Another advantage of a family-based design is its ability to avoid confounding due to population admixture or substructure. Population-based association studies can suffer from inflated false-positive rates as a result of population admixture or substructure, which is an even greater problem for rare variants.5 Rare variants are more likely to have more recent origins and are therefore more likely to be population specific than common variants, and there can be considerable differences in the rare-variant allelic spectrum, even between European ethnic groups. These differences can be more extreme in the study of admixed populations, such as African Americans and Hispanics. Family-based designs are robust against population admixture or substructure, and significant findings always imply association with the causal variant or association with a variant that is in linkage disequilibrium (LD) with the pathogenic variant.
A few tests have been proposed for family-based designs for the analysis of rare variants in sequence data. For example, the transmission disequilibrium test (TDT)6 has been extended to test rare-variant association by grouping information across multiple variants within a genomic region.7, 8 The extensions combine the benefits of rare-variant association analysis and family-based design, providing a robust and powerful approach to identifying and characterizing rare disease-susceptibility variants. However, these methods are not valid tests of LD for nuclear families with more than one affected child. Moreover, when extended pedigrees with multiple nuclear families and/or discordant sib-pairs are available, it is advantageous to include them in analysis because they also provide association information. The family-based association test (FBAT)9 has been extended to analyze sequence data in the rare-variant burden association test.10 A variance-component extension of FBAT has also been proposed, but the implemented software is applicable only to case-parent trio data.11 However, both FBAT extensions suffer from potential loss of power, which can be substantial for extended pedigrees, because these methods ignore parental phenotypes. Epstein et al. proposed a statistical approach for rare-variant association testing in affected sibships,12 which is less than optimal because it can be used only for analyzing affected sib-pairs in nuclear families (Michael Epstein, personal communication). Recently, Sul et al. proposed RareIBD for the analysis of large extended families of arbitrary structure.13 A main assumption of RareIBD is that only one founder in a family carries a rare variant in a given gene, which is often violated especially for extended pedigrees, and violation of this assumption will result in inflated type I error rates.
Here, we propose the rare-variant extension of the generalized disequilibrium test (GDT). The GDT utilizes genotype differences in all discordant relative pairs to assess associations within a family.14 The GDT has increased power by efficiently incorporating information beyond first-degree relatives. Moreover, quantitative or qualitative covariates, e.g., age, body mass index, and smoking status, can be incorporated in the analysis to control for confounding. The rare-variant extension of GDT (RV-GDT) aggregates a single-variant GDT statistic over a genomic region of interest, which is usually a gene. Additionally, the RV-GDT can incorporate weights that are based on allele frequencies or bioinformatics tools. Using simulated genetic data, we demonstrated that the RV-GDT method has well-controlled type I error rates, even when applied to admixed populations, populations with substructure, and pedigrees with family members missing genotype data. As a comparison, we also extended the pedigree disequilibrium test (PDT)15, 16 to analyze rare variants in general pedigrees. The PDT breaks a general pedigree into case-parent trios and discordant sib-pairs and then combines their contributions into a statistic that takes into account their non-independence. The rare-variant extension of the PDT (RV-PDT) is a weighted or unweighted combination of the single-variant PDT statistic over a genomic region of interest. The type I error was also evaluated in our simulated data for the RV-PDT and RareIBD. Although the RV-PDT had well-controlled type I error, the type I error for RareIBD was inflated, especially for extended pedigrees. The power of the RV-GDT was always substantially more powerful than that of Epstein’s affected-sib-pair (ASP) method and had similar or slightly higher power for nuclear families than the FBAT and RV-PDT under a variety of disease models. However, when applied to extended pedigrees and/or pedigrees in which family members were missing genotypes data, the RV-GDT was more powerful than these methods.
To further illustrate application of the proposed methods, we analyzed whole-genome sequence (WGS) data for 81 families affected by Alzheimer disease (AD [MIM: 104300]) from the Alzheimer Disease Sequencing Project (ADSP; dbGaP: phs000572.v6.p4). AD is a neurodegenerative disease characterized by dementia and typically begins with subtle and poorly recognized memory failure and slowly becomes more severe and incapacitating (see GeneReviews in Web Resources). AD is genetically heterogeneous and has an estimated heritability of 60%–80%.17 Although GWASs have successfully identified disease-associated loci, each locus accounts for only a small fraction of AD susceptibility, and a large proportion of AD heritability still remains unexplained.18 There is great interest in investigating the role of rare variants in the etiology of AD. Application of the RV-GDT identified suggestive associations between AD and rare variants in AXIN1 (MIM: 603816; GenBank: NM_003502.3) and TNK1 (MIM: 608076; GenBank: NM_001251902.1). An association between AD and a common variant in TNK1 was previously identified,19 and evidence of TNK1 involvement in AD has been further supported by experimental studies.20, 21 Although AXIN1 has not been previously shown to be associated with AD, experimental studies suggest that there might be a link between AXIN1 and AD.22, 23, 24, 25 These findings could provide new insights in the understanding of AD etiology.
Material and Methods
RV-GDT
The GDT utilizes the genotype differences in all discordant relative pairs within a family to assess the association.14 For the ith pedigree, ni is the total number of genotyped individuals, is the number of genotyped individuals who are affected, and is the number of genotyped individuals who are unaffected. The single-variant GDT statistic for the ith pedigree is defined as
where Xij and Xik are the numbers of minor alleles in the jth and kth unaffected individuals, respectively. Cijk is 1/ni if no covariates are included in the model; otherwise, it is given as
where and are the covariate vectors for the jth and kth unaffected individuals, respectively. Values in vector α are log odds ratios (ORs) for the association between the covariates and the trait, which are estimated from a logistic regression model that includes the phenotypes and covariates. The single-variant GDT statistic for a dataset with independent families is given as
which asymptotically follows a standard normal distribution under the null hypothesis of no association.14
It has been shown that an association test performed with individual rare variants (minor allele frequency [MAF] 1%) is underpowered26 given the small number of observed alternative alleles and the stringent multiple-testing correction. In order to increase power, it is advantageous to aggregate rare-variant information across a region. Similar to the burden of rare variants (BRV) method,27 here we aggregate the contributions of M variants across a region of interest, which is given as
where is the single-locus GDT statistic on the mth variant for the ith pedigree. In addition to aggregating the information across multiple variants, an alternative approach is to take a weighted sum of the contributions of each single variant, i.e.,
where wm is the weight assigned to the mth variant. The weights can be inferred from MAF in control28 or complete29 samples or from the predicted functionality of the variant,30 such as the C-score from the Combined Annotation Dependent Depletion (CADD) tool.31 The RV-GDT statistic is defined as
To infer its statistical significance, we apply a permutation procedure to derive empirical p values. We fix the genotypes and covariates for each pedigree and randomly shuffle the phenotypes among subjects within each pedigree. The vector α, a covariate adjustment, is also re-calculated after the phenotypes are shuffled. To reduce computational time, we use an adaptive permutation that evaluates the estimated p value at pre-defined checkpoints and stops further permutations for non-significant tests.
For rare-variant association tests, a common approach is to select a fixed MAF threshold and analyze only variants that meet the criterion. To determine whether the MAF of a variant is below the cutoff, one can obtain information on allele frequencies either from the sample or from public databases, e.g., the Exome Aggregation Consortium (ExAC) Browser. To avoid the implicit assumption about the relationship between allele frequency and variant functionality, we can use the variable threshold as an alternative approach to determine which variants should be analyzed.30 The intuition is that there exists an unknown threshold T for which variants with MAF < T are more likely to be functional than variants with MAF > T. In this approach, the RV-GDT score is calculated for each allele-frequency threshold, and the final RV-GDT statistic is defined as the maximum score. The p values must be inferred empirically for multiple-testing correction.
RV-PDT
The PDT takes into account the difference in the number of transmitted and non-transmitted minor alleles from parents to affected siblings and the difference in the number of minor alleles between affected and unaffected siblings.15, 16 For the ith pedigree, is the number of case-parent trios from informative nuclear families (at least one affected child, both parents genotyped at the marker, and at least one heterozygous parent), and nS is the number of informative discordant sib-pairs (at least one affected and one unaffected sibling with different marker genotypes). The single-variant PDT statistic for the ith pedigree is defined as
where Tik is the difference between the number of minor alleles transmitted and the number of minor alleles not transmitted from a heterozygous parent in the kth trio, and Sij is the difference between the number of minor alleles in affected siblings and those in unaffected siblings in the jth discordant sib-pair. Let N be the number of independent informative pedigrees (at least one informative nuclear family and/or discordant sibship); then, the single-variant PDT statistic is defined as
We can sum the contributions of each single variant across a region by
where is the single-variant PDT statistic on the mth variant in the ith pedigree, and M is the total number of variants in the region of interest. We can also consider the weighted sum of multiple variants, which is similar to the RV-GDT. The RV-PDT statistic is defined as
The p values can be inferred empirically via haplotype permutation, which is able to control the type I error in the TDT-based tests.7 For each pedigree, we fix the founders’ genotypes and obtain the genotypes of the non-founders by pairing a randomly selected paternal and maternal haplotype. Adaptive permutation can also be used to reduce computational time.
Simulation Framework
Generation of Family Data
To evaluate the performance of the RV-GDT, we compared it to other family-based association methods, including FBAT, RV-PDT, and Epstein’s ASP method, through simulating and analyzing family-based exome sequence data. Genotypes were simulated for autosomal genes across the genome on the basis of the observed variant sites and their corresponding MAFs obtained from the non-Finnish European and African and African American populations recorded in the ExAC Browser32 (17,987 autosomal genes for 33,370 subjects in the non-Finnish European population and 17,892 autosomal genes for 5,203 subjects in African and African American populations). Family data were generated with RarePedSim,33 which is able to effectively simulate sequence-based genotypes for any arbitrarily complex pedigree structure by conditioning on observed phenotypic data and incorporating a user-specified phenotype model and variant information. Using ExAC MAFs, we generated genotypes under linkage equilibrium (after assigning haplotypes to founders), which then segregated within the generated pedigrees.
Disease Model
The disease prevalence is assumed to be 1%, and the disease status for each subject is assigned on the basis of the multisite genotypes consisting of rare nonsense, missense, and splice-site variants (MAF ≤ 1% in its corresponding ExAC population). An OR of 2.5 is assigned to each variant that is deemed causal, and the disease probabilities of all variants within a gene are computed on the basis of a multiplicative mode of inheritance.33
Evaluation of Type I Error
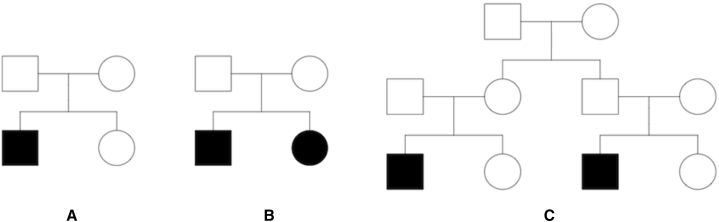
To evaluate the type I error rate of the RV-GDT and RV-PDT, we set the OR of the causal variant to 1 (no association between genetic variant and phenotype) and used the variant information from the ExAC non-Finnish European population to generate the family data. We considered four different types of family data: 1,000 nuclear families with one affected child and one unaffected child (discordant sib-pair; Figure 1A), 1,000 nuclear families with two affected children (affected sib-pair; Figure 1B), 1,000 extended pedigrees with two affected individuals in the third generation (extended pedigree; Figure 1C), and a mixture of these three pedigree structures (500 discordant sib-pairs, 250 affected sib-pairs, and 250 extended pedigrees). Genotype data were simulated for all autosomal genes across the genome, and genes with at least three informative variant sites were analyzed. Type I error rates were evaluated as the proportion of genes with a p value less than 0.05 and 0.005. The p values were obtained empirically via 100,000 permutations. Moreover, to demonstrate that the RV-GDT can appropriately handle pedigrees with missing data, we analyzed the data after removing genotype data from 50% of the founders.
Figure 1.
Pedigree Structures Used in the Simulation Studies
(A) Discordant nuclear sib-pair: the family contains parents, an affected child, and an unaffected child.
(B) Affected sib-pair: the nuclear family contains parents and two affected children.
(C) Extended three-generation pedigree.
We also evaluated the type I error rates of RV-GDT when there is population admixture or population substructure. Genotype data were simulated for 17,873 autosomal genes present in both the ExAC non-Finnish European and African and African American populations. To generate family data from an admixed population, we randomly generated the haplotypes of the founders in each pedigree from the European or African population with a probability of 20% or 80%, respectively. To generate family data with population substructure, we simulated 50% of the families with ExAC non-Finnish European variant information and simulated the other 50% of the families with ExAC African and African American variant information.
We also evaluated the type I error rates of RareIBD (version 1.1)13 in our simulation framework. We applied a maximum of 100,000 inheritance vector samplings to pre-compute the mean and standard deviation of the statistic and used 10,000 gene-dropping permutations to estimate the p values.
Power Evaluation
To evaluate the power of RV-GDT, we simulated 1,000 families of each pedigree structure shown in Figure 1 and a mixture of the three pedigree structures (500 discordant sib-pairs, 250 affected sib-pairs, and 250 extended pedigrees) by using ExAC non-Finnish European variant information. Genotype data were generated for autosomal genes across the genome when 75% of the rare nonsense, missense, and splice-site variants were randomly selected to be causal with an OR of 2.5. Genes with at least three informative variant sites were analyzed, and power was evaluated as the proportion of genes with a p value less than 0.05. To assess the influence of missing founder genotype data on power, we used a probability to determine which founders were missing all of their genotype data and considered three different probabilities (0%, 25%, and 50%). To further evaluate the power of RV-GDT for extended pedigrees in which family members are missing genotype data, we determined whether each parent, regardless of being a founder or non-founder (subjects in the first two generations), had a 0%, 25%, 50%, or 75% probability of missing all of their genotype data. Power was evaluated when 50%, 75%, and 100% of the randomly selected rare nonsense, missense, and splice-site variants were causal with an OR of 2.5.
We compared the power of the RV-GDT method to that of other family-based association tests, including Epstein’s ASP method (one-sided test),12 RV-PDT, and FBAT10 (version 2.0.4, with the “-v0” option to calculate unweighted rare-variant statistics). Epstein’s ASP method requires estimation of identity by descent (IBD) sharing between affected siblings. Because the IBD-sharing information is known in the simulated data, we used the exact IBD sharing in the power evaluation. The phase information generated during simulation of family data was used for haplotype permutation in the RV-PDT. Both Epstein’s ASP method and FBAT software report analytical p values. For RV-GDT and RV-PDT, we performed one-sided tests and obtained p values empirically by performing 2,000 permutations.
Application to AD Data
Description of the ADSP Data
The WGS data from 112 families were downloaded from dbGaP: phs000572.v6.p4. Study subjects with phenotypes coded as “definite AD,” “probable AD,” or “possible AD” were labeled as affected, and all other subjects were labeled as unaffected. The mean age of onset for AD was 72.63 years with a standard deviation of 8.46. In all 112 families selected for generation of WGS data, no more than 75% of affected members were positive for APOE4 (MIM: 107741), and no family members were homozygous for APOE4. Families in whom all sequenced subjects were affected were excluded from our analysis, which resulted in 81 families (21 nuclear and 60 extended), including 414 subjects with WGS data (316 affected and 98 unaffected; 167 male and 247 female) and 418 subjects without WGS data (22 affected and 396 unaffected; 221 male and 197 female). Their pedigrees and ethnicities (46 Dominican, 31 of European descent, 2 Puerto Rican, 1 Dutch isolate, and 1 African American) are shown in Figure S2 and Table S2, respectively.
Generation, Quality Control, Annotation, and Analysis of WGS Data
Genomic DNA were sequenced at the Broad Institute, Human Genome Sequencing Center at the Baylor College of Medicine, and McDonnell Genome Institute at Washington University. Reads were mapped to the GRCh37 reference genome assembly with the Burrows-Wheeler Aligner.34 BAM files from all three sequencing centers were collected, and genotype calling and primary quality control (QC) were performed by both the Broad Institute (Broad pipeline) and the Baylor College of Medicine Human Genome Sequencing Center (Baylor pipeline). The Broad and Baylor pipelines used the Genome Analysis Toolkit HaplotypeCaller35 and Atlas2 software,36 respectively, for genotype calling.
For the Broad pipeline, those variants that did not “pass” Variant Quality Score Recalibration were removed. For the Baylor pipeline, variants with a mapping score < 0.80 were deleted, and genotypes that did not “pass” the Sample Genotype Filter or that had a read depth < 10 or an out-of-range ratio of variant reads to total read depth (≤0.75 or ≥0.25) were deleted. For both pipelines, the following types of variants were excluded: monomorphic variants, those with a call rate ≤ 80%, those with excessive heterozygosity, and those with an average mean read depth > 500.
Once primary QC was completed for the Broad and Baylor pipelines, consensus genotypes were determined by keeping concordant variants and excluding variants in which a different alternative allele was called between the two pipelines. After consensus calling, a second round of variant-level QC was applied to remove any variants that were monomorphic, had >20% missing genotypes, or had an excessive number of heterozygous genotypes. QC was performed by the QC working group of the ADSP.
We used the RefGene database to select variants located in exon regions and included only single-nucleotide variants (SNVs) within the autosomal exome coding region in our analysis. Mendelian inconsistencies were identified and removed with PLINK software.37 Gene regions were assigned according to RefSeq definitions, and ANNOVAR was used to annotate variant sites.38 Variants within regions containing copy-number variants or pseudogenes were excluded, and variants that were either nonsynonymous or putative splice sites were included in the analysis. Only variants that were absent or had a MAF ≤ 2% in the ExAC Browser were analyzed. We used Variant Association Tools (VAT) to perform the variant-selection procedures described above.39 Only genes with at least three variant sites were analyzed, leaving 8,891 genes for analysis.
Results
Type I Error Rate
When the family data were generated under the null hypothesis of no association with ExAC non-Finish European variant information, type I error of the RV-GDT was well controlled for the family structures shown in Figure 1 and also for the mixed family structures. Type I error rates were evaluated at α = 0.05 and α = 0.005, the results of which are shown in Table 1. Additionally, the quantile-quantile plot of the −log10p values demonstrates that type I error was well controlled (Figure S1). The type I error was also well controlled for RV-PDT for all family structures (data not shown). When founders had a 50% probability of missing all of their genotype data, the RV-GDT method still had proper control of type I error rates for all pedigree and mixed family structures (Table 1 and Figure S1).
Table 1.
Type I Error Rate for RV-GDT at α Levels of 0.05 and 0.005
|
Discordant Sib-Pair |
Affected Sib-Pair |
Extended Pedigree |
Mixed Family Types |
|||||
|---|---|---|---|---|---|---|---|---|
| α = 0.05 | α = 0.005 | α = 0.05 | α = 0.005 | α = 0.05 | α = 0.005 | α = 0.05 | α = 0.005 | |
| RV-GDT | 0.047 | 0.0048 | 0.050 | 0.0051 | 0.051 | 0.0049 | 0.051 | 0.0048 |
| Each Founder Has a 50% Probability of Missing All Genotype Data | ||||||||
| RV-GDT | 0.051 | 0.0050 | 0.051 | 0.0049 | 0.049 | 0.0047 | 0.051 | 0.0047 |
| 80% African and 20% European Population Admixture | ||||||||
| RV-GDT | 0.051 | 0.0049 | 0.051 | 0.0048 | 0.048 | 0.0047 | 0.050 | 0.0051 |
| 50% African and 50% European Families | ||||||||
| RV-GDT | 0.050 | 0.0051 | 0.049 | 0.0053 | 0.048 | 0.0052 | 0.051 | 0.0048 |
We simulated 1,000 families for each pedigree structure shown in Figure 1 and mixed pedigree structures. Genotype data were generated for all autosomal genes across the genome with an OR of 1.0, and type I error rate was defined as the proportion of genes with a p value less than 0.05 or 0.005. We used variant information for 17,987 autosomal genes from the ExAC non-Finnish European population to generate family data when each founder had a 0% or 50% probability of missing genotype data. We used variant information for 17,873 autosomal genes present in both the ExAC non-Finnish European population and African and African American populations to generate family data with population admixture and substructure.
To demonstrate that the RV-GDT can adequately control for population admixture, we generated data for pedigrees with 80% African and 20% European admixture. We also evaluated whether type I error was well controlled in the presence of European-African substructure by simulating and analyzing data with 50% European pedigrees and 50% African pedigrees. For both scenarios, type I error was well controlled (Table 1 and Figure S1), suggesting that the RV-GDT is robust to population admixture and substructure.
We also evaluated the type I error rates of RareIBD by using simulated family data, and the results are shown in Table S1. We observed inflated type I error rates for discordant sib-pairs and extended pedigrees in each scenario evaluated (i.e., non-Finnish European population, missing founder data, African-European substructure, and African-European admixture) and deflated type I error rates for affected sib-pairs. For example, when data were generated with substructure (50% African and 50% European) and analyzed, the discordant sib-pairs and extended pedigrees had type I error rates of 0.063 and 0.477, respectively, at α = 0.05, whereas the affected sib-pairs had a type I error rate of 0.030.
Power Evaluation
We compared the power of RV-GDT with that of Epstein’s ASP method, RV-PDT, and FBAT for a variety of pedigree structures (Figure 1) when 75% of the rare variants were causal with an OR of 2.5. Additionally, we evaluated the effect of missing founder data on power by using different probabilities (0%, 25%, and 50%) to determine whether a founder was missing genotype data. The power of RV-GDT and other methods when founders were missing genotype data is shown in Table 2. When none of the founders were missing data, the differences in power between RV-GDT, RV-PDT, and FBAT were small for nuclear families (discordant and affected sib-pairs), but all methods were considerably more powerful than Epstein’s ASP method. When 1,000 discordant sib-pairs were analyzed, the power of FBAT, RV-PDT, and RV-GDT was 0.46, 0.51, and 0.53, respectively, whereas Epstein’s ASP method was unable to analyze these data because there were no affected sibships. When 1,000 affected sib-pairs were analyzed, the power of FBAT, RV-PDT, and RV-GDT was 0.80, 0.79, and 0.81, respectively, whereas the power of Epstein’s ASP method was 0.24. However, RV-GDT had considerably higher power than the other methods, i.e., FBAT and RV-PDT, which can analyze extended pedigrees. When 1,000 extended pedigrees were analyzed, the power of FBAT and RV-PDT was 0.42 and 0.48, respectively, whereas the power of RV-GDT was 0.64. Moreover, the power of RV-GDT was higher than that of the other methods when founders were missing genotype data. When ∼25% of the founders were missing their genotype data, the power of FBAT, RV-PDT, and RV-GDT for 1,000 extended pedigrees was 0.38, 0.45, and 0.62, respectively; when the missing probability was increased to 50%, the power of FBAT and RV-PDT was 0.34 and 0.41, respectively, whereas RV-GDT still had a power of 0.60.
Table 2.
Power Comparison of Epstein’s ASP Method, RV-PDT, FBAT, and RV-GDT when Founders Are Missing Different Proportions of Genotype Data
| Method |
Discordant Sib-Pair |
Affected Sib-Pair |
Extended Pedigree |
Mixed Family Types |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0%a | 25%a | 50%a | 0% | 25% | 50% | 0% | 25% | 50% | 0% | 25% | 50% | |
| Epstein’s ASPb | – | – | – | 0.23 | 0.23 | 0.23 | – | – | – | 0.10 | 0.10 | 0.10 |
| FBAT | 0.46 | 0.40 | 0.35 | 0.80 | 0.71 | 0.54 | 0.42 | 0.38 | 0.34 | 0.61 | 0.52 | 0.38 |
| RV-PDT | 0.51 | 0.45 | 0.42 | 0.79 | 0.70 | 0.52 | 0.48 | 0.45 | 0.41 | 0.63 | 0.55 | 0.43 |
| RV-GDT | 0.53 | 0.48 | 0.45 | 0.81 | 0.75 | 0.62 | 0.64 | 0.62 | 0.60 | 0.66 | 0.62 | 0.56 |
Genetic variant data were generated for 1,000 families of each pedigree structure shown in Figure 1 and mixed pedigree structures with the use of ExAC non-Finnish European variant information. Genotype data were generated for 17,987 autosomal genes across the genome when 75% of the rare nonsense, missense, and splice-site variants were randomly selected to be causal with an OR of 2.5, and power was evaluated as the proportion of genes with a p value < 0.05.
Probability that each founder is missing all genotype data.
Power was evaluated under the assumption that the exact IBD sharing between affected sib-pairs is known. Unknown IBD sharing and non-simulated data would reduce the power.
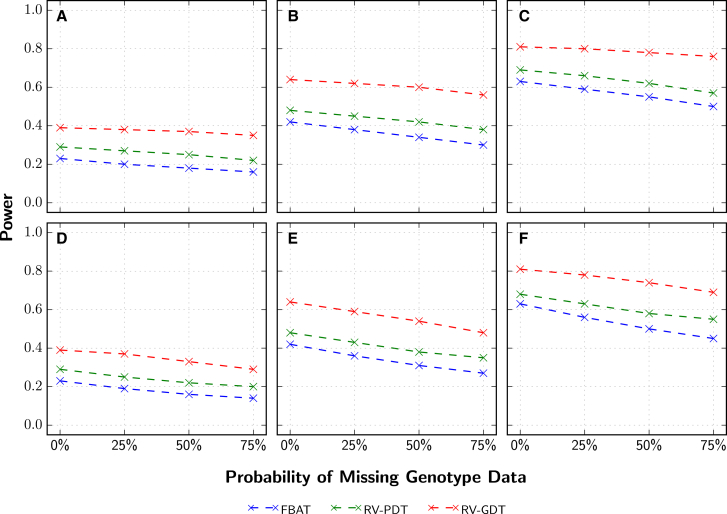
To further evaluate the power of RV-GDT when family members are missing genotype data, we simulated 1,000 extended pedigrees under different proportions of causal variants (50%, 75%, and 100%) with an OR of 2.5. When none of the pedigree members were missing genotype data, the power of the RV-GDT was higher than that of RV-PDT and FBAT (Figures 2A–2C). For example, when 100% of the rare nonsense, missense, and splice-site variants were causal and none of the pedigree members were missing genotype data, the power of FBAT, RV-PDT, and RV-GDT was 0.63, 0.69, and 0.81, respectively (Figure 2C). When founders had a 25%, 50%, or 75% probability of missing genotype data, the power decreased for each method as the percentage of founders missing genotype data increased (Figures 2A–2C). However, the RV-GDT still had considerably higher power than the other methods, not only because its initial power was higher than that of the other methods but also because it lost less power as the percentage of founders missing genotype data increased. For example, when 100% of the rare variants were causal and the probability that founders were missing genotype data was increased from 0 to 50% (Figure 2C), the FBAT and RV-PDT power was reduced by 12.70% (from 0.63 to 0.55) and 10.14% (from 0.69 to 0.62), respectively, whereas RV-GDT had a 3.84% (from 0.81 to 0.78) loss of power. Similar patterns of decreasing power were observed when family members in the first two generations had a 25%, 50%, or 75% probability of missing all of their genotype data, regardless of whether they were founders or non-founders (Figures 2D–2F). For the model in which 100% of the rare variants were causal (Figure 2F), the power of the FBAT and RV-PDT was reduced by 20.63% (from 0.63 to 0.50) and 15.94% (from 0.69 to 0.58), respectively, when the probability of individuals missing their genotype data was increased from 0% to 50%, whereas the power for RV-GDT was reduced by 8.64% (from 0.81 to 0.74).
Figure 2.
Power Comparisons of FBAT, RV-PDT, and RV-GDT for Extended Pedigrees with Family Members Missing Genotype Data
Genetic variant data were generated for 1,000 extended pedigrees with ExAC non-Finnish European variant information. Different proportions of the rare nonsense, missense, and splice-site variants were deemed to be causal: 50% (A and D), 75% (B and E), and 100% (C and F) with an OR of 2.5.
(A–C) Power comparisons when the probability that each founder was missing all genotype data ranged from 0% to 75%.
(D–F) Power comparisons when the probability that each parent (founder or non-founder) was missing all genotype data ranged from 0% to 75%.
Application to AD Data
We applied the RV-GDT method to analyze WGS data from the ADSP dataset. All pedigrees have at least one parental family member who is missing WGS data. Given the small sample size, application of the RV-GDT did not detect associations with exome-wide significance of 2.50 × 10−6 (Bonferroni correction for 20,000 genes). The most significant associations with AD were observed for MARCH10 (MIM: 613337; GenBank: NM_001100875.1; p value = 5.0 × 10−5), AMBN (MIM: 601259; GenBank: NM_016519.5; p value = 9.0 × 10−5), TCOF1 (MIM: 606847; GenBank: NM_000356.3; p value = 2.0 × 10−4), AXIN1 (p value = 2.5 × 10−4), and TNK1 (p value = 6.0 × 10−4). The ExAC MAFs of the variants within these genes, along with annotations from dbNSFP (version 2.9)40 (which include GERP,41 PhyloP,42 and CADD31 scores and Functional Analysis through Hidden Markov Models [fathmm],43 MutationTaster,44 PolyPhen-2,45 PROVEAN,46 and SIFT47 prediction), are shown in Table S3–S7.
Eight missense variants were observed in MARCH10 (Table S3). 53 alternative alleles were observed in family members with AD, whereas six were observed in unaffected individuals. Except for SNV rs13801568, which had the same number of alternative alleles in affected and unaffected family members, all other variant sites had higher alternative-allele counts in affected subjects than in unaffected family members. Five SNVs occurred at conserved nucleotides, and two SNVs were deemed to be deleterious by at least three of six bioinformatics tools (CADD, fathmm, MutationTaster, PolyPhen-2, PROVEAN, and SIFT). Eight nonsynonymous variants were observed in AMBN, and 41 and 4 alternative alleles of these variants were observed in affected and unaffected family members, respectively (Table S4). All SNVs except rs150017698 had a higher number of alternative alleles in affected family members than in unaffected family members. Four variants in AMBN were deemed to be conserved by both GERP and PhyloP and were predicted to be deleterious by at least three of six bioinformatics tools. 19 missense variants were observed in TCOF1—78 and 12 alternative alleles were observed in affected and unaffected family members, respectively (Table S5). Although only four variants were deemed to be conserved by both GERP and PhyloP, 12 variants were judged to be deleterious by at least three of six bioinformatics tools. AXIN1 included eight nonsynonymous variants; 34 alternative alleles were observed in affected family members, and one alternative allele was observed in an unaffected family member, and all variants had a higher number of alternative alleles in affected family members than in unaffected family members (Table S6). Six variants occurred at conserved nucleotides; however, none of the eight variants were deemed to be deleterious by at least three of six bioinformatics tools. Of the six variants observed in TNK1, 21 alternative alleles were observed in affected family members, and no alternative alleles were observed in unaffected family members (Table S7). Four variants were deemed conserved by both GERP and PhyloP, and four variants were deemed to be deleterious by at least three of six bioinformatics tools. rs201180891 occurs at a highly conserved residue and is predicted to be deleterious in all available bioinformatics results, and four affected family members were observed to be carriers of the alternative allele of this variant. The variant c.923T>A (p.Met308Lys) (GenBank: NM_003985.3) was not found in ExAC samples, and two individuals affected by AD are carriers of an alternative allele. This variant also occurs at a highly conserved residue and is predicted to be deleterious by five of six bioinformatics tools.
Discussion
We extended the family-based GDT to allow for the analysis of rare variants so that the method could be applied to association analysis of WES or exome sequence data. Our simulation studies demonstrated that the RV-GDT has well-controlled type I error rates, even when applied to admixed populations or populations with substructure. The RV-GDT has greater power than other family-based rare-variant association methods and is substantially more powerful when applied to extended pedigrees and/or pedigrees in which family members are missing genotype data. There are advantages to performing family-based association studies over employing population-based designs. Family-based studies can have higher power given an equivalent number of cases because they can involve more pathogenic susceptibility variants with larger effect sizes than those observed for sporadic disease.3 Additionally, many family-based association methods can control for population admixture and substructure on a local level, whereas for population-based designs, the inclusion of principal or multi-dimensional scaling components can control for population admixture and substructure only on a global level, which might not be sufficient for rare-variant association studies.48 However, family-based designs do have their drawbacks; compared with population-based studies, they require more resources for the recruitment of probands and their relatives. For family-based designs, genotype data are often missing because of unascertainable family members, e.g., non-paternity and deceased parents from late-onset disease. Usually, family data are composed of many different types of pedigree structures, and there are family members without genotype data, as observed in the 81 AD-affected families analyzed here (see Figure S2). The ability to analyze family data, including extended pedigrees and/or pedigrees in which family members are missing genotypes data, with minimal loss of power makes the RV-GDT an extremely valuable method for detecting associations and elucidating the genetic etiology of complex traits.
RareIBD has a main assumption that only one founder in each family carries a mutation for a specific rare variant. Violation of this assumption will result in an inflated test statistic.13 Complex traits, such as AD and coronary heart disease, have relatively high prevalences; therefore, a pedigree might have multiple affected individuals who do not have the same causal variants. Unlike for Mendelian diseases, the assumption that only one founder in a family carries the pathogenic susceptibility variant might not be valid for complex traits. Despite the fact that RareIBD (version 1.1) excludes variants violating this assumption, our simulations showed extremely inflated type I error rates for extended pedigrees. Our simulation framework is based on ExAC variant information for all genes, which represents exome sequence data more realistically than generating data for a single genomic region by using a population demographic model (the latter was previously used for evaluating type I error of RareIBD13).We also evaluated the power of RareIBD when 75% of rare nonsense, missense, and splice-site variants were randomly selected to be causal with an OR of 2.5. Although type I error was slightly inflated, the power of RareIBD was 0.32 when 1,000 discordant sib-pairs were analyzed, whereas the power of FBAT, RV-PDT, and RV-GDT was 0.46, 0.51, and 0.53, respectively. When 1,000 affected sib-pairs were analyzed, the power of RareIBD was 0.79, comparable to that of FBAT (0.80), RV-PDT (0.79), and RV-GDT (0.81). We did not evaluate power for extended pedigrees because it would not be valid given the extremely inflated type I error rates for RareIBD.
RareIBD (version 1.1) can analyze only families from single populations because of differences in the allelic spectrum between populations. Families of different ancestries need to be analyzed separately, and meta-analysis needs to be performed, which can lead to a loss of power. Genetic studies of complex traits are often composed of families ascertained from multiple populations. For example, ADSP includes both families of European descent and African American and Dominican families. Even the analysis of families of European descent can still be problematic, given that for rare variants, the allelic spectrum can differ greatly even between adjacent populations, e.g., Ashkenazi and other Eastern European populations.49
Neither RV-GDT nor FBAT requires haplotyping, IBD-sharing estimation, or imputation of missing genotypes, which avoids the potential decrease in power due to loss of information and/or inclusion of noise. For Epstein’s ASP method, IBD sharing between siblings must be estimated before statistical analysis. RV-PDT requires haplotype information in order to perform haplotype permutation, which is necessary to control type I error in the presence of LD between variants.7 Even though some algorithms can perform haplotyping and/or IBD-sharing estimation with acceptable accuracy, the potential loss of information and inclusion of noise can greatly jeopardize power. In our power evaluation of Epstein’s ASP method, we used exact IBD sharing in the simulated data. There would be a loss of power if the IBD-sharing information were inferred, for example, via MERLIN. Especially when founders are missing, it would introduce more uncertainties and lead to a further decrease in power. RareIBD requires family data without missing genotypes. Missing genotypes are imputed, and the most likely genotypes are analyzed. Although family-based imputation can reach relatively high accuracy, association testing with the most likely imputed genotype can lead to type I error inflation.50 More experiments are needed to investigate how imputed genotypes can be correctly incorporated in family-based association studies.
Adjustment for non-confounding covariates that are known to influence the trait can reduce spurious associations due to sampling artifacts or biases in study design.51 However, caution should be exercised for decisions about whether to incorporate covariates in the association analysis of binary traits. It has been shown for GWASs that including known covariates can reduce the power to identify associated variants when the disease prevalence is low. On the other hand, including non-confounding predictive covariates when disease prevalence is sufficiently high (>20%) will often lead to an increase in power.52 The RV-GDT can incorporate covariates in the analysis, but it does not provide an evaluation of covariate significance and also cannot be used for covariate selection. The FBAT can also incorporate covariates, whereas Epstein’s ASP method and RV-PDT have not been extended to adjust for covariates.
It was previously shown that integrating information on variant allele frequencies from population-based data into family-based studies can be useful for association studies of rare variants.53 Jiang et al. suggested incorporating population-control-based weights into the TDT framework to potentially up-weight pathogenic susceptibility variants, down-weight neutral variants, and also assign the direction of the effect for pathogenic variants.8 In our simulation, no improvement in power was observed for either RV-GDT or RV-PDT when weights were incorporated with data from population controls. In fact, the incorporation of weights from population controls led to slightly less power than not using any weights. Genetic data for 1,000 extended pedigrees were simulated with ExAC non-Finnish European variant information, and 75% of the rare nonsense, missense, and splice-site variants were randomly selected to be causal with an OR of 2.5. We also generated 20,000 population controls, and the weights inferred the control data, as suggested by Jiang et al. The power of the RV-GDT decreased from 0.81 to 0.78, and the power of the RV-PDT also decreased from 0.79 to 0.77. This is not surprising given that it has previously been shown that decreases in power can occur when weights are not optimal.54 In our simulations, the controls were generated from the same population as the family data, but because of random variability, they were not always optimal. The reduction in power could be even greater if controls are drawn from a different population. Moreover, how to handle variants that are not present in population controls is a practical problem that needs to be addressed. For example, 10.41% of variants analyzed in the AD pedigrees are not present in the ExAC Browser, which is one of the largest publicly available databases.
The application of the RV-GDT in the analysis of AD pedigrees highlights its applicability to family-based studies. The pedigree structures of this dataset are highly heterozygous, and each pedigree has at least one family member who has not been sequenced (see Figure S2). RV-GDT has fewer constraints on pedigree structure than Epstein’s ASP method and RV-PDT, and it can analyze any pedigree as long as it includes both affected and unaffected subjects. FBAT can analyze most pedigree structures, but those with missing parental data often cannot be analyzed, especially when there are no unaffected offspring. Epstein’s ASP method is only applicable to analyzing affected sibships in nuclear pedigrees, and it needs the estimated IBD sharing between sibships, which is problematic for pedigrees missing parental genotypes. Without parental genotypes, IBD sharing must be estimated from identity by state (IBS) sharing and variant allele frequencies. It is unclear how well Epstein’s ASP method performs in this situation. The majority of affected sibships in the AD pedigrees are missing both parental genotypes, and no single affected sibship has genotype data for both parents. The RV-PDT requires nuclear families with informative case-parent trios and/or discordant sib-pairs to detect association, but the haplotype permutation is necessary for the RV-PDT to control type I error,7 and the haplotype permutation needs the complete nuclear family or reconstructed haplotypes for the missing founders. No single AD pedigree has complete parental WGS data, and the RV-PDT hasn’t been extended to analyze family data with reconstructed haplotypes; therefore, it is not possible to analyze the AD pedigrees with the RV-PDT. The AD data could not be analyzed with RareIBD given the extreme inflation of type I error for extended pedigrees. Of the 81 AD-affected families, 25 cannot be analyzed by the FBAT (see Figure S2). Moreover, the FBAT returned “NaN” (not a number) when the genotype data for TCOF1 in the AD data were analyzed. The p values of FBAT for MARCH10, AMBN, AXIN1, and TNK1 were 0.06, 0.30, 0.04, and 0.04, respectively, and all p values were considerably less significant than those obtained from the RV-GDT.
The application of the RV-GDT on WGS data from 81 AD-affected families identified potential involvement of AXIN1 (16p13.3) and TNK1 (17p13.1) in this neurodegenerative disease. Previously, a SNP-based association study detected an association between rs1554948, which is within TNK1, and late-onset AD.19 Our study implicates multiple rare variants in TNK1 as a potential underlying cause of AD. We observed 21 alternative alleles in 21 affected family members and no alternative alleles in unaffected family members. Out of 21 family members carrying TNK1 alternative alleles, only two of them were APOE4 positive. The association between rare variants in TNK1 and AD is consistent with functional studies of its protein. TNK1 encodes a non-receptor tyrosine kinase and mediates intracellular signaling. Activated TNK1 has been reported to facilitate tumor necrosis factor alpha (TNFα)-induced apoptosis, which suggests its involvement in TNFα signaling and neuronal cell death.20 The involvement of TNK1 in AD pathogenesis could also be through its interaction with phospholipase C (PLC). TNK1 has been reported to be associated with PLC gamma 1,21 and multiple studies have observed aberrant PLC activity in AD brains.19 Identification of the involvement of TNK1 in AD etiology provides new insights that could be used for prevention and treatment. Although the association between AD and variants in AXIN1 has not been reported previously, the identification of AXIN1 is also consistent with experimental findings. AXIN1 encodes a scaffolding protein that plays a critical role in regulating GSK3-mediated phosphorylation of the protein tau.22 Axin negatively affects tau phosphorylation by GSK3,23 and phosphorylated tau has a decreased capacity to bind and stabilize microtubules.24 The abnormally phosphorylated tau has been observed in populations of AD-affected individuals at clinicopathological levels.25 These findings suggest that abnormal expression of AXIN1 might contribute to tau pathology in AD. No previous association studies have implicated MARCH10, AMBN, or TCOF1 in the etiology of AD. Additionally, no functional studies support the involvement of these genes in AD etiology. The associations between AD and MARCH10, AMBN, and TCOF1 might be false positives, and future replication studies will allow for the elucidation of whether these genes are involved in the etiology of AD.
The WGS data on the analyzed 81 families is part of the ADSP Discovery Phase, which will be followed by the Discovery Extension Phase and the Follow-Up Phase. The Discovery Extension Phase will include WGS data on 107 additional family members of the pedigrees studied in the Discovery Phase and additional WGS data on 213 family members from new pedigrees. The Follow-Up Phase will include more families undergoing whole-genome or exome sequencing, and individual investigators will share their sequence data with the ADSP. These additional sequence data will permit a replication study.
The RV-GDT method provides a robust and powerful way to use family-based sequence data to identify associations between complex disease and rare variants. Given its capability of adequately controlling for population admixture or substructure and its superior power over other methods for extended pedigrees and pedigrees with missing data, the RV-GDT is extremely beneficial in elucidating the involvement of rare variants in the etiology of complex traits. The RV-GDT is applicable to exome and genome sequence data and rare variants obtained from genotyping arrays. VAT is an all-in-one software pipeline package available for pre-processing data from various input formats, and the RV-GDT package is implemented to analyze rare-variant data exported by VAT. The RV-GDT is implemented in Python, and the software package and documentation are publicly available online.
Acknowledgments
We wish to thank the family members who participated in the Alzheimer Disease Sequencing Project and made this research possible. This work was supported by National Human Genome Research Institute grant R01 HG008972. Complete acknowledgments can be found in the Supplemental Acknowledgments.
Published: January 5, 2017
Footnotes
Supplemental Data include Supplemental Acknowledgments, two figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.12.001.
Web Resources
Epstein software, http://genetics.emory.edu/labs/epstein/software
ExAC Browser, http://exac.broadinstitute.org/
FreeBayes, https://github.com/ekg/freebayes
GeneReviews, Bird, T.D. (1993). Alzheimer Disease Overview, https://www.ncbi.nlm.nih.gov/books/NBK1161/
Genome Analysis Toolkit (GATK), https://www.broadinstitute.org/gatk/
OMIM, http://www.omim.org/
RV-GDT, https://statgen.research.bcm.edu/index.php/Main_Page#Statistical_Genetics_Software
Variant Association Tools (VAT), http://varianttools.sourceforge.net/Association/
Supplemental Data
References
- 1.Eichler E.E., Flint J., Gibson G., Kong A., Leal S.M., Moore J.H., Nadeau J.H. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cirulli E.T., Goldstein D.B. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat. Rev. Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 3.Ott J., Kamatani Y., Lathrop M. Family-based designs for genome-wide association studies. Nat. Rev. Genet. 2011;12:465–474. doi: 10.1038/nrg2989. [DOI] [PubMed] [Google Scholar]
- 4.Li M., Boehnke M., Abecasis G.R. Efficient study designs for test of genetic association using sibship data and unrelated cases and controls. Am. J. Hum. Genet. 2006;78:778–792. doi: 10.1086/503711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathieson I., McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat. Genet. 2012;44:243–246. doi: 10.1038/ng.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spielman R.S., McGinnis R.E., Ewens W.J. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am. J. Hum. Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 7.He Z., O’Roak B.J., Smith J.D., Wang G., Hooker S., Santos-Cortez R.L.P., Li B., Kan M., Krumm N., Nickerson D.A. Rare-variant extensions of the transmission disequilibrium test: application to autism exome sequence data. Am. J. Hum. Genet. 2014;94:33–46. doi: 10.1016/j.ajhg.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Y., Satten G.A., Han Y., Epstein M.P., Heinzen E.L., Goldstein D.B., Allen A.S. Utilizing population controls in rare-variant case-parent association tests. Am. J. Hum. Genet. 2014;94:845–853. doi: 10.1016/j.ajhg.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laird N.M., Horvath S., Xu X. Implementing a unified approach to family-based tests of association. Genet. Epidemiol. 2000;19(Suppl 1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 10.De G., Yip W.-K., Ionita-Laza I., Laird N. Rare variant analysis for family-based design. PLoS ONE. 2013;8:e48495. doi: 10.1371/journal.pone.0048495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ionita-Laza I., Lee S., Makarov V., Buxbaum J.D., Lin X. Family-based association tests for sequence data, and comparisons with population-based association tests. Eur. J. Hum. Genet. 2013;21:1158–1162. doi: 10.1038/ejhg.2012.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epstein M.P., Duncan R., Ware E.B., Jhun M.A., Bielak L.F., Zhao W., Smith J.A., Peyser P.A., Kardia S.L.R., Satten G.A. A statistical approach for rare-variant association testing in affected sibships. Am. J. Hum. Genet. 2015;96:543–554. doi: 10.1016/j.ajhg.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sul J.H., Cade B.E., Cho M.H., Qiao D., Silverman E.K., Redline S., Sunyaev S. Increasing Generality and Power of Rare-Variant Tests by Utilizing Extended Pedigrees. Am. J. Hum. Genet. 2016;99:846–859. doi: 10.1016/j.ajhg.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W.-M., Manichaikul A., Rich S.S. A generalized family-based association test for dichotomous traits. Am. J. Hum. Genet. 2009;85:364–376. doi: 10.1016/j.ajhg.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin E.R., Monks S.A., Warren L.L., Kaplan N.L. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am. J. Hum. Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin E.R., Bass M.P., Gilbert J.R., Pericak-Vance M.A., Hauser E.R. Genotype-based association test for general pedigrees: the genotype-PDT. Genet. Epidemiol. 2003;25:203–213. doi: 10.1002/gepi.10258. [DOI] [PubMed] [Google Scholar]
- 17.Van Cauwenberghe C., Van Broeckhoven C., Sleegers K. The genetic landscape of Alzheimer disease: clinical implications and perspectives. Genet. Med. 2016;18:421–430. doi: 10.1038/gim.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del-Aguila J.L., Koboldt D.C., Black K., Chasse R., Norton J., Wilson R.K., Cruchaga C. Alzheimer’s disease: rare variants with large effect sizes. Curr. Opin. Genet. Dev. 2015;33:49–55. doi: 10.1016/j.gde.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Grupe A., Abraham R., Li Y., Rowland C., Hollingworth P., Morgan A., Jehu L., Segurado R., Stone D., Schadt E. Evidence for novel susceptibility genes for late-onset Alzheimer’s disease from a genome-wide association study of putative functional variants. Hum. Mol. Genet. 2007;16:865–873. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- 20.Azoitei N., Brey A., Busch T., Fulda S., Adler G., Seufferlein T. Thirty-eight-negative kinase 1 (TNK1) facilitates TNFalpha-induced apoptosis by blocking NF-kappaB activation. Oncogene. 2007;26:6536–6545. doi: 10.1038/sj.onc.1210476. [DOI] [PubMed] [Google Scholar]
- 21.Felschow D.M., Civin C.I., Hoehn G.T. Characterization of the tyrosine kinase Tnk1 and its binding with phospholipase C-γ1. Biochem. Biophys. Res. Commun. 2000;273:294–301. doi: 10.1006/bbrc.2000.2887. [DOI] [PubMed] [Google Scholar]
- 22.Salahshor S., Woodgett J.R. The links between axin and carcinogenesis. J. Clin. Pathol. 2005;58:225–236. doi: 10.1136/jcp.2003.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoothoff W.H., Bailey C.D.C., Mi K., Lin S.-C., Johnson G.V.W. Axin negatively affects tau phosphorylation by glycogen synthase kinase 3beta. J. Neurochem. 2002;83:904–913. doi: 10.1046/j.1471-4159.2002.01197.x. [DOI] [PubMed] [Google Scholar]
- 24.Spittaels K., Van den Haute C., Van Dorpe J., Geerts H., Mercken M., Bruynseels K., Lasrado R., Vandezande K., Laenen I., Boon T. Glycogen synthase kinase-3beta phosphorylates protein tau and rescues the axonopathy in the central nervous system of human four-repeat tau transgenic mice. J. Biol. Chem. 2000;275:41340–41349. doi: 10.1074/jbc.M006219200. [DOI] [PubMed] [Google Scholar]
- 25.Kolarova M., García-Sierra F., Bartos A., Ricny J., Ripova D. Structure and pathology of tau protein in Alzheimer disease. Int. J. Alzheimers Dis. 2012;2012:731526. doi: 10.1155/2012/731526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorlov I.P., Gorlova O.Y., Sunyaev S.R., Spitz M.R., Amos C.I. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am. J. Hum. Genet. 2008;82:100–112. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auer P.L., Wang G., Leal S.M. Testing for rare variant associations in the presence of missing data. Genet. Epidemiol. 2013;37:529–538. doi: 10.1002/gepi.21736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madsen B.E., Browning S.R. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin D.Y., Tang Z.Z. A general framework for detecting disease associations with rare variants in sequencing studies. Am. J. Hum. Genet. 2011;89:354–367. doi: 10.1016/j.ajhg.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price A.L., Kryukov G.V., de Bakker P.I., Purcell S.M., Staples J., Wei L.J., Sunyaev S.R. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B., Wang G.T., Leal S.M. Generation of sequence-based data for pedigree-segregating Mendelian or Complex traits. Bioinformatics. 2015;31:3706–3708. doi: 10.1093/bioinformatics/btv412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Challis D., Yu J., Evani U.S., Jackson A.R., Paithankar S., Coarfa C., Milosavljevic A., Gibbs R.A., Yu F. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang G.T., Peng B., Leal S.M. Variant association tools for quality control and analysis of large-scale sequence and genotyping array data. Am. J. Hum. Genet. 2014;94:770–783. doi: 10.1016/j.ajhg.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X., Jian X., Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., NISC Comparative Sequencing Program Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shihab H.A., Gough J., Cooper D.N., Stenson P.D., Barker G.L.A., Edwards K.J., Day I.N.M., Gaunt T.R. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 45.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 48.Liu J., Lewinger J.P., Gilliland F.D., Gauderman W.J., Conti D.V. Confounding and heterogeneity in genetic association studies with admixed populations. Am. J. Epidemiol. 2013;177:351–360. doi: 10.1093/aje/kws234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atzmon G., Hao L., Pe’er I., Velez C., Pearlman A., Palamara P.F., Morrow B., Friedman E., Oddoux C., Burns E., Ostrer H. Abraham’s children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am. J. Hum. Genet. 2010;86:850–859. doi: 10.1016/j.ajhg.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y., Willer C., Sanna S., Abecasis G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bush W.S., Moore J.H. Chapter 11: Genome-wide association studies. PLoS Comput. Biol. 2012;8:e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pirinen M., Donnelly P., Spencer C.C.A. Including known covariates can reduce power to detect genetic effects in case-control studies. Nat. Genet. 2012;44:848–851. doi: 10.1038/ng.2346. [DOI] [PubMed] [Google Scholar]
- 53.He X., Sanders S.J., Liu L., De Rubeis S., Lim E.T., Sutcliffe J.S., Schellenberg G.D., Gibbs R.A., Daly M.J., Buxbaum J.D. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9:e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu D.J., Leal S.M. Estimating genetic effects and quantifying missing heritability explained by identified rare-variant associations. Am. J. Hum. Genet. 2012;91:585–596. doi: 10.1016/j.ajhg.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.