Abstract
Nephronophthisis (NPH), an autosomal-recessive tubulointerstitial nephritis, is the most common cause of hereditary end-stage renal disease in the first three decades of life. Since most NPH gene products (NPHP) function at the primary cilium, NPH is classified as a ciliopathy. We identified mutations in a candidate gene in eight individuals from five families presenting late-onset NPH with massive renal fibrosis. This gene encodes MAPKBP1, a poorly characterized scaffolding protein for JNK signaling. Immunofluorescence analyses showed that MAPKBP1 is not present at the primary cilium and that fibroblasts from affected individuals did not display ciliogenesis defects, indicating that MAPKBP1 may represent a new family of NPHP not involved in cilia-associated functions. Instead, MAPKBP1 is recruited to mitotic spindle poles (MSPs) during the early phases of mitosis where it colocalizes with its paralog WDR62, which plays a key role at MSP. Detected mutations compromise recruitment of MAPKBP1 to the MSP and/or its interaction with JNK2 or WDR62. Additionally, we show increased DNA damage response signaling in fibroblasts from affected individuals and upon knockdown of Mapkbp1 in murine cell lines, a phenotype previously associated with NPH. In conclusion, we identified mutations in MAPKBP1 as a genetic cause of juvenile or late-onset and cilia-independent NPH.
Keywords: nephronophthisis, ciliopathy, kidney, retinitis pigmentosa, digenism, mitotic spindle, MAP kinase, DNA damage, MAPKBP1, WDR62
Main Text
Nephronophthisis (NPH [MIM: 256100]) is an autosomal-recessive kidney disorder characterized by the development of massive interstitial fibrosis with abnormal thickness of the tubular basement membranes, atrophic and/or dilated tubules, and occasionally, formation of cysts mainly distributed at the cortico-medullary junction within normal sized or small kidneys. It is the main genetic cause of end-stage renal disease (ESRD) in the first two decades of life and three different forms have been clinically described based on the mean age at ESRD: infantile (<3 years), juvenile (mean 13 years), and late onset (mean 19 years).1
NPH can be either isolated or associated with different extra-renal manifestations (retinal dystrophy, liver fibrosis, skeleton dysplasia, etc.) in syndromic forms referred to hereafter as nephronophthisis-related ciliopathies (NPH-RCs). Indeed, the great majority of the 20 genes associated with NPH-RCs encode proteins (NPHP) that localize to primary cilia where they play key functions in the biogenesis of this organelle as well as in cilia-dependent signaling pathways.1, 2, 3 NPH has therefore been classified as a ciliopathy, in a growing family of genetic diseases linked to either primary and/or motile cilia dysfunctions.4 In addition, recent evidence established that NPH-RCs can also be linked to the DNA damage response (DDR) signaling pathway. Initially identified for NPHP14/ZNF423 (MIM: 614844) and NPHP15/CEP164 (MIM: 614848),5 enhanced DDR signaling was also associated with mutations in several other NPHP genes evidenced by mouse models of NPHP6/CEP2906 (MIM: 610142), NPHP9/NEK87, 8 (MIM: 613824), and NPHP10/SDCCAG89 (MIM: 613524). Altogether, these data suggest that increased DDR signaling may underlie progressive renal disease seen in NPH10 as well as a possible functional link between cilia and regulation of DDR signaling which remains to be uncovered. Except for ZNF423 (MIM: 604557), which was not implicated in direct ciliary function, all NPHPs that have been involved in DDR signaling do also play a crucial role at primary cilia.1, 10
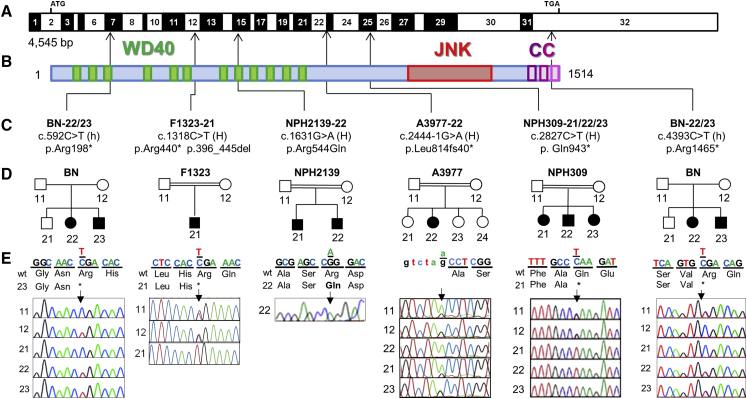
In order to identify additional genes mutated in NPH, we independently performed homozygosity mapping and/or whole-exome sequencing (WES) on affected individuals from our cohorts (Figure S1, Table S1). Blood samples and pedigrees were obtained from individuals with diagnosed NPH-RCs. Written informed consent was obtained from all individuals enrolled in this study and/or from parents and approved by the Comité de Protection des Personnes pour la Recherche Biomédicale Ile de France II, the Institutional Review Board (IRB) at the Boston Children’s Hospital (Boston), or the Regional Committee for Medical and Research Ethics, Western Norway (IRB no. 00001872). Affected individuals were included in WES upon either parental consanguinity or as member of a multiplex family. In total, WES was performed in 141 families, 85 with isolated NPH and 56 with syndromic NPH. Compound heterozygous nonsense (c.592C>T [p.Arg198∗]; c.4393C>T [p.Arg1465∗] in BN-23), homozygous nonsense (c.1318C>T [p.Arg440∗] in F1323-21; c.2827C>T [p.Gln943∗] in NPH309-21), as well as homozygous splice site mutations (c.2444−1G>A [p.Leu814fs40∗] in A3977-22) were identified in MAPKBP1 (GenBank: NM_001128608.1) in four affected individuals from four independent families (Figure 1 and Table 1). Additional competing biallelic missense variants were detected, but none of them are thought to possess an equally strong impact on the protein level, for either mild in silico prediction, occurrence in SNP databases (e.g., ExAC), or little evolutionary amino acid conservation (Table S2).
Figure 1.
Biallelic Mutations in MAPKBP1 Cause Nephronophthisis
(A) Exon structure of MAPKBP1 cDNA. Positions of start codon (ATG) and stop codon (TGA) are indicated.
(B) Domain structure of the protein. MAPKBP1 contains 12 WD repeats (green), located at the N terminus, a JNK-binding region located in the C-terminal part (red), and conserved coiled-coil domains at the C terminus (CC, purple).
(C) Relation of four homozygous (H) and two compound heterozygous mutations (h) to exons and protein domains is indicated by black arrows.
(D and E) Pedigrees (D) and chromatograms and segregation (E). Mutated nucleotides are shown above wild-type. In addition, individuals BN-21 and BN-23 also presented with retinitis pigmentosa (RP) linked to a homozygous mutation in PDE6A.
See Figure S5. Segregation of the MAPKBP1 mutations was not examined in family NPH2139 due to the unavailability of the DNA of the affected brother and the parents.
Table 1.
Mutations of MAPKBP1 in Five Families with Nephronophthisis
| Individuals | Ethnic Origin | Nucleotide Alterationsa | Deduced Protein Change | Exon, Zygosity | Parental Consanguinity | ESRD Years | RTX Years | Extrarenal Clinical Features |
|---|---|---|---|---|---|---|---|---|
| BN-22 | Norway | c.592C>T, c.4393C>T | p.Arg198∗, p.Arg1465∗ | 7, 32, Het | no | no (25) | – | loose patella, long fingers and feet |
| BN-23 | Norway | c.592C>T, c.4393C>T | p.Arg198∗, p.Arg1465∗ | 7, 32, Het | no | no (27) | – | RP,b CHD, severe meningomyelocele, statural growth delay |
| F1323-21 | Turkey | c.1318C>Tc, exon 12 skipping | p.Arg440∗, p.396_445del | 12, Hom | yes | 12 | 15 | facial dysmorphism |
| NPH2139-22 | Portugal | c.1631G>A | p.Arg544Glnd | 15, Hom | yes | no (23) | – | none |
| A3977-22 | Turkey | c.2444-1G>A, 5′ splice site | 5′ splice site | 22, Hom | yes | 15 | – | short stature |
| NPH309-21 | Italy | c.2827C>T | p.Gln943∗ | 25, Hom | yes | 22 | 22,39 | scoliosis |
| NPH309-22 | Italy | c.2827C>T | p.Gln943∗ | 25, Hom | yes | 25 | 27,38 | scoliosis, vesiculo-ureteral reflux, cholesteatoma, amyloid angiopathy |
| NPH309-23 | Italy | c.2827C>T | p.Gln943∗ | 25, Hom | yes | 20 | 21 | scoliosis, mild mental retardation, short stature, palate cleft |
Abbreviations are as follows: ESRD, end-stage renal disease; CHD, congenital heart disease; Het, heterozygous; Hom, homozygous; NPH, nephronophthisis; RP, retinitis pigmentosa; RTX, renal transplantation.
cDNA mutations are numbered according to human cDNA reference sequence GenBank: NM_001128608.1, isoform b (MAPKBP1), where +1 corresponds to the A of ATG start translation codon.
Individuals BN-23 and BN-21 also present with RP linked to homozygous mutation in PDE6A (Figure S5).
rs202001274: variant is listed in ExAC database, 2 of 121,258 are heterozygote. None of the others are currently listed.
PolyPhen-2 score, 1; MutationTaster, disease causing; SIFT deleterious/score: 0.
Additional screening of 342 unrelated affected individuals (108 with isolated NPH and 234 with syndromic NPH) using targeted exome sequencing2, 11 led to the identification in an affected individual (NPH2139-22) presenting late-onset isolated NPH of a homozygous missense variant (c.1631G>A [p.Arg544Gln]) never reported in ExAC database and predicted as damaging (PolyPhen-2 score, 1; SIFT, deleterious/score: 0; MutationTaster, disease causing). Interestingly, the Arg544 is part of a stretch of 12 aa that is highly conserved (almost identical) in both MAPKBP1 and WDR62 proteins from humans to Drosophila (Figure S2A). Furthermore, 3D structure analysis on equivalent residues in other WD40 domain-containing proteins indicates that Arg544 is located in a conserved site involved in the interaction of the WD40 domain with its partner(s) (Figures S2B and S2C), suggesting that the p.Arg544Gln variation may alter the binding of MAPKBP1 with potential and not yet identified partners.
Segregation of the identified mutations could be examined by Sanger sequencing for all the families except NPH2139. It showed that parents and unaffected siblings were heterozygous for the identified mutations (BN, F1323, NPH309, A3977) and that affected siblings were also presenting corresponding biallelic mutations in MAPKBP1 (BN, NPH309; Figure 1 and Table 1). We consequently identified mutations in MAPKBP1 in eight individuals from five families. They almost all presented with late-onset NPH (ESRD from 15 to >27 years) or juvenile NPH (for individual F1323-21 who presented ESRD at 12 years) (Table 1). Renal biopsies and ultrasounds from affected individuals (Figure 2) showed classical features of NPH characterized by atrophic tubules with thickening of the basement membranes, massive interstitial fibrosis, as well as the presence of interstitial infiltrate (BN-23 shown in Figure 2A, panels 1 and 2; NPH2139-21 shown in Figure 2C, panel 2; A3977-22 shown in Figure 2D, panel 1) as well as increased echogenicity and cysts (Figure 2D, panels 2 and 3).
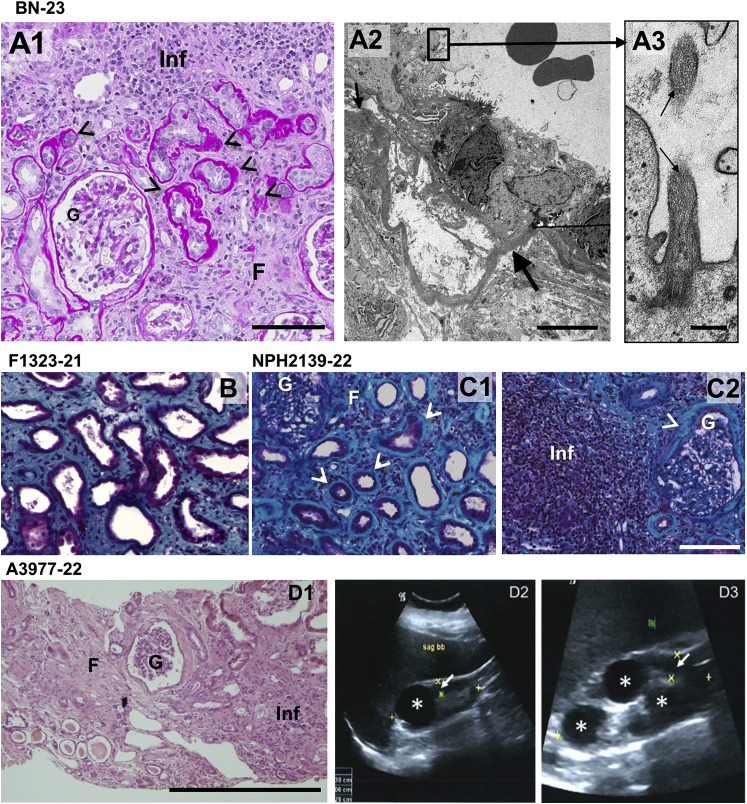
Figure 2.
Histological and Sonographic Kidney Lesions in Individuals with MAPKBP1 Mutations
(A1) Periodic acid-Schiff staining of a kidney biopsy from individual BN-23 showing tubular atrophy (arrowheads), interstitial fibrosis (F), focal interstitial infiltration (Inf), and normal glomerulus (G). Scale bar represents 100 μm.
(A2) Ultrastructure (transmission electron microscopy picture) from a tubule with abrupt transition from thin to thickened and disorganized basement membrane (arrow). Scale bar represents 5 μm.
(A3) Example of a primary cilium found at the apical membrane of a tubular epithelial cell (arrows). Scale bar represents 250 μm.
(B) Trichrome staining of a kidney biopsy from individual F1323-21 showing fibrosis, thickened basal membrane, as well as atrophic and dilated tubules.
(C) PAS trichrome staining of a kidney biopsy from individual NPH2139-21 showing tubular atrophy (arrowheads in C1), interstitial fibrosis (F) and focal interstitial infiltration (Inf, C2), and sclerotic glomeruli (G, C2) surrounded by a thickened capsular basement membrane (arrows, C1 and C2). Scale bar represents 100 μm.
(D1) Histological analysis of individual A3977-22 shows massive interstitial fibrosis and atrophic tubules. Scale bar represents 100 μm.
(D2 and D3) Upon renal ultrasound, kidneys present small with narrowed parenchyma (white arrow), increased echogenicity, and single cysts on both sides (white asterisk). D2 right kidney, D3 left kidney.
Among the three nonsense mutations, the homozygous c.1318C>T (p.Arg440∗) mutation in individual F1323-21 was investigated more closely. Indeed, in silico analysis of MAPKBP1 transcripts revealed an alternative splicing leading to the skipping of exon 12 (NCBI; GenBank: XM_011521383.1). This alternative transcript is predicted to lead to an in-frame deletion and a 1,475 amino acid protein (versus 1,514 for full-length MAPKBP1). Because c.1318C>T (p.Arg440∗) was located in the exon 12 (Figure S3A), we next analyzed the expression and ratio of the various transcripts in individual F1323-21. Amplification of the exon 11–13 region by RT-PCR from F1323-21 fibroblasts revealed the presence of two products: one product also detected in the unrelated affected and control individuals (BN-22 and BN-12) and a shorter product (Figure S3B). Sanger sequencing analysis confirmed that the longer product corresponds to the full-length exon 12 containing transcript presenting the c.1318C>T nonsense mutation while the shorter band corresponds to a transcript that effectively lacks exon 12 (Figures S2B and S2C). The high amount of transcript lacking exon 12 in individual F1323-21 may possibly be explained by the fact that the c.1318C>T mutation is predicted to affect the binding of serine-arginine-rich protein SF2/ASF to putative exonic splicing enhancers (ESEfinder 3.0;12 Figures S4A and S4B), which is then expected to affect the recruitment of splicing factors to splice sites and to lead to the skipping of exon 12 (Figure S4C). Skipping of exon 12 results in an in-frame deletion of 49 amino acids (aa 396 to 445) encompassing the sixth WD repeat out of the seven predicted to form a WD domain (Figure S3D). The resulting Δexon12 V5-tagged construct was poorly expressed upon transient transfection, with a migration profile in western blot experiments different from what was expected (Figure S3E, arrow), suggesting that the lack of exon 12 likely leads to misfolding of the N-terminal WD domain and decreased stability of the resulting protein. In conclusion, individual F1323-21 expressed two MAPKPB1 transcripts, one coding for the expected p.Arg440∗ protein and a shorter spliced form lacking exon 12, both of these proteins being likely non- or poorly functional (see below).
NPH in the affected individuals was not associated with other classically found extra-renal manifestations of NPH-RCs, except retinitis pigmentosa (RP [MIM: 613810]) found in two out of three siblings of the BN family, suggesting Senior-Loken syndrome (SLS).1 However, only individual BN-23 presented with both RP and NPH whereas his siblings presented with either NPH (BN-22) or RP (BN-21). Interestingly, while MAPKBP1 mutations segregated only with NPH but not with RP, WES of individual BN-23 revealed the co-occurrence of a homozygous mutation in PDE6A (c.2053G>A [p.Val685Met]), previously associated with RP13 (MIM: 180071), that segregated with RP in affected siblings but was not found in individual BN-22 (Figure S5). Altogether, these results show that RP and NPH in the BN siblings are caused by biallelic mutations in two different genes and that mutations in MAPKBP1 are associated with NPH without RP as observed in individual BN-22. Additional manifestations were observed in several affected individuals from different families including facial dysmorphism (F1323-21 and NPH309-23) and/or scoliosis (NPH309-21, -22, -23). Finally, congenital heart disease and meningomyelocele were observed in a BN family member (BN-23). It remains speculative at this point whether defects of MAPKBP1 are truly related to these conditions since none of them were observed in animal models (see below).
MAP-kinase binding protein 1 (MAPKBP1, also known as JNKBP1) is the paralog of WDR62,14 the product of a gene mutated in primary microcephaly (MCPH2 [MIM: 604317]),15, 16 which localizes at the mitotic spindle poles (MSPs) and has been involved in the orientation of the mitotic spindle.17 MAPKBP1 and WDR62 share the same structural organization with their N-terminal half characterized by the presence of WD40 repeats (12 for MAPKBP1, see Figure 1B). In addition, MAPKBP1,18 as well as WDR62,19 binds JNK family members through their C-terminal domain (aa 1,063–1,331 for MAPKBP1). It was shown to act as a scaffolding protein for JNK as well as for other signaling pathways.18, 20, 21 Finally, MAPKBP1 and WDR62 also share a highly conserved C-terminal coiled-coil containing region that is required for WDR62 homodimerization and some evidence indicates that it might also be involved in MAPKBP1-WDR62 heterodimerization.22 However, despite their similarity, no functional relationship has been characterized between the two proteins.
Expression of endogenous MAPKBP1 was tested in vitro and in vivo, using a commercial rabbit polyclonal antibody against MAPKBP1 that we validated by western blot and immunofluorescence experiments (Figures S6A–S6C). Analysis of the expression of MAPKBP1 in fibroblasts from both control subjects and affected individuals using the validated antibody led to the expected results (Figure S6D) with a slightly shorter protein for BN-22 (p.Arg1465∗) and no detectable protein for NPH309-21 (p.Gln943∗, loss of the epitope) and F1323-21 (p.Arg440∗, see Figure S3).
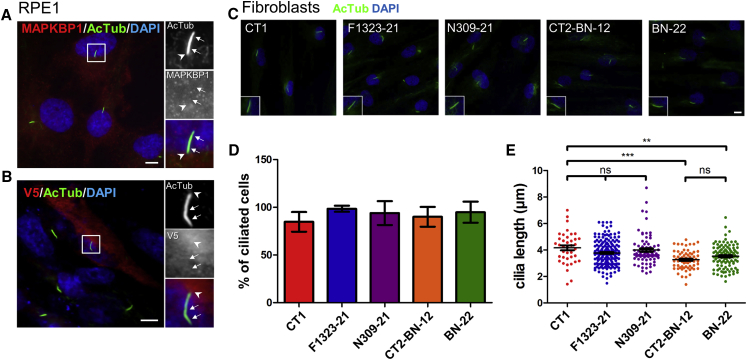
We then investigated the subcellular distribution of MAPKBP1 first in ciliated RPE1 cells. Endogenous (Figure 3A) and V5-tagged MAPKBP1 (Figure 3B) showed a similar diffuse or punctate cytoplasmic distribution and did not show any specific association with either the axoneme (acetylated α-tubulin, arrows) nor the basal body region (arrowhead) of primary cilia, an expected localization for a NPHP gene product. Similar results were obtained in ciliated human primary fibroblasts and IMCD3 kidney cells (Figure S7A) as well as in human adult kidney tubular cells in vivo, where MAPKBP1 showed a similar faint cytoplasmic staining not associated with cilia (Figure S8). The absence of MAPKBP1 from cilia and basal body was in agreement with proteomic and recent proximity-dependent biotinylation studies in which MAPKBP1 was not detected either at cilia or at centrosomes/basal bodies.23, 24, 25, 26, 27 Therefore, as expected, mutations in MAPKBP1 did not show any impact on ciliogenesis since fibroblasts obtained from affected individuals (NPH309-21, F1323-21, BN-22) formed cilia in similar proportion and with similar length as in cells from control individuals (Figures 3C–3E). Cilia with normal shape were also observed at the apical membrane of tubular epithelial cells from individual BN-23 (Figures 2A, panel 4, and S8). In addition, animal models that were generated, including zebrafish morphants (Figure S9) and mutant (Figures S10 and S11) as well as knock-out (Figure S12) mice, did not show any of the classical ciliopathy-associated phenotypes including body axis curvature, situs inversus, or pronephric cysts in the zebrafish, or situs inversus, polydactyly, or renal tubular cysts in the mouse. In agreement, in the zebrafish larvae, mapkbp1 was not expressed in the pronephros nor in the Kupffer vesicle and was mostly expressed in somites and in the brain (Figure S13), similarly as reported in mice.18 Altogether, these data show that MAPKBP1 is not directly nor indirectly involved in ciliary functions. Examination of 18-month-old fish, in which mapkbp1 is expressed at low level in the kidney (Figure S10B), revealed a slight proportion of mapkbp1 mutant with dilated tubules associated with fibrosis (1/8; Figure S11). This result, although anecdotal, may reflect a potential role for MAPKBP1 in kidney homeostasis in a long term. The lack of obvious kidney phenotype in mice models even in adults (up to 10 months; Figure S12) was already reported in other Nphp knockout mice, including Nphp1, Nphp4, and Nphp5,28, 29, 30 respectively mutated in juvenile isolated NPH1 and SLS-associated NPH.1, 31, 32 In the case of MAPKBP1, the lack of kidney phenotype in vertebrate models is possibly linked to either residual functional protein (zebrafish mutant) or compensatory effects by its paralog WDR62 or species- and tissue-specific differences in the function of MAPKBP1.
Figure 3.
MAPKBP1 Is Not Involved in Ciliogenesis
(A and B) Serum-starved (24 hr) nontransfected RPE1 cells (A) or RPE1 cells transiently transfected (FuGENE [Promega]) with V5-tagged MAPKBP1 encoding plasmid (B) were fixed (paraformaldehyde; PFA) and stained for acetylated α-tubulin (AcTub, green, cilia; mouse monoclonal antibody [6-11-B-1, Sigma], dil 1/10,000) and for endogenous MAPKBP1 (A) (red; rabbit polyclonal [HPA030832, Sigma], dil 1/300) or overexpressed MAPKBP1 (B) (red, V5 rabbit polyclonal antibody [Sigma V8137], dil 1/10,000). Insets on the right show higher magnifications of representative cilia indicated by a white square in the corresponding images. Arrows indicate cilia, arrowheads cilium base region.
(C–E) Serum-starved fibroblasts from controls (CT1 [unrelated healthy], CT2-BN-12 [mother of BN-22, heterozygous for the c.592C>T (p.Arg198∗) mutation]) or from the affected individuals (BN-22, F1323-21, NPH309-21) were stained for acetylated-tubulin (AcTub, green) to identify cilia (C). Insets on the right show higher magnifications of representative cilia. Ciliogenesis (% of ciliated cells [D]) and length of cilia (E) were analyzed and quantified by ImageJ from three independent experiments (n > 50 cells for each experiment). ns,∗p < 0.01, and ∗∗∗p < 0.0001 were calculated via Dunn’s multiple comparison test after the analysis of variance ANOVA test. Note that cilia length in cells from individual BN-22 is not significantly different from that of cilia from cells of his mother (CT2-BN-12). Cilia in cells from individuals F1323-21 and NPH309-21 were not significantly different than in control cells (CT1) but longer than in cells from individuals of the BN family. Scale bars represent 5 μm.
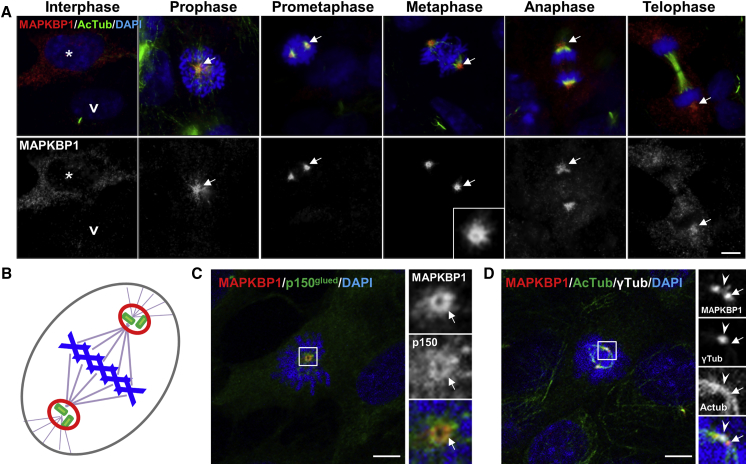
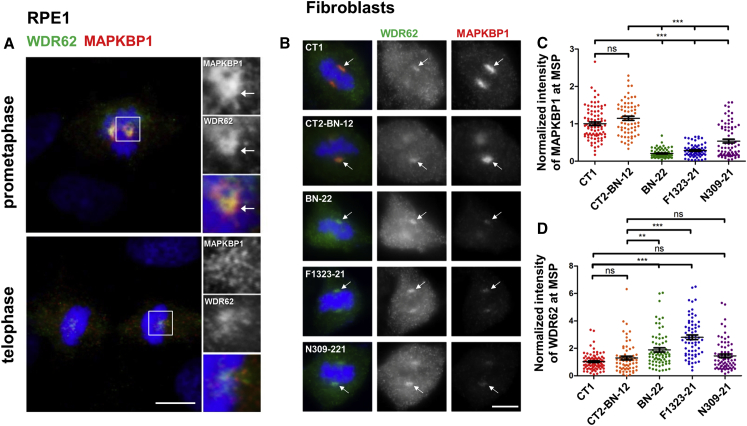
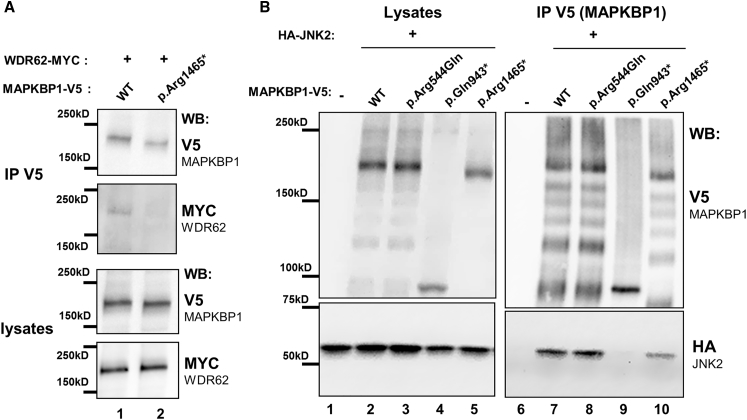
As MAPKBP1 was absent from cilia in quiescent cells where it showed non-specific cytoplasmic localization (Figures 3 and S7A), we analyzed its distribution in cycling cells. Interestingly, while MAPKBP1 staining was very faint in ciliated quiescent cells, it globally increased in the cytoplasm of non-ciliated cycling cells (Figure 4A, asterisk) and showed a strong accumulation around the poles of the mitotic spindle from prophase to anaphase in mitotic cells (Figure 4A, arrows). Interestingly, this distribution during mitosis was similar to the one reported for WDR62 that is recruited to MSP during early phases of mitosis.15, 17 The localization of MAPKBP1 at MSP was further confirmed by the fact that MAPKBP1 staining showed a typical ring-shaped organization (Figure 4A, metaphase, arrow) that colocalized with the MSP marker p150 glued (Figure 4C, arrows) and it surrounded centrioles as observed by confocal transverse sections (Figure 4D, arrows). MAPKBP1 also colocalized with WDR62 at MSP during early phases of mitosis (Figure 5A), as expected. We therefore investigated the impact of the mutations on the localization of MAPKBP1 and WDR62 during mitosis. As shown in Figure 5B, WDR62 was observed at MSP in mitotic fibroblasts from both control subjects and affected individuals (NPH309-21, F1323-21, BN-22), while MAPKBP1 staining at MSP was severely decreased in affected individuals compared to control subjects (arrows). Quantification of WDR62 and MAPKBP1 staining intensities at MSP confirmed these observations (Figures 5C and 5D). Similar results were obtained upon transient transfections of corresponding V5-tagged MAPKBP1 encoding constructs in cycling HeLa cells (Figure S14), showing that all variants of MAPKBP1, except the p.Arg544Gln, impair its recruitment to MSP. Interestingly, in fibroblasts from two out of three affected individuals (F1323-21 and BN-22, but not NPH309-21), the level of WDR62 staining at MSP was significantly increased compared to control subjects (Figure 5D) suggesting that WDR62 might compensate for the lack of MAPKBP1. In addition, the last conserved coiled-coil domain of MAPKBP1 (aa 1,488–1,514), which is lost in the p.Arg1465∗ protein (BN-22), was involved in WDR62-MAPKBP1 heterodimerization.22 As shown in Figures 6A and S15, WDR62 was efficiently co-immunoprecipitated by WT MAPKBP1 but not by the p.Arg1465∗ variant, confirming the interaction between the two proteins, the role of the last coiled-coil domain in MAPKBP1-WDR62 heterodimerization, and the negative impact of p.Arg1465∗ on this interaction. As expected, the interaction with WDR62 was also lost for the p.Gln943∗ shorter protein (NPH309-21). It was preserved for the p.Arg544Gln variant (NPH2139-22; Figure S15), a result in agreement also with the fact that this variation does not affect the WDR62 binding site and with the observation that its targeting to MSP was not affected (Figure S14). Altogether, these data indicate that most of the identified variations in MAPKBP1 affect its recruitment to MSP that is likely to occur through its interaction with WDR62.
Figure 4.
MAPKBP1 Is Recruited to Mitotic Spindle Poles during Early Stages of Mitosis
(A) Cycling RPE1 cells were fixed (PFA) and stained for MAPKBP1 (red) and for DNA with DAPI (blue). Arrows stress accumulation of MAPKBP1 staining at spindle poles in mitotic cells. The white arrowhead and star indicate representative ciliated and non-ciliated cells, respectively. Corresponding MAPKBP1 staining are shown in black and white in the lower panels.
(B) Scheme indicating mitotic spindle poles (MSP, red) surrounding centrosomes (green) at each pole of the mitotic spindle (pink) during metaphase (chromosomes in blue).
(C and D) Cycling RPE1 cells were fixed (PFA) and stained for MAPKBP1 (red) and either with p150 glued (MSP, green [C]; mouse monoclonal antibody [610473, BD Biosciences], dil 1/100) or acetylated α-tubulin (AcTub, spindle, green [D]) and γ-tubulin (γ-tub, centrioles, white [D]; goat polyclonal antibody [sc-7396, Santa Cruz], dil 1/2,000) together with DAPI (blue). Insets on the right show higher magnifications of representative MSP indicated by a white square in the corresponding images. Arrows and arrowheads point to MAPKBP1 staining at the MSP and to γ-tub, respectively. Scale bars represent 5 μm.
Figure 5.
Localization of MAPKBP1 at Mitotic Spindle Poles Is Disturbed in Fibroblasts from Affected Individuals
(A) Cycling RPE1 cells were fixed (PFA) and stained for endogenous MAPKBP1 (red) and WDR62 (green; mouse monoclonal [Clone 3G8, Sigma], dil 1/1,000) together with DAPI (blue). Representative pictures of cells in prometaphase and telophase are shown. Insets on the right show higher magnifications of representative MSP indicated by a white square in the corresponding images.
(B–D) Cycling fibroblasts from control subjects (CT1 and CT2-BN-12) or from affected individuals (BN-22, F1323-21, NPH309-21) were stained for both MAPKBP1 (red) and WDR62 (green). Representative pictures of cells in metaphase are shown in (B). Arrows stress MSPs. Intensity of MAPKBP1 (C) and WDR62 (D) stainings at the mitotic spindle poles was quantified with ImageJ (3 independent experiments, n > 65 cells). ns, ∗∗p < 0.001 and ∗∗∗p < 0.0001 were calculated via Dunn’s multiple comparison test after the analysis of variance ANOVA test. Scale bars represent 5 μm.
Figure 6.
Mutations in MAPKBP1 Affect JNK2 and/or WDR62 Binding
HEK293 cells were transiently transfected (lipofectamine [ThermoFischer]) with plasmids encoding V5-tagged WT and mutant forms of MAPKBP1 and Myc-tagged WDR62 (A) or HA-tagged JNK2 (B), as indicated. Cells were lysed and immunoprecipitated with the anti-V5 antibody (rabbit polyclonal [Sigma, V8137]). Lysates and immunoprecipitates (IP) were analyzed by western blot as indicated (WB; V5: Mouse monoclonal clone SV5-Pk1 [AbD Serotec MCA-1360], dil 1/5,000; HA: mouse monoclonal, clone 16B12 [Biolegend, 901502], dil 1/5,000; MYC: mouse monoclonal, Ab-2 clone 9E10.3 [Fisher Scientific MS-139-P1], dil 1/5,000).
The functional impact of the identified mutations was further evaluated by testing their ability to interact with JNK. As shown in Figure 6B, interaction with JNK2 was not affected by the p.Arg544Gln variation (Figure 6B, lane 8; NPH2139-22) as expected by the fact that the N-terminal WD repeat-containing domain is not required for JNK binding.18, 22 Interaction with JNK2 was lost for the p.Gln943∗ construct (Figure 6B, lane 9; NPH309-21), in agreement with the loss of the reported JNK binding site in this mutant18 (see Figures 1B and S6A). Moreover, the ability of the p.Arg1465∗ protein (BN-22) to interact with JNK2 was decreased compared to WT (Figure 6B, lane 10). This could be explained by the fact that the p.Arg1465∗ variant lacks the last C-terminal coiled-coil that was shown to be involved in dimerization and efficient binding to JNK in the context of WDR62.22 Despite the effect on JNK binding, MAPKBP1 mutations did not result in significant change in JNK signaling (phosphorylated JNK) in stable IMCD3 cells knockdown for Mapkbp1 expression nor in kidney biopsies from affected individual (Figure S16).
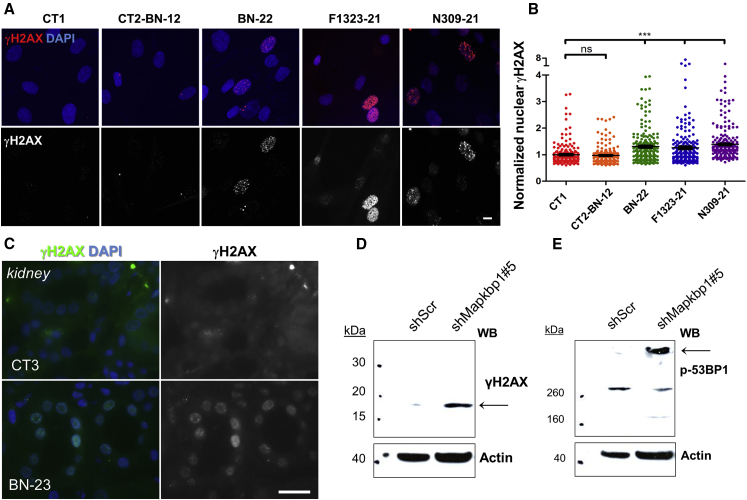
Increased DNA damage response (DDR) signaling was recently reported to be associated with NPH-RCs, so we tested whether this was also the case for NPH caused by mutations in MAPKBP1. Steady-state DDR signaling was first analyzed in control fibroblasts by immunofluorescence after phosphorylation of H2AX (γH2AX), a widely used marker of DDR signaling.33 As a positive control for the use of this assay, nuclear γH2AX staining was significantly increased in control fibroblasts treated with aphidicolin, an inhibitor of DNA replication known to induce DDR signaling,34 compared to untreated cells (Figures S17A and S17B). Steady-state DDR signaling was subsequently analyzed in fibroblasts from affected individuals with mutations in MAPKBP1. Interestingly, all mutant fibroblasts showed increased levels of nuclear γH2AX staining compared to healthy controls (Figures 7A and 7B), a phenotype also observed in kidney tubules from affected individual BN-23 (Figure 7C) and in two different murine cell lines (NIH 3T3 and IMCD3) that were knocked-down for Mapkbp1 expression (Figures 7D, 7E, S17E, and S17F). These data are in agreement with those recently reported in other NPH models (CEP290/NPHP6, NEK8/NPHP9, SDCCAG8/NPHP10, ZNF423/NPHP14, and CEP164/NPHP15)5, 6, 8, 9, 35 and suggest either direct or indirect function of MAPKBP1 in DDR signaling. The exact role of MAPKBP1 in DDR remains to be determined. Interestingly, JNK pathway positively regulates DDR signaling36 and defects in JNK regulation could then result in the accumulation of non-repaired DNA damage and then in steady-state increased DDR signaling (γH2AX). However, we could not detect a major general defect in JNK activation in mutant conditions, suggesting that MAPKBP1 might play a specific function in DDR independent of JNK.
Figure 7.
DNA Damage Response Is Increased in MAPKBP1 Mutant Cells
(A) Fibroblasts from control subjects (CT1; CT2-BN-12) and affected individuals (BN-22, F1323-21, NPH309-21) were fixed and stained for phosphorylated γH2AX (red; mouse monoclonal, clone JBW301 [MILLIPORE 05-636], dil 1/200) together with DAPI (blue). Scale bar represents 5 μm.
(B) The intensity of nuclear γH2AX staining was quantified from three independent experiments (n > 200). ns and ∗∗∗p < 0.0001 were calculated via Dunn’s multiple comparison test after the analysis of variance ANOVA test.
(C) Kidney biopsies from a control unrelated individual (CT3) and from the affected individual BN-23 were stained for γH2AX (green; rabbit polyclonal, dil 1/200). Scale bar represents 20 μm.
(D and E) NIH 3T3 cells (fibroblasts) were stably transduced with lentiviral vectors expressing either control scrambled (shScr) or Mapkbp1 targeting shRNA (shMapkbp1#5; Figures S16 and S17). Immunoblots show the expression of different components of DNA damage response signaling including γH2AX (Cell Signaling, rabbit polyclonal, dil 1/1,000); (D) and phospho-53BP1 (Cell Signaling, rabbit polyclonal, dil 1/1,000); (E).
In conclusion, the present study revealed biallelic mutations in MAPKBP1 in eight affected individuals from five unrelated families presenting with juvenile or late-onset NPH (from 12 to >27 years). We therefore propose NPHP20 (MIM: 617271) as an alias for MAPKBP1. Hypomorphic mutations in ciliary NPHP genes have been involved in late-onset NPH including NPHP3,37 WDR19,38 TTC21B,39 and NPHP5,40 whereas truncating or loss-of-function mutations of these genes were associated with early-onset NPH or even more severe cystic kidney disease or syndromic forms of ciliopathies41, 42, 43 in agreement with their key functions at cilia. Affected individuals described here did not share additional manifestations classically observed in ciliopathies, in agreement with the fact that MAPKBP1 is not present at cilia and does not seem to be involved in ciliogenesis. Scoliosis and/or facial dysmorphism were found in several but not all of the affected individuals; however, at this point, we cannot correlate mutations in MAPKBP1 with these manifestations that might be linked to other genetic causes in these consanguineous families. Interestingly, MAPKBP1 does not appear to be involved in cilia function in vitro and none of the generated animal models developed ciliopathy-like phenotypes. It rather seems that MAPKBP1 is likely the first member of a non-ciliary gene family for NPH that may account for non-syndromic forms of NPH for which causative mutated genes remain largely to be identified.
Acknowledgments
We are grateful to the families and studied individuals for their contribution. We would like to thank Hervé Enslen, Sylvie Legrand-Poels, and Ami Aronheim for their kind gifts of JNK2, MAPKBP1, and WDR62 encoding plasmids; Christelle Arrondel for her precious help for immunohistochemistry; Gérard Pivert (Pathology Department, Necker Hospital) for kidney biopsies; the bioinformatic Plateform (Université Paris Descartes, Institut Imagine) as well as Solenn Pruvost and Mohammed Zarhrate for their support in exome sequencing; Morgan Gallazzini for his kind help on Phospho-JNK blots; Marie-Claire Gubler for her precious help with interpretation of kidney biopsies; Jorunn Skeie Bringsli for genotyping; Torbjørn Leivestad at the Norwegian Renal Registry; and Joseph Szustakowski for his contribution in the early part of the project. The Mapkbp1 mutant mice were obtained from the Medical Research Council centre for mouse genetics (MRC-Harwell) which distributes these mice on behalf of the European Mouse Mutant Archive. The MRC-Harwell is also a member of the International Mouse Phenotyping Consortium (IMPC), which funded the generation of the Mapkbp1 mutant mice. This work was supported by grants from the Regional Health Authority Western Norway (grants no. 911688 and 911466 to C.B. and 911746 to E.R.), from the NIH (DK068306 to F.H.), from the Fondation pour la Recherche Médicale (FRM; DEQ20130326532 to S.S.), and the foundation GIS-Maladies Rares (FONDATION_HTS-RD; I201302013 to S.S.). The Imagine Institute is supported by an ANR grant ANR-A0-IAHU-01 and from Fondation ARC (EML20110602384) for the purchase of the LEICA SP8 confocal microscope.
Published: January 12, 2017; corrected online February 2, 2017
Footnotes
Supplemental Data include 17 figures and 2 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.12.011.
Contributor Information
Sophie Saunier, Email: sophie.saunier@inserm.fr.
Friedhelm Hildebrandt, Email: friedhelm.hildebrandt@childrens.harvard.edu.
Accession Numbers
The phenotype presented in this paper has been named NPHP20 with the MIM accession number 61721.
Web Resources
Ensembl Genome Browser, http://www.ensembl.org/index.html
ESEfinder3.0, http://krainer01.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
Infrafrontier, https://www.infrafrontier.eu/
International Mouse Phenotyping Consortium, http://www.mousephenotype.org/data/genes/
Jalview, http://www.jalview.org/
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Wolf M.T.F. Nephronophthisis and related syndromes. Curr. Opin. Pediatr. 2015;27:201–211. doi: 10.1097/MOP.0000000000000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bizet A.A., Becker-Heck A., Ryan R., Weber K., Filhol E., Krug P., Halbritter J., Delous M., Lasbennes M.-C., Linghu B. Mutations in TRAF3IP1/IFT54 reveal a new role for IFT proteins in microtubule stabilization. Nat. Commun. 2015;6:8666. doi: 10.1038/ncomms9666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schueler M., Braun D.A., Chandrasekar G., Gee H.Y., Klasson T.D., Halbritter J., Bieder A., Porath J.D., Airik R., Zhou W. DCDC2 mutations cause a renal-hepatic ciliopathy by disrupting Wnt signaling. Am. J. Hum. Genet. 2015;96:81–92. doi: 10.1016/j.ajhg.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hildebrandt F., Benzing T., Katsanis N. Ciliopathies. N. Engl. J. Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaki M., Airik R., Ghosh A.K., Giles R.H., Chen R., Slaats G.G., Wang H., Hurd T.W., Zhou W., Cluckey A. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell. 2012;150:533–548. doi: 10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slaats G.G., Saldivar J.C., Bacal J., Zeman M.K., Kile A.C., Hynes A.M., Srivastava S., Nazmutdinova J., den Ouden K., Zagers M.S. DNA replication stress underlies renal phenotypes in CEP290-associated Joubert syndrome. J. Clin. Invest. 2015;125:3657–3666. doi: 10.1172/JCI80657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otto E.A., Trapp M.L., Schultheiss U.T., Helou J., Quarmby L.M., Hildebrandt F. NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J. Am. Soc. Nephrol. 2008;19:587–592. doi: 10.1681/ASN.2007040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grampa V., Delous M., Zaidan M., Odye G., Thomas S., Elkhartoufi N., Filhol E., Niel O., Silbermann F., Lebreton C. Novel NEK8 mutations cause severe syndromic renal cystic dysplasia through YAP dysregulation. PLoS Genet. 2016;12:e1005894. doi: 10.1371/journal.pgen.1005894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Airik R., Slaats G.G., Guo Z., Weiss A.-C., Khan N., Ghosh A., Hurd T.W., Bekker-Jensen S., Schrøder J.M., Elledge S.J. Renal-retinal ciliopathy gene Sdccag8 regulates DNA damage response signaling. J. Am. Soc. Nephrol. 2014;25:2573–2583. doi: 10.1681/ASN.2013050565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slaats G.G., Giles R.H. Are renal ciliopathies (replication) stressed out? Trends Cell Biol. 2015;25:317–319. doi: 10.1016/j.tcb.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Failler M., Gee H.Y., Krug P., Joo K., Halbritter J., Belkacem L., Filhol E., Porath J.D., Braun D.A., Schueler M. Mutations of CEP83 cause infantile nephronophthisis and intellectual disability. Am. J. Hum. Genet. 2014;94:905–914. doi: 10.1016/j.ajhg.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corton M., Blanco M.J., Torres M., Sanchez-Salorio M., Carracedo A., Brion M. Identification of a novel mutation in the human PDE6A gene in autosomal recessive retinitis pigmentosa: homology with the nmf28/nmf28 mice model. Clin. Genet. 2010;78:495–498. doi: 10.1111/j.1399-0004.2010.01487.x. [DOI] [PubMed] [Google Scholar]
- 14.Pervaiz N., Abbasi A.A. Molecular evolution of WDR62, a gene that regulates neocorticogenesis. Meta Gene. 2016;9:1–9. doi: 10.1016/j.mgene.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholas A.K., Khurshid M., Désir J., Carvalho O.P., Cox J.J., Thornton G., Kausar R., Ansar M., Ahmad W., Verloes A. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 2010;42:1010–1014. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu T.W., Mochida G.H., Tischfield D.J., Sgaier S.K., Flores-Sarnat L., Sergi C.M., Topçu M., McDonald M.T., Barry B.J., Felie J.M. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat. Genet. 2010;42:1015–1020. doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogoyevitch M.A., Yeap Y.Y.C., Qu Z., Ngoei K.R., Yip Y.Y., Zhao T.T., Heng J.I., Ng D.C.H. WD40-repeat protein 62 is a JNK-phosphorylated spindle pole protein required for spindle maintenance and timely mitotic progression. J. Cell Sci. 2012;125:5096–5109. doi: 10.1242/jcs.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koyano S., Ito M., Takamatsu N., Shiba T., Yamamoto K., Yoshioka K. A novel Jun N-terminal kinase (JNK)-binding protein that enhances the activation of JNK by MEK kinase 1 and TGF-beta-activated kinase 1. FEBS Lett. 1999;457:385–388. doi: 10.1016/s0014-5793(99)01084-4. [DOI] [PubMed] [Google Scholar]
- 19.Wasserman T., Katsenelson K., Daniliuc S., Hasin T., Choder M., Aronheim A. A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation. Mol. Biol. Cell. 2010;21:117–130. doi: 10.1091/mbc.E09-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaguchi T., Miyashita C., Koyano S., Kanda H., Yoshioka K., Shiba T., Takamatsu N., Ito M. JNK-binding protein 1 regulates NF-kappaB activation through TRAF2 and TAK1. Cell Biol. Int. 2009;33:364–368. doi: 10.1016/j.cellbi.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Lecat A., Di Valentin E., Somja J., Jourdan S., Fillet M., Kufer T.A., Habraken Y., Sadzot C., Louis E., Delvenne P. The c-Jun N-terminal kinase (JNK)-binding protein (JNKBP1) acts as a negative regulator of NOD2 protein signaling by inhibiting its oligomerization process. J. Biol. Chem. 2012;287:29213–29226. doi: 10.1074/jbc.M112.355545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen-Katsenelson K., Wasserman T., Darlyuk-Saadon I., Rabner A., Glaser F., Aronheim A. Identification and analysis of a novel dimerization domain shared by various members of c-Jun N-terminal kinase (JNK) scaffold proteins. J. Biol. Chem. 2013;288:7294–7304. doi: 10.1074/jbc.M112.422055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 24.Jakobsen L., Vanselow K., Skogs M., Toyoda Y., Lundberg E., Poser I., Falkenby L.G., Bennetzen M., Westendorf J., Nigg E.A. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011;30:1520–1535. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa H., Thompson J., Yates J.R., 3rd, Marshall W.F. Proteomic analysis of mammalian primary cilia. Curr. Biol. 2012;22:414–419. doi: 10.1016/j.cub.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mick D.U., Rodrigues R.B., Leib R.D., Adams C.M., Chien A.S., Gygi S.P., Nachury M.V. Proteomics of primary cilia by proximity labeling. Dev. Cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta G.D., Coyaud É., Gonçalves J., Mojarad B.A., Liu Y., Wu Q., Gheiratmand L., Comartin D., Tkach J.M., Cheung S.W.T. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell. 2015;163:1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang S.-T., Chiou Y.-Y., Wang E., Lin H.-K., Lee S.-P., Lu H.-Y., Wang C.-K.L., Tang M.-J., Li H. Targeted disruption of Nphp1 causes male infertility due to defects in the later steps of sperm morphogenesis in mice. Hum. Mol. Genet. 2008;17:3368–3379. doi: 10.1093/hmg/ddn231. [DOI] [PubMed] [Google Scholar]
- 29.Won J., Marín de Evsikova C., Smith R.S., Hicks W.L., Edwards M.M., Longo-Guess C., Li T., Naggert J.K., Nishina P.M. NPHP4 is necessary for normal photoreceptor ribbon synapse maintenance and outer segment formation, and for sperm development. Hum. Mol. Genet. 2011;20:482–496. doi: 10.1093/hmg/ddq494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronquillo C.C., Hanke-Gogokhia C., Revelo M.P., Frederick J.M., Jiang L., Baehr W. Ciliopathy-associated IQCB1/NPHP5 protein is required for mouse photoreceptor outer segment formation. FASEB J. 2016;30:3400–3412. doi: 10.1096/fj.201600511R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaki M., Hoefele J., Allen S.J., Ramaswami G., Janssen S., Bergmann C., Heckenlively J.R., Otto E.A., Hildebrandt F. Genotype-phenotype correlation in 440 patients with NPHP-related ciliopathies. Kidney Int. 2011;80:1239–1245. doi: 10.1038/ki.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halbritter J., Porath J.D., Diaz K.A., Braun D.A., Kohl S., Chaki M., Allen S.J., Soliman N.A., Hildebrandt F., Otto E.A., GPN Study Group Identification of 99 novel mutations in a worldwide cohort of 1,056 patients with a nephronophthisis-related ciliopathy. Hum. Genet. 2013;132:865–884. doi: 10.1007/s00439-013-1297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogakou E.P., Nieves-Neira W., Boon C., Pommier Y., Bonner W.M. Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J. Biol. Chem. 2000;275:9390–9395. doi: 10.1074/jbc.275.13.9390. [DOI] [PubMed] [Google Scholar]
- 34.Ikegami S., Taguchi T., Ohashi M., Oguro M., Nagano H., Mano Y. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature. 1978;275:458–460. doi: 10.1038/275458a0. [DOI] [PubMed] [Google Scholar]
- 35.Choi H.J.C., Lin J.-R., Vannier J.-B., Slaats G.G., Kile A.C., Paulsen R.D., Manning D.K., Beier D.R., Giles R.H., Boulton S.J., Cimprich K.A. NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol. Cell. 2013;51:423–439. doi: 10.1016/j.molcel.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaukat Z., Liu D., Hussain R., Khan M., Gregory S.L. The role of JNK signalling in responses to oxidative DNA damage. Curr. Drug Targets. 2016;17:154–163. doi: 10.2174/1389450116666150126111055. [DOI] [PubMed] [Google Scholar]
- 37.Olbrich H., Fliegauf M., Hoefele J., Kispert A., Otto E., Volz A., Wolf M.T., Sasmaz G., Trauer U., Reinhardt R. Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat. Genet. 2003;34:455–459. doi: 10.1038/ng1216. [DOI] [PubMed] [Google Scholar]
- 38.Bredrup C., Saunier S., Oud M.M., Fiskerstrand T., Hoischen A., Brackman D., Leh S.M., Midtbø M., Filhol E., Bole-Feysot C. Ciliopathies with skeletal anomalies and renal insufficiency due to mutations in the IFT-A gene WDR19. Am. J. Hum. Genet. 2011;89:634–643. doi: 10.1016/j.ajhg.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huynh Cong E., Bizet A.A., Boyer O., Woerner S., Gribouval O., Filhol E., Arrondel C., Thomas S., Silbermann F., Canaud G. A homozygous missense mutation in the ciliary gene TTC21B causes familial FSGS. J. Am. Soc. Nephrol. 2014;25:2435–2443. doi: 10.1681/ASN.2013101126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otto E.A., Loeys B., Khanna H., Hellemans J., Sudbrak R., Fan S., Muerb U., O’Toole J.F., Helou J., Attanasio M. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat. Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 41.Tory K., Rousset-Rouvière C., Gubler M.-C., Morinière V., Pawtowski A., Becker C., Guyot C., Gié S., Frishberg Y., Nivet H. Mutations of NPHP2 and NPHP3 in infantile nephronophthisis. Kidney Int. 2009;75:839–847. doi: 10.1038/ki.2008.662. [DOI] [PubMed] [Google Scholar]
- 42.Coussa R.G., Otto E.A., Gee H.-Y., Arthurs P., Ren H., Lopez I., Keser V., Fu Q., Faingold R., Khan A. WDR19: an ancient, retrograde, intraflagellar ciliary protein is mutated in autosomal recessive retinitis pigmentosa and in Senior-Loken syndrome. Clin. Genet. 2013;84:150–159. doi: 10.1111/cge.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis E.E., Zhang Q., Liu Q., Diplas B.H., Davey L.M., Hartley J., Stoetzel C., Szymanska K., Ramaswami G., Logan C.V., NISC Comparative Sequencing Program TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat. Genet. 2011;43:189–196. doi: 10.1038/ng.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.