Abstract
In 2015, the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) published updated standards and guidelines for the clinical interpretation of sequence variants with respect to human diseases on the basis of 28 criteria. However, variability between individual interpreters can be extensive because of reasons such as the different understandings of these guidelines and the lack of standard algorithms for implementing them, yet computational tools for semi-automated variant interpretation are not available. To address these problems, we propose a suite of methods for implementing these criteria and have developed a tool called InterVar to help human reviewers interpret the clinical significance of variants. InterVar can take a pre-annotated or VCF file as input and generate automated interpretation on 18 criteria. Furthermore, we have developed a companion web server, wInterVar, to enable user-friendly variant interpretation with an automated interpretation step and a manual adjustment step. These tools are especially useful for addressing severe congenital or very early-onset developmental disorders with high penetrance. Using results from a few published sequencing studies, we demonstrate the utility of InterVar in significantly reducing the time to interpret the clinical significance of sequence variants.
Keywords: InterVar, variant annotation, variant interpretation, clinical interpretation, ClinVar, genetic diagnosis, ACMG, ANNOVAR
Introduction
With the continued development and deployment of massively parallel next-generation sequencing (NGS) technologies, clinical and molecular laboratories are now rapidly adopting NGS in genetic testing and human genetics research. Although it is becoming easier and more affordable for individual laboratories to generate NGS data, the major hurdle in utilizing these data lies in how to interpret the genotype-phenotype relationships, especially in genomic medicine settings.1, 2 The process of identifying disease-causing or disease-contributing variants among the thousands of genetic variants within an individual’s genome generally involves a number of steps, such as variant annotation, variant filtering, in silico prediction, and clinical interpretation by human experts.3 Each of these steps can involve the use of specific computational and bioinformatics tools.
Several tools and databases have been developed to assist laboratories and clinicians with understanding the functional significance of genetic variants with respect to their potential effects on genes and diseases. They generally fall into several categories. First, a number of annotation tools, such as ANNOVAR,4, 5 VAAST,6 SeattleSeq,7 SNPeff,8 and VEP,9 can predict how genetic variants affect transcript structure or coding sequences. They can classify variants into intronic, intergenic, splice, and exonic variants, and for exonic variants, they can compute how amino acid sequences are affected. Second, for coding variants, a variety of tools can predict whether the variant is deleterious to protein function or structure by using evolutionary information, context within the protein sequence, and biochemical properties. These in silico methods include individual scoring systems, such as SIFT,10 PolyPhen-2,11 CADD,12 FATHMM,13 and MutationTaster,14 as well as meta-predictors, such as Condel15 and MetaSVM.16 Many have a similar theoretical basis, but they also have known limitations, such as moderate accuracy, low specificity, and over-prediction.17, 18 Third and finally, public disease-specific and gene-specific databases, such as the Human Gene Mutation Database (HGMD),19 ClinVar,20 and various locus-specific databases,21 can document functionally or clinically validated genetic variants that are pathogenic for particular diseases. The HGMD is a comprehensive collection of germline mutations in nuclear genes that underlie, or are associated with, human inherited disease and is compiled primarily from the published literature.19 ClinVar20 archives the clinical significance of variants reported directly from submitters. However, these databases often contain variants that are incorrectly classified without a primary review of evidence, and they sometimes have contradictory records on the assessment of pathogenicity. The NIH began the ClinGen initiative22 to build an authoritative central resource that defines the clinical relevance of genes and variants for use in precision medicine and research. To improve the accuracy of variant interpretations, ClinGen uses a ranking system to denote the quality associated with each submission to ClinVar. Despite the existence of a variety of resources, a more systematic way to evaluate the pathogenicity of genetic variants observed in sequencing studies is needed to facilitate clinical evaluation of variants and to enable the precise implementation of genomic medicine.
To standardize the clinical interpretation of genetic variants, the American College of Medical Genetics and Genomics (ACMG) recommended standards for the interpretation of sequence variations and offered a decision-tree algorithm for variant interpretation in 2000 and 2007.23, 24 With the rapid development and adoption of NGS, variant interpretation has become more complex, and new challenges in the clinical interpretation of Mendelian and complex diseases have emerged. To address these challenges and to provide more concrete guidelines, the ACMG and the Association for Molecular Pathology (AMP) published updated guidelines for the interpretation of sequence variants in May of 2015.25 This new report describes updated standards and guidelines for classifying sequence variants by using criteria informed by expert opinion and experience. To better describe the causality of variants identified in genes associated with Mendelian diseases, the ACMG and AMP recommend a widely used five-tiered categorization system—pathogenic, likely pathogenic, uncertain significance, likely benign, and benign—for classifying variants. The system uses a total of 28 criteria based on different sources of data, such as population data, in silico data, functional data, and segregation data. The ACMG and AMP also propose a set of scoring rules, which combine criteria to give the five-tier classification system for genetic variants.
Although the ACMG-AMP guidelines were developed to enable consistent and reliable interpretation of genetic variants, application of the ACMG-AMP criteria still involves some discrepancies between intra- and inter-laboratory settings. Some efforts have been taken to reduce inter-laboratory inconsistencies,26 but >66% of variant classifications are still discordant in inter-laboratory classifications. There could be several reasons for the discordances. For many clinical labs, implementing the variant scoring rules into a standardized workflow is difficult with available informatics tools. For example, the ACMG and AMP recommend using 28 criteria during the interpretation process; however, gathering information on each of the criteria is quite complicated and might not be easily accomplished by individual interpreters or might not be reproducible by the same interpreter at different times. Furthermore, the ACMG and AMP provide only general guidelines on how to assess each criterion but do not offer specific algorithms for implementing these guidelines (for example, which databases to use); different researchers might prefer to use different algorithms, making the results less reproducible between different human interpreters. Finally, although a variety of databases (such as ClinVar and the 1000 Genomes Project) or in silico tools (such as SIFT and PolyPhen-2) are available online and easily accessible to the average user, there is a lack of tools that combine all of these databases together to offer a one-stop shop for human interpreters to derive a final score for genetic variants. Addressing these challenges will require easy-to-use yet automated computational tools and web services that can generate versioned and reproducible criteria for every variant and help human interpreters quickly understand the clinical significance of genetic variants. In this study, we present such a tool, InterVar (Clinical Interpretation of Genetic Variants), to fill these unmet needs on the basis of the 2015 ACMG-AMP guidelines and user-supplied domain knowledge.
Material and Methods
Generation of Variant Annotation
The required input for InterVar is a simple tab-delimited file including a list of variants that are already annotated with a set of required information, such as amino acid changes and allele frequency. Users can generate this input file themselves by using an in-house variant analysis workflow; alternatively, InterVar can take a VCF file, call the ANNOVAR software (a powerful and widely used annotation tool), and generate the required input data. The following is an example command line for running ANNOVAR: “perl table_annovar.pl input.vcf humandb/ -buildver hg19 -remove -out output -protocol refGene,esp6500siv2_all,1000g2015aug_all,avsnp144,dbnsfp30a,clinvar_20160302,exac03,dbscsnv11,dbnsfp31a_interpro,rmsk,ensGene,knownGene -operation g,f,f,f,f,f,f,f,f,r,g,g -nastring. -vcfinput.” The description for these databases is given below: “esp6500siv2_all” is a database for allele frequency in the NHLBI Exome Sequencing Project (ESP6500), “refGene” is a database for gene annotation from RefSeq, “1000 g2015aug_all” is a database for alternative allele frequency (AAF) in the 1000 Genomes Project27 (version August 2015), “exac03” is a database for AAF in the Exome Aggregation Consortium (ExAC) Browser28 (version 0.3), “dbnsfp30a” is a database for various functional deleteriousness prediction scores from dbNSFP29, 30 (version 3.0a), “clinvar_20160302” is for the variants reported in ClinVar20 (version 20160302), “avsnp144” is for the ANNOVAR-compiled dbSNP (version 144), “ensGene” is for gene annotation from Ensembl, “knownGene” is for gene annotation from UCSC Known Genes, “dbnsfp31a_interpro” is a database of the domain information from dbNSFP29, 30 and InterPro31 (which integrates information about protein families, domains, and functional sites), “dbscsnv11” is a database for predicting the splicing impact by Ada Boost and Random Forest,32 and “rmsk” is a database on the repeat masking track from the UCSC Genome Browser. These databases might be updated to new versions when they become available.
Criteria and Scoring System
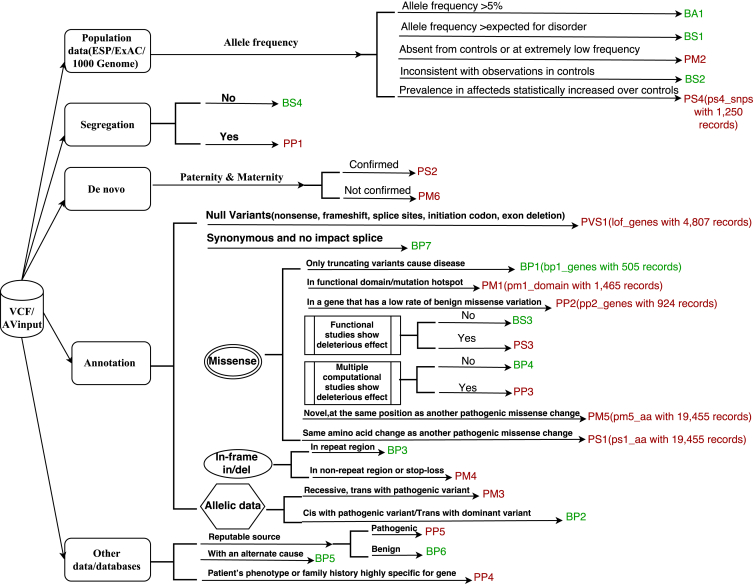
Based on the 2015 ACMG-AMP guidelines, the criteria fall into two sets: pathogenic or likely pathogenic (P/LP) and benign or likely benign (B/LB), whereas “uncertain significance” is assigned to variants for which the criteria for P/LP and B/LB are contradictory or not met. There are a total of 28 criteria: the 16 criteria for the P/LP criterion are very strong (PVS1), strong (PS1–PS4), moderate (PM1–PM6), or supporting (PP1–PP5); whereas the 12 criteria for the B/LB criterion are stand-alone (BA1), strong (BS1–BS4), or supporting (BP1–BP7). If a criterion is positive, InterVar will assign 1; otherwise, InterVar will assign 0. For these 28 criteria, InterVar can automatically generate predictions on 18 (PVS1, PS1, PS4, PM1, PM2, PM4, PM5, PP2, PP3, PP5, BA1, BS1, BS2, BP1, BP3, BP4, BP6, and BP7) according to the current annotation datasets, yet the rest (PS2, PS3, PM3, PM6, PP1, PP4, BS3, BS4, BP2, and BP5) require user input in the manual adjustment step. Below, we describe the details on how to assign these criteria from various sources of annotation information.
PVS1 by Automated Scoring
The null variants include nonsense variants, frameshift indels, and canonical splice variants, which often lead to loss of function (LOF). From ANNOVAR annotations, these LOF variants are represented as frameshift indel, stop-gain, stop-loss, and splicing variants in canonical transcripts. We first filtered ClinVar (version 20160302) by taking those variants shown in MedGen and then removing common variants (allele frequencies > 5%) and variants with conflicting annotations. The variants in ClinVar were annotated by ANNOVAR with RefGene definitions, and we identified 1,988 genes harboring at least one LOF variant that is “pathogenic” in ClinVar. Recently, the ExAC analyzed high-quality exome (protein-coding region) DNA sequence data for 60,706 individuals and identified 3,230 genes as LOF intolerant.28 We combined these two gene sets from ClinVar and the ExAC Browser and generated 4,807 genes as our final LOF-intolerant gene list. Null variants in the canonical transcripts for these 4,807 genes were assigned a PVS1 of 1. However, on the basis of the canonical rules for nonsense-mediated mRNA decay,33 we did not consider nonsense variants that are downstream of or within 50 nucleotides of the final exon-junction complex.
PS1 and PM5 by Automated Scoring
Generally speaking, if one missense variant is pathogenic, then a different nucleotide change that results in the same amino acid alteration should also be pathogenic for PS1. However, if a different nucleotide change results in a different amino acid change, then it suggests moderate evidence of pathogenicity by PM5. We first filtered ClinVar (subject to the same data-cleaning procedure described above), picked out all missense variants annotated as pathogenic, and stored the amino acid changes in an InterVar-specific database. We also inferred the splicing impact of these exonic missense variants by ANNOVAR from the “dbscsnv11” database to assess the possibility that they act through splicing disruption rather than amino acid changes. If a variant supplied by the user results in the same amino acid change, the PS1 value will be assigned as 1. However, if a variant supplied by the user results in a different amino acid change, then PM5 will be assigned as 1.
PS2 and PM6 by Manual Scoring
The de novo status of the variants gives strong support for the pathogenic status for PS2 if both maternity and paternity can be confirmed; if maternity or paternity is not confirmed, then moderate evidence of pathogenicity should be applied to PM6. Because InterVar cannot directly annotate the de novo status of the user’s input variants, PS2 and PM6 are treated as user-supplied values in the second step (manual adjustment) of InterVar.
PS3 and BS3 by Manual Scoring
If in vitro or in vivo functional studies are supportive of a damaging effect on the gene or gene product, PS3 should be assigned as 1. If in vitro or in vivo functional studies show no damaging effect on protein function or splicing, BS3 should be assigned as 1. InterVar does not have the information on functional studies, so by default these values are 0 and can be overridden by users. In the future, we might establish a database with validated genetic variants that are known to affect the function of genes or gene products.
BA1, BS1, BS2, PS4, and PM2 by Automated Scoring
The AAFs in control populations are useful for scoring the pathogenicity of variants, given that frequently occurring variants in the population are unlikely to cause rare diseases. We retrieved information on disease prevalence from OrphaNet and translated OrphaNet identifiers into OMIM identifiers. Here, we used three datasets to assess the variant frequency: the NHLBI Exome Sequencing Project (ESP6500), 1000 Genomes Project, and ExAC Browser. If any of the AAFs in any database is >5%, BA1 will be assigned as 1. If the AAF in the ExAC Browser is great than expected for the disorder caused by mutations in the corresponding gene, BS1 will be assigned as 1 (here, we set a default cutoff as 1% for rare disease, but users can specify their own cutoff in the configuration file of InterVar). If a variant is observed in a healthy adult in the 1000 Genomes Project as a homozygote (for diseases defined as recessive in OMIM) or as a heterozygote otherwise, then BS2 will be applied. We manually removed known major adult-onset disorders from consideration. We did not use the ExAC Browser or ESP6500 here because these datasets can contain variants from individuals with various diseases.
Variants that are absent or are present at extremely low frequencies in a large control cohort could represent moderate evidence for pathogenicity. If a variant that is responsible for dominant diseases is absent in all control subjects from ESP6500, 1000 Genomes Project, and the ExAC Browser, PM2 will be applied. If the variant causes recessive diseases and has a very low frequency with AAF < 0.5%, then PM2 can also be applied. Information on the gene-disease relationship, such as dominance or recessiveness, is obtained from OMIM.
In some cases, pathogenic variants have a significantly higher frequency in affected subjects than in control subjects. To handle these variants, we also cataloged all variants with an odds ratio (OR) > 5.0 from GWASdb34 version 2. For these variants, PS4 will be applied. For some rare variants where case-control studies might not reach statistical significance, PS4 also can be downgraded to a moderate level during the manual adjustment step.
PM1 by Automated Scoring
Many protein domains play essential roles for protein function, so missense variants in these domains tend to be pathogenic. The domain information can be inferred from dbNSFP by ANNOVAR through the “dbnsfp31a_interpro” database. We first annotated all ClinVar variants (subject to the same data-cleaning procedure described above) with protein-domain information and then compiled a list in which domains contained only pathogenic or likely pathogenic variants without benign or common (allele frequency > 5%) variants. This list is provided within the InterVar package and will be updated regularly. If the user’s input variants are located in these domains, then PM1 will be applied.
PM3 and BP2 by Manual Scoring
The pathogenicity of a variant also needs to be evaluated on the basis of whether variants with known pathogenicity exist in cis or trans with it. InterVar does not know the cis/trans status for variants, so this needs to be provided by users in the second step (manual adjustment) of InterVar. For two heterozygous variants that are present in a gene associated with recessive disorders, if one is pathogenic and the other is located in trans, then moderate evidence of PM3 will be applied. If more than two variants are observed in trans, then moderate evidence for pathogenicity can be upgraded to strong. If the variants are present in a gene associated with dominant diseases, yet one variant is pathogenic and the other is located in trans, then supporting evidence of benign status will be applied to BP2 for the other variant. Regardless of models of disease inheritance, for two variants, if one is pathogenic and the other is observed in cis, then BP2 will be applied for the other variant.
PM4 and BP3 by Automated Scoring
Indels and stop losses can change the length of proteins and disrupt protein function. We annotated the repeat region by using the “rmsk” database from the UCSC Genome Browser. This database was created by the RepeatMasker program, which screens DNA sequences for interspersed repeats and low-complexity DNA sequences. When the variants are “non-frameshift insertion,” “non-frameshift deletion” in the non-repeat region, or stop-loss variants, PM4 will be applied. If the variants are “non-frameshift insertion” or “non-frameshift deletion” in the repeat region, BP3 will be applied.
PP1 and BS4 by Manual Scoring
Familial segregation of a variant with a disease is an important sign for linking the variant to the disease. If segregation is found in multiple affected family members and if this gene is definitively known to be associated with this disease, then PP1 will be applied. When there is a lack of segregation in affected members of a family, then the benign supporting evidence of BS4 will be applied. Because InterVar does not know the information on segregation, this piece of evidence can be provided by users in the second step (manual adjustment) of InterVar.
PP2 and BP1 by Automated Scoring
For many genes, the spectrum or distribution of pathogenic and benign variants can be informative for the pathogenicity status. For a given gene, if the missense variants are common causes of the disorder and the gene also has very few benign variants, then a missense variant in this gene can be supporting evidence for pathogenicity, and PP2 will be applied. However, if the truncating variants are major causes of the disease, then a missense variant in this gene can be supporting evidence for a benign status, and BP1 will be applied.
We annotated all variants in ClinVar (subject to the same data-cleaning procedure described above). For a given gene, if most of the pathogenic variants (>80% and at least one variant) are missense, and if a small proportion (<10% and less than one variant) of missense variants are benign, then for missense variants, PP2 will be assigned as 1. The treatment for BP1 is similar to that for PP2, but we assess whether most of pathogenic variants (>80% and at least one variant) are truncating variants. The truncating variants are defined as stop-gain, stop-loss, frameshift indel, or those disrupting splice sites. If the user’s variants are missense in this gene, BP1 will be assigned as 1.
PP3 and BP4 by Automated Scoring
When multiple pieces of computational evidence support a deleterious effect on the gene or gene product (conservation, evolutionary, splicing impact, etc.), then the supporting pathogenic evidence of PP3 will be assigned as 1. In comparison, when multiple pieces of computational evidence suggest no impact on the gene or gene product, then supporting benign evidence of BP4 will be assigned as 1. All sets of in silico results must agree when PP3 or BP4 is assigned.
These multiple pieces of computational evidence can be provided by ANNOVAR from the “dbnsfp30a” database, where the MetaSVM score16 is used for deleteriousness prediction and GERP++ is used for evolutionary conservation. The splicing impacts can be inferred by ANNOVAR from the “dbscsnv11” database. For the evidence of PP3 and BP4, we set the cutoff to 0.0 for MetaSVM scores (greater scores indicate more likely deleterious effects), 2.0 for GERP++_RS (smaller scores indicate less conservation), and 0.6 for adaptive boosting (ADA) and random forest (RF) scores of dbscSNV as splicing impact (larger scores indicate more likely splice altering).
PP4 by Manual Scoring
For a given gene, if the individual’s phenotype or family history is highly specific to the disorder associated with the gene, then it is supporting evidence for pathogenicity; in such a case, PP4 should be applied. This information needs to be provided by the user in the second step (manual adjustment) of InterVar.
PP5 and BP6 by Automated Scoring
If a reputable source has already reported a variant as pathogenic but the evidence is not provided for independent evaluation, then PP5 will be applied. When a reputable source has already reported a benign variant but without detailed evidence, then BP6 will be applied. In InterVar, we used the ClinVar dataset (subject to the same data-cleaning procedure described above) to perform this analysis by default, but users can select to use HGMD or other proprietary databases for this analysis.
BP5 by Manual Scoring
If a disease has an alternate molecular basis (caused by more than one gene) and if a variant is observed in a gene related to the disease, then it will be supporting evidence for a benign status, and BP5 will be assigned as 1. Note that this criterion is stronger for a gene associated with a dominant disorder than for a gene associated with a recessive disorder. Because of the multiple exceptions for this criterion, as described before,25 users can adjust this criterion by using their own knowledge in the manual adjustment step.
BP7 by Automated Scoring
If a synonymous (silent) variant has no effect on splicing and if the nucleotide position is not highly conserved, then we can classify this variant as likely benign and assign BP7 as 1. The prediction on the effect on splicing can be extracted by ANNOVAR with the “dbscSNV” database. Both scores dbscSNV_RF_SCORE and dbscSNV_ADA_SCORE should be <0.6 when the variant is predicted to have no impact on splicing. The conservation information is retrieved from the “dbnsfp30a” database, where a GERP++ score > 2 indicates that the nucleotide is highly conserved.
InterVar and wInterVar
InterVar is a command-line-driven software written in Python and can be used as a standalone application on a variety of operating systems—including Windows, Linux, and MacOS—where Python is installed. The source code of InterVar is available from GitHub (see Web Resources).
InterVar takes either pre-annotated files in tab-delimited formats or unannotated input files in VCF format or ANNOVAR input format, where each line corresponds to one genetic variant. If the input files are unannotated, InterVar will call ANNOVAR to generate necessary annotations. Users can also use software tools other than ANNOVAR to generate pre-annotated files. The execution of InterVar mainly consists of two major steps: (1) automatically interpreting the variant by using the criteria outlined above and (2) manually adjusting specific criteria to re-interpret the clinical significance. However, users can also specify their own evidence file for a subset of the criteria and import it into InterVar by using the argument “-evidence_file” so that one single step is sufficient to generate the final results. In the output, on the basis of all 28 pieces of criteria that are either automatically generated or manually supplied by the user, each variant will be assigned as pathogenic, likely pathogenic, uncertain significance, likely benign, or benign by rules specified in the 2015 ACMG-AMP guidelines.25
We also developed a web server called wInterVar, which offers a graphical user interface for InterVar (see Web Resources). Users can directly input their missense variants into wInterVar by chromosomal position, by dbSNP identifier, or by gene name with the nucleic acid change. The wInterVar server will provide full details on the variants, including all automatically generated criteria, most of the supportive evidence, and sub-population information. Users then have the ability to manually adjust these criteria and resubmit to the server to perform re-interpretation. We scanned all exons, and for each position we generated all three possible nucleotide changes. If the mutation was non-synonymous, we kept it in our database. The human genome contains approximately 80,000,000 non-synonymous variants, and we pre-computed the 18 criteria for all of them. Therefore, the execution of wInterVar is very fast, typically less than 1 s to obtain the result on a variant. However, the wInterVar server cannot process other types of variants (such as indels), and the user will need to rely on InterVar instead.
Results
Summary of the Interpretation Procedure
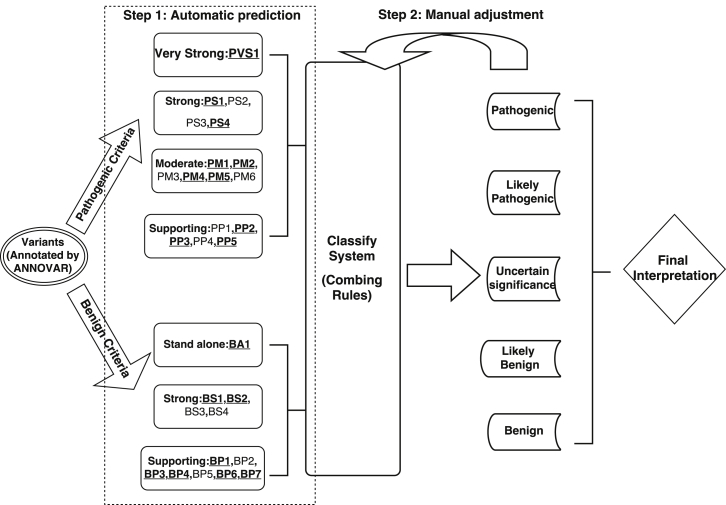
A flowchart for InterVar is given in Figure 1. InterVar mainly consists of two major steps: (1) automated scoring on each of the 18 pieces of criteria and (2) manual review and adjustment on specific criteria to arrive at a final interpretation. During the first step, InterVar calls an annotation software, such as ANNOVAR,5 to obtain necessary annotation information on variants and then uses its own internal annotation database to supplement additional annotations. Using these annotations on variants and genes, InterVar performs a preliminary interpretation of the variant and presents all relevant evidence for manual review. Currently, 18 pieces of criteria can be automatically generated and used in the first step. During the second step, the user can manually adjust each of the criteria on the basis of prior information (such as a variant’s de novo status) or his or her own domain knowledge to reach a final interpretation. We emphasize here that automated scoring is based on default parameters and that users are advised to examine detailed evidence and use prior knowledge on ethnicity and/or disease to perform manual adjustments. A detailed explanation of these 28 criteria is given in Figure 2.
Figure 1.
Flowchart of the Two-Step Procedure of InterVar
Underlined and bold fonts denote automated criteria.
Figure 2.
Illustration of the 28 Criteria from the 2015 ACMG-AMP Guidelines
For some criteria, the name of the internal database and its size are denoted within parentheses.
For example, consider missense variant chr12: 52,093,447T>C (GRCh37 coordinate) in exon 7 of SCN8A (MIM: 600702), which causes early infantile epileptic encephalopathy type 13 (MIM: 614558). We recently reported this variant as a de novo mutation in a 4-year-old female who, at 5 months of age, exhibited symptoms of epilepsy that progressed to a severe condition with very little movement, including the inability to sit or walk on her own.35 We illustrate the scoring logic for this variant. This variant is located in a protein domain called the ion transport domain. This domain does not have any benign variants in public databases compiled by InterVar, so we assigned PM1 as 1. In addition, this variant is not present in the 1000 Genomes Project, ExAC Browser, or ESP6500, so PM2 was assigned as 1. For SCN8A, all known pathogenic variants are missense, so PP2 was assigned as 1. According the 2015 ACMG-AMP rules, the variant falls into the class of “uncertain significance.” In the second step, if we manually adjust the criteria by providing de novo information as PS2 = 1, then the clinical significance will change to “likely pathogenic” on the basis of “1 strong (PS1–PS4) and 1–2 moderate (PM1–PM6).” This procedure illustrates how to use automated interpretation and manual adjustment to derive a final interpretation for genetic variants.
Interpretation of De Novo Variants in Neurodevelopmental Disorders
We compiled a dataset of 9,305 de novo variants from 12 published trio-based exome sequencing studies on autism spectrum disorders,36, 37 developmental disorders,38 schizophrenia,39, 40, 41, 42 epileptic encephalopathies,43 and intellectual disability.44, 45, 46, 47 Among them, 8,346 variants were detected from affected subjects (n = 6,515), and 959 were detected from control subjects (n = 900). Among these 8,346 variants from affected subjects, 4,526 were non-synonymous, resulting in coding sequence changes in 3,462 genes, whereas 616 non-synonymous variants were present in 592 genes from control subjects.
We next performed automated variant interpretation by InterVar on all of these variants by using default options in the program and setting expected prevalence for these disorders as 1% (Table 1). Given that each published exome sequencing study used Sanger sequencing to validate the de novo status of the variants, we assigned PM6 as 1, indicating that these variants were assumed to be de novo without confirmed paternity or maternity. Among these variants, 4,459 (53.4%) and 493 (51.4%) were interpreted as having uncertain significance in affected and control subjects, respectively. Among affected subjects, 430 (5.1%) and 1,666 (20.0%) variants were interpreted as pathogenic and likely pathogenic, respectively. Among control subjects, 10 (1.0%) and 206 (21.5%) variants were interpreted as pathogenic and likely pathogenic, respectively.
Table 1.
Illustration of Automated Interpretation of De Novo Variants from Individuals with Several Different Diseases and Control Subjects
| Interpretation | DD | SCZ | ASD | EE | ID | Affected Subjects | Control Subjects |
|---|---|---|---|---|---|---|---|
| Benign | 7 | 3 | 52 | 0 | 0 | 62 | 0 |
| Likely benign | 288 | 241 | 1,085 | 59 | 56 | 1,729 | 250 |
| Uncertain significance | 819 | 466 | 2,869 | 180 | 125 | 4,459 | 493 |
| Likely pathogenic | 339 | 199 | 967 | 81 | 80 | 1,666 | 206 |
| Pathogenic | 125 | 26 | 226 | 17 | 36 | 430 | 10 |
| Total | 1,578 | 935 | 5,199 | 337 | 297 | 8,346 | 959 |
| Benign and likely benign | 295 | 244 | 1,137 | 59 | 56 | 1,791 | 250 |
| Pathogenic and likely pathogenic | 464 | 225 | 1,193 | 98 | 116 | 2,096 | 216 |
| p value (compared to control subjects)a | 4.71E−7 | 0.65 | 0.06 | 0.00061 | 2.07E−6 | 0.0022 | – |
| OR and 95% CI | 0.55 (0.44–0.69) | 0.94 (0.72–1.21) | 0.82 (0.67–1.00) | 0.52 (0.35–0.75) | 0.42 (0.29–0.60) | 0.74 (0.61–0.90) | – |
Abbreviations are as follows: DDD, developmental disorder; SCZ, schizophrenia; ASD, autism spectrum disorder; EE, epileptic encephalopathy; ID, intellectual disability; OR, odds ratio; and CI, confidence interval.
p values were calculated with a two-sided Fisher’s exact test.
We next combined variants with a benign or likely benign interpretation as one category (B/LB) and those with pathogenic or likely pathogenic as another category (P/LP) and compared their frequency between affected and control subjects. (Please note that we do not have access to individual-level data, so our analysis below focused on comparing detected variants between affected and control subjects.) Using Fisher’s exact test, we detected a strong enrichment of P/LP variants among de novo variants in affected subjects (p = 0.0022) on the basis of automated interpretation. This result confirms that de novo variants that might be pathogenic are more prevalent in subjects with neurodevelopmental disorders than in control subjects. Please note that this analysis leveraged results only from automated interpretation (step 1) and did not account for manual adjustment (step 2) based on additional domain knowledge of the variants, genes, phenotypes, or diseases.
In comparison, we also predicted the pathogenicity of these variants by using SIFT and PolyPhen-2 scores on a subset of the variants for which the scores were available (Table 2). SIFT predicted 2,242 (26.8%) of 8,346 variants as deleterious (SIFT < 0.05 as the cutoff) for the subjects with neurodevelopmental disorders and predicted 283 (29.5%) of 959 variants as deleterious for control subjects. PolyPhen-2 predicted 3,157 (37.8%) of 8,346 variants as probably damaging or possibly damaging (PolyPhen-2_HDIV > 0.453 as the cutoff) for affected subjects and predicted 403 (42.0%) of 959 variants as probably damaging or possibly damaging for control subjects. Comparing affected and control subjects (Table 2), we did not observe a strong enrichment of P/LP variants with these two methods (p = 0.64 for SIFT and p = 0.08 for PolyPhen-2_HDIV). These results demonstrate that in silico predictions alone might not be sufficient to identify P/LP variants in exome sequencing studies.
Table 2.
Analysis of De Novo Variants by SIFT and PolyPhen-2
| Interpretation |
SIFT |
PolyPhen-2 |
||
|---|---|---|---|---|
| Affected Subjects | Control Subjects | Affected Subjects | Control Subjects | |
| Benign or tolerated | 2,608 (31.2%) | 343 (35.7%) | 1,426 (17.1%) | 214 (22.3%) |
| Deleterious, probably damaging, or possibly damaging | 2,242 (26.8%) | 283 (29.5%) | 3,157 (37.8%) | 403 (42.0%) |
| Unknown | 3,496 (42.0%) | 333 (34.8%) | 3763 (45.1%) | 342 (35.7%) |
| Total | 8,346 | 959 | 8,346 | 959 |
| p value (compared to control subjects)a | 0.64 | 0.08 | ||
p values were calculated with a two-sided Fisher’s exact test.
Comparative Analysis on ClinVar
Although variant databases such as HGMD, ClinVar, and OMIM have been very helpful for cataloging genetic variants known to be associated with human diseases, they also have known limitations, e.g., that a portion of benign variants are incorrectly classified as pathogenic variants.48, 49 For example, Dorschner et al.50 manually examined primary literature for 239 unique variants reported as pathogenic in HGMD and confirmed that only 7.5% are actually pathogenic from the original publication. The discrepancy in variant clinical significance between HGMD and clinical labs also highlights the lack of standards in interpreting a variant as pathogenic or likely pathogenic in the literature. Similarly, Bell et al.51 found that 27% of the pathogenic variants cited in the literature are common polymorphisms or misannotated, underscoring the need for better mutation databases. Interestingly, we recently sequenced a personal genome and identified two variants reported as pathogenic in ClinVar, but manual examination of the cited publication indicated that neither was reported as pathogenic in the original publication.52 This problem has been increasingly recognized in recent years,48 suggesting that “known” pathogenic variants in various databases should not be taken at face value and instead deserve more detailed re-examination. Here, we analyzed the entire ClinVar dataset and compared their annotations with the automated interpretation (step 1) by InterVar to assess the concordance rates and examine sources of discordance. We recognized that because InterVar compiled some of its internal databases from ClinVar, its interpretation might be slightly biased toward being more similar to that of ClinVar.
We retrieved ClinVar version 2016-03-02 and selected all non-conflicting nonsynonymous variants categorized as one of the following: (1) benign or likely benign and (2) pathogenic or likely pathogenic. We then re-interpreted these variants by using the automated interpretation function in InterVar (Table 3). For the benign category in ClinVar, InterVar also classified 4,898 (80.6%) variants as benign or likely benign, suggesting that InterVar is largely consistent with ClinVar on this category of variants. However, for variants in the pathogenic category, InterVar and ClinVar have large differences. In fact, InterVar classified only 2,058 (13.9%) variants in the category as likely pathogenic yet none as pathogenic. Obviously, we acknowledge that all of these interpretations by InterVar were based on only 18 pieces of criteria in step 1, and none of them were subjected to manual examination; yet, additional information such as familial segregation, family history, and de novo status could move some variants with uncertain significance into a more deleterious category (likely pathogenic or pathogenic).
Table 3.
Illustration of Automated Interpretation of Pathogenic and Benign Variants Annotated in ClinVar
| InterVar (Automated Interpretation) |
ClinVar |
|
|---|---|---|
| Pathogenic or Likely Pathogenic | Benign or Likely Benign | |
| Benign | 65 (0.4%) | 1,505 (24.8%) |
| Likely benign | 448 (3.0%) | 3,393 (55.9%) |
| Uncertain significance | 12,207 (82.6%) | 1,173 (19.3%) |
| Likely pathogenic | 2,058 (13.9%) | 0 (0%) |
| Pathogenic | 0 (0%) | 0 (0%) |
| Sum of five tiers | 14,778 | 6,071 |
| Benign and likely benign | 513 (3.5%) | 4,898 (80.6%) |
| Pathogenic and likely pathogenic | 2,058 (13.9%) | 0 (0%) |
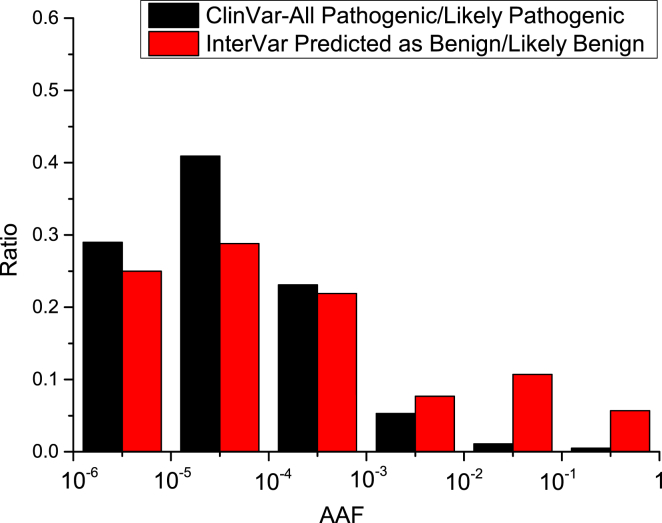
Given the differences between ClinVar annotation and InterVar prediction, we performed a more detailed analysis on the 513 (3.5%) variants that were classified as pathogenic by ClinVar but predicted as benign or likely benign by InterVar. First, we plotted the distribution of the maximum AAF of these variants in three databases (1000 Genomes Project, ExAC Browser, and NHLBI ESP6500; Figure 3). From this analysis, we found that there were >10% variants with AAF > 0.01 and 5% variants with AAF > 0.1. Clearly, >10% variants might be merely genetic polymorphisms that were incorrectly cataloged as pathogenic in ClinVar. Nevertheless, we also confirmed that in ClinVar, more than half of the pathogenic or likely pathogenic variants were very rare with an AAF < 0.0001, and >85% pathogenic variants had an AAF < 0.001, which fits our expectations. For manual examination of these variants, the cutoff of disease prevalence could be essential for assigning benign criteria such as BS1.
Figure 3.
AAF Distribution of Pathogenic or Likely Pathogenic ClinVar Variants Predicted to Be Benign or Likely Benign by InterVar and All Pathogenic or Likely Pathogenic ClinVar Variants
Analysis on Previously Reported Clinically Actionable Variants
Clinical exome and genome sequencing are likely to uncover “incidental findings” that are unrelated to the indication for ordering the sequencing tests but are of clinical significance.53 The ACMG has recommended returning incidental findings from a minimum set of 56 actionable genes,53 but many researchers have used an expanded list of genes selected according to domain knowledge. Several studies have examined incidental findings from large-scale genome or exome sequencing projects, so here we investigated how InterVar classifies clinically actionable genetic variants reported in previous studies.
Amendola et al.54 previously examined exome sequencing data on 4,300 European Americans and 2,203 African Americans as part of NHLBI ESP6500 and reported 616 variants in 112 actionable genes (Table 4). These 616 variants were classified as actionable and pathogenic on the basis of HGMD annotations. Amendola et al. re-classified these 616 variants by using their own classification criteria, such as rules based on allele frequency, segregation, de novo status, function data, etc. They found only 70 (11.4%) as pathogenic or likely pathogenic, yet most of them (66.4%) were classified as variants of uncertain significance. Automated prediction (step 1) from InterVar classified only 33 (5.4%) variants as pathogenic or likely pathogenic, whereas most of the variants (43.2%) were classified as benign or likely benign. Please note that during variant classification, Amendola et al. leveraged information such as segregation and de novo status, but we did not have access to these pieces of information. Therefore, the number of pathogenic variants classified by InterVar in step 2 (manual adjustment) could increase significantly given additional information. Nevertheless, these results already suggest that the interpretation of InterVar is consistent with the manual interpretation by Amendola et al., who concluded that the vast majority of variants annotated as pathogenic in HGMD are probably not really pathogenic. This analysis confirms that incorrect classification of the pathogenic variant, even in ACMG actionable genes, represents a substantial issue when HGMD is the only criterion used for variant interpretation.
Table 4.
Interpretation of 616 HGMD-Classified Pathogenic Variants from NHLBI ESP6500
| Clinical Significance | InterVar (Automated Interpretation) | ESP6500 Team (Manual Interpretation) | Concordant |
|---|---|---|---|
| Benign | 5 | 0 | 0 |
| Likely benign | 261 | 137 | 77 |
| Likely pathogenic | 30 | 38 | 2 |
| Pathogenic | 3 | 32 | 0 |
| Uncertain significance | 317 | 409 | 234 |
| Sum of five tiers | 616 | 616 | 313 |
| Benign or likely benign | 266 | 137 | 79 |
| Pathogenic or likely pathogenic | 33 | 70 | 6 |
Comparative Analysis with CLINVITAE
CLINVITAE (see Web Resources) is a database of clinically observed genetic variants aggregated from public sources and is operated and made freely available by INVITAE. Although the vast majority of the variants were collected from public databases, 11,696 variants were detected and classified by the INVITAE team. Unlike ClinVar and HGMD, which compile information from diverse sources, CLINVITAE potentially represents a more homogeneous collection of variants interpreted by a consistent set of institution-specific rules. Among these 11,696 variants, 5,405 (46.2%) and 717 (6.1%) were classified as benign or likely benign and pathogenic or likely pathogenic, respectively. Among them, 4,226 (36.1%) benign or likely benign variants were also classified as benign or likely benign by InterVar, whereas only 227 (1.9%) pathogenic or likely pathogenic variants were classified as pathogenic or likely pathogenic by InterVar (Table 5). This analysis again demonstrates that the concordance between automated interpretation of InterVar and expert-compiled classification is higher for benign or likely benign variants than for pathogenic or likely pathogenic variants.
Table 5.
Comparison of Variant Interpretation by CLINVITAE and Automated Interpretation by InterVar
| Clinical Significance | InterVar (Automated Interpretation) | CLINVITAE | Concordant |
|---|---|---|---|
| Benign | 242 | 2,407 | 230 |
| Likely benign | 6,593 | 2,998 | 2,428 |
| Likely pathogenic | 286 | 106 | 11 |
| Pathogenic | 137 | 611 | 132 |
| Uncertain significance | 4,438 | 5,574 | 3,047 |
| Sum of five tiers | 11,696 | 11,696 | 5,848 |
| Benign or likely benign | 6,835 | 5,405 | 4,226 |
| Pathogenic or likely pathogenic | 423 | 717 | 227 |
wIntervar: Web Version of InterVar to Facilitate Manual Interpretation
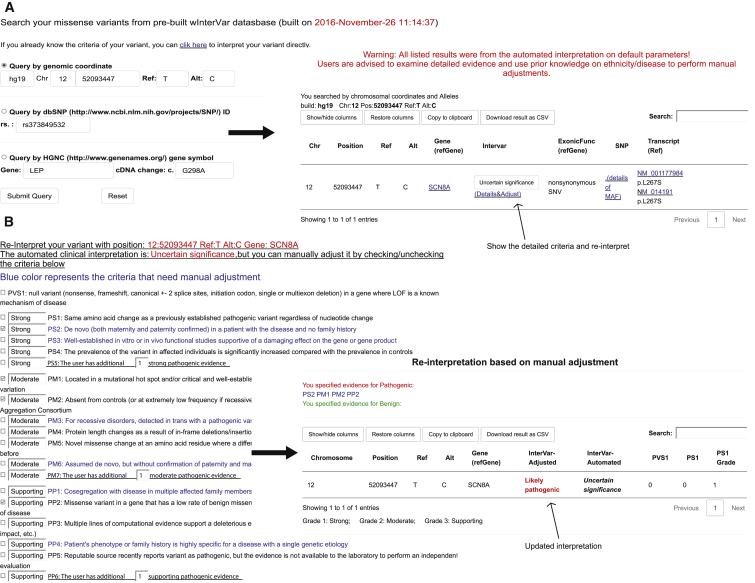
wInterVar (see Web Resources) is a web implementation of InterVar so that users can use an online web server to perform interpretation on individual variants without using command-line tools. The wInterVar server has two steps for assessing and adjusting the clinical significance of variants: users first input a variant to obtain pre-computed, automated interpretation (Figure 4A). After reviewing the results of automated interpretation, users can then click the “adjust” button to perform the manual adjustment step by selecting and de-selecting appropriate criteria according to additional information and domain knowledge. The wInterVar server will then perform the final interpretation with the two-step procedure (Figure 4B).
Figure 4.
Illustration of wInterVar
(A) Automatic interpretation of genetic variants, which can be entered by several means.
(B) Once users click “adjust,” the full list of criteria is shown for manual adjustment, after which the final results are given.
We assessed the speed of InterVar and wInterVar. Using a machine with 16 GB of memory and two Intel Xeon X5650 (2.67 GHz) CPUs, the InterVar pipeline takes approximately 40 min to annotate 3,000,000 variants from a whole genome. The runtime can be greatly reduced to <5 min (∼0.1 ms per variant) if an existing ANNOVAR annotation file is already available. For the wInterVar server, all annotation results for all possible non-synonymous variants were already pre-computed and imported into MongoDB, a NoSQL database system. Therefore, users can quickly search specific variants and receive an almost immediate response (<1 s for a variant). In addition, users can manually adjust the criteria and re-submit to wInterVar to obtain the final interpretation with an almost immediate response.
Discussion
In this article, we have presented two computational tools, InterVar and wInterVar, for performing evidence-based clinical interpretation of genetic variants according to the 2015 ACMG-AMP guidelines. To the best of our knowledge, we are not aware of software tools that are freely available to the academic community and perform similar functionalities. We wish to emphasize that although InterVar is a computational tool, it requires human input to derive accurate results with a two-step design: in the first step, InterVar performs automated interpretation with preliminary results, yet in the second step, InterVar takes additional information provided by human experts to adjust the criteria and provide a final interpretation. The two-step procedure allows InterVar to leverage automated information retrieval as much as possible, yet also allows additional input by human experts, to obtain the most accurate interpretations for genetic variants.
We applied InterVar to annotate and interpret de novo variants in subjects with neurodevelopmental disease and control subjects and observed a strong enrichment of pathogenic or likely pathogenic variants in affected subjects. In comparison, simple deleteriousness prediction algorithms such as SIFT and PolyPhen-2 failed to differentiate affected from control subjects. This observation suggests that one should compile multiple sources of criteria (in this case, up to 28 criteria), including deleteriousness prediction algorithms, to assess the potential pathogenicity of genetic variants rather than rely on deleteriousness prediction algorithms only.
Currently, a number of public databases, such ClinVar and HGMD, document the clinical significance of genetic variants, which are mostly provided by submitters or manually compiled from scientific literature. Because different submitters or different authors can have very different criteria to assess the pathogenicity of genetic variants, the quality of entries in these databases can be highly heterogeneous. As a result, it is expected that a proportion of pathogenic variants in these databases might simply be false positives that are misclassified.48, 49, 50, 51 Several studies have demonstrated that after manual re-interpretation, many of the pathogenic variants are indeed benign or have uncertain significance.55, 56, 57 Our results in the current study further support the observation that a very large proportion of documented pathogenic or likely pathogenic variants are indeed polymorphisms segregating in the population and are unlikely to contribute significantly to disease risk. These observations further support the importance of efforts, such as ClinGen, to compile high-quality, gold-standard datasets with confidence scores to be used by the community for more accurate interpretation of genetic variants.
InterVar has several limitations that we wish to discuss here. First, InterVar needs a variant knowledgebase for accurate interpretation, so some variants in some genes might be more accurately interpreted than others. For example, well-studied genes tend to have more entries in clinical databases and are more likely to be interpreted accurately. Second, InterVar is designed to interpret genetic variants that are likely to cause Mendelian diseases or are highly penetrant for Mendelian diseases (OR > 5) and cannot handle alleles that increase susceptibility to common and complex traits. Therefore, we caution that the current interpretation is appropriate only for Mendelian diseases or Mendelian forms of complex diseases. Third, although we provide a set of default databases to help implement 18 of the 2015 ACMG-AMP criteria, it is expected that different users or groups might want to use their own versions of these criteria. Therefore, we designed InterVar to be highly flexible in taking user-supplied annotations for each of the criteria to accommodate a variety of users with different needs.
Another issue we wish to emphasize is that the 2015 ACMG-AMP guidelines use 28 criteria with equal weights. One underlying rationale might be that it is difficult to quantify the contribution of each criterion given the complexity of interpreting genetic evidence.25 Another potential reason is that equal weighting is intuitively easier to understand and implement by clinicians and researchers. However, it is expected that different types of criteria might have different contributions and weights for the classification of the pathogenicity or quantitative prediction of pathogenicity. If we can accumulate very large datasets of true positives and true negatives, it is possible to apply machine-learning approaches in the future for more accurate prediction and quantitative assessment of pathogenicity for genetic variants.
One important caveat that we wish to stress is that InterVar is better suited to addressing the variant-interpretation problem for severe congenital or very early-onset developmental disorders with nearly 100% penetrance, but it might work less well for late-onset or recessive diseases. For example, amyotrophic lateral sclerosis (ALS) is a fatal, progressive neurodegenerative disease, and the non-canonical IκB kinase family member TANK binding kinase 1 (TBK1 [MIM: 604834]) was recently identified as an ALS-related gene in whole-exome sequencing of 2,874 ALS individuals and 6,405 control individuals.58 InterVar classified all TBK1 variants reported in the study as benign or having uncertain significance. Another example is TREM2 (MIM: 605086), associated with Alzheimer disease, from a recent sequencing study on a heterogeneous population of 1,092 affected and 1,107 control subjects.59 Rare variants in TREM2 (especially SNP rs75932628, which has the strongest association) were reported in their study. However, none of these variants were predicted to be pathogenic by InterVar. One main reason is that databases such as the ExAC Browser and ESP6500 were used in compiling the criteria, but they are technically not appropriate control databases because they are actually composed of many adult individuals with diseases. In comparison, the 1000 Genome Project is probably a more appropriate source of general control subjects, but its sample size is too small to enable adequate evaluation of rare variants. In any case, when databases such as the ExAC Browser and ESP6500 are used, it could be tricky to assign BS1 and BS2 to adult-onset or late-onset disorders, and some user-specific adjustments might be necessary for these diseases.
In summary, we have developed InterVar, a computational tool, and wInterVar, a web server, for the clinical interpretation of genetic variants according to the 2015 ACMG-AMP guidelines. InterVar can automatically generate the preliminary interpretations for 18 criteria and then allow manual adjustment of additional criteria to arrive at the final interpretation. InterVar can be easily used by researchers and clinicians and will greatly facilitate our understanding of the functional consequences of genetic variants in human diseases.
Acknowledgments
The authors thank Dr. Fan Xia (Baylor College of Medicine) and Dr. Rong Mao (ARUP Laboratories) for reading the manuscripts and offering valuable suggestions on the web server. We thank three anonymous reviewers for their valuable comments, which helped improve the manuscript substantially. We also want to thank members of the K.W. lab for testing the InterVar and wInterVar tools and providing feedback. This study was supported by NIH grants HG006465 and MH108728. K.W. was previously a board member and stock holder of Tute Genomics, a bioinformatics software company.
Published: January 26, 2017
Web Resources
1000 Genomes Project, http://www.1000genomes.org/
CLINVITAE, http://clinvitae.invitae.com/
Ensembl, http://www.ensembl.org/
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org
GWASdb, http://jjwanglab.org/gwasdb
HGMD, http://www.hgmd.org
InterVar, https://github.com/WGLab/InterVar
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://omim.org/
OrphaNet, http://www.orpha.net/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2
RepeatMasker, http://www.repeatmasker.org/
SIFT, http://sift.jcvi.org/
UCSC Genome Browser, http://genome.ucsc.edu
wIntervar, http://wintervar.wglab.org/
References
- 1.McPherson J.D. Next-generation gap. Nat. Methods. 2009;6(11, Suppl):S2–S5. doi: 10.1038/nmeth.f.268. [DOI] [PubMed] [Google Scholar]
- 2.Lyon G.J., Wang K. Identifying disease mutations in genomic medicine settings: current challenges and how to accelerate progress. Genome Med. 2012;4:58. doi: 10.1186/gm359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quintáns B., Ordóñez-Ugalde A., Cacheiro P., Carracedo A., Sobrido M.J. Medical genomics: The intricate path from genetic variant identification to clinical interpretation. Appl. Transl. Genomics. 2014;3:60–67. doi: 10.1016/j.atg.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang X., Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J. Med. Genet. 2012;49:433–436. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yandell M., Huff C., Hu H., Singleton M., Moore B., Xing J., Jorde L.B., Reese M.G. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21:1529–1542. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 11.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shihab H.A., Gough J., Cooper D.N., Day I.N., Gaunt T.R. Predicting the functional consequences of cancer-associated amino acid substitutions. Bioinformatics. 2013;29:1504–1510. doi: 10.1093/bioinformatics/btt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 15.González-Pérez A., López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thusberg J., Olatubosun A., Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum. Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 18.Thompson B.A., Greenblatt M.S., Vallee M.P., Herkert J.C., Tessereau C., Young E.L., Adzhubey I.A., Li B., Bell R., Feng B. Calibration of multiple in silico tools for predicting pathogenicity of mismatch repair gene missense substitutions. Hum. Mutat. 2013;34:255–265. doi: 10.1002/humu.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horaitis O., Talbot C.C., Jr., Phommarinh M., Phillips K.M., Cotton R.G. A database of locus-specific databases. Nat. Genet. 2007;39:425. doi: 10.1038/ng0407-425. [DOI] [PubMed] [Google Scholar]
- 22.Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., Nussbaum R.L., ClinGen ClinGen--the Clinical Genome Resource. N. Engl. J. Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazazian H.H., Boehm C.D., Seltzer W.K. ACMG recommendations for standards for interpretation of sequence variations. Genet. Med. 2000;2:302–303. [Google Scholar]
- 24.Richards C.S., Bale S., Bellissimo D.B., Das S., Grody W.W., Hegde M.R., Lyon E., Ward B.E., Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 25.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amendola L.M., Jarvik G.P., Leo M.C., McLaughlin H.M., Akkari Y., Amaral M.D., Berg J.S., Biswas S., Bowling K.M., Conlin L.K. Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am. J. Hum. Genet. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum. Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X., Jian X., Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T.K., Bateman A., Bernard T., Binns D., Bork P., Burge S. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jian X., Boerwinkle E., Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42:13534–13544. doi: 10.1093/nar/gku1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis B.P., Green R.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M.J., Liu Z., Wang P., Wong M.P., Nelson M.R., Kocher J.P., Yeager M., Sham P.C., Chanock S.J., Xia Z., Wang J. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2016;44(D1):D869–D876. doi: 10.1093/nar/gkv1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malcolmson J., Kleyner R., Tegay D., Adams W., Ward K., Coppinger J., Nelson L., Meisler M.H., Wang K., Robison R., Lyon G.J. SCN8A mutation in a child presenting with seizures and developmental delays. Cold Spring Harb Mol Case Stud. 2016;2:a001073. doi: 10.1101/mcs.a001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deciphering Developmental Disorders S., Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Girard S.L., Gauthier J., Noreau A., Xiong L., Zhou S., Jouan L., Dionne-Laporte A., Spiegelman D., Henrion E., Diallo O. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat. Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 40.Xu B., Ionita-Laza I., Roos J.L., Boone B., Woodrick S., Sun Y., Levy S., Gogos J.A., Karayiorgou M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gulsuner S., Walsh T., Watts A.C., Lee M.K., Thornton A.M., Casadei S., Rippey C., Shahin H., Nimgaonkar V.L., Go R.C., Consortium on the Genetics of Schizophrenia (COGS) PAARTNERS Study Group Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fromer M., Pocklington A.J., Kavanagh D.H., Williams H.J., Dwyer S., Gormley P., Georgieva L., Rees E., Palta P., Ruderfer D.M. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen A.S., Berkovic S.F., Cossette P., Delanty N., Dlugos D., Eichler E.E., Epstein M.P., Glauser T., Goldstein D.B., Han Y., Epi4K Consortium. Epilepsy Phenome/Genome Project De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamdan F.F., Srour M., Capo-Chichi J.M., Daoud H., Nassif C., Patry L., Massicotte C., Ambalavanan A., Spiegelman D., Diallo O. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10:e1004772. doi: 10.1371/journal.pgen.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rauch A., Wieczorek D., Graf E., Wieland T., Endele S., Schwarzmayr T., Albrecht B., Bartholdi D., Beygo J., Di Donato N. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 46.de Ligt J., Willemsen M.H., van Bon B.W., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 47.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 48.(2016). Improving databases for human variation. Nat. Methods 13, 103. [DOI] [PubMed]
- 49.MacArthur D.G., Manolio T.A., Dimmock D.P., Rehm H.L., Shendure J., Abecasis G.R., Adams D.R., Altman R.B., Antonarakis S.E., Ashley E.A. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dorschner M.O., Amendola L.M., Turner E.H., Robertson P.D., Shirts B.H., Gallego C.J., Bennett R.L., Jones K.L., Tokita M.J., Bennett J.T., National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am. J. Hum. Genet. 2013;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bell C.J., Dinwiddie D.L., Miller N.A., Hateley S.L., Ganusova E.E., Mudge J., Langley R.J., Zhang L., Lee C.C., Schilkey F.D. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci. Transl. Med. 2011;3:65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi L., Guo Y., Dong C., Huddleston J., Yang H., Han X., Fu A., Li Q., Li N., Gong S. Long-read sequencing and de novo assembly of a Chinese genome. Nat. Commun. 2016;7:12065. doi: 10.1038/ncomms12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green R.C., Berg J.S., Grody W.W., Kalia S.S., Korf B.R., Martin C.L., McGuire A.L., Nussbaum R.L., O’Daniel J.M., Ormond K.E., American College of Medical Genetics and Genomics ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amendola L.M., Dorschner M.O., Robertson P.D., Salama J.S., Hart R., Shirts B.H., Murray M.L., Tokita M.J., Gallego C.J., Kim D.S. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res. 2015;25:305–315. doi: 10.1101/gr.183483.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue Y., Chen Y., Ayub Q., Huang N., Ball E.V., Mort M., Phillips A.D., Shaw K., Stenson P.D., Cooper D.N., Tyler-Smith C., 1000 Genomes Project Consortium Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am. J. Hum. Genet. 2012;91:1022–1032. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shearer A.E., Eppsteiner R.W., Booth K.T., Ephraim S.S., Gurrola J., 2nd, Simpson A., Black-Ziegelbein E.A., Joshi S., Ravi H., Giuffre A.C. Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am. J. Hum. Genet. 2014;95:445–453. doi: 10.1016/j.ajhg.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tabor H.K., Auer P.L., Jamal S.M., Chong J.X., Yu J.H., Gordon A.S., Graubert T.A., O’Donnell C.J., Rich S.S., Nickerson D.A., Bamshad M.J., NHLBI Exome Sequencing Project Pathogenic variants for Mendelian and complex traits in exomes of 6,517 European and African Americans: implications for the return of incidental results. Am. J. Hum. Genet. 2014;95:183–193. doi: 10.1016/j.ajhg.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cirulli E.T., Lasseigne B.N., Petrovski S., Sapp P.C., Dion P.A., Leblond C.S., Couthouis J., Lu Y.F., Wang Q., Krueger B.J., FALS Sequencing Consortium Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S.K., Younkin S., Alzheimer Genetic Analysis Group TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]