Abstract
Diagnosis at the edges of our knowledge calls upon clinicians to be data driven, cross-disciplinary, and collaborative in unprecedented ways. Exact disease recognition, an element of the concept of precision in medicine, requires new infrastructure that spans geography, institutional boundaries, and the divide between clinical care and research. The National Institutes of Health (NIH) Common Fund supports the Undiagnosed Diseases Network (UDN) as an exemplar of this model of precise diagnosis. Its goals are to forge a strategy to accelerate the diagnosis of rare or previously unrecognized diseases, to improve recommendations for clinical management, and to advance research, especially into disease mechanisms. The network will achieve these objectives by evaluating patients with undiagnosed diseases, fostering a breadth of expert collaborations, determining best practices for translating the strategy into medical centers nationwide, and sharing findings, data, specimens, and approaches with the scientific and medical communities. Building the UDN has already brought insights to human and medical geneticists. The initial focus has been on data sharing, establishing common protocols for institutional review boards and data sharing, creating protocols for referring and evaluating patients, and providing DNA sequencing, metabolomic analysis, and functional studies in model organisms. By extending this precision diagnostic model nationally, we strive to meld clinical and research objectives, improve patient outcomes, and contribute to medical science.
Keywords: rare diseases, diagnosis, National Institutes of Health, cooperative behavior, phenotyping, high-throughput nucleotide sequencing
Main Text
Introduction
Rare, novel, and undiagnosed disorders challenge patients, their families, and clinicians. The National Institutes of Health (NIH) Office of Rare Diseases Research noted that 6% of individuals seeking their assistance had an undiagnosed disease; for those finally diagnosed, 15% had experienced persistent symptoms without a diagnosis for at least 5 years.1 Long diagnostic odysseys are expensive, and they involve repeated and duplicative diagnostic efforts with risks of invasive procedures and false diagnoses.
The Undiagnosed Diseases Network (UDN) was designed to forge a paradigm for diagnosing rare and previously unrecognized diseases, improving the management of these conditions, and accelerating research on rare diseases. The UDN exists to increase the national capacity to assess those with mysterious medical conditions, to create a network with broad collaborative expertise, and to determine ways to extend this model to other medical centers. Our work products include findings, data, specimens, and best practices, which we share broadly with the research, clinical, and patient communities. This commentary offers the lessons we have learned in building the UDN with the goal of stimulating dissemination of this precision diagnostic model.
Building the Network
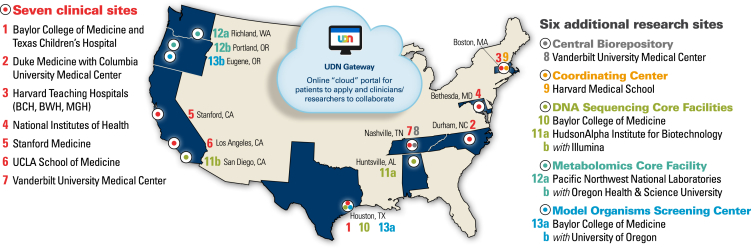
The UDN is an extension of the Undiagnosed Diseases Program (UDP), pioneered in 2008, at the NIH Clinical Center in Bethesda.2, 3, 4 The UDP initially was funded by the NIH Office of Rare Diseases Research. Based upon its success, planning for the UDN began in 2012. The UDN has a 5-year budget of $120,000,000 from the NIH Common Fund: the National Human Genome Research Institute (NHGRI) serves as the lead administrator. Currently, the UDN consists of seven clinical sites, two sequencing cores, a metabolomics core, a model-organism screening center, a central biorepository, and a coordinating center (Figure 1). The first patient was seen in September 2015. The large number of patients with rare and newly recognized conditions, the breadth of the expertise in the network, and the highly collaborative nature of the UDN represent a signal opportunity to accelerate discovery about health and disease while developing best practices for the diagnosis of challenging cases.
Figure 1.
The Undiagnosed Diseases Network
The Undiagnosed Diseases Network (UDN) includes seven clinical sites, two DNA sequencing cores, a metabolomics core, a model-organism screening center, a central biorepository, and a coordinating center. Technologically, the sites are linked via the UDN Gateway, through which participants also apply. Abbreviations are as follows: BCH, Boston Children's Hospital; BWH, Brigham and Women's Hospital; and MGH, Massachusetts General Hospital. (Figure modified from the UDN website, maintained by the coordinating center.)
The UDN coordinating center, based at the Department of Biomedical Informatics at Harvard Medical School (Boston), opened in January 2014. In July 2014, six additional clinical sites (beyond the NIH UDP) were announced: Baylor College of Medicine (Houston), Duke University Medical Center (Durham) in collaboration with Columbia University (New York), Harvard Teaching Hospitals (Boston Children’s Hospital, Brigham and Women’s Hospital, and Massachusetts General Hospital in Boston), Stanford University (Stanford), the University of California, Los Angeles (Los Angeles), and Vanderbilt University (Nashville). In September 2014, two DNA sequencing cores were named: Baylor College of Medicine and HudsonAlpha (Huntsville)—initially based at the Medical College of Wisconsin (Milwaukee)—in collaboration with Illumina Laboratory (San Diego). The UDN’s capabilities were further extended in September 2015 through a central biorepository at Vanderbilt University Medical Center, a model-organism screening center at the Baylor College of Medicine and the University of Oregon (Eugene), and a metabolomics core at Battelle Pacific Northwest Laboratories (Richland) and the Oregon Health and Science University (Portland).
An integrated network is emerging as a result of close interactions among the investigators and NIH scientists, a mutual enthusiasm for the mission, and shared governance, which is laid out in the UDN Manual of Operations. The primary governing body is the Steering Committee. UDN operations take place in the context of working groups and committees (e.g., Billing, Biosamples and Biorepository, Case Review Committee, Clinical Protocols, Genetic Counseling and Testing, Metabolomics, Model Organisms, Publications and Research Committee, and Sequencing), which will evolve over time to meet the changing needs of the network. For example, the Clinical Protocols working group has two subgroups: Utility and Utilization and Site Operations. Patients and their families are integral to the network, for which reason we have formalized a Patient Engagement Group. An independent group of external scientific advisors also reviews the UDN and makes recommendations to the NIH.
The Protocol
Referral
Patients and their families are the center of the UDN. They simultaneously are patients, volunteer research subjects, and partners. Patients or their referring clinicians begin the UDN application process through an online gateway (“UDN Gateway”) developed and maintained by the coordinating center. When submitted online, the application is routed to a clinical site, typically the closest one geographically.
Professionals at the clinical site decide whether to offer a UDN evaluation on the basis of review of the online application as well as medical records submitted to the site. A network review committee meets regularly to advise the clinical sites on case selection and offer suggestions. In general, patients are accepted after earlier extensive evaluations fail to reach a diagnosis that explains objective findings or when phenotypic manifestations are novel and suggestive of an unrecognized disorder or relatives are similarly affected. By 2017, after a scale-up interval, we anticipate that the network will accept 450 patients with a wide range of phenotypes each year—150 at the NIH Clinical Center and 50 at each of the six other clinical sites.
Diagnostic Protocol
The UDN diagnostic process emphasizes a highly collaborative and coordinated clinical evaluation, detailed and standardized documentation of patient phenotype, and tightly coupled bedside and bench collaborations. The process does not have a fixed termination date. Among those diagnosed at the NIH UDP from 2008 to 2015, the length of time to arrive at a diagnosis after the on-site evaluation ranged from 1 week to 4 years.2, 3, 4
Clinical Evaluation
A signature feature of the NIH UDP, and now the UDN, is the compression of a patient’s clinical evaluation into the equivalent of five inpatient days, which requires rigorous coordination of activities at the clinical site. The evaluation involves detailed clinical consultations by multiple specialists, laboratory testing, diagnostic procedures, and imaging studies, all tailored to the patient’s individual presentation. While focusing on the specific patient, the clinical sites follow standardized protocols for phenotyping and specimen collection to facilitate later comparisons. Samples, including plasma, serum, peripheral-blood mononuclear cells, DNA, urine, and sometimes skin biopsies, are collected and stored both at the local clinical site and in a central repository to facilitate specimen backup, access, and sharing.
Phenotyping
The term “phenotype” can have various meanings. In the UDN, the term refers to an individual’s morphologic, physiologic, and behavioral characteristics with a particular emphasis on deviations from normal. The systematic documentation of phenotype is fundamental to the diagnosis of elusive conditions. Phenotyping has been described as “…a surprising and embarrassing secret [… because] the final determination of whether an individual mutation is responsible for disease in an individual patient remains fundamentally impressionist.”5 In other words, the potential precision of genomics is starkly contrasted with the subjectiveness of the clinical assessments, for which reason the National Research Council emphasized the need for new phenotype characterization and disease taxonomy.6 Thus, a detailed and standardized phenotype is an essential feature of the UDN diagnostic evaluation.
The use of a standardized, computer-parsable classification system (ontology) is the first step toward improving documentation of signs and symptoms. The UDN encodes phenotypes with Human Phenotype Ontology (HPO) terms,7 which are used worldwide by numerous projects and are listed on the HPO website. Some examples of projects include FORGE Canada8 and DECIPHER. The HPO has approximately 11,000 computer- and human-interpretable terms. Primary HPO terms do not capture quantitative data; however, terms can be associated with metadata, which allow for the inclusion of quantitative data, age of onset, severity, periodicity, and other modifiers. HPO terms are internally arranged in acyclic, hierarchical graphs, allowing for similar terms to be related to one another in a computable manner. Similar relatedness procedures can be used to match patients to patients, patients to disease-associated genes, and patients to model organisms with orthologous gene-phenotype associations. The UDN uses PhenoTips software to collect and analyze phenotypic information; it provides a flexible interface for selecting HPO terms and recording associated metadata.9
The focused effort required for encoding phenotypes into standardized terms pays dividends in at least six ways:
-
•
Delineation of the phenotype of novel conditions.
-
•
Identification of additional cases: capturing phenotypic features in a systematic and standardized way enables the use of tools, such as those present in PhenomeCentral, for identifying individuals affected by the same unnamed disorder through a phenotype-matching algorithm.10
-
•
Analysis of genome sequence data: one example of the added value of phenotyping to the analysis of sequence data is Exomiser, a tool that accounts for phenotypic relevance to prioritize variants.11
-
•
Expansion of the phenotype of rare conditions: some individuals diagnosed by the NIH UDP had previously defined rare conditions, and a number of these individuals presented with novel variations of the classic phenotype.2
-
•
Genetic heterogeneity: the other side of the phenotypic-expansion coin is that a detailed map of molecular and phenotypic data helps to untangle convergent and overlapping phenotypes by identifying phenotypes that might have different genetic bases. It would also allow for the identification of underlying genetic heterogeneity of specific phenotypes.
-
•
Identification of model-organism phenotypes that are potentially related to the participant’s phenotype.
Sequencing and Interpretation
Most UDN participants are candidates for exome or genome sequencing. Filtering the large number of variants that sequencing identifies is a challenge. Variant classification is facilitated by the accumulation of curated information on factors such as population frequency, segregation according to a proposed genetic model, and predicted impact on gene function.
Many diagnostic successes have used clues that were present before sequencing, such as linkage analysis,12 regions of homozygosity,13 the presence of non-physiologic metabolites,14 and clinical resemblance to known syndromes. Application of sequencing techniques to the UDN cohort is especially challenging because of the dearth of pre-sequencing clues. Many patients have unique presentations, no history of consanguinity, and no similarly affected relatives, making traditional linkage methods unfeasible. Especially in the context of undiagnosed diseases, the successful interpretation of sequencing data relies on collaboration among clinicians, bioinformaticians, and other scientists familiar with the phenotypic features of the patients. The UDN reflects this synergy in three ways: (1) the emphasis on in-depth phenotyping at clinical sites, (2) the integration of sequencing core investigators into the UDN team and activities, and (3) the fact that interpretations of a participant’s sequencing results most often take place at both a sequencing core and a clinical site, whose combined efforts lead to the final clinical report. Through this work, the UDN will contribute to the understanding of what conditions predict the success of DNA sequencing as a first-line approach for patients whose initial conventional testing fails to give an answer.
Going Beyond Phenotype and Genotype
Metabolomics
Additional inborn errors of metabolism surely remain to be discovered, and the NIH UDP, in its early years, identified two previously unrecognized congenital disorders of glycosylation, IIb (MOGS-CDG)15and Iz,16 and deficiency of cytosolic phosphoenolpyruvate carboxylate.17 Hence, a core metabolomics laboratory provides analyses for selected subjects, especially when autosomal-recessive inheritance is likely, the clinical history suggests a metabolic disorder, and routine biochemical assays, e.g., urinary and serum amino acids and organic acids, are not diagnostic. Through the UDN metabolomics core, the network is able to identify and quantitate non-traditional metabolites through targeted and untargeted assays.
Model Systems and Functional Studies
Findings in model systems help discriminate among candidate variants identified by sequencing in a genetically and environmentally controlled background. Further, functional studies in model systems provide insight because they allow repeated observations of pathogeneses of rare genetic disorders and the patients’ clinical courses, thus enabling the study of early phases of disorders that might be subclinical in humans, allowing access to tissues that are difficult to obtain from humans, and providing a platform for developing therapeutics.18 The UDN incorporates these studies in two ways: (1) at the model-organism screening center and (2) though supplements and grant awards for research on gene function.
Environmental Determinants
The search for the etiologies of rare conditions often focuses on the genome, and the diagnoses achieved by the NIH UDP have largely been based on genetic variants. Nevertheless, we subscribe to a disease model in which each person diverts from a path of health as a result of a unique combination of intrinsic genetic variations that interact within cells and the person’s environment, a lifetime of extrinsic factors.19, 20, 21 Most human carcinogens and teratogens have been discovered by alert practitioners who have probed deeply into the histories and pathologies of individual patients with unusual or rare cancers or congenital defects.22 Hence, all patients accepted into the UDN are asked to complete a wide-ranging environmental questionnaire so the UDN can identify potentially significant exposures.
The UDN is also equipped to investigate other axes of the diagnostic matrix, including immune and inflammatory contributors.
Practical Considerations in Implementing the UDN
Over its 2 years of existence, the UDN has successfully established governance procedures and regulatory milestones and has resolved a number of challenges relevant to emerging efforts in precision medicine.
Central IRB and Reliance Agreements
The NIH has encouraged the use of single institutional review boards (IRBs) for NIH-funded multi-site studies to assure productivity and uniformity without compromising ethical principles or protections. In a dynamic project such as the UDN, such an arrangement is certainly more efficient than requiring multiple IRB approvals. This efficiency came at the cost of an 11-month investment in negotiating reliance agreements between the NHGRI IRB and 13 other site IRBs; both prime awardees and subcontractors had to sign reliance agreements. In parallel, the NHGRI IRB acted as the central IRB and reviewed the initial UDN clinical protocol. Together with the NIH Office of Human Subjects Research Protections, the UDN central IRB worked closely with the coordinating center to complete the reliance agreement and IRB review process. The coordinating center now serves as the IRB liaison for the UDN and maintains the protocol.
Financial Assistance and Billing Procedures
The UDN strives to permit access to all patients irrespectively of their socioeconomic or health insurance status. The only way to ensure that patients would not be burdened with co-payments and deductibles would be to draw upon grant funds to cover the entire evaluation. Some clinical sites have chosen this approach for all participants. This is made feasible because those sites have substantial institutional discounts for patient care performed as part of research studies. Other clinical sites have chosen a hybrid model in which insurance is billed for clinically indicated evaluations and procedures and grant funding is used for research investigations. To enable the offset of co-payments and deductibles of patients at sites that bill insurance, the UDN has partnered with the National Organization for Rare Disorders to create a UDN patient assistance fund, made possible through the generous contributions of the Running for Rare Diseases Team. A collaboration with Mercy Medical Angels assists with travel expenses for those with financial need. We intend to assess the financial implications for the sustainability and dissemination of the UDN strategy as part of our internal review process.
UDN Data Sharing and Use Agreement
Achieving our combined clinical and research mission depends upon the exchange of identifiable patient data. A network-wide data sharing and use agreement enables such an exchange, which is reflected in the IRB protocol, the informed-consent process, and the UDN Manual of Operations. The data sharing and use agreement was the result of 8 months of negotiations among the participating institutions, which were usually represented by a member of their technology transfer offices.
External Data Sharing
In 2013, the White House Office of Science and Technology Policy released a memorandum entitled “Increasing Access to the Results of Federally Funded Scientific Research.”23 In response, the NIH redoubled its efforts to increase access to publications and digital data from the research it funds, culminating in both the publication of the “Plan for Increasing Access to Scientific Publications and Digital Scientific Data from NIH Funded Scientific Research”24 and added funds to advance data discovery.25
There are important ethical, legal, and social considerations associated with data sharing. The UDN illustrates many of the points raised in the Institute of Medicine Report, “Sharing Clinical Trial Data: Maximizing Benefits, Minimizing Risk.”26 A prime objective is respecting participant privacy. By definition, UDN participants have rare conditions, which challenges anonymization. Furthermore, in the context of “N of 1” clinical research, any perturbations of the data to increase anonymity could hamper their validity and utility. At the same time, although data sharing in many contexts is unlikely to benefit the participants themselves, UDN data sharing might do so by leading to the identification of additional cases that would clinch a definitive diagnosis. As the NIH UDP has done, the UDN shares data with PhenomeCentral, a secure data-sharing repository that encourages global scientific collaboration within the rare disease community while respecting the privacy of profiled patients. Users contribute records that consist of an anonymized description of observed phenotypes, along with the genetic variants, specified as either DNA sequencing results or a list of candidate genes. For a given patient record, users are shown limited information about other patients, including phenotypic similarities with potential genetic causes. PhenomeCentral is, in turn, a part of the MatchMaker Exchange collaborative effort.27, 28 Anonymized UDN data will also be deposited in dbGaP (dbGaP: phs001232.v1.p1), the NIH’s curated repository of studies that investigate genotype-phenotype interactions.29
Not only can UDN researchers share data with those outside of the network, but UDN patients and families can also lead their own data-sharing efforts, as illustrated by the work of Matthew Might, who is a family advisor to the UDN coordinating center.30
Collaborative Clinical Sites
To extend the reach of the UDN approach, we welcome the participation of investigators who are interested in collaborating with the network. A number of international sites have expressed interest in learning from the UDN experience, so as a first step, we announced the opportunity to participate as a UDN International Collaborative Clinical Site. To be considered, sites must meet certain criteria, including preparation of an application that summarizes their plans for clinical evaluation and laboratory tests (including sequencing), their adherence to the UDN publications policy, a data-sharing plan, IRB approvals, and evidence of independent funding. Sites that are accepted are expected to contribute significantly to the UDN mission and goals through their substantial intellectual contributions and participation in UDN activities. Likewise, international interest in coordinated sharing of data from undiagnosed cases is reflected by the establishment of the UDN International and substantial efforts in 12 countries.31
The UDN and Beyond
The lessons that the UDN holds for medicine extend well beyond the individuals we evaluate. The overwhelming majority of our patients come to us after misdiagnoses or no unifying diagnosis at all. In the general population, many common diseases, such as cardiovascular conditions, cancer, and infections—not just rare diseases—are often misdiagnosed.32 Most Americans experience a diagnostic error—a delayed diagnosis, missed diagnosis, or misdiagnosis—which can cause harm because of delayed or inappropriate tests and treatments.33 The Institute of Medicine’s report considers this fact a call-to-arms to reform the US healthcare system by strengthening teamwork and building health information technology that supports the diagnostic process, both central elements of the UDN.
Coda: Distinguishing Clinical from Research Activities
A decade ago, F. Miller argued for the tradition of distinct conceptual and moral boundaries between research and clinical care: “Medical care has a personalized focus. It is directed to helping a particular person in need of expert medical attention. Clinical research essentially lacks this purpose of personalized help for particular individuals. . . . The distinctive purpose of clinical research [is] to develop generalizable knowledge.”34
Currently, the search for the causes of undiagnosed diseases necessarily incorporates both clinical and research activities. This dichotomy carries with it three assumptions that do not hold in precision medicine efforts such as the UDN: (1) research presents less net clinical benefit and greater overall risk, (2) research introduces clinically irrelevant burdens and risks, and (3) research protocols dictate which interventions a patient receives.35 Most of the patient evaluation that takes place at a UDN clinical site is clinical care (e.g., consultations, procedures, specimen and collection) according to accepted medical practice. Few procedures are performed for purely research purposes (e.g., skin biopsies to culture fibroblasts), but even this formal designation as “research” might underemphasize the role that these activities could play in identifying the causes of these conditions that extend beyond current clinical knowledge.
The distinction between clinical and research purposes is not merely academic, for it directly influences both UDN operations and the definition of activities that are subject to formal ethical oversight. Apart from the NIH UDP, all UDN clinical sites are based at Health Information Portability and Accountability Act (HIPAA)-covered entities. At those clinical sites, the consent forms include authorization to disclose protected health information, as required by HIPAA. As mentioned above, exchange within the network is enabled by a data sharing and use agreement. An alternative could have been to frame the UDN as primarily engaged in clinical work but with business associate agreements in place to underpin the exchange of data under HIPAA (45 CFR 164.5).
It is increasingly recognized that “research” and “practice” are becoming less useful as shorthand for our fundamental moral concerns. Indeed, artificially enforcing this distinction could be detrimental in addressing undiagnosed diseases, the learning healthcare system,36 and precision medicine.37 In the UDN, the collaborative integration—the so-called virtuous cycle—of the bedside and the bench simultaneously serves the patient and biomedical science, given that each patient presents with compelling clinical needs and important research questions.
Consortia
Members of the Undiagnosed Diseases Network include David R. Adams, Christopher J. Adams, Mercedes E. Alejandro, Patrick Allard, Euan A. Ashley, Mashid S. Azamian, Carlos A. Bacino, Ashok Balasubramanyam, Hayk Barseghyan, Alan H. Beggs, Hugo J. Bellen, David Bernick, Jonathan A. Bernstein, Anna Bican, David P. Bick, Camille L. Birch, Braden E. Boone, Lauren C. Briere, Donna M. Brown, Catherine A. Brownstein, Matthew Brush, Elizabeth A. Burke, Lindsay C. Burrage, Katherine R. Chao, Gary D. Clark, Joy D. Cogan, Cynthia M. Cooper, William J. Craigen, Mariska Davids, Jyoti G. Dayal, Esteban C. Dell’Angelica, Shweta U. Dhar, Katrina M. Dipple, Laurel A. Donnell-Fink, Naghmeh Dorrani, Daniel C. Dorset, David D. Draper, Annika M. Dries, Rachel Eastwood, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Cecilia Esteves, Tyra Estwick, Paul G. Fisher, Trevor S. Frisby, Kate Frost, William A. Gahl, Valerie Gartner, Rena A. Godfrey, Mitchell Goheen, Gretchen A. Golas, David B. Goldstein, Mary “Gracie” G. Gordon, Sarah E. Gould, Jean-Philippe F. Gourdine, Brett H. Graham, Catherine A. Groden, Andrea L. Gropman, Mary E. Hackbarth, Melissa Haendel, Rizwan Hamid, Neil A. Hanchard, Lori H. Handley, Isabel Hardee, Matthew R. Herzog, Ingrid A. Holm, Ellen M. Howerton, Brenda Iglesias, Howard J. Jacob, Mahim Jain, Yong-hui Jiang, Jean M. Johnston, Angela L. Jones, Alanna E. Koehler, David M. Koeller, Isaac S. Kohane, Jennefer N. Kohler, Donna M. Krasnewich, Elizabeth L. Krieg, Joel B. Krier, Jennifer E. Kyle, Seema R. Lalani, Lea Latham, Yvonne L. Latour, C. Christopher Lau, Jozef Lazar, Brendan H. Lee, Hane Lee, Paul R. Lee, Shawn E. Levy, Denise J. Levy, Richard A. Lewis, Adam P. Liebendorder, Sharyn A. Lincoln, Carson R. Loomis, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Thomas C. Markello, Casey Martin, Paul Mazur, Alexandra J. McCarty, Allyn McConkie-Rosell, Alexa T. McCray, Thomas O. Metz, Matthew Might, Paolo M. Moretti, John J. Mulvihill, Jennifer L. Murphy, Donna M. Muzny, Michele E. Nehrebecky, Stan F. Nelson, J. Scott Newberry, John H. Newman, Sarah K. Nicholas, Donna Novacic, Jordan S. Orange, J. Carl Pallais, Christina G.S. Palmer, Jeanette C. Papp, Loren D.M. Pena, John A. Phillips III, Jennifer E. Posey, John H. Postlethwait, Lorraine Potocki, Barbara N. Pusey, Rachel B. Ramoni, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Sarah Sadozai, Katherine E. Schaffer, Kelly Schoch, Molly C. Schroeder, Daryl A. Scott, Prashant Sharma, Vandana Shashi, Edwin K. Silverman, Janet S. Sinsheimer, Ariane G. Soldatos, Rebecca C. Spillmann, Kimberly Splinter, Joan M. Stoler, Nicholas Stong, Kimberly A. Strong, Jennifer A. Sullivan, David A. Sweetser, Sara P. Thomas, Cynthia J. Tifft, Nathanial J. Tolman, Camilo Toro, Alyssa A. Tran, Zaheer M. Valivullah, Eric Vilain, Daryl M. Waggott, Colleen E. Wahl, Nicole M. Walley, Chris A. Walsh, Michael F. Wangler, Mike Warburton, Patricia A. Ward, Katrina M. Waters, Bobbie-Jo M. Webb-Robertson, Alec A. Weech, Monte Westerfield, Matthew T. Wheeler, Anastasia L. Wise, Lynne A. Wolfe, Elizabeth A. Worthey, Shinya Yamamoto, Yaping Yang, Guoyun Yu, and Patricia A. Zornio.
Conflicts of Interest
E.A.A. (Stanford) is an advisor to and stockholder in Personalis Inc. D.P.B. (HudsonAlpha) is a founder and chief medical officer of Envision Genomics, medical director of Smith Family Clinic LLC, assistant director of Clinical Services Laboratory LLC, and a scientific advisory board member of Genomics England. C.M.E. (BCM-Seq) is a full-time faculty member of Baylor College of Medicine and provides services as chief medical officer and chief quality officer to Baylor Miraca Genetics Laboratories through a professional services agreement. P.G.F. (Stanford) is an associate editor of the Journal of Pediatrics (Elsevier) and a paid consultant. D.B.G. (Columbia) owns equity in two precision medicine companies, Pairnomix and Clarus/EpiPM. J.E.P. (Baylor) is an employee of the Department of Molecular and Human Genetics at Baylor College of Medicine, which has entered a joint venture with Baylor Genetics Diagnostic Laboratory. J.A.R. (Baylor) is a member of the Department of Molecular and Human Genetics at Baylor College of Medicine, which derives revenue from clinical genetic testing offered by Baylor Genetics. M.S.C. (HudsonAlpha) is a laboratory director for the HudsonAlpha Clinical Services Lab LLC, which performs fee-for-service clinical laboratory testing. D.A.S. (Baylor) is a member of the Clinical Advisory Board of Baylor Genetics and works for the Department of Molecular and Human Genetics at Baylor College of Medicine, which derives revenue from genetic analyses offered through Baylor Genetics. E.K.S. (Harvard) received honoraria and consulting fees from Merck, grant support and consulting fees from GlaxoSmithKline, and honoraria and travel support from Novartis. P.A.W. (BCM-Seq) is a contract employee for Baylor Genetics, a clinical laboratory that derives income from whole-exome sequencing and other genetic testing.
Acknowledgments
Rachel Eastwood of the Undiagnosed Diseases Network Coordinating Center prepared Figure 1. We are grateful for the participation of patients and family members and their referring clinicians. This work was supported in part by the Intramural Research Program of the National Human Genome Research Institute and the NIH Common Fund through the Office of Strategic Coordination and Office of the NIH Director. Research reported in this manuscript was supported by the NIH Common Fund through the Office of Strategic Coordination and Office of the NIH Director under award numbers U01HG007530, U01HG007674, U01HG007703, U01HG007709, U01HG007672, U01HG007690, U01HG007708, U01HG007942, U01HG007943, U54NS093793, and U01TR001395. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Web Resources
Clinical Sites for an Undiagnosed Diseases Network (UDN) (U01), RFA-RM-13-004, http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-13-004.html
Coordinating Center for an Undiagnosed Diseases Network (UDN) (U01), RFA-RM-12-020, http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-12-020.html
DECIPHER, https://decipher.sanger.ac.uk/about
DNA Sequencing Core for an Undiagnosed Diseases Network (UDN) (U01), RFA-RM-13-018, http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-13-018.html
Gene Function Studies to Investigate Rare and Undiagnosed Diseases (Admin Supp), PA-13-076, http://grants.nih.gov/grants/guide/pa-files/PA-13-076.html
Human Phenotype Ontology, http://human-phenotype-ontology.github.io/
Mercy Medical Angels, http://mercymedical.org/
Metabolomics Core for the Undiagnosed Diseases Network (UDN) (U01), RFA-RM-15-001, http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-15-001.html
Model Organisms Screening Center for the Undiagnosed Diseases Network (UDN) (U54), RFA-RM-14-016, http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-14-016.html
National Human Genome Research Institute website for the UDN, https://www.genome.gov/27550959/undiagnosed-diseases-network-udn/
NIH Common Fund website for the UDN, https://commonfund.nih.gov/Diseases/index
NIH, Request for Comments on the Draft NIH Policy on the Use of a Single Institutional Review Board for Multi-Site Research, http://grants.nih.gov/grants/guide/notice-files/NOT-OD-15-026.html
NIH, Final NIH Policy on the Use of a Single Institutional Review Board for Multi-Site Research, https://grants.nih.gov/grants/guide/notice-files/NOT-OD-16-094.html
Running for Rare Disease Team, http://running4rare.org/about/
Undiagnosed Diseases Gene Function Research (R21), RFA-RM-14-005, http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-14-005.html
Undiagnosed Diseases Gene Function Research (R21), RFA-RM-15-004, http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-15-004.html
Undiagnosed Diseases Network (UDN), https://undiagnosed.hms.harvard.edu
Undiagnosed Diseases Network International (UDNI), http://www.udninternational.org
UDN Manual of Operations, http://undiagnosed.hms.harvard.edu/resources/for-researchers/
UDN online gateway, http://undiagnosed.hms.harvard.edu/apply/
References
- 1.National Technical Information Services (1989). Report of the National Commission on Orphan Diseases. Office of the Assistant Secretary for Health, Public Health Service, US Department of Health and Human Services, February 1989, p. 17. https://rarediseases.info.nih.gov/files/report_of_the_national_commission_on_orphan_diseases_february_1989.pdf.
- 2.Gahl W.A., Markello T.C., Toro C., Fajardo K.F., Sincan M., Gill F., Carlson-Donohoe H., Gropman A., Pierson T.M., Golas G. Genet. Med. 2012;14:51–59. doi: 10.1038/gim.0b013e318232a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gahl W.A., Wise A.L., Ashley E.A. JAMA. 2015;314:1797–1798. doi: 10.1001/jama.2015.12249. [DOI] [PubMed] [Google Scholar]
- 4.Gahl W.A., Mulvihill J.J., Toro C., Markello T.C., Wise A.L., Ramoni R.B., Adams D.R., Tifft C.J., UDN Mol. Genet. Metab. 2016;117:393–400. doi: 10.1016/j.ymgme.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrovski S., Goldstein D.B. Sci. Transl. Med. 2014;6:254fs35. doi: 10.1126/scitranslmed.3010272. [DOI] [PubMed] [Google Scholar]
- 6.National Research Council Committee on a Framework for Developing a New Taxonomy of Disease . Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. National Academies Press; 2011. The National Academies Collection: Reports funded by National Institutes of Health.https://www.ncbi.nlm.nih.gov/books/NBK4119/ [PubMed] [Google Scholar]
- 7.Köhler S., Doelken S.C., Mungall C.J., Bauer S., Firth H.V., Bailleul-Forestier I., Black G.C., Brown D.L., Brudno M., Campbell J. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beaulieu C.L., Majewski J., Schwartzentruber J., Samuels M.E., Fernandez B.A., Bernier F.P., Brudno M., Knoppers B., Marcadier J., Dyment D., FORGE Canada Consortium Am. J. Hum. Genet. 2014;94:809–817. doi: 10.1016/j.ajhg.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girdea M., Dumitriu S., Fiume M., Bowdin S., Boycott K.M., Chénier S., Chitayat D., Faghfoury H., Meyn M.S., Ray P.N. Hum. Mutat. 2013;34:1057–1065. doi: 10.1002/humu.22347. [DOI] [PubMed] [Google Scholar]
- 10.Buske O.J., Girdea M., Dumitriu S., Gallinger B., Hartley T., Trang H., Misyura A., Friedman T., Beaulieu C., Bone W.P. Hum. Mutat. 2015;36:931–940. doi: 10.1002/humu.22851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson P.N., Köhler S., Oellrich A., Wang K., Mungall C.J., Lewis S.E., Washington N., Bauer S., Seelow D., Krawitz P., Sanger Mouse Genetics Project Genome Res. 2014;24:340–348. doi: 10.1101/gr.160325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rehman A.U., Morell R.J., Belyantseva I.A., Khan S.Y., Boger E.T., Shahzad M., Ahmed Z.M., Riazuddin S., Khan S.N., Riazuddin S., Friedman T.B. Am. J. Hum. Genet. 2010;86:378–388. doi: 10.1016/j.ajhg.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh T., Shahin H., Elkan-Miller T., Lee M.K., Thornton A.M., Roeb W., Abu Rayyan A., Loulus S., Avraham K.B., King M.C., Kanaan M. Am. J. Hum. Genet. 2010;87:90–94. doi: 10.1016/j.ajhg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rios J., Stein E., Shendure J., Hobbs H.H., Cohen J.C. Hum. Mol. Genet. 2010;19:4313–4318. doi: 10.1093/hmg/ddq352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadat M.A., Moir S., Chun T.W., Lusso P., Kaplan G., Wolfe L., Memoli M.J., He M., Vega H., Kim L.J. N. Engl. J. Med. 2014;370:1615–1625. doi: 10.1056/NEJMoa1302846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng B.G., Wolfe L.A., Ichikawa M., Markello T., He M., Tifft C.J., Gahl W.A., Freeze H.H. Hum. Mol. Genet. 2015;24:3050–3057. doi: 10.1093/hmg/ddv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams D.R., Yuan H., Holyoak T., Arajs K.H., Hakimi P., Markello T.C., Wolfe L.A., Vilboux T., Burton B.K., Fajardo K.F. Mol. Genet. Metab. 2014;113:161–170. doi: 10.1016/j.ymgme.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson P.N., Webber C. PLoS Genet. 2014;10:e1004268. doi: 10.1371/journal.pgen.1004268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bearn A.G. Clarendon Press; 1993. Archibald Garrod and the Individuality of Man. [Google Scholar]
- 20.Childs B. Johns Hopkins University Press; 1999. Genetic Medicine: A Logic of Disease. [Google Scholar]
- 21.Mulvihill J.J. J. Natl. Cancer Inst. 1976;57:3–7. doi: 10.1093/jnci/57.1.3. [DOI] [PubMed] [Google Scholar]
- 22.Miller R.W. Princess Takamatsu Symp. 1987;18:3–12. [PubMed] [Google Scholar]
- 23.Holden, J.P. (2013). Increasing Access to the Results of Federally Funded Scientific Research. Memorandum for the Heads of Executive Departments and Agencies from the Office of Science and Technology Policy, Executive Office of the President, February 22, 2013. https://www.whitehouse.gov/sites/default/files/microsites/ostp/ostp_public_access_memo_2013.pdf.
- 24.National Institutes of Health (2015). National Institutes of Health Plan for Increasing Access to Scientific Publications and Digital Scientific Data from NIH Funded Scientific Research. February 2015. https://grants.nih.gov/grants/NIH-Public-Access-Plan.pdf.
- 25.Ohno-Machado, L., Alter, G., Fore, I., Martone, M., Sansone, S.-A., and Xu, H. (2015). bioCADDIE White Paper: Vision and Implementation. http://dx.doi.org/10.6084/m9.figshare.1362572.
- 26.Institute of Medicine of the National Academies . National Academies Press; 2015. Sharing Clinical Trial Data: Maximizing Benefits, Minimizing Risks. [PubMed] [Google Scholar]
- 27.Brownstein C.A., Holm I.A., Ramoni R., Goldstein D.B., Members of the Undiagnosed Diseases Network Hum. Mutat. 2015;36:985–988. doi: 10.1002/humu.22840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., Brunner H.G., Buske O.J., Carey K., Doll C. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tryka K.A., Hao L., Sturcke A., Jin Y., Wang Z.Y., Ziyabari L., Lee M., Popova N., Sharopova N., Kimura M., Feolo M. Nucleic Acids Res. 2014;42:D975–D979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Might M., Wilsey M. Genet. Med. 2014;16:736–737. doi: 10.1038/gim.2014.23. [DOI] [PubMed] [Google Scholar]
- 31.Taruscio D., Groft S.C., Cederroth H., Melegh B., Lasko P., Kosaki K., Baynam G., McCray A., Gahl W.A. Mol. Genet. Metab. 2015;116:223–225. doi: 10.1016/j.ymgme.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Singh H., Graber M.L. N. Engl. J. Med. 2015;373:2493–2495. doi: 10.1056/NEJMp1512241. [DOI] [PubMed] [Google Scholar]
- 33.National Academies of Sciences, Engineering, and Medicine . National Academies Press; 2015. Improving Diagnosis in Health Care. [Google Scholar]
- 34.Miller F.G. APA Newsletters: Newsletter on Philosophy and Medicine. 2006;5:10–14. [Google Scholar]
- 35.Kass N.E., Faden R.R., Goodman S.N., Pronovost P., Tunis S., Beauchamp T.L. Hastings Cent. Rep. 2013;43:S4–S15. doi: 10.1002/hast.133. [DOI] [PubMed] [Google Scholar]
- 36.Faden R.R., Kass N.E., Goodman S.N., Pronovost P., Tunis S., Beauchamp T.L. Hastings Cent. Rep. 2013;43:S16–S27. doi: 10.1002/hast.134. [DOI] [PubMed] [Google Scholar]
- 37.Collins F.S., Varmus H. N. Engl. J. Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]