Dynein-mediated movement of microtubules is required for chromosome movement; its absence leads to aberrant segregation. Alleva et al. show that FKB-6, a cochaperone of Hsp-90, is required for proper chromosome movement through down-regulation of resting time between movements.
Abstract
In meiotic prophase I, homologous chromosome pairing is promoted through chromosome movement mediated by nuclear envelope proteins, microtubules, and dynein. After proper homologue pairing has been established, the synaptonemal complex (SC) assembles along the paired homologues, stabilizing their interaction and allowing for crossing over to occur. Previous studies have shown that perturbing chromosome movement leads to pairing defects and SC polycomplex formation. We show that FKB-6 plays a role in SC assembly and is required for timely pairing and proper double-strand break repair kinetics. FKB-6 localizes outside the nucleus, and in its absence, the microtubule network is altered. FKB-6 is required for proper movement of dynein, increasing resting time between movements. Attenuating chromosomal movement in fkb-6 mutants partially restores the defects in synapsis, in agreement with FKB-6 acting by decreasing chromosomal movement. Therefore, we suggest that FKB-6 plays a role in regulating dynein movement by preventing excess chromosome movement, which is essential for proper SC assembly and homologous chromosome pairing.
Introduction
The halving of meiocyte ploidy to form gametes occurs via two cellular divisions in a process known as meiosis. The first meiotic cellular division segregates homologous chromosome pairs by means of a reductional division, reducing ploidy by half. The second meiotic division then segregates sister chromatids, leading to the formation of haploid gametes. Meiosis is essential for the inheritance of genetic information in offspring, as well as creating genetic diversity through recombination and subsequent crossover formation, which occurs in meiotic prophase I. Disturbance of the recombination and subsequent crossover formation process leads to improper segregation and thus aneuploidy.
Formation of crossovers requires the establishment of pairing interactions that bring homologous chromosomes into close proximity. Pairing interactions are stabilized by the synaptonemal complex (SC), a protein structure that connects homologous chromosomes throughout their length during prophase I of meiosis. In Caenorhabditis elegans meiosis, SC assembly is required for the formation of all crossover events and proceeds independently of recombination (Dernburg et al., 1998; MacQueen et al., 2002). In C. elegans, proper assembly of the SC is dependent on the initiation of pairing interactions at the pairing centers located at a single end of each chromosome (MacQueen et al., 2005). Homologous pairing requires chromosome movement via dynein-mediated forces of the microtubule cytoskeleton, which is connected to the pairing centers of the chromosomes through the linker of nucleoskeleton and cytoskeleton (LINC) complex (in C. elegans, SUN-1–ZYG-12) that spans the nuclear envelope (Penkner et al., 2009; Sato et al., 2009). The SC forms after the establishment of pairing interactions; lateral element proteins (HTP-1/2/3 and HIM-3) that are assembled on the chromosomal axis at meiotic entry are connected by central region proteins (SYP-1/2/3/4) along the full length of the pairs of homologous chromosomes (Zetka et al., 1999; MacQueen et al., 2002; Colaiácovo et al., 2003; Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005; Smolikov et al., 2007b, 2009; Goodyer et al., 2008). Formation of double-strand breaks (DSBs), which initiates recombination, requires the lateral element protein HTP-3 but does not require a fully assembled SC; thus DSBs concurrently form with the initiation of pairing interactions (Goodyer et al., 2008). However, these DSBs are repaired to form crossovers only when the SC is fully assembled and pairing interactions are stabilized along the entire chromosome pair (MacQueen et al., 2002).
Regulation of the assembly of the SC occurs at multiple levels: the SC is regulated to form only between homologous chromosomes and is inhibited from assembling between nonhomologous chromosomes, sister chromatids, or separately from chromosomes. Another layer of SC regulation prevents polycomplex (PC) formation, an ordered protein aggregate of SC proteins (typically central region proteins). Central region SC proteins have been shown to form a PC when misexpressed or overexpressed, suggesting that these proteins are capable of precocious self-assembly when not properly regulated (Sym and Roeder, 1995; Costa et al., 2005; Öllinger et al., 2005; Merritt and Seydoux, 2010). In C. elegans, PCs have been observed at elevated temperatures (>26°C), suggesting that protein misfolding may contribute to PC formation (Bilgir et al., 2013). Posttranslational modification pathways are also involved in preventing PC formation (Brockway et al., 2014). Lastly, chromosome movement can affect PC formation in C. elegans—preventing LINC complex attachment to the cytoskeleton, inhibiting dynein motor activity, or removing all pairing center proteins leads to PC formation (Sato et al., 2009; Labella et al., 2011). However, some defects in chromosome movement do not lead to PC formation, such as hindering the movement of a single chromosome or preventing the assembly of any one of the central region proteins (MacQueen et al., 2005; Phillips et al., 2005; Phillips and Dernburg, 2006; Labella et al., 2011). Similarly, reduction, but not elimination, of chromosomal movement (preventing dimerization of ZYG-12 or reducing the level of the SUN-1–ZYG-12 complex) leads to nonhomologous synapsis without PC formation (Penkner et al., 2007; Sato et al., 2009). Collectively, these findings suggest that preventing chromosome movement of all chromosomes leads to pairing defects and consequently PC formation.
FK506 binding proteins (FKBPs) are a family of conserved cochaperones. FKBPs contain a catalytic domain with peptidylprolyl cis-trans isomerase (PPIase) activity (Erlejman et al., 2014). This catalytic domain alters the structure of a client protein, assisting in chaperone function (Kang et al., 2008). Some of the FKBPs contain additional domains, such as the tetratricopeptide repeat (TPR) protein–protein interaction domain. FKBP52 is a well-studied FKBP cochaperone of HSP90, with a role in cellular trafficking and microtubule dynamics (Storer et al., 2011). FKBP52 was shown to promote nuclear transport of various targets, such as glucocorticoid receptors and p53 (Silverstein et al., 1999). FKBP52 function in transport requires its PPIase domain (but not PPI catalytic activity), as well as the ability to bind to dynein and nuclear pores (Silverstein et al., 1999). FKBP52 not only interacts with microtubules in transport, but also has an active role in microtubule dynamics. FKBP52 was shown to inhibit microtubule formation by tau in vitro, and its depletion led to reorganization of the microtubule network (Chambraud et al., 2007).
Of the 13 FKBP proteins in mammals, homologues for eight are found in C. elegans (Pemberton and Kay, 2005). FKB-6 (also known as FKBP-48) is considered to be the ortholog of the human cochaperone FKBP52/FKBP4 and the yeast FPR1; it is the only TPR-containing FKBP in C. elegans (Galat, 2000; Pemberton and Kay, 2005). FKB-6 contains two N-terminal FKB/PPIase domains and three C-terminal TPR domains (Richardson et al., 2007). FKB-6 is expressed in all developmental stages and in various tissues, such as neuronal, hypodermal, and somatic (Richardson et al., 2007; Fasseas et al., 2012). As expected from studies of the human homologue, C. elegans FKB-6 interacts physically with DAF-21, a germline-expressed HSP90 (Richardson et al., 2007). Before our work, no biological function for FKB-6 had been identified in C. elegans.
Here we show that FKB-6 plays a role in meiotic chromosome behavior by regulating chromosomal movements. In fkb-6 mutants, SYP proteins form PCs at the entry to meiosis, and the PCs persist up to midpachytene (MP). This delay in proper SC assembly leads to defects in DSB repair and elevated apoptosis, despite the formation of most meiotic crossovers. fkb-6 genetically interacts with zyg-12 and dynein and localizes to the cytoplasmic space. We show that the defects observed in fkb-6 mutants are associated with more frequent movement of DHC-1 patches, compared with wild type. Before our work, positive regulation of SUN-1–ZYG-12 complex movement was shown to support synapsis and pairing defects (Penkner et al., 2007, 2009; Sato et al., 2009). Our findings are intriguing because they suggest that down-regulating the SUN-1–ZYG-12 complex movement is important in facilitating proper synapsis and homologous chromosome pairing.
Results
Identification of fkb-6 as a gene required for down-regulation of PC complex formation
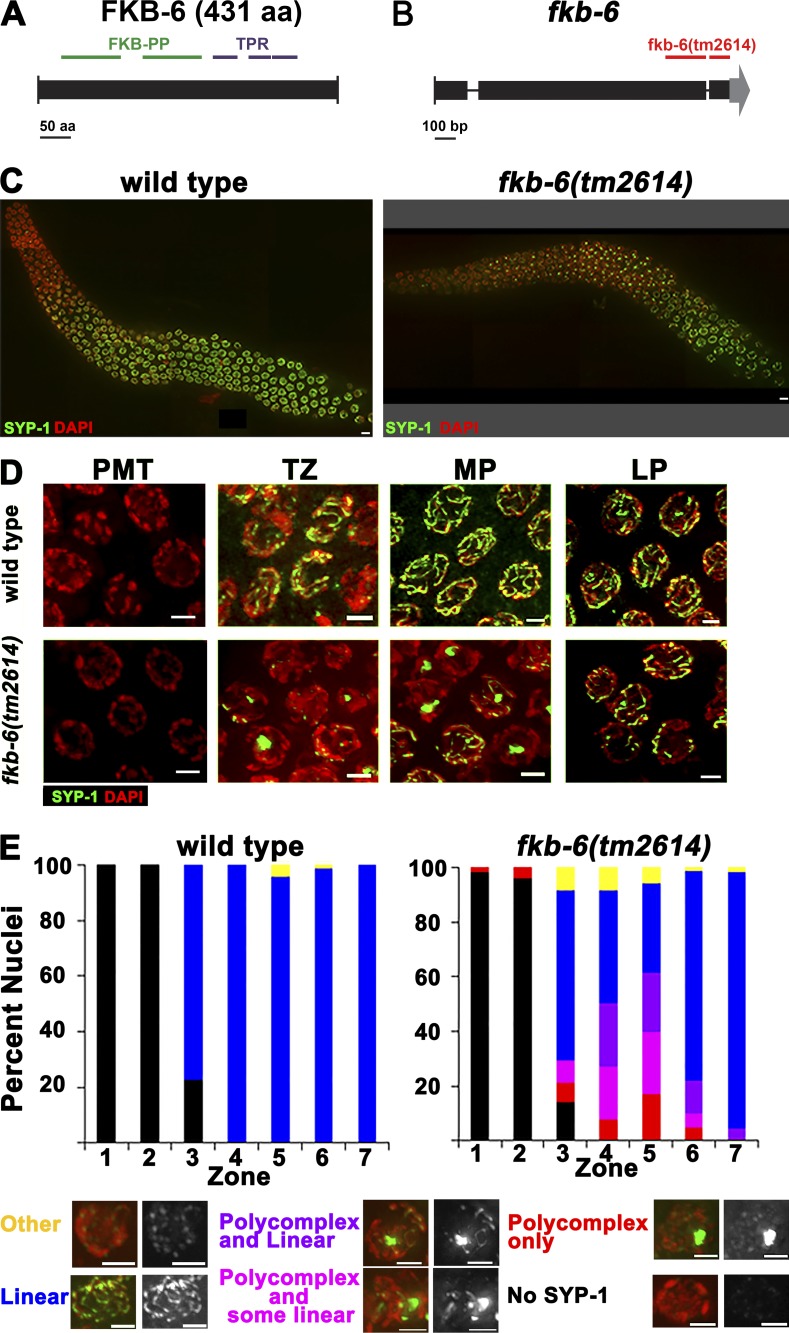
FKB-6 is a DAF-21/Hsp90 cochaperone with homology to FKBP52 from mouse/human. It contains two N-terminal PPIase catalytic domains and three C-terminal TPR repeats (Fig. 1 A). We isolated fkb-6 as a gene that, when knocked down, induces PC formation in akir-1(gk528) mutants (see Materials and methods). fkb-6(RNAi) had no noticeable phenotypes when performed in a wild-type background, likely because of incomplete depletion of the fkb-6 mRNA. To test whether a null mutant conferred a phenotype on an otherwise wild-type background, we analyzed a deletion mutant of fkb-6 predicted to remove two of the TPR repeats of the FKB-6 protein (Fig. 1 B). fkb-6(tm2614) homozygous mutants exhibited PCs when stained with the SC central region protein SYP-1 (Fig. 1, C and D). We also generated two N-terminal deletions, creating early out-of-frame mutations. Both of these mutants showed PC formation, a phenotype indistinguishable from that of fkb-6(tm2614) (Fig. S1, A and B). We were unable to obtain an fkb-6(tm2614);akir-1(gk528) double mutant, which suggests a synthetic lethal relationship between the alleles. akir-1(RNAi) increased the number of nuclei containing PCs in fkb-6(tm2614) mutants (Fig. S1 C), consistent with our findings of fkb-6(RNAi);akir-1(gk528) mutants. We did not find evidence for the germline HSP90 homologue acting in the same pathway, as daf-21(RNAi) did not lead to PC formation and null daf-21 mutants are lethal at the larval stage (Fig. S1 D; Birnby et al., 2000).
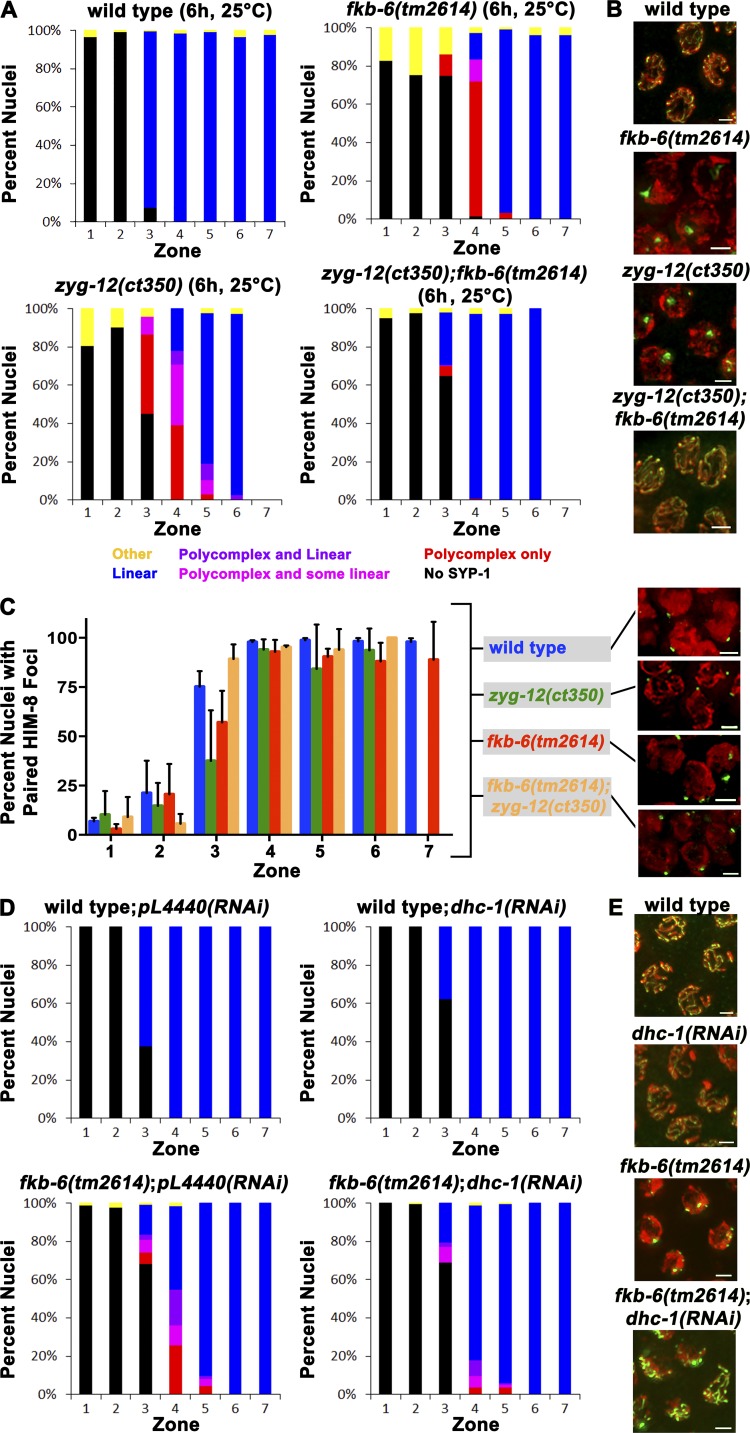
Figure 1.
PC formation of SYP-1 proteins in fkb-6(tm2614) mutant germline. (A) C. elegans FKB-6 protein mapped with its protein domains. (B) Schematics of the fkb-6 gene with the position of the tm2614 deletion. (C) Images of full-length gonad stained with SYP-1 (central region, green) and DAPI (false-colored red) of the genotypes indicated. Images are z-stack projections of half the germline. (D) Representative images of SYP-1 staining in PMT, TZ, MP, and LP. Images are projections of a z-stack halfway through the nuclei. (E) Quantification of SYP-1 staining of wild-type and fkb-6 mutants. Scoring was divided into six categories, as indicated at the bottom. Representation of zones in terms of meiotic stage are as follows: zones 1 and 2, PMT (mitotic nuclei); zone 3, mostly TZ and some PMT; zone 4, mostly EP; zones 5 and 6, MP; and zone 7, LP. n nuclei: wild type = 657 and fkb-6 = 621. Bars: (A) 50 aa; (B) 100 bp; (C) 5 µm; (D and E) 2 µm.
In C. elegans, the meiotic nuclei are positioned sequentially in the germline; therefore, meiotic events can be examined as a time-course analysis in a single germline. To further define the phenotype of fkb-6(tm2614), we performed quantitative 3D analysis throughout the germline of wild-type and fkb-6(tm2614) mutants via SYP-1 and DAPI staining (Brockway et al., 2014; Materials and methods). Similar to wild type, SYP-1 was absent from mitotically proliferating nuclei of fkb-6(tm2614) mutants (zones 1 and 2). At the entry to meiotic prophase I (zone 3, transition zone [TZ], leptotene/zygotene), in wild-type gonads, SYP-1 localization initiates in short dots and stretches (the width of a fully assembled SC), whereas SYP-1 localization in fkb-6(tm2614) mutants exhibited PC formation (Fig. 1 E). PCs were also observed throughout early to midpachytene, the meiotic stage in which chromosomes are fully synapsed in wild-type gonads. The mean width of a PC was 0.95 ± 0.2 µm (n = 82), which is about four times wider than that of a typical SC (wild type, 0.19 ± 0.04 µm; n = 16, P < 0.0001 by Mann–Whitney U test [MW]). Nuclei with PCs had 1.23 ± 0.57 PCs/nucleus (n = 82), indicating that a PC formation event occurs a mean of once per nucleus. Overall, in fkb-6 mutants, 35% of the meiotic nuclei (all stages combined) contained a PC. In the mid-to-late pachytene transition (zone 6) of fkb-6 mutants, the percentage of nuclei with PCs diminished, and they were almost completely absent by the end of pachytene (zone 7). No PCs were observed in diplotene and diakinesis oocytes, the final stage of meiotic prophase I. Chaperones and cochaperones are required at higher temperatures, where protein folding is challenged; the fraction of nuclei with PCs and the duration of PC aggregation is increased with exposure to elevated temperatures in fkb-6(tm2614) mutants (Fig. S1 E).
To examine whether the PCs consist of only central region proteins, we performed immunostaining with SYP-2 and SYP-4, two central region proteins, as well as HTP-3, HIM-3, and LAB-1, lateral element–associated proteins, in wild-type and fkb-6(tm2614) mutants (Fig. S2; Zetka et al., 1999; Colaiácovo et al., 2003; Smolikov et al., 2007b, 2009; de Carvalho et al., 2008; Goodyer et al., 2008). In wild-type pachytene nuclei, all of the indicated proteins localized continuously between homologous chromosomes. In fkb-6(tm2614) mutants, HTP-3, HIM-3, and LAB-1 localized to chromosomes in a linear manner. However, SYP-2 and SYP-4 formed PCs similarly to SYP-1. We therefore conclude that the defects in SC assembly in fkb-6(tm2614) mutants are specific to central region proteins of the SC.
fkb-6 mutants show defects in chromosome pairing
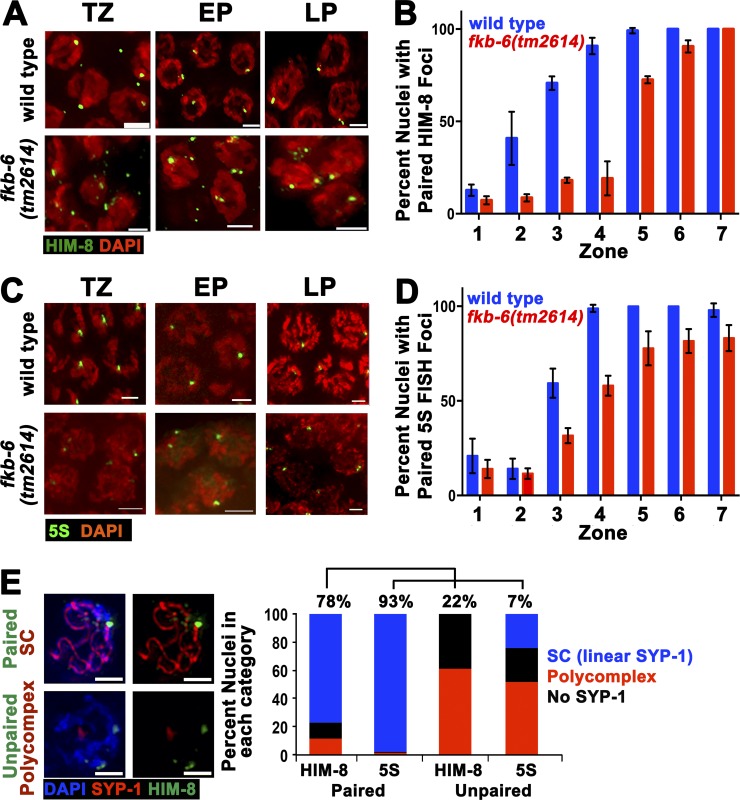
The SC is essential for the stabilization of pairing interactions during meiosis. Because PC formation has been shown to affect pairing stabilization and vice versa, we examined whether pairing is affected in fkb-6(tm2614) mutants. We performed immunofluorescent staining for the pairing center protein HIM-8 (localizes to the X chromosome pairing center [Phillips et al., 2005]) and assayed the percentage of paired versus unpaired HIM-8 foci in each of the seven zones (acquired in the same manner as the data in Fig. 1 E). In wild-type nuclei, HIM-8 foci pair at the entrance to meiosis and remain paired throughout pachytene (Fig. 2, A and B). fkb-6(tm2614) mutants showed a decrease in percentage of paired foci (e.g., 26% and 21% of wild type in zones 3 and 4, respectively; Fig. 2 B). To examine whether pairing of autosomes is also affected, we performed FISH analysis for the 5S rDNA locus located on chromosome V. In agreement with HIM-8 analysis, we observed a delay in pairing for the 5S locus (Fig. 2, C and D). The defects in pairing correlated with SC assembly defects; zones with high levels of PC formation (Fig. 1 E) showed lower levels of pairing (Fig. 2, B and D).
Figure 2.
fkb-6(tm2614) mutants show defects in pairing of the X-chromosome and chromosome V. Representative images of HIM-8 (A) antibody staining or 5S FISH (C) in the genotypes indicated in TZ/zone 3, EP/zone 4, and LP/zone 7. Stained with HIM-8 or FISH (green) and DAPI (red). Images are z-stack projections halfway through the nuclei. Bars, 2 µm. (B) Quantification of percentage of nuclei with paired HIM-8 foci. n nuclei: wild type = 790 and fkb-6 = 614. (D) Quantification of percentage of nuclei with paired 5S foci. n nuclei: wild type = 1,113 and fkb-6 = 1,277. Zones in B and D in terms of meiotic stage are as indicated in Fig. 1 D. Error bars are SD. (E) Representative images of common classes and division into categories of SYP-1 staining with HIM8 or 5S FISH probe (n = 743, 1,028 nuclei). Bars, 2 μm.
The number of nuclei with linear SC was higher than the number of paired nuclei (e.g., in zone 4, 65% linear SYP-1, whereas 38% and 58% paired with HIM-8 and 5S, respectively). This may be caused by some nonhomologous synapsis or a more successful synapsis of autosomes not examined here. To test for nonhomologous synapsis, we performed costaining of nuclei with HIM-8 and SYP-1 and immuno-FISH (5S and SYP-1 costaining). In both analyses, we observed nuclei with unpaired chromosomes as more likely to contain a PC (Fig. 2 E). However, 24% of unpaired chromosome V contained a SYP-1 stretch surrounded by DAPI that colocalized with at least one 5S focus, suggestive of low levels of nonhomologous synapsis.
fkb-6 mutants show perturbation of DSB repair but form oocytes with bivalents
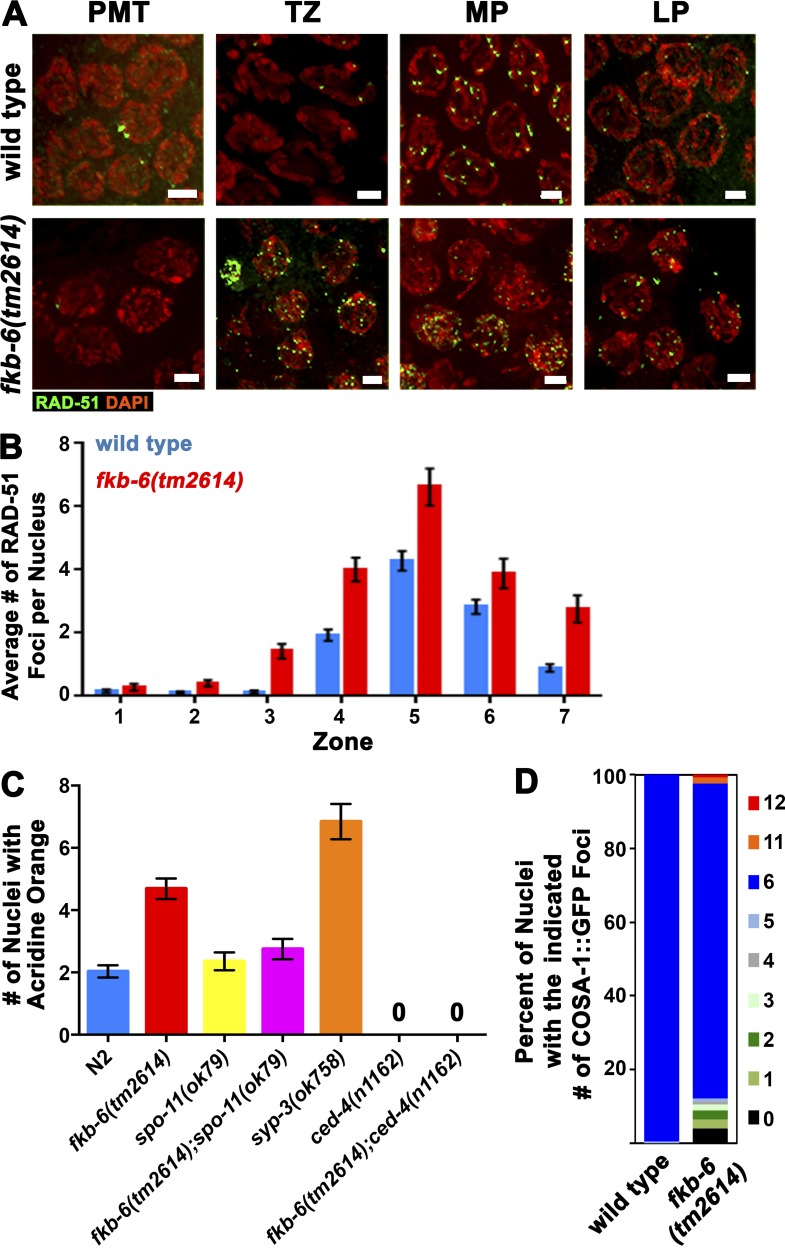
Lack of homologous synapsis and pairing stabilization is known to lead to defects in the repair of meiotic DSBs (Colaiácovo et al., 2003). To address whether DSB repair is affected in fkb-6(tm2614) mutants, we performed analysis of RAD-51 localization (Fig. 3 A). RAD-51 is a single-stranded DNA-binding protein that is essential for DSB repair. A RAD-51 focus indicates that a DSB was formed, resected to form single-stranded DNA, and was coated with RAD-51 protein (Colaiácovo et al., 2003). In fkb-6(tm2614) mutants, the overall levels of RAD-51 foci increased, in comparison to a wild-type background, at all meiotic stages examined (from 1.28 to 1.67 foci/nucleus; Fig. 3 B), but the overall kinetics (timing of RAD-51 appearance and disappearance) remained the same as in wild type. This phenotype is consistent with a delay in the repair of DSBs caused by a delay in SC assembly. The timing of the decrease in RAD-51 foci correlates with that of the resolution of PCs (zone 6), in agreement with the dependence of meiotic DSB repair on proper SC formation.
Figure 3.
fkb-6 mutants accumulate recombination intermediates. (A) Representative images of RAD-51 antibody staining in wild-type and fkb-6 mutants in PMT/zone 1, TZ/zone 3, EP/zone 4, and LP/zone 7. Stained with RAD-51 (green) and DAPI (red). Images are a z-stack projection halfway through the nuclei. Bars, 2 µm. (B) Quantification of RAD-51 immunostaining. Representation of zones in terms of meiotic stage are as indicated in Fig. 1 E. Error bars are SEM. n nuclei: wild type = 718 and fkb-6 = 610. (C) Apoptotic nuclei per gonadal arm in the genotypes indicated. Error bars are SEM. n gonads: wild type = 125, fkb-6 = 74, syp3 = 57, spo-11 = 42, fkb-6;spo-11 = 56, ced-4 = 22, and fkb-6;ced-4 = 21. (D) Distribution of the number of COSA-1 foci per gonad in LP nuclei. n nuclei: wild type = 168 and fkb-6 = 123.
Delay in the repair of meiotic DSBs can induce the DNA damage checkpoint in late pachytene (LP) and result in an increase in apoptosis (Smolikov et al., 2007a). fkb-6(tm2614) mutants exhibit 4.7 apoptotic nuclei/gonad (n = 74), compared with 1.8 in wild type (Fig. 3 C; P < 0.0001, MW). The levels of apoptosis observed in fkb-6(tm2614) mutants were significantly lower than those found in synapsis-defective mutants (syp-3; Fig. 3 C; P < 0.001, MW). The elevated levels of apoptosis in fkb-6(tm2614) mutants were suppressed by removal of meiotic DSBs (Fig. 3 C; fkb-6 to spo-11;fkb-6, P < 0.0001; spo-11 to spo-11;fkb-6, P = 0.5662, MW) and eliminated by removing a gene required for apoptosis, ced-4(n1162) (Fig. 3 C; P < 0.0001, MW). These data indicate that apoptosis is triggered by DNA damage in fkb-6(tm2614) mutants.
Our data indicate that in fkb-6(tm2614) mutants, PC formation leads to defects in DSB repair. However, because PCs are resolved by the end of pachytene, it is possible that nuclei may have a narrow window of time in which functional SC can support the repair of DSBs. In C. elegans, each pair of homologous chromosomes is joined by a single crossover event to form a bivalent. In wild-type LP nuclei, COSA-1 localizes to the six interfering crossovers (Yokoo et al., 2012). We used COSA-1::GFP foci counts as a marker to analyze the number of crossovers in LP and diakinesis DAPI body counts (bivalents/univalents) as a measure of crossover formation in nuclei progressing to form oocytes. In fkb-6(tm2614) mutants, 85% of the nuclei contained six COSA-1 foci, compared with 99% in wild type (Fig. 3 D; P = 0.007, MW). Prevalence of crossovers is in agreement with our DAPI body counts; 5.85 ± 0.87 in fkb-6(tm2614) mutants compared with 5.77 ± 0.43 in wild type (n = 26 and 20; P = 0.85, MW). The absence of severe crossover formation defects was not caused by preferential elimination of non-crossover nuclei by apoptosis, as ced-4;fkb-6(tm2614) double mutants did not show an increase in DAPI body number (5.78 ± 0.47, n = 42; P = 0.76, MW). However, when fkb-6(tm2614) mutants were grown at high temperatures, leading to PC formation at LP, oocytes with 11 or more univalents were readily observed (31%). Our data are consistent with the idea that nuclei can resolve recombination intermediates into crossovers as long as they reach LP with assembled SC.
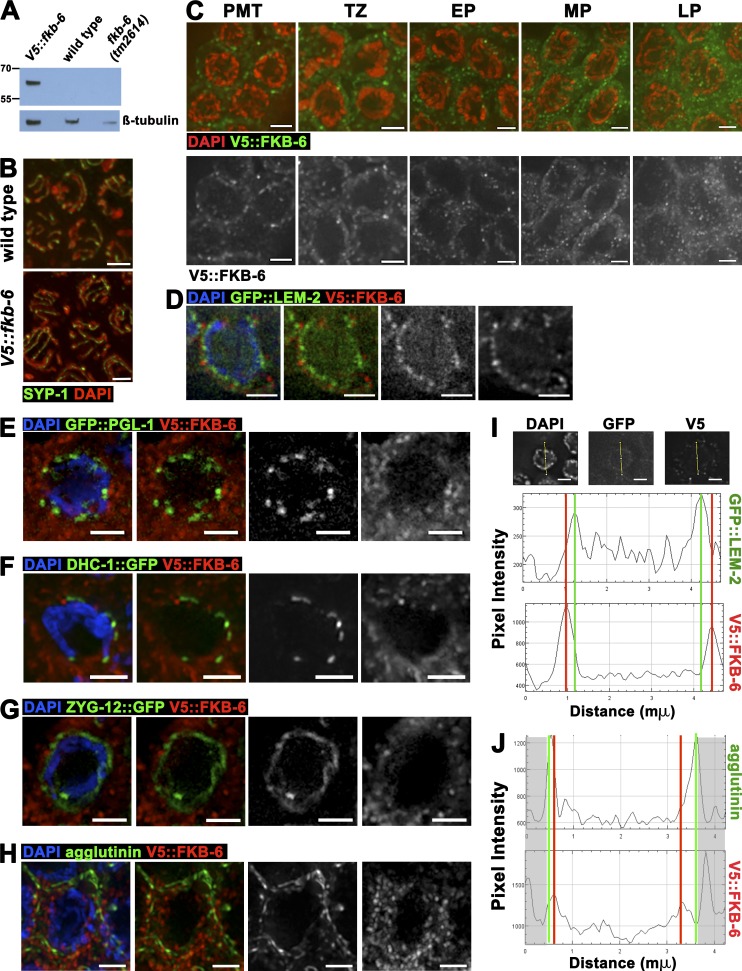
FKB-6 localizes to the cytoplasmic space
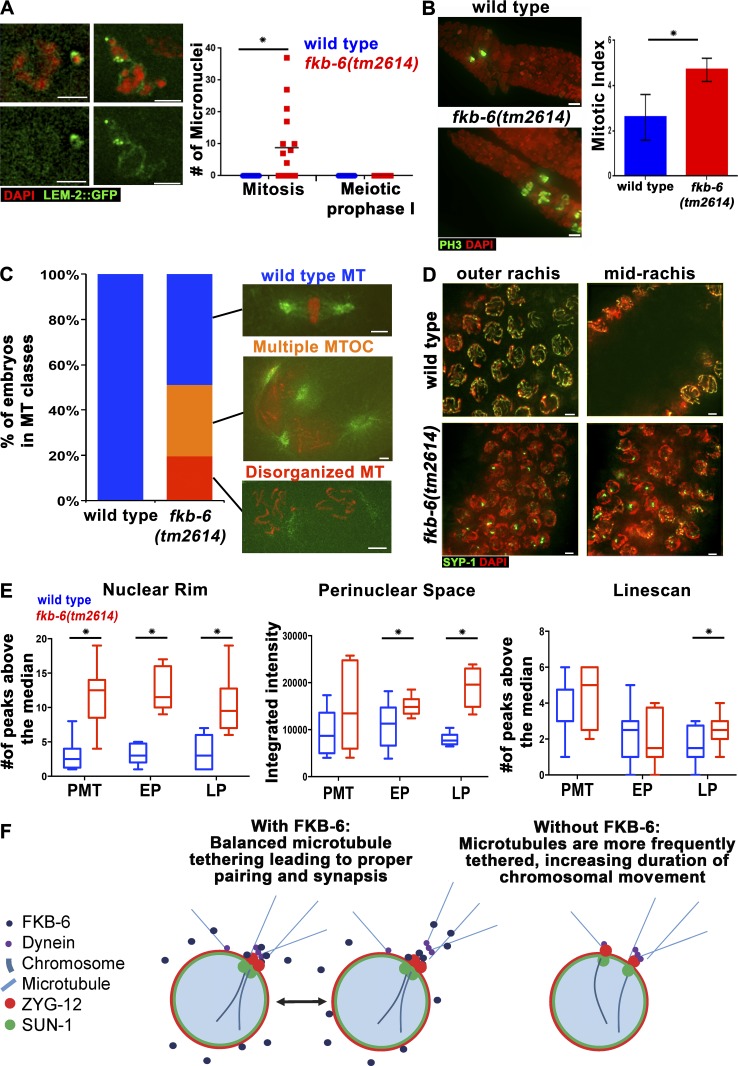
The localization of FKB-6 may be suggestive of how it regulates SC assembly and homologous chromosome pairing. We used the CRISPR/Cas9 system to generate a V5 N-terminally tagged FKB-6 line (Fig. 4 A). V5::fkb-6 worms are viable and fertile and have a functional germline, based on DAPI and SYP-1 staining (Fig. 4 B). Immunolocalization of FKB-6, using an antibody for V5, revealed a punctate, circular localization pattern surrounding the DAPI-stained region of each nucleus; this pattern is found throughout the germline, from premeiotic tip (PMT) to diakinesis (Fig. 4 C). FKB-6 was found in proximity surrounding, but not colocalizing to, a protein found next to the inner nuclear envelope (LEM-2; Fig. 4, D and I). FKB-6 staining slightly overlapped with proteins found at the outer nuclear envelope (Fig. 4, E–G) and was internal to plasma membrane stains (Fig. 4, H and J). These data suggest that FKB-6 localizes to the cytoplasm. The localization of nuclear envelope–associated proteins LEM-2 and SUN-1 and P-granules were not affected in fkb-6(tm2614) mutants (Fig. S3, A–C).
Figure 4.
FKB-6 localizes to the cytoplasmic space throughout the germline. (A) Western blot of V5 antigen (top) and β-tubulin (bottom) as loading control of the genotypes indicated. (B) Projections of a z-stack halfway through the nuclei of V5::fkb-6 transgenic worms. Gonad stained with SYP-1 (green) and DAPI (red). (C) Representative images of V5 antibody staining in V5::fkb-6 strain in PMT, TZ, EP, MP, and LP. FKB-6 (via V5; green) and DAPI (red). Images on the bottom are V5::FKB-6 channel (white) of the images shown on the left. (D–H) Early pachytene images of V5::fkb-6 strain stained for V5 (red); in green, GFP::LEM-2 (D), GFP::PGL-1 (E), DHC-1::GFP (F), ZYG-12::GFP (G), and agglutinin (H). (I and J) Intensity plots of the LEM-2::GFP (I) or agglutinin (H) and V5::fkb-6 strain. Shaded boxes are part of the adjacent nucleus. Bars, 2 µm.
FKB-6 counteracts the action of proteins involved in chromosomal movement
The localization of FKB-6 next to the external nuclear envelope suggests that its effect on chromosome pairing and PC formation of SC proteins is through chromosome movement. The LINC complex is embedded in the nuclear membrane and connects the chromosomes to microtubules forming the cytoskeleton, which permits motor-driven movement of the chromosomes. This movement is required for pairing of homologous chromosomes during the early stages of meiotic prophase I.
The zyg-12(ct350) temperature-sensitive mutant perturbs both chromosomal movement and dimerization of the LINC protein ZYG-12, which leads to PC formation, a phenotype similar to that observed for fkb-6 mutants (Malone et al., 2003; Sato et al., 2009; Zhou et al., 2009). We therefore explored the genetic interaction between the two mutants. Temperature shift of zyg-12(ct350) mutants at the restrictive temperature (25°C) for 6 h led to PC formation (zone 3, 51%; zone 4, 78%; Fig. 5 A), as previously reported (Sato et al., 2009). fkb-6(tm2614) mutants also showed PCs under these conditions (zone 3, 11%; zone 4, 82%; Fig. 5 A). Although individually both mutants displayed similar phenotypes, the zyg-12(ct350);fkb-6(tm2614) double mutants had a suppressed phenotype, with only a small fraction of the nuclei showing PCs (zone 3, 6%; zone 4, 1%; Fig. 5, A and B). Pairing defects were also suppressed in the double mutant compared with the single mutants (Fig. 5 C; zone 3: zyg-12(ct350) vs. double mutant, P < 0.0001; fkb-6(tm2614) vs. double mutant, P < 0.0001, Fisher’s exact test).
Figure 5.
fkb-6(tm2614) suppresses pairing and SC assembly defects in mutants with defective chromosomal movement. (A and D) Quantification of SYP-1 staining of the indicated genotypes and conditions. Representation of zones in terms of meiotic stage and classes of scoring are as indicated in Fig. 1 E. fkb-6(tm2614) suppresses the SC assembly defects in zyg-12(ct350) and dhc-1(RNAi). (B, E, and C inset) Representative images of zone 4 of A, D, and C, respectively. SYP-1/HIM-8 (green) and DAPI (red). (C) Quantification of HIM-8 pairing. Representation of zones in terms of meiotic stage are as indicated in Fig. 1 E. Error bars are SD. n nuclei: (A and C) wild type = 1,261, fkb-6 = 1,162, zyg-12 = 628, fkb-6;zyg-12 = 545, wild type;pl4440(RNAi) = 834, wild type;dhc-1(RNAi) = 1,126, fkb-6;pl4440(RNAi) = 899, and fkb-6;dhc-1(RNAi) = 1,832; (B) wild type = 818, fkb-6 = 706, zyg-12 = 533, and fkb-6;zyg-12 = 415. Bars, 2 μm.
dhc-1 encodes for the dynein heavy chain, which is required for chromosome movement via the LINC complex and, in turn, for SC elongation (Gönczy et al., 1999; Sato et al., 2009; Rog and Dernburg, 2015). We used dhc-1(RNAi), which led to partial depletion of DHC-1 and was not sufficient to induce a notable PC formation phenotype (Fig. 5, D and E). This is consistent with previous findings indicating that dhc-1(RNAi) at 20°C is partially penetrant (Sato et al., 2009). Similarly to what was observed for zyg-12(ct350);fkb-6(tm2614), knockdown of dhc-1 was able to suppress the PC formation phenotype observed in fkb-6(tm2614) mutants (Fig. 5 D). This suppression was not observed in severe depletion conditions using dhc-1(or195) (temperature sensitive mutant; grown at 25°C for 24 h), in which both alleles showed PC formation (Fig. S4 A). Depletion of two genes required for LINC movement (dhc-1(RNAi);zyg-12(ct350) double mutant) enhanced their individual phenotypes, leading to severe defects in SC assembly (Fig. S4 B), a phenotype strikingly different from that of zyg-12(ct350);fkb-6(tm2614) double mutants. These data indicate that FKB-6 has an opposing role to that of ZYG-12 and DHC-1 within the same mechanism.
FKB-6 regulates the movement of DHC-1 patches
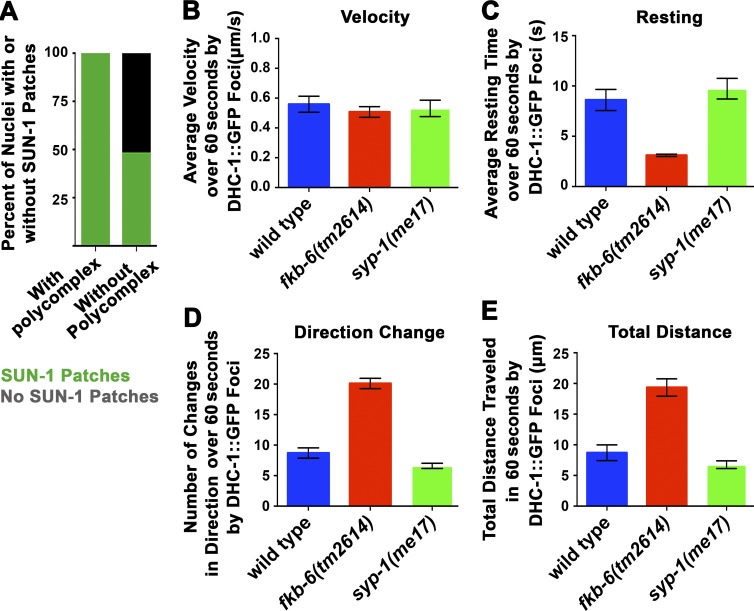
Lack of chromosome movement leads to pairing defects accompanied by various synapsis defects, some of which lead to PC formation (zyg-12(ct350), dhc-1;dlc-1 double mutants, deletion of all pairing center proteins [Sato et al., 2009; Labella et al., 2011]). FKB-6 may interact with LINC complex members SUN-1–ZYG-12 and the motor DHC-1 by affecting their function or their localization. We examined the localization of DHC-1, ZYG-12, and SUN-1 in fkb-6(tm2614) mutants and found these proteins to be localized to the nuclear envelope and form patches, as found in wild type (Fig. S3, B and F). These patches were found in early meiotic prophase I (TZ) and dispersed once pairing interactions were stabilized. In fkb-6(tm2614) mutants, DHC-1–SUN-1–ZYG-12 patches were temporally extended. We quantified this for SUN-1 and found that these patches were more frequently found on nuclei with PCs (Fig. 6 A), as was found in other mutants with defects in synapsis, despite forming an axis (Penkner et al., 2009).
Figure 6.
fkb-6(tm2614) mutants show decreased resting time of DHC-1 foci between movements. (A) Percentage of nuclei with SUN-1 patches in nuclei that do and do not show PC formation in fkb-6(tm2614) mutants. n nuclei: fkb-6(tm2614) mutants = 387. (B) Mean velocity of DHC-1 patches. (C) Resting time of DHC-1 patches. (D) Change in direction of DHC-1 patches. (E) Total distance traveled by DHC-1 patches in wild type, fkb-6(tm2614), and syp-1 mutants. Error bars in B–E are SEM; n nuclei = 10 for all.
To examine whether the meiotic defects found in fkb-6(tm2614) mutants stem from defects in movement of SUN-1–ZYG-12 patches, we performed live imaging of the movement of DHC-1::GFP and ZYG-12::GFP patches in wild-type and fkb-6(tm2614) mutants (DHC-1 localizes to SUN-1–ZYG-12 patches; Sato et al., 2009). Quantification of DHC-1::GFP patch movement revealed that the mean velocity of the patch is not different in fkb-6(tm2614) mutants compared with wild type (Fig. 6 B; n = 10; P = 0.4357, MW). However, other parameters of DHC-1 movement were altered in fkb-6(tm2614) mutants (Videos 1–3 and Fig. S5 A). DHC-1 patches had significantly reduced resting time between movements in fkb-6(tm2614) mutants (Fig. 6 C; n = 10; P = 0.0005, MW). This reduced resting time is likely to account for the increase in other events examined: the number of times DHC-1 patches changed direction and the total distance traveled (Fig. 6, D and E; n = 10; P < 0.0001, MW). Similar data were obtained using ZYG-12::GFP (Fig. S5 B). As expected, synapsis-defective mutants (syp-1) had no effect on DHC-1 movement (Fig. 6, B–E). These data indicate that FKB-6 regulates the resting time between episodes of chromosomal movement, and this method of regulation is key for proper chromosomal pairing and synapsis.
FKB-6 affects mitotic divisions of the germline nuclei and the microtubule network
The role of FKB-6 in negative regulation of DHC-1 movement, which is known to be microtubule driven, suggests that FKB-6 may be involved in other cellular events requiring microtubule dynein–driven function. The distal nuclei of the germline (PMT) proliferate mitotically to create the population of nuclei that enter meiosis. We have found evidence for defects in mitotic divisions and nuclear positioning, both of which require microtubule function. In fkb-6(tm2614) mutants, mitotic chromosomes missegregate, leading to the formation of micronuclei (Fig. 7 A; n = 10; P = 0.007, t test with Welch’s correction). We also observed an increased mitotic index in fkb-6(tm2614) mutants that can be attributed to a metaphase arrest (Fig. 7 B; n = 8; P = 0.005, MW). These defects in germline mitotic divisions led us to examine the mitotic divisions in fkb-6(tm2614) mutant embryos. We observed a high proportion of embryos with spindle defects in fkb-6(tm2614) mutants: 31% of the embryos showed more than two spindles in a cell, and 19% showed abnormal/absent microtubule organization centers, whereas none showed such defects in wild type (Fig. 7 C; wild type, n = 69; fkb-6, n = 51). These defects were associated with abnormal chromosome morphology (71%) and lagging chromosomes (12%). No embryos past the 40-cell stage were observed in fkb-6(tm2614) mutants. These defects were likely the cause of the high embryonic lethality observed in fkb-6(tm2614) mutants, which laid 33 ± 17 eggs/hermaphrodite as opposed to 225 ± 80 eggs/hermaphrodite for wild type: 100% of the eggs laid by fkb-6(tm2614) mutants were nonviable, compared with 1.4% in wild type (wild type, n = 675; fkb-6, n = 267).
Figure 7.
fkb-6(tm2614) mutants show defects typical to microtubule organization defects in the germline. (A) Micronuclei in fkb-6(tm2614) mutants (grown at 25°C for 6 h). LEM-2 (nuclear envelope, green) and DAPI (red). Images are a z-stack projection halfway through the nuclei. n gonads: wild type = 16 and fkb-6(tm2614) mutants = 16. (B) Metaphase nuclei are found in higher proportion in fkb-6(tm2614) mutants. PH-3 to mark metaphase nuclei (green) and DAPI (red). Images are a z-stack projection through the nuclei at the PMT. Error bars are SD. n gonads: wild type = 8 and fkb-6(tm2614) mutants = 8. (C) Microtubule defects found in embryos, as indicated in the images, distributed to categories in wild-type (n = 68) and fkb-6(tm2614) mutants (n = 31). (D) Nuclear positioning defects in fkb-6(tm2614) mutants, SYP-1 (red) and DAPI (blue). Images are a z-stack projection of the top and bottom layer. Bars, 2 μm. (E) Quantification of microtubule density around the nucleus by three methods (ImageJ). Box-and-whisker plots are made with minimum to maximum values. n nuclei: wild type = 12 and fkb-6 = 12 for all. *, P < 0.05. (F) Model.
In addition to defects specific to mitosis, the positioning of nuclei in the germline was also perturbed, with the distance between nuclei in fkb-6(tm2614) mutants showing a wider spread than observed in wild type, as well as nuclei found in the midrachis (Fig. 7 D). These defects in nuclear organization in fkb-6(tm2614) mutants were also indicated by cell membrane, tbg-1::GFP, and F-actin staining; this staining formed a honeycomb pattern in wild type; however, in fkb-6(tm2614) mutants, it was misshapen (Fig. S3, D and E). Similar nuclear organization defects were reported for zyg-12 mutants (Zhou et al., 2009). If FKB-6 destabilizes microtubule polymerization at the nuclear envelope, then in its absence, microtubules should be found more frequently next to the nuclear envelope. We performed microtubule staining intensity analysis, measuring intensity surrounding germline nuclei of the PMT (mitotic) region, early pachytene (EP), and LP (corresponding to zones 1, 4, and 7). Peaks of microtubule staining were more frequently found next to the nucleus in fkb-6(tm2614) mutants compared with wild type in all analyses for LP and for two analyses in EP (Figs. 7 E and S5 C; nuclear rim: PMT, P = 0.0001; EP, P = 0.0008; LP, P = 0.0160; adjacent cytoplasmic space: PMT, P = 0.0839; EP, P = 0.0100; LP, P < 0.0001; linescans: PMT, P = 0.1254; EP, P = 0.8873; LP, P = 0.0223). This evidence altogether suggests that FKB-6 has a role in microtubule dynamics throughout the germline.
Discussion
Here, we have identified a novel function for FKB-6, a relatively unstudied protein in C. elegans: regulation of chromosome pairing and synapsis. We have shown that in fkb-6 mutants, SC central region proteins form PCs and homologous chromosome pairing is delayed. These defects are associated with perturbed DSB repair and increased apoptosis. Despite these defects, crossovers are formed and bivalents are observed. FKB-6 localizes outside the nucleus and is required to down-regulate chromosomal movements by opposing the function of dynein and ZYG-12. To our knowledge, this is the first study to show the importance of negative regulation of chromosome movement in SC assembly and pairing between homologous chromosomes. This indicates that precise control of chromosome movement is imperative for the success of these processes.
The importance of chromosome movement and pauses in movement for chromosome synapsis
Proper movement of chromosomes in meiosis is key for forming pairing interactions and proper SC assembly between homologous chromosomes. Studies in C. elegans have established that chromosome movement in meiosis is controlled by a microtubule network that is attached to the chromosomes via the LINC complex, containing SUN (SUN-1) and KASH (ZYG-12) proteins. It has been shown that chromosome movement is critical for pairing in other organisms as well: the LINC complex connects chromosomes to the nuclear envelope in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and mouse meiosis (Niwa et al., 2000; Ding et al., 2007; Conrad et al., 2008; Morimoto et al., 2012; Boateng et al., 2013). Similarly to what was found in C. elegans, the homology search requires dynein-mediated movement of microtubules in Mus musculus, S. pombe, and Drosophila melanogaster (Yamamoto et al., 1999; Horn et al., 2013; Christophorou et al., 2015). It was proposed that the main function of these movements is to perturb nonhomologous interactions between chromosomes.
Despite the clear importance of chromosome movement for pairing and synapsis, previous studies had not examined the limit of this positive effect of movement for pairing and synapsis. Could chromosome movement be restrained to allow proper synapsis and pairing interactions? Can increased movement be deleterious to chromosome synapsis and pairing? Our work suggests that the answer to these questions is yes. We have identified FKB-6 as a protein in whose absence the LINC complex moves more frequently and both pairing and synapsis are perturbed. We propose that resting phases between bursts of chromosomal movement are required to allow for synapsis to proceed past the initiation point. Therefore, whereas chromosomal movement acts to perturb nonhomologous synapsis, we propose that the pauses are required to allow for homologous synapsis to take place. The opposing roles of FKB-6 to that of ZYG-12 and DHC-1 is supported by the suppression of PC formation in fkb-6 mutants by down-regulating movement (zyg-12 or dhc-1 mild knockdown) as opposed to fully abrogating movement (zyg-12 or dhc-1 strong loss of function). In our model, both FKB-6 and LINC complex contribute to the balance between movement (dynein) and pausing movement (FKB-6); this balance is required for the fine-tuning of pairing and synapsis events, prohibiting promiscuous interactions between nonhomologous chromosomes, yet also creating an environment proper for homologous chromosomes to interact (Fig. 7 F).
FKB-6 and FKBP52 may share similar roles
FKBP52, the mammalian homologue of FKB-6, may share a similar mechanism of action to that of FKB-6, as both proteins affect processes related to microtubule dynamics. A meiotic role for FKBP52 is not known, but what is known about its biochemical function may suggest a mechanism of action for FKBP52 in meiosis. FKBP52 physically interacts with dynein to regulate microtubule dynamics and was shown to be involved in microtubule depolymerization in vitro (Chambraud et al., 2007). If this function is conserved in FKB-6, one mechanism by which FKB-6 can regulate the LINC complex movement is by depolymerizing microtubules. In fkb-6 mutants, resting time between movements is shorter. This resting time is likely the time spent between detachments of one microtubule fiber to the attachment of the next. This switch of microtubules results in changes in the directionality of movement. If FKB-6 reduces the chance of attachment for a new microtubule filament to ZYG-12 via dynein, then in fkb-6 mutants, the switch between microtubule fibers will frequently lead to a decrease in resting time and more frequent directionality changes, as we observed. If FKB-6 function is affecting only microtubule attachment to LINC, but not stability, then removal of FKB-6 is not expected to result in loss of microtubules but changes in their organization at the vicinity of the nucleus, as we have found. A role for FKB-6 in microtubule dynamics is also supported by nuclear positioning and mitotic division defects that are found in fkb-6 mutants. As expected, these defects are not as severe as the ones observed when microtubule movement is lost, as a result of FKB-6 modulating this activity and not eliminating it.
FKB-6 and other FKBPs
FKBP52 has not been shown to act in meiosis; however, meiotic roles were identified for two other FKBP proteins: Fkbp6 in mice and Fpr3 in yeast. Fkbp6 is required for pairing during spermatogenesis (Crackower et al., 2003). Interestingly, Fpr3 deletion suppresses PC formation that is induced by removal of DSBs (Macqueen and Roeder, 2009). This phenotype is the opposite from that observed in fkb-6 mutants. In both cases, the mechanisms suggested for these other FKBP proteins are different from those identified here for FKB-6, as well as likely to require them to act inside the nucleus. We cannot exclude a direct role for FKB-6 in SC assembly (e.g., via transport of SC proteins), but because of the effect we have observed on chromosome movement and microtubule-related phenotypes, it is more parsimonious to suggest that FKB-6 acts on the SC indirectly by controlling chromosomal movement.
Control of SC central region protein PC formation and chromosome movement
The SC is a dynamic structure that is able to reshape and acquire different structures: aggregated form/PC versus linear morphology. It is also a structure capable of self-assembly. The most striking evidence for this comes from mammalian tissue culture studies. Expression of central region proteins in mitotically dividing cells is sufficient to assemble long filaments of central region proteins (Costa et al., 2005; Öllinger et al., 2005). These filaments are not connected to DNA, suggesting that the importance of meiotic proteins is not to facilitate SC assembly but to regulate it so that they assemble between homologous chromosomes in an elongated manner. Growth of the central region of the SC in an unregulated manner, in multiple directions, may be the cause of PC formation, whereas restricting growth to one direction may lead to linear polymerization. As evidence, EM analysis of the SC shows protein dimers of the central region subunit forming a single structure that is repeated multiple times in one direction longitudinal to the chromosome axis (Sym and Roeder, 1995; MacQueen et al., 2002). Some models suggest thickness to the SC, but growth in this direction is limited in wild-type SC (Page and Hawley, 2004; Costa et al., 2005). EM analysis of PCs shows a different organization: multiple SC-like structures are attached side by side, allowing for the growth of these structures in multiple directions (Sym and Roeder, 1995; Jeffress et al., 2007).
This study, and others, establish that chromosome movement plays a key role in promoting the longitudinal assembly of the SC (Labella et al., 2011). In the absence of such movement, longitudinal polymerization is blocked, leading to PC formation. PC formation may therefore be a default state: if polymerization is blocked in one direction, it will proceed in others, creating a PC. This model assumes that accurate polymerization is the favorable state, as long as the mechanisms for longitudinal assembly are in place. If chromosome movement is attenuated, but not blocked, a wrong partner will be chosen (nonhomologous synapsis). In this situation, polymerization of the SC will be longitudinal, and PCs will not be formed. If only one chromosome loses movement and polymerization is blocked, SC components that were transported into the nucleus have the option to assemble on other chromosomes that are engaged in longitudinal SC assembly. However, if movement is perturbed on all chromosomes, the central region proteins do not associate with chromosomes, and hence form PCs.
Another key aspect of SC biology, supported by our studies, is that the SC is dynamic and can be reshaped, even after a PC has formed. Despite the presence of PCs in fkb-6 mutants in early stages of prophase, these PCs can be resolved as meiosis progresses. PCs found in early prophase are associated with impaired SC function: delay in the repair of DSBs, as indicated by accumulation of recombination intermediates (RAD-51). This delay in repair has deleterious effects on the germline, as apoptosis is increased. However, because functional SC structure is restored in late prophase, recombination intermediates can be resolved in time to produce crossovers. These findings suggest that functional SC is crucial for crossover formation only from mid- to late pachytene and is dispensable in earlier stages (as long as HTP-3 loads and DSBs are made). The fact that SC assembly defects are not permanent when chromosome movement is perturbed corroborates what was found for other mutants affecting chromosomal movement that show PC formation in early prophase. Mutants that have severe meiotic defects also have other anomalies in the germline (short germline and severely disorganized nuclei), making it difficult to dissect the cause for the extended duration of PC formation.
In some cases, the dynamic property of the SC may be jeopardized: high temperatures (>26°C) lead to PC formation in wild type (Bilgir et al., 2013), and milder heat shock conditions (25°C) enhance PC formation in fkb-6 mutants. These findings indicate that although the SC is dynamic, its ability to be dynamic is temperature sensitive, similar to what is found for chaperone client proteins. This evidence suggests that chaperones may contribute directly to folding of SC proteins, preventing PC formation (like Fpr3), as well as indirectly, by controlling other processes, such as microtubule movement (as found for FKB-6). The collaboration between these two modes of action of chaperones is likely needed to ensure proper SC assembly.
Materials and methods
Strains
Most C. elegans strains were cultured under standard conditions at 20°C. Several strains were cultured at 15°C (indicated by an asterisk in the strain list) and experimentally cultured at 25°C (Figs. 4, 5, and S1). N2 Bristol strain was used as the wild-type background. The following mutations and chromosome rearrangements were used: LGI: syp-3(ok758), akir-1(gk528), dhc-1(or195),* hT2[bli-4(e937) let-?(q782) qIs48] (I;III); LGII: zyg-12(ct350),* zyg-12(or577),* pch-2(tm1458), mIn1[mIs14 dpy-10(e128)] (II); LGIII: ced-4(n1162); LGIV: syp-1(me17), spo-11(ok79), dpy-20(e1282) IV, nT1[qIs51] (IV;V); and LGV: fkb-6(tm2614), fkb-6(iow3), fkb-6(iow4).
The following transgenic lines were used: meIs9[unc-119(+)pie-1promoter::gfp::SYP-3];unc-119(ed3) III, meIs8[pie-1p::GFP::cosa-1+unc-119(+)] II, V5::fkb-6 V, qaIs3507[unc-119(+)+pie-1p::GFP::lem-2], bnIs1[pie-1::GFP::pgl-1+unc-119(+)], dhc-1(ie28[dhc-1::degron::GFP]) I, ojIs9[zyg-12(all)::GFP + unc-119(+)], ddIs6[tbg-1::GFP + unc-119(+)] V, itIs37 [pie-1p::mCherry::H2B::pie-1 3′UTR + unc-119(+)], ltIs24 [(pAZ132) pie-1p::GFP::tba-2 + unc-119(+)], ieSi38 IV, oxTi633 V, and oxTi1014 IV.
RNAi screen leading to the identification of FKB-6
To identify novel genes involved in the regulation of SC assembly, we conducted an RNAi-based screen for genes required for PC formation of SYP-3 proteins in pachytene, a meiotic stage in which the SC is normally fully assembled and in which there are no PCs observed (Smolikov et al., 2009). A PC was defined as SYP-3 localized in a globular manner with a width of at least three times that of wild-type SC, by qualitative inspection. The screen was performed in the akir-1(gk528) mutant background (Clemons et al., 2013) because previous work in our laboratory has shown that these mutants can be used as a sensitized background for isolating genes that affect SC assembly (Brockway et al., 2014). We hypothesized that chaperones and cochaperones may regulate SC assembly by preventing PC formation. Therefore, we performed the screen using RNAi for a small subset of C. elegans genes that included predicted and/or known chaperones and cochaperones. In this screen, we exposed akir-1(gk528) mutants to RNAi by feeding for two generations, then performed ethanol whole worm fixation and observed SYP-3::GFP to identify enhancers leading to SYP-3::GFP aggregation. The phenotype was confirmed by staining for SYP-1 (central region protein of the SC; MacQueen et al., 2002).
CRISPR-Cas9
To generate deletion mutants and the V5 tag insertion, we used CRISPR-Cas9 technology. Single guide RNA (sgRNA) plasmid targeting the sequence 5′-GTTGAAACTCATCAAGAAGGAGG-3′ was cloned in PU6::sgRNA plasmid (Friedland et al., 2013). Selection methods used were: dyp-10 coconversion for V5 tag insertion (Arribere et al., 2014) and coselection from unc-22 mutations for deletion mutants (Kim et al., 2014). V5 tag sequence used was as reported in Paix et al. (2014).
RT-PCR
To determine the genetic landscape of fkb-6(tm2614), we performed RT-PCR analysis. This was done using the Superscript III OneStep RT PCR kit (12574-026; Thermo Fisher Scientific) and primers 5′-CTTGAAGCGCTTCTTGTCACGC-3′ and 5′-CCAACCACTGGAACGACCGTG-3′. Our analysis revealed a deletion of the last 119 amino acids of the 431-aa protein. There was also a simultaneous insertion of 52 amino acids. This deletion removes the third TPR domain and part of the second TPR domain. Our analysis also showed that transcript levels are not different from a wild-type background. RT-PCR of other fkb-6 mutations, created through CRISPR-Cas9, was performed. Using the technique described earlier, we found that fkb-6(iow4) is a 230-bp deletion that contains a 166-bp deletion of the promoter and 64-bp of the first exon; this deleted region encodes for the first 79 amino acids of the FKB-6 protein, which includes the first PP domain. Another small deletion mutation leading to sterility, fkb-6(iow3), was isolated and had a frameshift around the PAM site of the sgRNA (5′-GTTGAAACTCATCAAGAAGGAGG-3′). Both of these mutants were examined by DAPI and SYP-1 staining and were shown to exhibit a phenotype indistinguishable from that of the fkb-6(tm2614) allele.
Fecundity assay
Stage L4 worms were moved to individual NGM plates and allowed to lay eggs for 24 h. These single adult worms were then moved to new NGM plates and allowed to lay eggs again. Adult worms were moved a total of three times. These experiments were performed with three different worms for N2 and fkb-6(tm2614) strains.
Immunofluorescent staining and microscopy
Adult hermaphrodites were dissected 20–24 h after L4 to release gonads. Antibodies used for immunostaining as in Colaiácovo et al. (2003) are as follows: SYP-1 (1:500), RAD-51 (1:10,000; ModEncod), SYP-2 (1:100; gift from A. Villeneuve, Stanford University, Stanford, CA), SYP-4 (1:100), HTP-3 (1:500; gift from A. Dernburg, University of California, Berkeley, Berkeley, CA), HTP-1/2 (1:500; gift from A. Dernburg, McGill University, Montreal, Canada), HIM-3 (1:500; gift from M. Zetka, University of Iowa, Iowa City, IA), LAB-1 (1:300; gift from M. Zetka), agglutinin (1:10, FL-1071; Vector Laboratories), phalloidin (1:10, A12380; Molecular Probes), KT3 (1:10, PGL-3; Developmental Studies Hybridoma Bank), SUN-1 (1:500; Novus Biologicals), and HIM-8 (1:1,000; Sdix). Secondary antibodies used were α-goat Alexa Fluor 549, α-rabbit Alexa Fluor 488, α-guinea pig Alexa Fluor 488, and α-mouse Alexa Fluor 488. HIM-8 (1:500; gift from A. Dernburg) staining was done as in Nabeshima et al. (2004). Secondary antibody was α-guinea pig Alexa Fluor 488. V5 staining was performed with the V5 antibody (1:200; Novus Biologicals) and was performed as in Reid et al. (2014), with a few minor changes. Methanol fix after dissection was limited to 1 min, and acetone fix was removed. Formaldehyde fix was lowered to 2 h from 12 h. Secondary antibody was α-rabbit Cy3 (gift from J. Weiner). Staining for phosphorylated histone 3 (PH3) was done as in Hsu et al. (2000). PH3 antibody (1:100; EMD Millipore) staining was performed with adult hermaphrodites dissected 20–24 h after L4. The secondary antibody used was α-rabbit Alexa Fluor 555. Microtubule staining was done as in Dennis et al. (2012). 12G10 (1:10; Developmental Studies Hybridoma Bank) antibody staining was done with adult hermaphrodites dissected 20–24 h after L4. Secondary antibody used was α-mouse Alexa Fluor 488. All gonads were stained with DAPI (1:2,000 dilution of a 5-mg/ml DAPI stock in PBS-Tween) for 10 min.
FISH
The 5S FISH probe generation and homologous pairing was monitored as in MacQueen et al. (2002). To generate the 5S rDNA probe, primers (5′-TACTTGGATCGGAGACGGCC-3′ and 5′-CTAACTGGACTCAACGTTGC-3′) amplifying the 1-kb region were used and then digested. The region of interest was labeled fluorescently by terminal deoxynucleotidyl transferase. Freeze cracking method was used (see antibody staining), except 7.4% PFA was added before freezing. SSC-Tween was used as wash buffer. Hybridization was performed at 94°C for 90 s. Nuclei were scored from three germlines per genotype. SYP-1 staining and FISH were performed as in Martinez-Perez and Villeneuve (2005).
Microscope image acquisition
Imaging medium used for fixed samples was Vectashield. Images were acquired using the DeltaVision wide-field fluorescence microscope system (Applied Precision Ltd.) with Olympus 100×/1.40-NA lenses. Optical sections were collected at 0.20-µm increments with a coolSNAPHQ camera (Photometrics) and softWoRx 5.0.0 software (Applied Precision Ltd.) and deconvolved using softWoRx. Images are projections through 3D data stacks of whole nuclei (15 to 30 0.2-µm slices/stack). Imaging was performed at RT. Images were processed using Adobe Photoshop CC. Images were adjusted after assembly using the levels function; images of wild type and mutants in the same panel were manipulated identically and simultaneously.
Sample size for figures
n reflects the number of gonads or nuclei counted. In Fig. 3 C, apoptosis: wild type = 96, fkb-6 = 74, syp3 = 57, spo-11 = 42, and fkb-6;spo-11 = 56 gonads. In Fig. 7 A, micronuclei: wild type = 16 and fkb-6(tm2614) mutants = 16 gonads. In Fig. 1 E, SYP-1: wild type = 657 and fkb-6 = 621 nuclei from three independent gonads each. In Fig. 2 B, HIM-8: wild type = 790 and fkb-6 = 614 nuclei from three independent gonads each. In Figs. 2 D and 5S, wild type = 1,113 and fkb-6 = 1,277 nuclei from three independent gonads each. In Fig. 2 E, HIM-8: 743 and fkb-6 = 1,028 nuclei. In Fig. 3 B, RAD-51: wild type = 718 and fkb-6 = 610 nuclei from three independent gonads each. In Fig. 3 D, COSA-1: wild type = 168 and fkb-6 = 123 nuclei from three independent gonads each. In Fig. 5 (A and D), SYP-1: wild type = 1,261, fkb-6 = 1,162, zyg-12 = 628, fkb-6;zyg-12 = 545, wild type;pl4440(RNAi) = 834, wild type;dhc-1(RNAi) = 1,126, fkb-6;pl4440(RNAi) = 899, and fkb-6;dhc-1(RNAi) = 1,832 nuclei from three independent gonads each. In Fig. 5 C, HIM-8: wild type = 818, fkb-6 = 706, zyg-12 = 533, and fkb-6;zyg-12 = 415 nuclei from three independent gonads each. In Fig. 6 A: SUN-1 fkb-6(tm2614) mutants = 387 nuclei from five gonads. In Fig. 6 (B–E), DHC-1: wild type = 10 and fkb-6 = 10 nuclei from four gonads each. In Fig. 7 E, microtubule analysis: wild type = 12 and fkb-6 = 12 nuclei from three gonads each. In Fig. S1, SYP-1: wild type;pl4440(RNAi) = 834, wild type;akir-1(RNAi) = 958, fkb-6;pl4440(RNAi) = 899, and fkb-6;akir-1(RNAi) = 989, pL4440(RNAi) 25°C for 6 h = 1,261, daf-21(RNAi) 25°C for 6 h = 1,466, pL4440(RNAi) 25°C for 24 h = 1,339, daf-21(RNAi) 25°C for 24 h = 1,301 nuclei from three gonads each. In Fig. S4, SYP-1: wild type = 1339, fkb-6 = 742, dhc-1 = 1,377, fkb-6;dhc-1 = 795, zyg-12(ct350) = 398, fkb-6;zyg-12(ct350) = 316, zyg-12(or577) = 1,242, fkb-6;zyg-12(or577) = 916, wild type;pL4440(RNAi) = 1,551, wild type;dhc-1(RNAi) = 1,749, zyg-12(ct350);pL4440(RNAi) = 398, zyg-12(ct350);dhc-1(RNAi) = 1,140 nuclei from three gonads each; ZYG-12: wild type = 12 and fkb-6 = 12 nuclei from four gonads each.
SYP-1 localization analysis
SYP-1 localization pattern was assigned to each nucleus qualitatively using the following categories: linear (found in elongated stretches), PC and linear (PC in addition to mostly elongated stretches, as found in wild-type pachytene nuclei), PC and some linear (PC in addition to short stretches, discontinuous or less than a length of a wild-type stretch in pachytene, predominantly), PC only (all the SYP-1 signal is in a PC), no SYP-1 localization, and other (not included in the previous categories).
HIM-8 foci and 5S FISH analysis
The gonads, of all genotypes analyzed, were divided into seven zones, as in SYP-1 localization analysis, and nuclei were scored for HIM-8/5S FISH foci pairing. Scoring was done similarly to FISH analysis in MacQueen et al. (2002), with 0.7-µm distance between HIM-8/5S FISH foci used as the cutoff for paired homologous chromosomes.
Nonhomologous synapsis analysis
Quantification of nonhomologous synapsis was done in an fkb-6 mutant background with HIM-8/SYP-1 and 5S FISH/SYP-1 double staining. For HIM-8/SYP-1, nuclei with SYP-1 staining were examined for paired or nonpaired HIM-8 foci. Nonpaired HIM-8 foci were determined to be either on or not on SYP-1 linear staining. If a single HIM-8 focus was on a SYP-1 linear strand, the nucleus was determined to have nonhomologous synapsis of the X chromosome. Nonhomologous synapsis quantification for 5S/SYP-1 was performed as for HIM-8/SYP-1 with squashed nuclei.
RAD-51 foci analysis
Quantification of RAD-51 foci was performed for all seven zones composing the PMT to LP regions of the germline as in Colaiácovo et al. (2003). RAD-51 foci were counted for each nucleus. Three gonads were scored per genotype. The mean was determined from each zone across the three gonads analyzed. Statistical comparisons between genotypes were performed using two-tailed MW.
COSA-1 foci analysis
COSA-1::GFP was quantified as in Yokoo et al. (2012) in zone 7 (end of LP) of wild-type and fkb-6 mutant gonads. COSA-1 foci were counted per nucleus within zone 7. Three gonads were scored per genotype. Statistical comparisons were performed using two-tailed MW.
Apoptosis assay
Germ cell corpses were analyzed in adult hermaphrodites 20–24 h after L4 stage, as described in Kelly et al. (2000). 22–24 h after L4, worms were incubated in 25 µg/ml of acridine orange for 2 h. Worms were placed on 1.5% agarose pads and examined on the DeltaVision system described earlier. The number of acridine orange–positive nuclei was counted in each gonad near the bend region. Statistical comparisons between genotypes were performed using two-tailed MW.
RNAi
fkb-6 was identified in an RNAi screen on an akir-1 mutant background and GFP::SYP-3 meIs9[unc-119(+)pie-1promoter::gfp::SYP-3] transgenic line for mutants affecting SC morphogenesis. Synchronized L1 larvae were placed on nemotode growth medium, ampicillin, and IPTG plates seeded with RNAi bacteria from the Ahringer C. elegans RNAi library (Kamath et al., 2003) or pL4440 empty vector control. 4-d-old worms grown on the RNAi were cytologically analyzed and examined for PC formation (as described in Results). RNAi clones were inoculated for 6 h to overnight in lysogeny broth and ampicillin (50 ng/µl). Cultures were then seeded onto IPTG plates and grown for at least 12 h (Kamath et al., 2003). Synchronized L1 or L4 larvae were placed on the seeded IPTG plates and developed to adults. These adults or the next generation of adults were cytologically analyzed.
SUN-1 analysis
Using the zone division system as described earlier, analysis of SUN-1 staining was examined at zone 5 (end of MP). Scoring of nuclei was performed by scoring SYP-1 PC formation and SUN-1 foci around the nucleus. Nuclei were divided into four categories: nuclei with both SYP-1 PC and SUN-1 foci, nuclei with SYP-1 PC and without SUN-1 foci, nuclei without SYP-1 PC and with SUN-1 foci, or nuclei without either SYP-1 PC or SUN-1 foci. Analysis was performed on N2 and fkb-6 mutant strains. Statistical analysis was performed using Fisher’s exact test.
Live imaging
DHC-1::GFP or ZYG-12::GFP transgenic worms, on an N2, fkb-6, and syp-1 background, were immobilized as in Rog and Dernburg (2015). 22–24 h after L4, worms were placed on 7.5% agarose pads and immobilized with Polybead 0.1-µm polystyrene beads (#00876; Polysciences). Images were taken with the DeltaVision system described earlier. Images were taken at increments of 0.5 s (a single 0.2-µm slice) at the start of the visualization of DHC-1::GFP and ZYG-12::GFP foci in each germline imaged.
DHC-1::GFP and ZYG-12::GFP movement analysis
Analysis of DHC-1::GFP transgenic strains in the N2 and fkb-6 mutant background was performed in ImageJ. The plugin Manual Tracking was used to determine distance and velocity traveled by DHC-1::GFP foci for 60 s. Data points were taken at the onset and conclusion of each directional movement. These data points allowed for calculation of total distance traveled, mean velocity, number of times foci changed direction, and time between movements of foci. Statistical analysis was performed using unpaired t test with Welch’s correction.
Western blot
100 adult homozygote worms were lysed 20–24 h after L4 stage selection, per genotype. Lysis buffer contained SDS urea and 2-mercaptoethanol and was flash frozen with liquid nitrogen. Samples were boiled for 5 min and run on 4–20% gradient Expressplus PAGE (#M42012; GenScript). PBS-Tween (1×) was used as a wash buffer. The V5 (1:200; Invitrogen) antibody was used as primary antibody. Mouse α-tubulin (1:1,000; Developmental Studies Hybridoma Bank) was used as the loading control. The secondary antibody used was α-mouse antibody conjugated to HRP (1:10,000). PBS-Tween (1×)–5% milk was used for incubation and blocking. WesternBright ECL (#K-12045-D20; Advansta) was used for HRP detection and blot development.
PH3 Analysis
Analysis of PH3 staining was performed in N2 and fkb-6 mutants. The number of PH3-positive nuclei was quantified. Statistical analysis was performed using two-tailed MW.
Microtubule staining analysis
Analysis of microtubule staining was performed in N2 and fkb-6 mutant background using ImageJ in three different methods. Nuclear rim analysis: The circle tool was used to place an ellipsis (5 pixels wide) around a nucleus (DAPI channel). A plot profile of the intensity around the ellipsis was taken. The median was taken between the highest and lowest peaks, and the number of peaks greater than the median was quantified as the number of microtubule–filament connections. Adjacent cytoplasmic space analysis: A second ellipsis analysis was used by placing an ellipsis (1 pixel wide) inside the nuclear space near the edge of DAPI staining. A second ellipsis was placed directly outside the nucleus in such a way that space was seen between DAPI and the ellipsis on all sides. The integrated intensity was calculated for each ellipsis, and the inner ellipsis integrated intensity was subtracted from the outer integrated intensity. Linescan analysis: Linescans at a 5-pixel width were taken from each nuclear center radiating outward in six different directions. A plot profile was taken of each linescan. Linescans were examined by determining the highest and lowest points of intensity, 3 µm from the edge of DAPI staining. The midpoint was determined, and the number of peaks above the midpoint was counted as a microtubule strand (cytosolic network analysis). Microtubule connections to the nucleus were determined by whether the intensity within a 5-pixel length outside of the nucleus exceeded the midpoint. Statistical analyses were performed using unpaired t test with Welch’s correction.
Micronuclei analysis
Analysis of micronuclei was performed using GFP::lem-2 in wild-type and fkb-6(tm2614) mutant background. Worms were dissected 20–24 h after L4 and a 6-h incubation at 25°C. Gonads were visually analyzed for micronuclei. Micronuclei were defined as DAPI bodies at least half the size of wild-type nuclei and surrounded by GFP. Analysis was divided into two zones, PMT and meiosis, differentiated by the appearance of SYP-1 staining. Statistical analysis was performed using unpaired t test with Welch’s correction.
Microtubule embryo analysis
Mitotic division in wild-type and fkb-6 mutant embryos were analyzed by tba-2::GFP and DAPI morphology. 20–24 h after L4, worms were dissected, stained with DAPI, and fixed with ethanol. Using the DeltaVision system, embryos were qualitatively analyzed. DAPI staining and TBA-2::GFP were both examined for abnormal appearance and structure in fkb-6 mutants compared with wild type.
Online supplemental material
Fig. S1, A and B, show SYP-1 staining of early frameshift mutants of fkb-6 showing PC, similar to that described for fkb-6(tm2614); Fig. S1 C shows SYP-1 localization quantification in single and double mutants, with akir-1 showing an additive effect with fkb-6(tm2614); in Fig. S1 D, down-regulation of daf-21 does not show PC formation; and Fig. S1 E shows increased PC formation in fkb-6 mutants at 25°C. In Fig. S2, staining of various SC components shows that the defects in fkb-6(tm2614) mutants are specific to SC central region proteins. Fig. S3 shows staining of various nuclear envelope and membrane proteins with normal localization in fkb-6(tm2614) mutants. In Fig. S4 A, SYP-1 localization quantification in single and double mutants shows aggravated SYP-1 PC formation in long exposure to high temperature; Fig. S4 B shows enhancement of PC formation in zyg-12 mutants exposed to dhc-1 RNAi. Fig. S5 A explains methods; Fig. S5 B shows movement defects of ZYG-12::GFP patches; and Fig. S5 C shows microtubule staining in fkb-6 mutants. Videos 1–3 are examples of live imaging of DHC-1 in all genotypes examined.
Supplementary Material
Acknowledgments
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). We thank the National Bioresource Project for the Experimental Animal “Nematode C. elegans,” Japan, for providing the tm2614 allele. We thank M. Zetka for the HIM-3 and HTP-3 antibodies, A. Dernburg for the HIM-8 and HTP-1/2 antibodies, M. Colaiacovo for the LAB-1 antibody, J. Wiener for help with reagents, and D. Houston for his help with membrane and nuclear envelope staining reagents. We are grateful to R. Malone, Y. Tzur, and members of the Smolikove laboratory for critical reading of this manuscript and for T. Tootle and V. Prahlad for discussion.
This work was supported by the National Science Foundation grant MCB-1515551 (to S. Smolikove).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- DSB
- double-strand break
- EP
- early pachytene
- FKBP
- FK506 binding protein
- LINC
- linker of nucleoskeleton and cytoskeleton
- LP
- late pachytene
- MP
- midpachytene
- MW
- Mann–Whitney U test
- PC
- polycomplex
- PH3
- phosphorylated histone 3
- PMT
- premeiotic tip
- PPIase
- peptidylprolyl cis-trans isomerase
- SC
- synaptonemal complex
- sgRNA
- single guide RNA
- TPR
- tetratricopeptide repeat
- TZ
- transition zone
References
- Arribere J.A., Bell R.T., Fu B.X.H., Artiles K.L., Hartman P.S., and Fire A.Z.. 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 198:837–846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgir C., Dombecki C.R., Chen P.F., Villeneuve A.M., and Nabeshima K.. 2013. Assembly of the synaptonemal complex is a highly temperature-sensitive process that is supported by PGL-1 during Caenorhabditis elegans meiosis. G3 (Bethesda). 3:585–595. 10.1534/g3.112.005165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnby D.A., Link E.M., Vowels J.J., Tian H., Colacurcio P.L., and Thomas J.H.. 2000. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans. Genetics. 155:85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boateng K.A., Bellani M.A., Gregoretti I.V., Pratto F., and Camerini-Otero R.D.. 2013. Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev. Cell. 24:196–205. 10.1016/j.devcel.2012.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway H., Balukoff N., Dean M., Alleva B., and Smolikove S.. 2014. The CSN/COP9 signalosome regulates synaptonemal complex assembly during meiotic prophase I of Caenorhabditis elegans. PLoS Genet. 10:e1004757 10.1371/journal.pgen.1004757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambraud B., Belabes H., Fontaine-Lenoir V., Fellous A., and Baulieu E.E.. 2007. The immunophilin FKBP52 specifically binds to tubulin and prevents microtubule formation. FASEB J. 21:2787–2797. 10.1096/fj.06-7667com [DOI] [PubMed] [Google Scholar]
- Christophorou N., Rubin T., Bonnet I., Piolot T., Arnaud M., and Huynh J.-R.. 2015. Microtubule-driven nuclear rotations promote meiotic chromosome dynamics. Nat. Cell Biol. 17:1388–1400. 10.1038/ncb3249 [DOI] [PubMed] [Google Scholar]
- Clemons A.M., Brockway H.M., Yin Y., Kasinathan B., Butterfield Y.S., Jones S.J.M., Colaiácovo M.P., and Smolikove S.. 2013. Akirin is required for diakinesis bivalent structure and synaptonemal complex disassembly at meiotic prophase I. Mol. Biol. Cell. 24:1053–1067. 10.1091/mbc.E12-11-0841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiácovo M.P., MacQueen A.J., Martinez-Perez E., McDonald K., Adamo A., La Volpe A., and Villeneuve A.M.. 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 5:463–474. 10.1016/S1534-5807(03)00232-6 [DOI] [PubMed] [Google Scholar]
- Conrad M.N., Lee C.-Y., Chao G., Shinohara M., Kosaka H., Shinohara A., Conchello J.-A., and Dresser M.E.. 2008. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 133:1175–1187. 10.1016/j.cell.2008.04.047 [DOI] [PubMed] [Google Scholar]
- Costa Y., Speed R., Öllinger R., Alsheimer M., Semple C.A., Gautier P., Maratou K., Novak I., Höög C., Benavente R., and Cooke H.J.. 2005. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J. Cell Sci. 118:2755–2762. 10.1242/jcs.02402 [DOI] [PubMed] [Google Scholar]
- Couteau F., and Zetka M.. 2005. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 19:2744–2756. 10.1101/gad.1348205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower M.A., Kolas N.K., Noguchi J., Sarao R., Kikuchi K., Kaneko H., Kobayashi E., Kawai Y., Kozieradzki I., Landers R., et al. . 2003. Essential role of Fkbp6 in male fertility and homologous chromosome pairing in meiosis. Science. 300:1291–1295. 10.1126/science.1083022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho C.E., Zaaijer S., Smolikov S., Gu Y., Schumacher J.M., and Colaiácovo M.P.. 2008. LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes Dev. 22:2869–2885. 10.1101/gad.1691208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis S., Sheth U., Feldman J.L., English K.A., and Priess J.R.. 2012. C. elegans germ cells show temperature and age-dependent expression of Cer1, a Gypsy/Ty3-related retrotransposon. PLoS Pathog. 8:e1002591 10.1371/journal.ppat.1002591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A.F., McDonald K., Moulder G., Barstead R., Dresser M., and Villeneuve A.M.. 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 94:387–398. 10.1016/S0092-8674(00)81481-6 [DOI] [PubMed] [Google Scholar]
- Ding X., Xu R., Yu J., Xu T., Zhuang Y., and Han M.. 2007. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell. 12:863–872. 10.1016/j.devcel.2007.03.018 [DOI] [PubMed] [Google Scholar]
- Erlejman A.G., Lagadari M., Harris D.C., Cox M.B., and Galigniana M.D.. 2014. Molecular chaperone activity and biological regulatory actions of the TPR-domain immunophilins FKBP51 and FKBP52. Curr. Protein Pept. Sci. 15:205–215. 10.2174/1389203715666140331113753 [DOI] [PubMed] [Google Scholar]
- Fasseas M.K., Dimou M., and Katinakis P.. 2012. The Caenorhabditis elegans parvulin gene subfamily and their expression under cold or heat stress along with the fkb subfamily. Biochem. Biophys. Res. Commun. 423:520–525. 10.1016/j.bbrc.2012.05.157 [DOI] [PubMed] [Google Scholar]
- Friedland A.E., Tzur Y.B., Esvelt K.M., Colaiácovo M.P., Church G.M., and Calarco J.A.. 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods. 10:741–743. 10.1038/nmeth.2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galat A. 2000. Sequence diversification of the FK506-binding proteins in several different genomes. Eur. J. Biochem. 267:4945–4959. 10.1046/j.1432-1327.2000.01509.x [DOI] [PubMed] [Google Scholar]
- Gönczy P., Pichler S., Kirkham M., and Hyman A.A.. 1999. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 147:135–150. 10.1083/jcb.147.1.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyer W., Kaitna S., Couteau F., Ward J.D., Boulton S.J., and Zetka M.. 2008. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev. Cell. 14:263–274. 10.1016/j.devcel.2007.11.016 [DOI] [PubMed] [Google Scholar]
- Horn H.F., Kim D.I., Wright G.D., Wong E.S.M., Stewart C.L., Burke B., and Roux K.J.. 2013. A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J. Cell Biol. 202:1023–1039. 10.1083/jcb.201304004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.Y., Sun Z.W., Li X., Reuben M., Tatchell K., Bishop D.K., Grushcow J.M., Brame C.J., Caldwell J.A., Hunt D.F., et al. . 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 102:279–291. 10.1016/S0092-8674(00)00034-9 [DOI] [PubMed] [Google Scholar]
- Jeffress J.K., Page S.L., Royer S.K., Belden E.D., Blumenstiel J.P., Anderson L.K., and Hawley R.S.. 2007. The formation of the central element of the synaptonemal complex may occur by multiple mechanisms: the roles of the N- and C-terminal domains of the Drosophila C(3)G protein in mediating synapsis and recombination. Genetics. 177:2445–2456. 10.1534/genetics.107.078717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. . 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421:231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Kang C.B., Hong Y., Dhe-Paganon S., and Yoon H.S.. 2008. FKBP family proteins: Immunophilins with versatile biological functions. Neurosignals. 16:318–325. 10.1159/000123041 [DOI] [PubMed] [Google Scholar]
- Kelly K.O., Dernburg A.F., Stanfield G.M., and Villeneuve A.M.. 2000. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics. 156:617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ishidate T., Ghanta K.S., Seth M., Conte D. Jr., Shirayama M., and Mello C.C.. 2014. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics. 197:1069–1080. 10.1534/genetics.114.166389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella S., Woglar A., Jantsch V., and Zetka M.. 2011. Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev. Cell. 21:948–958. 10.1016/j.devcel.2011.07.011 [DOI] [PubMed] [Google Scholar]
- Macqueen A.J., and Roeder G.S.. 2009. Fpr3 and Zip3 ensure that initiation of meiotic recombination precedes chromosome synapsis in budding yeast. Curr. Biol. 19:1519–1526. 10.1016/j.cub.2009.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Colaiácovo M.P., McDonald K., and Villeneuve A.M.. 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16:2428–2442. 10.1101/gad.1011602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Phillips C.M., Bhalla N., Weiser P., Villeneuve A.M., and Dernburg A.F.. 2005. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 123:1037–1050. 10.1016/j.cell.2005.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.J., Misner L., Le Bot N., Tsai M.C., Campbell J.M., Ahringer J., and White J.G.. 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 115:825–836. 10.1016/S0092-8674(03)00985-1 [DOI] [PubMed] [Google Scholar]
- Martinez-Perez E., and Villeneuve A.M.. 2005. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 19:2727–2743. 10.1101/gad.1338505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., and Seydoux G.. 2010. The Puf RNA-binding proteins FBF-1 and FBF-2 inhibit the expression of synaptonemal complex proteins in germline stem cells. Development. 137:1787–1798. 10.1242/dev.050799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto A., Shibuya H., Zhu X., Kim J., Ishiguro K., Han M., and Watanabe Y.. 2012. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J. Cell Biol. 198:165–172. 10.1083/jcb.201204085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A.M., and Hillers K.J.. 2004. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics. 168:1275–1292. 10.1534/genetics.104.030700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O., Shimanuki M., and Miki F.. 2000. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 19:3831–3840. 10.1093/emboj/19.14.3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öllinger R., Alsheimer M., and Benavente R.. 2005. Mammalian protein SCP1 forms synaptonemal complex-like structures in the absence of meiotic chromosomes. Mol. Biol. Cell. 16:212–217. 10.1091/mbc.E04-09-0771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S.L., and Hawley R.S.. 2004. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20:525–558. 10.1146/annurev.cellbio.19.111301.155141 [DOI] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H.E., Lee C.Y.S., Calidas D., Lu T., Smith J., Schmidt H., Krause M.W., and Seydoux G.. 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics. 198:1347–1356. 10.1534/genetics.114.170423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton T.J., and Kay J.E.. 2005. Identification and comparative analysis of the peptidyl-prolyl cis/trans isomerase repertoires of H. sapiens, D. melanogaster, C. elegans, S. cerevisiae and Sz. pombe. Comp. Funct. Genomics. 6:277–300. 10.1002/cfg.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A., Tang L., Novatchkova M., Ladurner M., Fridkin A., Gruenbaum Y., Schweizer D., Loidl J., and Jantsch V.. 2007. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell. 12:873–885. 10.1016/j.devcel.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Penkner A.M., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T., Woglar A., Csaszar E., Pasierbek P., Ammerer G., Gruenbaum Y., and Jantsch V.. 2009. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 139:920–933. 10.1016/j.cell.2009.10.045 [DOI] [PubMed] [Google Scholar]
- Phillips C.M., and Dernburg A.F.. 2006. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell. 11:817–829. 10.1016/j.devcel.2006.09.020 [DOI] [PubMed] [Google Scholar]
- Phillips C.M., Wong C., Bhalla N., Carlton P.M., Weiser P., Meneely P.M., and Dernburg A.F.. 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 123:1051–1063. 10.1016/j.cell.2005.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A., Yücel D., Wood M., Llamosas E., Kant S., Crossley M., and Nicholas H.. 2014. The transcriptional repressor CTBP-1 functions in the nervous system of Caenorhabditis elegans to regulate lifespan. Exp. Gerontol. 60:153–165. 10.1016/j.exger.2014.09.022 [DOI] [PubMed] [Google Scholar]
- Richardson J.M., Dornan J., Opamawutthikul M., Bruce S., Page A.P., and Walkinshaw M.D.. 2007. Cloning, expression and characterisation of FKB-6, the sole large TPR-containing immunophilin from C. elegans. Biochem. Biophys. Res. Commun. 360:566–572. 10.1016/j.bbrc.2007.06.080 [DOI] [PubMed] [Google Scholar]
- Rog O., and Dernburg A.F.. 2015. Direct visualization reveals kinetics of meiotic chromosome synapsis. Cell Reports. 10:1639–1645. 10.1016/j.celrep.2015.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Isaac B., Phillips C.M., Rillo R., Carlton P.M., Wynne D.J., Kasad R.A., and Dernburg A.F.. 2009. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 139:907–919. 10.1016/j.cell.2009.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein A.M., Galigniana M.D., Kanelakis K.C., Radanyi C., Renoir J.M., and Pratt W.B.. 1999. Different regions of the immunophilin FKBP52 determine its association with the glucocorticoid receptor, hsp90, and cytoplasmic dynein. J. Biol. Chem. 274:36980–36986. 10.1074/jbc.274.52.36980 [DOI] [PubMed] [Google Scholar]
- Smolikov S., Eizinger A., Hurlburt A., Rogers E., Villeneuve A.M., and Colaiácovo M.P.. 2007a Synapsis-defective mutants reveal a correlation between chromosome conformation and the mode of double-strand break repair during Caenorhabditis elegans meiosis. Genetics. 176:2027–2033. 10.1534/genetics.107.076968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Eizinger A., Schild-Prüfert K., Hurlburt A., McDonald K., Engebrecht J., Villeneuve A.M., and Colaiácovo M.P.. 2007b SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics. 176:2015–2025. 10.1534/genetics.107.072413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Schild-Prüfert K., and Colaiácovo M.P.. 2009. A yeast two-hybrid screen for SYP-3 interactors identifies SYP-4, a component required for synaptonemal complex assembly and chiasma formation in Caenorhabditis elegans meiosis. PLoS Genet. 5:e1000669 10.1371/journal.pgen.1000669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storer C.L., Dickey C.A., Galigniana M.D., Rein T., and Cox M.B.. 2011. FKBP51 and FKBP52 in signaling and disease. Trends Endocrinol. Metab. 22:481–490. 10.1016/j.tem.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M., and Roeder G.S.. 1995. Zip1-induced changes in synaptonemal complex structure and polycomplex assembly. J. Cell Biol. 128:455–466. 10.1083/jcb.128.4.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., West R.R., McIntosh J.R., and Hiraoka Y.. 1999. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 145:1233–1249. 10.1083/jcb.145.6.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo R., Zawadzki K.A., Nabeshima K., Drake M., Arur S., and Villeneuve A.M.. 2012. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 149:75–87. 10.1016/j.cell.2012.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka M.C., Kawasaki I., Strome S., and Müller F.. 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13:2258–2270. 10.1101/gad.13.17.2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Rolls M.M., Hall D.H., Malone C.J., and Hanna-Rose W.. 2009. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J. Cell Biol. 186:229–241. 10.1083/jcb.200902101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.