The kinetochore complex is conserved across many eukaryotes, but the protozoan lineage Kinetoplastida builds kinetochores from components without apparent homology to models. D’Archivio and Wickstead describe a new family of proteins with homology to outer kinetochore proteins Ndc80 and Nuf2 that defines the outer kinetochore of trypanosomes, suggesting that all eukaryotes have divergent versions of a universal kinetochore machine.
Abstract
Kinetochores are multiprotein complexes that couple eukaryotic chromosomes to the mitotic spindle to ensure proper segregation. The model for kinetochore assembly is conserved between humans and yeast, and homologues of several components are widely distributed in eukaryotes, but key components are absent in some lineages. The recent discovery in a lineage of protozoa called kinetoplastids of unconventional kinetochores with no apparent homology to model organisms suggests that more than one system for eukaryotic chromosome segregation may exist. In this study, we report a new family of proteins distantly related to outer kinetochore proteins Ndc80 and Nuf2. The family member in kinetoplastids, KKT-interacting protein 1 (KKIP1), associates with the kinetochore, and its depletion causes severe defects in karyokinesis, loss of individual chromosomes, and gross defects in spindle assembly or stability. Immunopurification of KKIP1 from stabilized kinetochores identifies six further components, which form part of a trypanosome outer kinetochore complex. These findings suggest that kinetochores in organisms such as kinetoplastids are built from a divergent, but not ancestrally distinct, set of components and that Ndc80/Nuf2-like proteins are universal in eukaryotic division.
Introduction
During cell division, genetic material must be faithfully transmitted to daughter cells. In eukaryotes, this is achieved by coupling the movement of spindle microtubules to replicated chromosomes via a multiprotein attachment complex called the kinetochore. In most organisms, kinetochores are built around a site of specialized chromatin that is distinguished by the presence of the histone H3 variant CENP-A. This centromeric DNA recruits a set of ∼16 proteins known as the constitutive centromere-associated network (CCAN), which forms the core of the inner kinetochore (Cheeseman and Desai, 2008; Westhorpe and Straight, 2013). In human cells, the CCAN is associated with centromeres throughout the cell cycle (Foltz et al., 2006; Okada et al., 2006). From late G2 onwards, components of the outer kinetochore are recruited to the exterior side of the CCAN, in particular a set of three protein complexes (Knl-1, Mis12, and Ndc80), which together form the KMN network (Cheeseman and Desai, 2008). This network mediates the interaction of the kinetochore with the spindle, and the Ndc80 complex—consisting of a heterotetramer of Ndc80 (also known as HEC1), Nuf2, Spc25, and Spc24—forms a long rod with microtubule-binding globular domains distal from the centromere (Wigge and Kilmartin, 2001; Ciferri et al., 2005; Wei et al., 2005; Cheeseman et al., 2006; DeLuca et al., 2006; Alushin et al., 2010). These domains in Ndc80 and Nuf2 have the same calponin homology (CH) fold (Wei et al., 2007; Ciferri et al., 2008), and the overall architecture of the proteins is also similar, implying that they diverged from a single ancestor that most likely formed a homodimer (Schou et al., 2014).
The above model for kinetochore assembly is conserved between humans and yeast, and homologues of several components are found in diverse eukaryotes. In spite of this, components are not universally identifiable, and a lineage of flagellate protozoa called the kinetoplastids build kinetochores from components without apparent homology to models, suggesting there may be alternative systems. The Kinetoplastida are a group of protozoa that diverged from the animal-yeast lineage very early in evolution (Hampl et al., 2009; Rogozin et al., 2009; He et al., 2014). Several kinetoplastid organisms cause important diseases of humans and other animals, and the African trypanosome Trypanosoma brucei is the causative agent of human sleeping sickness. Trypanosomes undergo a closed mitosis based around an intranuclear spindle, and electron-dense plaques very similar in ultrastructure to vertebrate kinetochores have been observed in dividing nuclei (Ogbadoyi et al., 2000). However, they have an unusual genome architecture (Daniels et al., 2010), including ∼100 small linear chromosomes, each of which is segregated with fidelity (Wickstead et al., 2003). Moreover, when the genome of T. brucei and two other kinetoplastids were sequenced, they were found to encode no readily identifiable homologues of kinetochore proteins in other systems (Berriman et al., 2005; Akiyoshi and Gull, 2013), including the centromere-specific histone CENP-A (Lowell and Cross, 2004). This has been reinforced by the recent identification of 20 kinetochore proteins in trypanosomes (KKT1–20), defining an “unconventional” kinetochore (Akiyoshi and Gull, 2014; Nerusheva and Akiyoshi, 2016). KKTs associate to kinetochore-like nuclear foci, are involved in chromosome segregation, and at least two (KKT2 and KKT3) are highly enriched at trypanosome centromeres, but none has clear orthology to proteins in nonkinetoplastid lineages. As a result, it has been proposed that the Kinetoplastida build kinetochores from a set of proteins distinct from other lineages and perhaps representing an ancestral set (Akiyoshi and Gull, 2014).
An ancestrally distinct kinetochore in trypanosomes would support a controversial rooting of the eukaryotic tree in which the Euglenozoa (kinetoplastids, euglenids, and diplonemids) are the earliest branching extant line (Cavalier-Smith, 2010). However, although kinetoplastids are exceptional in possessing no obvious conventional kinetochore components, they are not unique in lacking key components (Meraldi et al., 2006; Westermann and Schleiffer, 2013). As a result, it is unclear whether any eukaryotic kinetochores are truly distinct or if they have diverged from a kinetochore composed of canonical components. This also raises the question of which proteins are performing key functions in which clear orthologues are missing. In this study, we describe a new family of proteins with homology to Ndc80 and Nuf2, which define the outer kinetochore of trypanosomes. These proteins are present in organisms lacking Ndc80/Nuf2, and we present data to show that all eukaryotes have divergent versions of the same universal kinetochore machine.
Results
All eukaryotes encode at least one protein with similarity to Ndc80 and Nuf2
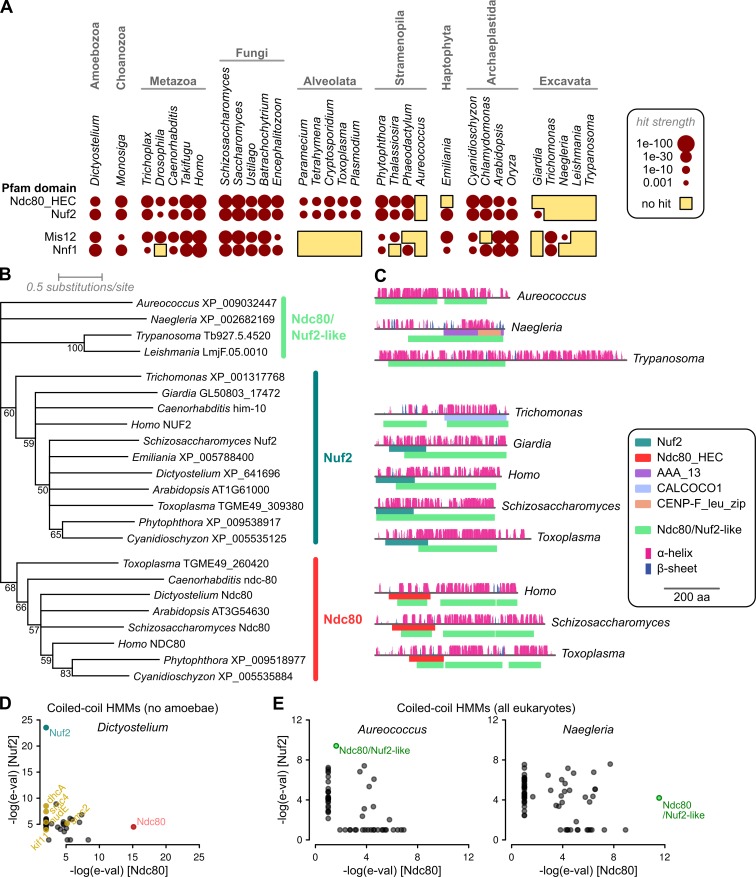
Ndc80 and Nuf2 are among the most highly conserved kinetochore components in terms of sequence similarity (Meraldi et al., 2006). Domain models covering the N-terminal regions of Ndc80/HEC1 and Nuf2 are included in the Pfam database (PF03801 and PF03800, respectively). In spite of these regions being conserved in previously identified homologues of Ndc80 and Nuf2, predicted proteins with significant similarity to the Pfam domains are not encoded in the genomes of several eukaryotes in addition to the Kinetoplastida (Fig. 1 A). Most noticeably, there are no readily identifiable homologues in Naegleria gruberi, which is a close relative of the kinetoplastids, the more distantly related excavate Trichomonas vaginalis or the golden alga Aureococcus anophagefferens. A similar situation exists for the two most highly conserved components of the Mis12 complex, Mis12 and Nnf1, readily identifiable homologues of which are absent from Giardia, Aureococcus, and all aveolates, in addition to kinetoplastids (Fig. 1 A). This patchy distribution of identifiable homologues may be the product of genuine loss or ancestral difference, but it could also represent divergence of protein sequences in some lineages, leading to difficult detection of true homologues.
Figure 1.
Identification of Ndc80- and Nuf2-like sequences across eukaryotes. (A) Most excavates and the golden alga A. anophagefferens lack proteins matching either Ndc80_HEC or Nuf2 Pfam domains (e-value ≤ 0.001). A similar, but not identical, distribution exists for Mis12 and Nnf1 domains, with notable additional absence in Alveolata. (B) Maximum likelihood phylogeny of Ndc80/Nuf2-like protein sequences. Tree represents a majority consensus from 500 bootstrapped inferences based on 406 aligned residues. Numbers show bootstrap support for nodes. (C) Predicted protein architectures (secondary structure features and Pfam domains) of example sequences, showing also position of hits to a pan-Ndc80/Nuf2 HMM. (D) The coiled-coil tails of both Ndc80 and Nuf2 contain information specific to these families; HMMs built only from regions of Ndc80 or Nuf2 homologues outside of the CH fold, and excluding all sequences from Amboebae, specifically identify Ndc80 and Nuf2 from the predicted proteome of Dictyostelium discoideum. (E) Similar models including all eukaryotes specifically identify the Ndc80/Nuf2-like sequences detected by the pan-Ndc80/Nuf2 HMM.
We reasoned that if all eukaryotic kinetochores are based on a common machinery with divergent components in some lineages, the most constrained components would likely be in the outer kinetochore, where the kinetochore interfaces with microtubules. To this end, we undertook a sensitive search for Ndc80 and Nuf2 homologues using an iterative hidden Markov model (HMM)–based approach working from clear homologues to more divergent lineages (see Materials and methods for details). HMMs were first constructed for Ndc80 and Nuf2 families separately using all alignable residues (including those outside of CH domains). These models readily identify a previously unidentified Nuf2-like protein in T. vaginalis.
Ndc80 and Nuf2 share evolutionary ancestry (Schou et al., 2014). Profile–profile comparisons between these the two HMMs showed significant similarity (e-value = 2 × 10−13), and the protein families are alignable across both N-terminal CH and C-terminal tail domains (Fig. S1). Alignable residues from the full proteins were combined to form a pan-Ndc80/Nuf2 HMM. This identified proteins with Ndc80/Nuf2-like properties encoded in both N. gruberi and A. anophagefferens. Significantly, new proteins with apparent Ndc80/Nuf2 homology were identified in organisms otherwise lacking both Ndc80 and Nuf2 proteins, with no additional hits in organisms containing obvious homologues, suggesting that our combined model is specifically identifying divergent members of this family and not other classes of protein containing either CH folds or coiled-coil regions. To give the greatest sensitivity for searching in kinetoplastida, we compared our pan-Ndc80/Nuf2 HMM to alignments of all orthologue groups from a selection of kinetoplastid species, thereby weighting our comparison for residues conserved in both sets (Table S1). The best hit, orthologue group OG5_141718, contains proteins from kinetoplastids that group in phylogenies with Naegleria and Aureococcus Ndc80/Nuf2-like proteins (Fig. 1 B). They also have architectures reminiscent of both Ndc80 and Nuf2, with a large quantity of predicted α-helices across the whole protein, but no detectable Ndc80 or Nuf2 Pfam domains (Fig. 1 C). Alignments of these proteins with clear Ndc80 and Nuf2 sequences shows there are alignable residues at the C-terminal end of the Ndc80/Nuf2 domain, but the major contribution to detection is from coiled-coils. These regions are much more similar to each other than to nonorthologous coiled-coil proteins, and HMMs built from these regions contain sufficient specific information to clearly identify Ndc80 and Nuf2 sequences encoded in the Dictyostelium genome with very little cross-reaction to other proteins, even with no sequence from the CH domains or from any Amoebae in the models (Fig. 1 D). They also identify the same Ndc80/Nuf2-like proteins in Naegleria and Aureococcus as the pan-family HMM (Fig. 1 E).
Trypanosome Ndc80/Nuf2-like protein, KKT-interacting protein 1 (KKIP1), is part of the kinetochore
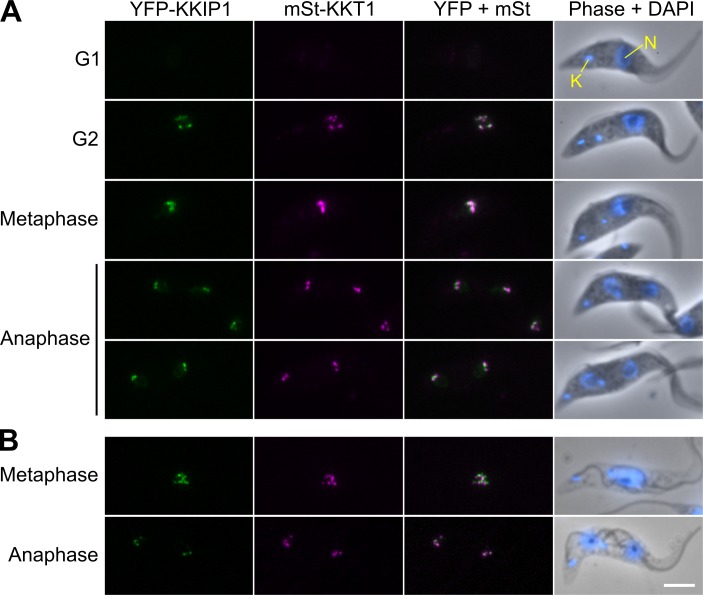
Our expanded HMM protocol identified new Ndc80/Nuf2-like sequences in all eukaryotes lacking conventional Ndc80 or Nuf2 (and nowhere else). However, these proteins lack clear similarity across the CH region, and this distant relationship alone provides little evidence that the predicted homologues are involved in kinetochore function. Those from the kinetoplastids are the most divergent in sequence, sharing only 9–12% identity across aligned residues (Fig. S1). To test if divergent Ndc80/Nuf2-like molecules are true kinetochore components, we tagged the identified protein in T. brucei by integration of coding sequence for YFP at the N terminus of the endogenous gene (Tb927.5.4520). In agreement with our prediction, the protein showed a clear kinetochore-like localization (Fig. 2 A). Location and movement of the protein in the nucleus were similar to those seen for the first identified kinetochore component, KKT1 (Fig. 2 A), and the temporal behavior through the cell cycle is similar to that seen for several other KKT proteins (Akiyoshi and Gull, 2014). Levels of the tagged protein are undetectable in G1 cells, with visible foci forming around S phase. These foci strengthen in signal through G2 before congression to the middle of the nucleus at metaphase and then movement to the spindle poles at anaphase. The protein is stably associated with detergent-extracted cytoskeleton preparations (Fig. 2 B), but was not identified as part of the KKT set. To reflect this behavior and the alternative method for identification, we have named the trypanosomal protein KKIP1.
Figure 2.
KKIP1 is a new kinetoplastid kinetochore-associated protein. (A) Micrographs of insect midgut-form (procyclic) cells expressing YFP-KKIP1 and mStrawberry-KKT1. Counterstaining with the DNA stain DAPI is also shown. (B) Stable association of fluorescently tagged proteins with detergent-extracted cytoskeleton preparations. Bar, 4 µm. K, kinetoplast; N, nucleus.
KKIP1 is essential for chromosome segregation and spindle function
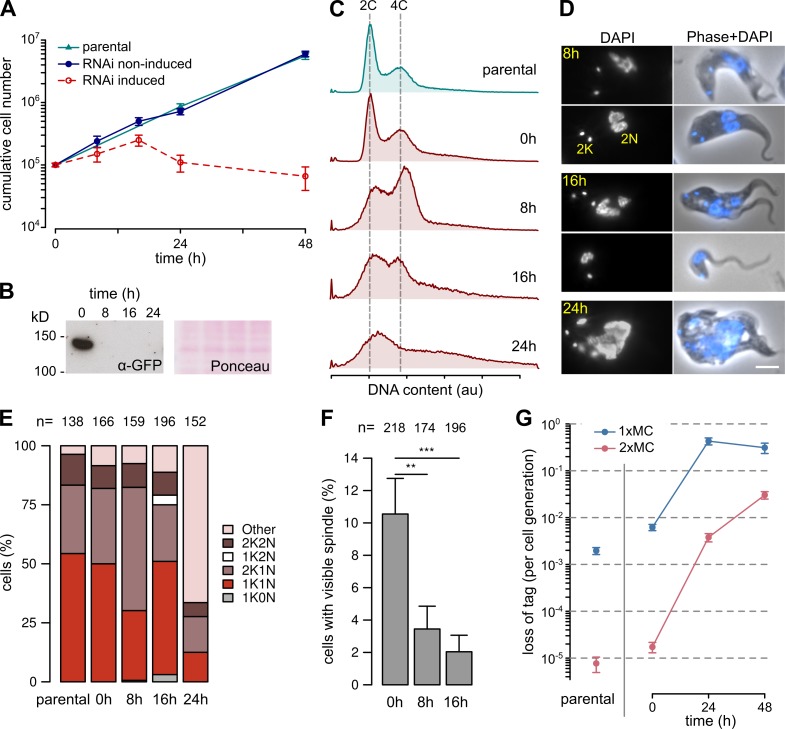
Depletion of at least six KKTs resulted in some disruption of trypanosome mitosis, although growth of cell populations was not greatly affected (Akiyoshi and Gull, 2014). Depletion of KKIP1 levels by inducible RNAi resulted in a rapid and severe defect in cell growth (Fig. 3, A and B). This defect was apparent within the first cell cycle after RNAi induction with a large accumulation of cells with 4C DNA content (Fig. 3 C). By 16 h after induction, these cells start to be replaced by cells with DNA content outside of the normal range, consistent with cell division without correct partitioning of nuclear DNA (Fig. 3, C and D). A cell cycle defect was also apparent when observing the division of the kinetoplast (mitochondrial DNA) and nuclei. These organelles have distinct segregation timing and can be used to morphologically follow progression of trypanosome cells through the cell cycle (Woodward and Gull, 1990). RNAi against KKIP1 for 8 h (just over one cell generation time) resulted in ∼50% of cells with a morphology found in S/G2 cells (2K1N), followed by release and accumulation of cells with aberrant numbers of DNA-containing organelles (Fig. 3 E).
Figure 3.
KKIP1 is essential for chromosome segregation and spindle function. (A) Cell growth in cultures after RNAi-induced ablation of KKIP1. Error bars show SEM (n = 3; P < 0.001 for induced versus noninduced cell numbers at points after 20 h; Student's t test). (B) Immunoblots of cells expressing YFP-KKIP1 showing depletion of protein. Protein loading is shown by Ponceau S stain. (C) Flow cytometry showing disruption of DNA content caused by KKIP1 depletion (representative data from two repeats shown). (D) Phenotypic changes to cells upon RNAi. Bar, 4 µm. (E) Morphological analysis of cell cycle on RNAi shows a buildup of undivided cells with late morphology (2K1N), then formation of cells with aberrant numbers of kinetoplasts (K) and nuclei (N). (F) Loss of visible spindles on KKIP1 depletion, judged by immunofluorescence against β-tubulin (**, P < 0.01; ***, P < 0.001; Student's t test). (G) RNAi-stimulated loss of one or two minichromosomes (MC) from cells. Error bars represent standard error estimates from counts of resistant cells (n = 15–44; P < 0.001 for 24 and 48 h vs. 0 h; Z-test). au, arbitrary units.
Cells depleted of KKIP1 are unable to correctly assemble or maintain the spindle, with a greater than fourfold decrease in visible spindles at 8 or 16 h after induction (P = 0.006 and <0.001, respectively, Z-test; Fig. 3 F). This is in spite of the majority of cells having G2/M DNA content by 8 h after induction, suggesting that in trypanosomes failure to attach kinetochores to the spindle causes destabilization of the spindle itself or initiation of an assembly checkpoint.
To test for chromosome loss on KKIP1 depletion, we integrated a negative selection marker (a gene encoding herpes simplex virus thymidine kinase) on either one or two individual minichromosomes. These small chromosomes are segregated with fidelity but are not required for growth in culture, allowing loss to be monitored after reversal of RNAi induction (removal of tetracycline). In parental cells, the rate of loss of a single marked chromosome was 0.002 per cell generation (Fig. 3 G), in line with previous estimates of minichromosome loss (Wickstead et al., 2003). Loss rates in noninduced cells were slightly above this rate, suggesting some level of RNAi in the absence of induction, but this was greatly increased by 24-h induction to 70 and 220 times these levels for loss of one or two chromosomes, respectively (Fig. 3 G). Minichromosomes were lost independently both in the presence and absence of RNAi induction (combined rate of loss of both chromosomes being approximately the product of two single rates), suggesting that detachment of individuals from the spindle does not influence segregation of others.
KKIP1 interacts with KKTs and identifies new kinetochore components
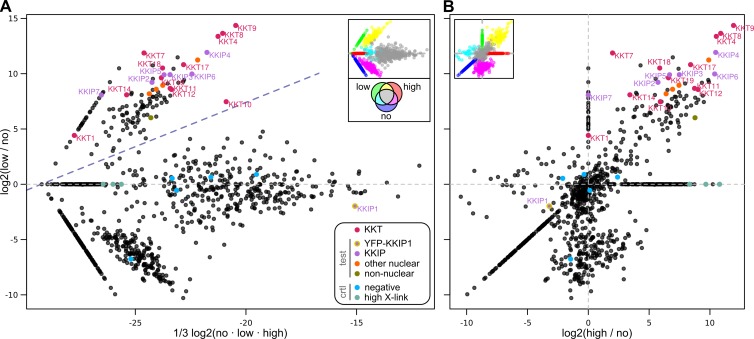
KKIP1 did not copurify with any of the trypanosome KKTs (Akiyoshi and Gull, 2014). In agreement with this observation, affinity purification of YFP-KKIP1 under similar conditions did not identify KKTs as copurifying proteins (except KKT10 at levels near background; Table S3). However, only KKIP1 itself was found above background, suggesting that the KKIP1–kinetochore interaction is unstable under standard conditions used for immunoprecipitation. To identify interacting proteins for this potentially labile association, we used a proximity-based approach of affinity purification after limiting, reversible cross-linking. This was combined with label-free semiquantitative mass spectrometry (Trudgian et al., 2011) to estimate enrichment after stabilization of complexes under conditions of low or high formaldehyde cross-linking (one and five times approximate molar ratio to available reactive groups, respectively) relative to controls without cross-linking. Samples were compared by integrated spectral intensities to identify proteins enriched under specific conditions (Fig. 4 and see Materials and methods for details). Spectral intensities and enrichment data for all 935 nonredundant trypanosome proteins detected in these experiments are presented in Table S3.
Figure 4.
Reversible cross-linking shows that KKIP1 interacts with KKTs. (A) Label-free semiquantitative mass spectrometry showing relative enrichment of proteins under conditions of “low” cross-linking against total intensity across all samples. (B) Enrichment under “low” versus “high” cross-linking. Signals from KKTs as well as 11 test and 8 control “hypothetical” proteins localized in this study are highlighted. For display, intensity of proteins not detected for a specific condition are set to an arbitrary minimum value. Insets show positions in the main plot of sets found under specific combinations of conditions, colored as demonstrated by the inset Venn diagram. Intensities and relative enrichment for all 935 nonredundant trypanosome proteins detected are presented in Table S3.
In agreement with the observed KKIP1 localization, several KKTs (namely, KKT4, 8, 9, 10, 11, 12, 14, 17, 18, and 19) copurify with YFP-KKIP1 in low and high cross-linked samples, with KKT1 being additionally detected in low cross-linking only (Fig. 4). The set of proteins enriched on cross-linking also contained the Aurora B kinase homologue, TbAUK1 (Fig. S2), which interacts with kinetochores until anaphase (Li et al., 2009). They are also significantly enriched in trypanosomal nucleoporins, including TbNup92/Mlp2, which is present at nuclear pores at interphase, but associates with the spindle at mitosis (Holden et al., 2014). In contrast, KKT13, which reaches peak levels at S phase, and KKT2 and KKT3, which are thought to interact closely with the centromere, were not detected.
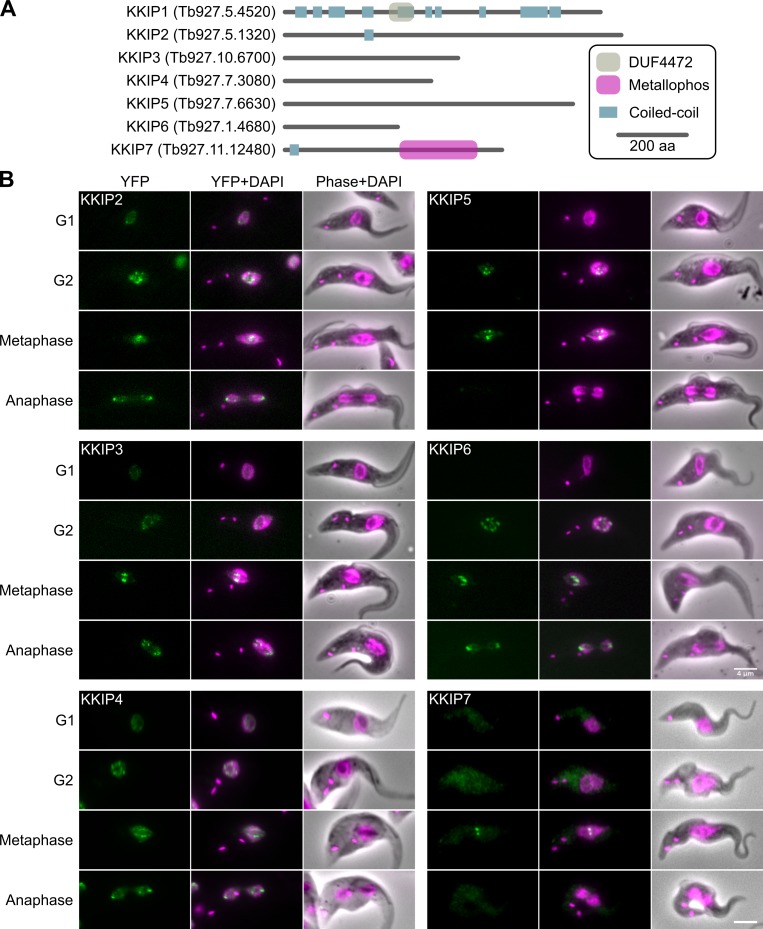
To validate our proteomic approach, we tagged 12 proteins of unknown function that were enriched in immunoprecipitates on cross-linking, plus several controls (Table S2). Of 11 with detectable signal, 10 were nuclear, with 6 localized to the kinetochore. We have named these new kinetochore proteins KKIP2–7 to follow from KKIP1 (Figs. 4 and 5 A). In contrast, no kinetochore association was seen for controls taken from sets of hypothetical proteins that were either: (a) not enriched (five proteins); or (b) enriched only under high cross-linking (three proteins). As expected, all of the high cross-link controls were other nuclear proteins, but none localized to the kinetochore.
Figure 5.
KKIP1-interacting proteins include new kinetochore components. (A) Predicted protein architectures for newly identified KKIP proteins. Regions of coiled-coil were predicted using Ncoils (Lupas et al., 1991). (B) Micrographs of procyclic cells expressing KKIPs tagged with YFP at the N termini. Counterstaining with DAPI is also shown. Bars, 4 µm.
KKIP2–7 are novel kinetoplastid kinetochore-associated proteins and can be grouped by their patterns of expression/localization through the cell cycle. KKIP2, 3, and 6 have a similar temporal pattern to KKIP1 (and KKT1, 5, 6, 7, 16, 17, and 18), loading gradually from S phase onwards and being unloaded/degraded at the end of mitosis (Fig. 5 B). RNAi against KKIP2 and KKIP3 caused defects in DNA segregation and population growth, although not with such rapid or large an effect as seen when depleting KKIP1 (Fig. S3). KKIP4 is also loaded to kinetochores in S/G2, although it is present in the nucleus throughout the cell cycle (Fig. 5 B). In contrast, KKIP5 signal is rapidly lost from kinetochores at the onset of anaphase (as seen for KKT8–12 and 19). Knockdown of KKIP4 and 5 had little effect on growth in culture (Fig. S3).
As for KKTs, KKIPs possess no uniquely defining predicted domains in current Pfam profiles (Fig. 5 A). Excepting the very sensitive method described in the first section of the Results, homologues for KKIP1–6 are not readily identified in eukaryotes outside of the Kinetoplastida (Fig. S4). KKIP7 is a predicted protein phosphatase. This is the first phosphatase to be localized to the kinetoplastid kinetochore and is suggestive of possible antagonism with the four kinases in the KKTs or TbAUK1. It is a member of the phosphoprotein phosphatase group of Ser/Thr phosphatases, which include the PP1 and PP2A families that coordinate mitotic progression and exit in fission yeast, but the protein has been shown to be part of a kinetoplastid-specific subfamily (Brenchley et al., 2007). Nonetheless, KKIP7 is present only on metaphase kinetochores (Fig. 5) in a manner reminiscent of PPA2-B56 (Kitajima et al., 2006), and we speculate that this phosphatase may be performing a similar role in attachment biorientation, perhaps in association with an as-yet-unidentified shugoshin-like molecule.
KKIP1 defines a trypanosome outer kinetochore complex
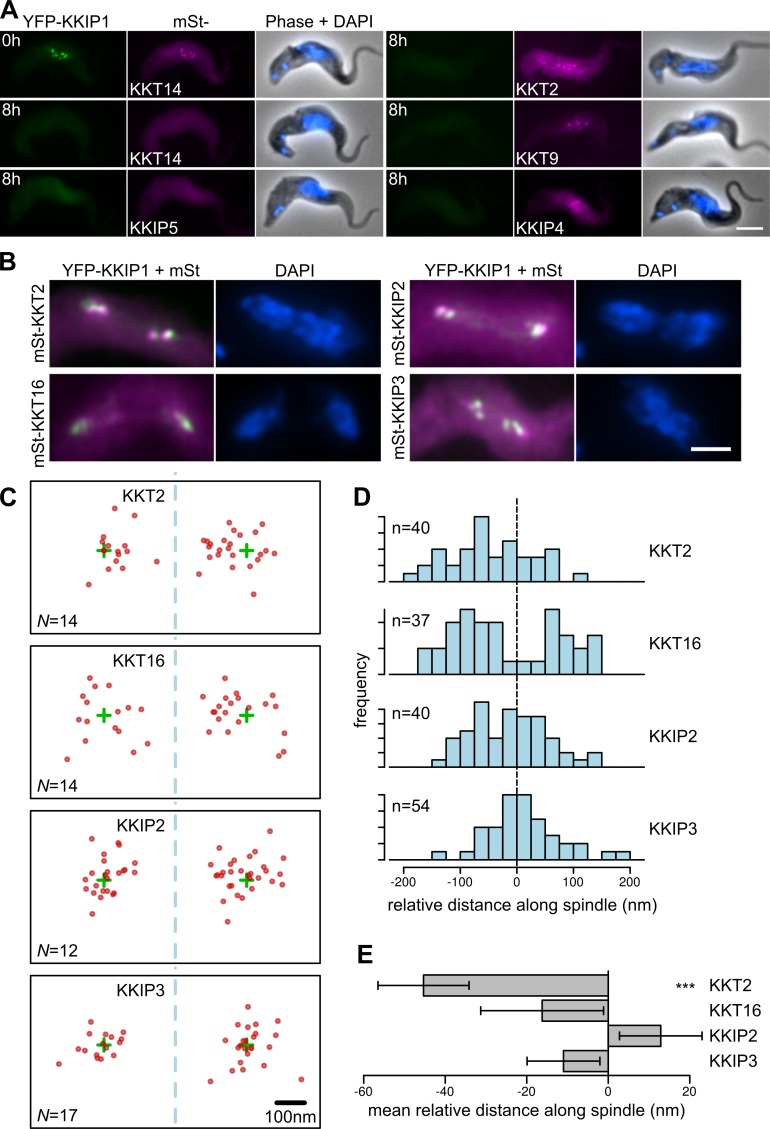
The Ndc80 complex is part of the outer kinetochore KMN network. In vertebrate cells, it is one of the last complexes recruited to the kinetochore before mitosis, requiring the presence of more centromere-proximal components for recruitment (Screpanti et al., 2011; Schleiffer et al., 2012; Gascoigne and Cheeseman, 2013; Nishino et al., 2013; Rago et al., 2015). To test for the biochemical position of KKIP1 in recruitment of proteins to the trypanosome kinetochore, we looked at the levels of other components in cells depleted of KKIP1. This was done for representatives from each of the potential complexes in the KKT set (excepting KKT13, which is present only in S phase), by tagging KKT2, 7, 9, 14, and 16 and also for KKIP2–5. Consistent with a centromere-distal role, KKIP1 is not required for recruitment of KKT2, 7, 9, 16 kinetochore-like foci, demonstrating that the majority of KKT complexes are upstream of KKIP1 (Fig. 6 A and Fig. S5). This was also observed for the newly identified kinetochore component KKIP4. In contrast, KKIP2, 3, and 5 and also KKT14 are dependent on KKIP1 for localization, demonstrating that even some proteins stably associating with the KKTs require outer kinetochore components for recruitment and/or persistence at the kinetochore.
Figure 6.
KKIP1 is an outer kinetochore protein. (A) Localization of KKT and KKIP components in cells at 0 or 8 h after induction of KKIP1 RNAi. (B) YFP-KKIP1 shows incomplete colocalization with predicted inner kinetochore components. (C) Subpixel positions of peak intensity for mStrawberry-tagged KKTs and KKIPs in anaphase cells. Positions of foci in individual cells (red dots) are shown relative to YFP-KKIP1 peak (green crosses) with spindle axis aligned to x. N gives number of independent spindles contributing to positions shown. (D) Distribution of relative positions along spindle axis. Negative values represent positions toward the mid-spindle. (E) Mean relative positions. Error bars show SEM (***, P < 0.001; Student's t test). Bars, 4 µm.
To directly observe the locations of KKT and KKIP components in the kinetochore, we used two-color fluorescence microscopy to determine relative subpixel positions of tagged proteins (Joglekar et al., 2009; Wan et al., 2009). We performed this analysis with cells in anaphase, to derive distance measurements for kinetochores that are not under metaphase tension-induced stretch, and measurements were made of positions of focus peak signals relative to the major axis of the spindle (Fig. 6, B and C). Owing to the shape of dividing trypanosomes, cells in anaphase adhere to slides with the mitotic spindle well aligned to the xy plane. Movement relative to this plane that maintains both poles in the same focal plane (less than ∼1-µm difference along z axis) equates to <3% change in measured distance along a typical 4-µm spindle because of projection into the xy plane, meaning that the system can be reasonably approximated to 2D without substantial underestimation of distances along this axis.
KKT2 is constitutively present at trypanosome kinetochores and thought to be one of the components closest to the centromere (Akiyoshi and Gull, 2014). The distribution of peak intensities for KKT2 foci relative to KKIP1 was significantly skewed away from the spindle poles (Fig. 6 D), with a mean distance of 45 ± 11 nm (Fig. 6 E; P = 0.0002, Student's t test). This is in agreement with the thickness of the kinetochore-like plaques (∼50 nm along the spindle axis) observed in trypanosome nuclei in ultrastructural studies (Ogbadoyi et al., 2000). In contrast, the mean relative distances for KKIP2 and KKIP3 were not significantly different from 0 (Fig. 6 E). This was also the case for KKT16, which has a similar temporal behavior to KKIP1–3. However, this was the result of having a distribution different to KKIP2 (P = 0.009; Kolmogorov–Smirnov test) with modes both on the interior and polar sides of KKIP1 signal, suggesting that this protein is part of a complex that does not associate along the main spindle axis (Fig. 6 D).
Discussion
KKIP1 is a kinetoplastid protein that we have shown to be a highly diverged member of the Ndc80/Nuf2 family of kinetochore proteins. Homology was very difficult to infer from sequence information, but the protein is recruited to the outer kinetochore in trypanosomes, and its depletion from cells impacts on DNA segregation in a manner similar to temperature-sensitive or degron mutants of Nuf2 in budding yeast (Osborne et al., 1994; McCleland et al., 2003) and RNAi against Nuf2 in human cells (DeLuca et al., 2002; Cheeseman et al., 2008). This defect is not because of disruption of the core kinetochore structure because most KKT components tested remain associated upon KKIP1 depletion (Fig. 6 and Fig. S5). Together, these data strongly suggest that KKIP1 is both evolutionarily related to Ndc80 and Nuf2 and performing the same function. This is the first demonstration that the kinetochore of kinetoplastids is related at the level of individual components to canonical kinetochores.
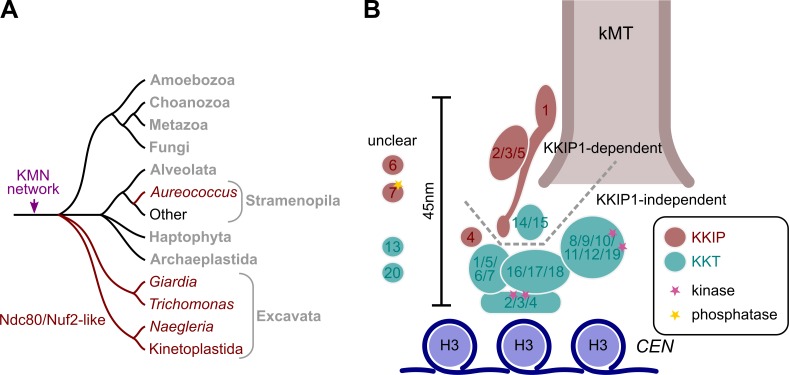
How does the discovery of Ndc80/Nuf2-like molecules in kinetoplastids, and other excavates, affect the view of kinetochore evolution? The identification of 20 proteins forming part of an unconventional kinetochore in kinetoplastids was an apparent synapomorphy separating Euglenozoa (kinetoplastids, euglenids, and diplonemids) away from the rest of the eukaryotic line, in agreement with a hypothesis placing this group as the earliest branch of extant eukaryotes (Cavalier-Smith, 2010; Akiyoshi and Gull, 2014). However, phylogenetic data do not support this branching order. Kinetoplastids are members of the potential eukaryotic “supergroup” Excavata. Monophyly of excavates is still somewhat unclear, but the consensus strongly supports that Euglenozoa and Percolozoa (including Naegleria) are part of a single clade (Hampl et al., 2009; Derelle and Lang, 2012; He et al., 2014). This means that a common ancestor gave rise to the kinetochores in the Naegleria and kinetoplastids. Significantly, the Naegleria genome encodes an identifiable homologue of Mis12 (Akiyoshi and Gull, 2014; Fig. 1 A), and we show in this study that it possesses an Ndc80/Nuf2-like sequence in the same family as KKIP1. To date, we have been unable to identify clear orthologues of Mis12 or Nnf1 in kinetoplastids using similar iterative procedures to that used to identify KKIP1. However, this is also the case for Aureococcus, Giardia, or any of several alveolates, suggesting that these proteins are either more commonly lost than for Ndc80/Nuf2 or at least have diverged in these lineages to be undetectable with even these sensitive methods. Similarly, we have yet to identify good candidates for Knl1 in Naegleria or kinetoplastids, but this protein is less constrained in sequence (Meraldi et al., 2006), making detection more difficult. Nonetheless, our data show that a KMN network containing at least Ndc80 and Mis12 complexes was present in the ancestor of kinetoplastids and Naegleria, reuniting the eukaryotic outer kinetochore around a universal protein set with common ancestry (Fig. 7 A).
Figure 7.
The evolution of trypanosome kinetochores. (A) Schematic representation of the likely relationships between some of the major groups of eukaryotes showing the presence of Ndc80/Nuf2-like sequences. (B) Model for trypanosome kinetochore architecture based on interactions and temporal loading seen in Akiyoshi and Gull (2014) and work in this study. kMT, kinetochore microtubule.
Fig. 7 B shows our current model for the kinetochore structure in trypanosomes. The distance along the spindle axis between an inner kinetochore component, KKT2, and a tag on the N terminus of KKIP1 was ∼45 nm for cells in anaphase. Because the Ndc80/Nuf2-like region of KKIP1 is toward the N terminus, it is expected that this end of the protein will be outermost, and this is in good agreement with the distance between the inner and outer kinetochore plaques seen by electron microscopy in trypanosomes (Ogbadoyi et al., 2000). It is also the distance along the same axis between Cse4p (CENP-A) and the Ndc80 N terminus observed for budding yeast kinetochores in anaphase (Joglekar et al., 2009). In spite of this spatial similarity, the primary sequence of KKIP1 is considerably longer than typical Ndc80 or Nuf2 (Fig. 1), and the structure of the rest of the protein is currently unclear. It is noteworthy that we identified only one Ndc80/Nuf2-like sequence in organisms lacking easily identifiable Ndc80 and Nuf2 (Fig. 1). It may be that a second, even more dissimilar homologue exists in these lines. However, immunopurification of KKIP1 without stabilization did not identify a partner protein, suggestive that divergent Ndc80/Nuf2-like proteins may be homodimers or even monomers. A homodimer was the ancestral state for the Ndc80 complex, but the presence of a single Ndc80/Nuf2-like protein in the golden alga A. anophagefferens means that if these proteins are homodimers, it is as likely a derived as ancestral characteristic.
The kinetochores of kinetoplastid organisms remain dissimilar to the canonical arrangement in several important ways. The lack of CENP-A at the centromeres (Lowell and Cross, 2004) is intriguing, given the wide distribution of this central component and the relative ease of detection because of the sequence constraints imposed on histones by their roles. However, lack of CENP-A/CenH3 is also a feature of some insect kinetochores, in which it is associated with a transition to holocentricity (Drinnenberg et al., 2014). Similarly, the lack of identifiable Mis12 and Nnf1 homologues is not a unique feature, as these proteins are also undetectable in Giardia, Aureococcus, and alveolates (Fig. 1 A). The presence of four kinases as structural components in the KKT set is a clear contrast with well-studied model kinetochores (Akiyoshi and Gull, 2014). Two of these kinases are likely to be very close to the centromere, which, together with the lack of CENP-A, reveals a very different structure at the inner kinetochore. Our data suggest that the KKT proteins (perhaps with the exception of KKT14/15) may represent a kinetoplastid CCAN set, with at least some outer kinetochore components being unstable under the conditions of immunopurification. Although the kinetoplastid CCAN has changed beyond recognition, with replacement of at least some components, the KMN network is still present, although with highly divergent sequence. There are striking biological parallels between this hypothesis and kinetochore evolution in Drosophila melanogaster and Caenorhabditis elegans, in which the kinetochores have been hugely simplified by the widespread loss of CCAN subunits in an extremely short evolutionary time, but with conservation of the KMN network (Meraldi et al., 2006; Przewloka et al., 2007; Westermann and Schleiffer, 2013). This plasticity in components as well as sequence has made understanding the evolution of the kinetochore a substantial challenge, and there are still important questions to address, but the work in this study shows that although the kinetoplastid kinetochores are highly diverged from models at the sequence level, no eukaryotic line thus far identified is ancestrally different.
Materials and methods
Bioinformatic analyses
All searches were based on predicted protein datasets for 46 diverse eukaryotes for which complete or near-complete genome sequence data are publicly available. Organisms selected were based on those used in (Wickstead et al., 2010b), with the inclusion of data from the haptophyte Emiliania huxleyi (Read et al., 2013). Initial profiles for Ndc80 and Nuf2 (PF03801.9 and PF03800.10, respectively) were taken from Pfam (Finn et al., 2010). HMMER3 (Eddy, 2009) was used to find similar sequences in a database made of all predicted proteomes. Hits were aligned with MAFFTv6.925b, adopting the accurate L-INS-i strategy involving local pairwise alignment with iterative refinement (Katoh et al., 2005), trimmed to conserved regions with trimAl (Capella-Gutiérrez et al., 2009), and used to create new profiles. These steps of identification of homologues, alignment, and refinement of models were then iterated until no new sequences were identified (two iterations). To search the pan-Ndc80/Nuf2 profile against kinetoplastids, proteins from T. brucei TREU927, T. cruzi CL Brener, Leishmania major Friedlin, and Crithidia fasciculata CfCl were aligned by orthologue group (http://www.tritrypdb.org), trimmed, and converted to profiles. Profile–profile comparisons were performed using HH-suite (Söding, 2005) and alignments were made using the “–profile” option of MAFFT (L-INS-i). For phylogenetic inference, alignments were trimmed to conserved regions and used to infer maximum likelihood phylogenies as implemented by the program PhyML3.0 (Guindon et al., 2010) using the WAG substitution matrix with a gamma-distributed variation in substitution rate approximated to five discrete categories (shape parameter estimated from the data). Tree shown is a majority-rule consensus of 500 bootstrap replicates. Protein domain architectures were predicted with PSIPRED (McGuffin et al., 2000), COILS (Lupas et al., 1991), and the Pfam database (Finn et al., 2010).
Cell lines and cell culture
All work was performed in SmOxP427 or SmOxB427 cells (in the case of procyclic- or bloodstream-form cells, respectively), which are Lister 427–based lines expressing transgenic T7 RNA polymerase and Tet-repressor protein from the tubulin locus (Poon et al., 2012). Procyclic cells were grown at 28°C in SDM79 medium (Brun and Schönenberger, 1979) supplemented with 10% FBS. Bloodstream-form cells were grown in HMI-9 medium supplemented with 15% FBS at 37°C and 5% CO2 (Hirumi and Hirumi, 1989).
For N-terminal tagging, constructs encoding fluorescent proteins were integrated at the 5′ end of the endogenous coding sequence for the protein of interest. All constructs were derived from pEnNY0, pEnNmSt0-B, or pEnNmSt0-N, which encode YFP (pEnNY0) or mStrawberry (pEnNmSt0-B/N), in addition to hygromycin (pEnNY0), blasticidin (pEnNmSt0-B), or neomycin (pEnNmSt0-N) resistance markers, and were made by modifications to the pEnG0 previously developed by the laboratory (Wickstead et al., 2010a). Targeting sequences comprised ∼200 bp from the N-terminal end of the coding sequence and ∼200 bp of upstream sequence, cloned downstream of the fluorescent protein coding sequence, along with a linearization site between the targeting sequences. All primers used for cloning are available in Table S4. For inducible RNA interference, ∼400-bp fragments of coding sequence were cloned into p2T7-177 (Wickstead et al., 2002), which integrates into 177-bp repeats found on minichromosomes. Plasmids were linearized with NotI and transfected into trypanosomes by electroporation as described in Schumann Burkard et al. (2011). Stable transfectants were selected with 50 µg/ml hygromycin, 10 µg/ml blasticidin, 2.5 µg/ml G418, or 5 µg/ml phleomycin in the case of SmOxP427 cells or 5 µg/ml hygromycin, 2 µg/ml blasticidin, or 2.5 µg/ml phleomycin for SmOxB427. RNAi was induced by the addition of 1 µg/ml tetracycline to the growth medium. Primers used to generate RNAi fragments are available in Table S4.
The HSV-TK coding sequence was obtained from pCIHDAdGT8-3 (Addgene), fused to blasticidin or neomycin resistance genes, cloned downstream of a T7 polymerase promoter into a construct integrating into 177-bp repeats of minichromosomes to generate pMC-T7-HSVTK-B and pMC-T7-HSVTK-N plasmids. To assess the loss rate of individual minichromosomes, cell lines containing either one or two minichromosomes tagged with HSV-TK were generated. Minichromosome loss rate per generation was determined by growth for 48 h without selection (with or without induction of RNAi) followed by quantification of the proportion of cells resistant to 100 µg/ml ganciclovir.
Protein localization
For analysis of localization of tagged proteins by native fluorescence, cells were harvested from mid-log phase cultures, washed twice in PBS (137 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4), and allowed to settle onto glass (procyclic cells) or glutaraldehyde-derivatized silanized slides (bloodstream-form cells). Cells were fixed for 10 min in 2% (wt/vol) formaldehyde, permeabilized in −20°C methanol for at least 2 min, and rehydrated in PBS before mounting in a solution containing a DNA stain and photostabilizing agent (1% wt/vol 1,4-diazabicyclo[2.2.2]octane, 90% vol/vol glycerol, 50 mM sodium phosphate, pH 8.0, and 200 ng/ml 4′,6-diamidino-2-phenylindole). For immunolocalization, samples prepared as for native fluorescence were incubated for 1 h with mouse anti–β-tubulin mAb KMX-1 (Birkett et al., 1985; 1:10 dilution of hybridoma culture supernatant). Slides were subsequently washed extensively, then incubated with 1:200 TRITC-conjugated goat anti-mouse IgG (Stratech Scientific Ltd), and mounted as above. For cytoskeleton preparations, cells were settled as above and then detergent extracted by the addition of 0.2% vol/vol NP-40 (Sigma-Aldrich) in PEME buffer (100 mM Pipes, pH 6.9, 2 mM EGTA, 1 mM MgSO4, and 0.1 mM EDTA) for 2 min. Cytoskeletons were then fixed for 5 min in 2% wt/vol formaldehyde/PBS, followed by washing twice in PBS, and mounting as above.
Images were captured on a BX51 microscope equipped with a 100× UPlanApo objective (NA 1.35; Olympus) and CoolSnap-HQ (Photometrics) or Retiga R1 (QImaging) CCD cameras. All images of fluorescent proteins were captured at RT with equal exposure settings and no prior illumination. Images for level comparison were also processed in parallel with the same alterations to minimum and maximum display levels. Image acquisition was controlled by µManager open source software (Edelstein et al., 2014). Analysis was performed in ImageJ (Schneider et al., 2012) and the statistical programming package R (http://www.r-project.org). For analysis of relative positions of kinetochore components, locations were captured corresponding to the subpixel peak of signal for individual foci of fluorescence at anaphase for both YFP-KKIP1 and mStrawberry-tagged kinetochore components. Only cells with at least one focus clearly visible at both ends of the spindle and in both channels were considered. The major axis of the spindle was defined by the line between the mean xy positions of all foci visible at each pole and the data transformed such that this lay along the x axis. Relative positions of the mStrawberry and YFP peak signals were then taken from each transformed focus. Full scripts used for transformation are available from the authors on request. Because mitotic trypanosomes tend to settle with the spindle axis aligned to the xy plane, no correction was made for components of the spindle axis in z. Elevation of one pole of a typical 4-µm spindle by up to 1 µm in z (sufficient for the foci to move out of the focal plane) would lead to an underestimate of the distances by <3%.
Flow cytometry
For quantitative analysis of DNA content by flow cytometry, ∼5 × 106 cells were harvested by centrifugation and resuspended in 0.25% (wt/vol) formaldehyde in PBS. After 5 min, fixed cells were again pelleted and resuspended in 500 ml PBS containing 0.4% Triton X-100, 100 µg/ml RNaseA, and 25 μg/ml propidium iodide. These samples were incubated at 37°C for 30 min before analysis.
Immunopurification
Immunopurification was performed essentially as described in Daniels et al. (2012), with the addition of limited cross-linking before purification. In brief, 3 × 109 procyclic form cells expressing YFP-KKIP1 were harvested by centrifugation from actively dividing cultures. Cells were washed once in ice-cold HKMEG (150 mM KCl, 150 mM glucose, 25 mM Hepes, pH 7.8, 4 mM MgCl2, and 1 mM EGTA) and then with HKMEG containing 5 µM E64-d. Cells were treated with 1.5 ml of 0, 0.1, or 0.5% formaldehyde for 5 min, quenched with 1.5 ml of 1 M glycine, and lysed in HKMEG containing 1% (vol/vol) NP-40, 1 mM DTT, and protease inhibitors (2 mM 1,10-phenanthroline, 0.5 mM PMSF, 50 µM leupeptin, 7.5 µM pepstatin A, and 5 µM E64-d). Lysate was sonicated for 2 min at 20% intensity applied for 20% of the cycle and cleared by centrifugation at 20,000 g for 30 min. Cleared lysate was allowed to bind for 2 h on ice with gentle agitation to approximately five times molar excess of affinity-purified rabbit anti-GFP polyclonal antibodies that had been covalently attached to paramagnetic beads (Dynabeads Protein G; Invitrogen) by dimethyl pimelimidate treatment (Unnikrishnan et al., 2012). Beads were washed extensively in HKMEG containing 0.1% (vol/vol) NP-40, 0.5 mM DTT, and bound complex subsequently eluted by the incubating the beads in 100 mM glycine, pH 2.7.
Mass spectrometry and label-free quantitation
Immunopurified samples were desalted by precipitation with acetone at −20°C, washed in cold acetone, and solubilized in Laemmli sample buffer before treatment with 10 mM iodoacetamide for alkylation of cysteines. Samples were encapsulated in a polyacrylamide matrix by running a short distance into an SDS-PAGE gel, followed by staining with Coomassie and excision of gel fragment. Gel fragments were washed with 50% acetonitrile in 50 mM NH4HCO3, pH 8.5, dehydrated in 100% acetonitrile, and air-dried. Proteins were digested for 16 h with 20 µg/ml trypsin (Promega) in 25 mM NH4HCO3, pH 8.5, at 37°C. Mass spectrometry was performed on an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) at the University of Oxford Central Proteomics Facility (http://www.proteomics.ox.ac.uk).
Label-free quantitation was performed from mzXML data files using the Central Proteomics Facilities Pipeline (http://www.proteomics.ox.ac.uk). Data were searched with X!Tandem and OMSSA engines against a custom, nonredundant protein database of predicted protein sequences from TREU927/4 strain (http://www.tritrypdb.org) with the inclusion of exogenous protein sequence (including fluorescent proteins, drug selection markers, and exogenous proteins expressed in the parental cells) and common contaminating peptides. Possible modification of peptides by N-terminal acetylation, carbamidomethylation (C), oxidation (M), and deamidation (N/Q) was permitted in searches. Peptide identifications were validated with PeptideProphet and ProteinProphet (Nesvizhskii et al., 2003) and lists compiled at the peptide and protein level. iProphet was used to combine search engine identifications and refine identifications and probabilities. Normalized spectral index quantitation (SINQ) was applied to the grouped metasearches to give protein-level quantitation between labeled samples and controls, as described in Trudgian et al. (2011), and implemented by the Central Proteomics Facilities Pipeline at the University of Oxford. SINQ values are summed intensities of matched fragment ions for all spectra assigned to a peptide (identified by ProteinProphet), normalized for differences in protein loading between datasets and for individual protein length. A probability cutoff corresponding to 1% false discovery rate relative to a target-decoy database (reversed sequences) was applied. Data in Fig. 4 are presented as log2 enrichment (ratio of SINQ value in cross-linked sample versus non–cross-linked control) against log2 of the geometric mean intensity across all experiments. Processed data for all 935 nonredundant trypanosome proteins detected in these experiments are presented in Table S3, and raw mzXML data files are available on request.
Online supplemental material
Fig. S1 shows alignment of Ndc80- and Nuf2-like sequences from various eukaryotic models. Fig. S2 shows label-free semiquantitative proteomic data highlighting sequences from protein sets associated with specific cellular functions. Fig. S3 shows growth and DNA content of cells depleted of KKIP2, 3, 4, or 5 by induction of RNAi. Fig. S4 shows the distribution across eukaryotes of easily detected orthologues to trypanosomal kinetochore proteins. Fig. S5 shows changes in the overall levels and localization of KKT and KKIP components upon depletion of KKIP1. Table S1 shows top hits between kinetoplastid orthologue groups and an HMM of diverse Ndc80 and Nuf2 sequences. Table S2 shows KKIP1-interacting proteins and controls localized in trypanosomes. Table S3 provides data from label-free semiquantitative mass spectrometry of KKIP1-interacting proteins. Table S4 shows primer sequences used in the generation of constructs for endogenous locus tagging and RNAi.
Supplementary Material
Acknowledgments
The authors thank Tom Richards, Liz Sockett, and Dick McIntosh for discussion and comments on the manuscript; Catarina Gadelha and Ben Thomas for assistance with semiquantitative analysis of mass spectrometry data; and Keith Gull for providing the KMX-1 reagent. The authors also thank Stephen Beverley and the Genome Institute, Washington University School of Medicine (St. Louis, MO), for making genomic data for C. fasciculata available via TriTrypDB (http://tritrypdb.org) in advance of publication.
This research was funded by the Biotechnology and Biological Sciences Research Council (BB/J01477X/1).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CCAN
- constitutive centromere-associated network
- CH
- calponin homology
- HMM
- hidden Markov model
- KKIP
- KKT-interacting protein
- SINQ
- spectral index quantitation
References
- Akiyoshi B., and Gull K.. 2013. Evolutionary cell biology of chromosome segregation: Insights from trypanosomes. Open Biol. 3:130023 10.1098/rsob.130023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B., and Gull K.. 2014. Discovery of unconventional kinetochores in kinetoplastids. Cell. 156:1247–1258. 10.1016/j.cell.2014.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin G.M., Ramey V.H., Pasqualato S., Ball D.A., Grigorieff N., Musacchio A., and Nogales E.. 2010. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 467:805–810. 10.1038/nature09423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science. 309:416–422. 10.1126/science.1112642 [DOI] [PubMed] [Google Scholar]
- Birkett C.R., Foster K.E., Johnson L., and Gull K.. 1985. Use of monoclonal antibodies to analyse the expression of a multi-tubulin family. FEBS Lett. 187:211–218. 10.1016/0014-5793(85)81244-8 [DOI] [PubMed] [Google Scholar]
- Brenchley R., Tariq H., McElhinney H., Szöor B., Huxley-Jones J., Stevens R., Matthews K., and Tabernero L.. 2007. The TriTryp phosphatome: Analysis of the protein phosphatase catalytic domains. BMC Genomics. 8:434 10.1186/1471-2164-8-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R., and Schönenberger M.. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 36:289–292. [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., and Gabaldón T.. 2009. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25:1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2010. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol. Lett. 6:342–345. 10.1098/rsbl.2009.0948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46. 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., and Desai A.. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594. 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C., De Luca J., Monzani S., Ferrari K.J., Ristic D., Wyman C., Stark H., Kilmartin J., Salmon E.D., and Musacchio A.. 2005. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J. Biol. Chem. 280:29088–29095. 10.1074/jbc.M504070200 [DOI] [PubMed] [Google Scholar]
- Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J.G., De Wulf P., et al. 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 133:427–439. 10.1016/j.cell.2008.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels J.-P., Gull K., and Wickstead B.. 2010. Cell biology of the trypanosome genome. Microbiol. Mol. Biol. Rev. 74:552–569. 10.1128/MMBR.00024-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels J.-P., Gull K., and Wickstead B.. 2012. The trypanosomatid-specific N terminus of RPA2 is required for RNA polymerase I assembly, localization, and function. Eukaryot. Cell. 11:662–672. 10.1128/EC.00036-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Moree B., Hickey J.M., Kilmartin J.V., and Salmon E.D.. 2002. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 159:549–555. 10.1083/jcb.200208159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., and Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Derelle R., and Lang B.F.. 2012. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol. Biol. Evol. 29:1277–1289. 10.1093/molbev/msr295 [DOI] [PubMed] [Google Scholar]
- Drinnenberg I.A., deYoung D., Henikoff S., and Malik H.S.. 2014. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLife. 3 10.7554/eLife.03676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform. 23:205–211. [PubMed] [Google Scholar]
- Edelstein A.D., Tsuchida M.A., Amodaj N., Pinkard H., Vale R.D., and Stuurman N.. 2014. Advanced methods of microscope control using μManager software. J. Biol. Methods. 1:e10 10.14440/jbm.2014.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Mistry J., Tate J., Coggill P., Heger A., Pollington J.E., Gavin O.L., Gunasekaran P., Ceric G., Forslund K., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38(Database):D211–D222. 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E.T., Black B.E., Bailey A.O., Yates J.R. III, and Cleveland D.W.. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469. 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- Gascoigne K.E., and Cheeseman I.M.. 2013. CDK-dependent phosphorylation and nuclear exclusion coordinately control kinetochore assembly state. J. Cell Biol. 201:23–32. 10.1083/jcb.201301006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., and Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hampl V., Hug L., Leigh J.W., Dacks J.B., Lang B.F., Simpson A.G.B., and Roger A.J.. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc. Natl. Acad. Sci. USA. 106:3859–3864. 10.1073/pnas.0807880106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Fiz-Palacios O., Fu C.-J., Fehling J., Tsai C.-C., and Baldauf S.L.. 2014. An alternative root for the eukaryote tree of life. Curr. Biol. 24:465–470. 10.1016/j.cub.2014.01.036 [DOI] [PubMed] [Google Scholar]
- Hirumi H., and Hirumi K.. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75:985–989. 10.2307/3282883 [DOI] [PubMed] [Google Scholar]
- Holden J.M., Koreny L., Obado S., Ratushny A.V., Chen W.-M., Chiang J.-H., Kelly S., Chait B.T., Aitchison J.D., Rout M.P., and Field M.C.. 2014. Nuclear pore complex evolution: A trypanosome Mlp analogue functions in chromosomal segregation but lacks transcriptional barrier activity. Mol. Biol. Cell. 25:1421–1436. 10.1091/mbc.E13-12-0750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bloom K., and Salmon E.D.. 2009. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19:694–699. 10.1016/j.cub.2009.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., and Miyata T.. 2005. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518. 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima T.S., Sakuno T., Ishiguro K., Iemura S., Natsume T., Kawashima S.A., and Watanabe Y.. 2006. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 441:46–52. 10.1038/nature04663 [DOI] [PubMed] [Google Scholar]
- Li Z., Umeyama T., and Wang C.C.. 2009. The aurora kinase in Trypanosoma brucei plays distinctive roles in metaphase-anaphase transition and cytokinetic initiation. PLoS Pathog. 5:e1000575 10.1371/journal.ppat.1000575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell J.E., and Cross G.A.M.. 2004. A variant histone H3 is enriched at telomeres in Trypanosoma brucei. J. Cell Sci. 117:5937–5947. 10.1242/jcs.01515 [DOI] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., and Stock J.. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- McCleland M.L., Gardner R.D., Kallio M.J., Daum J.R., Gorbsky G.J., Burke D.J., and Stukenberg P.T.. 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17:101–114. 10.1101/gad.1040903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin L.J., Bryson K., and Jones D.T.. 2000. The PSIPRED protein structure prediction server. Bioinformatics. 16:404–405. 10.1093/bioinformatics/16.4.404 [DOI] [PubMed] [Google Scholar]
- Meraldi P., McAinsh A.D., Rheinbay E., and Sorger P.K.. 2006. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7:R23 10.1186/gb-2006-7-3-r23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerusheva O.O., and Akiyoshi B.. 2016. Divergent polo box domains underpin the unique kinetoplastid kinetochore. Open Biol. 6:150206 10.1098/rsob.150206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A.I., Keller A., Kolker E., and Aebersold R.. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658. 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- Nishino T., Rago F., Hori T., Tomii K., Cheeseman I.M., and Fukagawa T.. 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32:424–436. 10.1038/emboj.2012.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbadoyi E., Ersfeld K., Robinson D., Sherwin T., and Gull K.. 2000. Architecture of the Trypanosoma brucei nucleus during interphase and mitosis. Chromosoma. 108:501–513. 10.1007/s004120050402 [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R. III, Desai A., and Fukagawa T.. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457. 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- Osborne M.A., Schlenstedt G., Jinks T., and Silver P.A.. 1994. Nuf2, a spindle pole body-associated protein required for nuclear division in yeast. J. Cell Biol. 125:853–866. 10.1083/jcb.125.4.853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon S.K., Peacock L., Gibson W., Gull K., and Kelly S.. 2012. A modular and optimized single marker system for generating Trypanosoma brucei cell lines expressing T7 RNA polymerase and the tetracycline repressor. Open Biol. 2:110037 10.1098/rsob.110037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka M.R., Zhang W., Costa P., Archambault V., D’Avino P.P., Lilley K.S., Laue E.D., McAinsh A.D., and Glover D.M.. 2007. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS One. 2:e478 10.1371/journal.pone.0000478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rago F., Gascoigne K.E., and Cheeseman I.M.. 2015. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr. Biol. 25:671–677. 10.1016/j.cub.2015.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read B.A., Kegel J., Klute M.J., Kuo A., Lefebvre S.C., Maumus F., Mayer C., Miller J., Monier A., Salamov A., et al. 2013. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature. 499:209–213. 10.1038/nature12221 [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Basu M.K., Csürös M., and Koonin E.V.. 2009. Analysis of rare genomic changes does not support the unikont-bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol. Evol. 1:99–113. 10.1093/gbe/evp011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffer A., Maier M., Litos G., Lampert F., Hornung P., Mechtler K., and Westermann S.. 2012. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 14:604–613. 10.1038/ncb2493 [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schou K.B., Andersen J.S., and Pedersen L.B.. 2014. A divergent calponin homology (NN-CH) domain defines a novel family: Implications for evolution of ciliary IFT complex B proteins. Bioinformatics. 30:899–902. 10.1093/bioinformatics/btt661 [DOI] [PubMed] [Google Scholar]
- Schumann Burkard G., Jutzi P., and Roditi I.. 2011. Genome-wide RNAi screens in bloodstream form trypanosomes identify drug transporters. Mol. Biochem. Parasitol. 175:91–94. 10.1016/j.molbiopara.2010.09.002 [DOI] [PubMed] [Google Scholar]
- Screpanti E., De Antoni A., Alushin G.M., Petrovic A., Melis T., Nogales E., and Musacchio A.. 2011. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21:391–398. 10.1016/j.cub.2010.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics. 21:951–960. 10.1093/bioinformatics/bti125 [DOI] [PubMed] [Google Scholar]
- Trudgian D.C., Ridlova G., Fischer R., Mackeen M.M., Ternette N., Acuto O., Kessler B.M., and Thomas B.. 2011. Comparative evaluation of label-free SINQ normalized spectral index quantitation in the central proteomics facilities pipeline. Proteomics. 11:2790–2797. 10.1002/pmic.201000800 [DOI] [PubMed] [Google Scholar]
- Unnikrishnan A., Akiyoshi B., Biggins S., and Tsukiyama T.. 2012. An efficient purification system for native minichromosome from Saccharomyces cerevisiae. Methods Mol. Biol. 833:115–123. 10.1007/978-1-61779-477-3_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.-T., Yen T.J., McEwen B.F., et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell. 137:672–684. 10.1016/j.cell.2009.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R.R., Sorger P.K., and Harrison S.C.. 2005. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. USA. 102:5363–5367. 10.1073/pnas.0501168102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R.R., Al-Bassam J., and Harrison S.C.. 2007. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 14:54–59. 10.1038/nsmb1186 [DOI] [PubMed] [Google Scholar]
- Westermann S., and Schleiffer A.. 2013. Family matters: Structural and functional conservation of centromere-associated proteins from yeast to humans. Trends Cell Biol. 23:260–269. 10.1016/j.tcb.2013.01.010 [DOI] [PubMed] [Google Scholar]
- Westhorpe F.G., and Straight A.F.. 2013. Functions of the centromere and kinetochore in chromosome segregation. Curr. Opin. Cell Biol. 25:334–340. 10.1016/j.ceb.2013.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B., Ersfeld K., and Gull K.. 2002. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 125:211–216. 10.1016/S0166-6851(02)00238-4 [DOI] [PubMed] [Google Scholar]
- Wickstead B., Ersfeld K., and Gull K.. 2003. The mitotic stability of the minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 132:97–100. 10.1016/j.molbiopara.2003.08.007 [DOI] [PubMed] [Google Scholar]
- Wickstead B., Carrington J.T., Gluenz E., and Gull K.. 2010a The expanded kinesin-13 repertoire of trypanosomes contains only one mitotic kinesin indicating multiple extra-nuclear roles. PLoS One. 5:e15020 10.1371/journal.pone.0015020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B., Gull K., and Richards T.A.. 2010b Patterns of kinesin evolution reveal a complex ancestral eukaryote with a multifunctional cytoskeleton. BMC Evol. Biol. 10:110 10.1186/1471-2148-10-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A., and Kilmartin J.V.. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349–360. 10.1083/jcb.152.2.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward R., and Gull K.. 1990. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J. Cell Sci. 95:49–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.