Peroxisomes and the ER exchange lipids for various metabolic and anabolic reactions. In this study, Hua et al. show that the interaction between the ER-resident VAPs with the peroxisomal protein ACBD5 tethers peroxisomes to the ER. This tether is required for the exchange of lipids, including cholesterol, between the two organelles.
Abstract
Lipid exchange between the endoplasmic reticulum (ER) and peroxisomes is necessary for the synthesis and catabolism of lipids, the trafficking of cholesterol, and peroxisome biogenesis in mammalian cells. However, how lipids are exchanged between these two organelles is not understood. In this study, we report that the ER-resident VAMP-associated proteins A and B (VAPA and VAPB) interact with the peroxisomal membrane protein acyl-CoA binding domain containing 5 (ACBD5) and that this interaction is required to tether the two organelles together, thereby facilitating the lipid exchange between them. Depletion of either ACBD5 or VAP expression results in increased peroxisome mobility, suggesting that VAP–ACBD5 complex acts as the primary ER–peroxisome tether. We also demonstrate that tethering of peroxisomes to the ER is necessary for peroxisome growth, the synthesis of plasmalogen phospholipids, and the maintenance of cellular cholesterol levels. Collectively, our data highlight the importance of VAP–ACBD5–mediated contact between the ER and peroxisomes for organelle maintenance and lipid homeostasis.
Introduction
Peroxisomes are unique among the endomembrane organelles because of their semiautonomous nature. They are capable of importing newly synthesized peroxisomal matrix (lumen) proteins directly from the cytosol and multiply in numbers by the growth and division of existing peroxisomes (Agrawal and Subramani, 2016; Hua and Kim, 2016; Schrader et al., 2016). However, like other organelles of the endomembrane system, peroxisomes depend on the ER for their lipid composition and also receive some of their membrane proteins from the ER (Hettema et al., 2014). Similarly, the ER receives lipid precursors required for the biosynthesis of specialized lipids from peroxisomes. For example, the synthesis of plasmalogens, a class of ether phospholipids that represent ∼20% of the total phospholipid mass in humans, is initiated in the peroxisomes and completed in the ER (Braverman and Moser, 2012). Furthermore, cholesterol is not only trafficked through peroxisomes, but peroxisomes may also synthesize precursors for cholesterol biosynthesis (Kovacs et al., 2007; Chu et al., 2015).
Several models have been proposed as to how lipids and/or proteins are exchanged between the ER and peroxisomes. The earliest model is the shuttling of specific lipids to peroxisomes through the cytosol by lipid binding proteins such as carnitine transporters (Antonenkov and Hiltunen, 2012; Hunt et al., 2014). A vesicular model has recently been popularized with the discovery of peroxisomal protein–containing vesicles in various yeasts (Lam et al., 2010; Agrawal et al., 2011) that transport proteins from the ER to peroxisomes (Agrawal and Subramani, 2016). Another mechanism of peroxisome interface with the ER may be through the ER–peroxisome membrane contact sites or tethers (Schrader et al., 2015; Shai et al., 2016). In budding yeast Saccharomyces cerevisiae and the filamentous Ascomycetes, ER–peroxisome tethers are required for proper peroxisome inheritance (Ng et al., 2009; Knoblach et al., 2013). Contact sites mediated by two proteins, Pex30p and Pex31p, are reported to play a role in peroxisome growth in a number of yeast (Raychaudhuri and Prinz, 2008; Yan et al., 2008; David et al., 2013). However, no known homologue of Pex30p or Pex31p has been identified in mammalian cells. In mammalian cells, contacts between peroxisomes and lysosomes were shown to be necessary for cholesterol trafficking from lysosomes to peroxisomes (Chu et al., 2015). However, whether peroxisomes and the ER exchange lipids in mammalian cells is not known.
Previously, we have shown that a key peroxisomal biogenesis protein, PEX16, initially targets to the ER before being trafficked to peroxisomes (Kim et al., 2006; Aranovich et al., 2014; Hua et al., 2015). We also showed that PEX16 recruits peroxisomal membrane proteins to the ER before being trafficked to existing peroxisomes, suggesting that PEX16 may be interacting with components involved in trafficking of proteins and lipids between the two organelles. To understand the mechanisms by which the ER and peroxisomes communicate with each other, we performed a screen to identify both proximal and interacting proteins of PEX16. Along with peroxisomal membrane proteins, we identified the VAMP-associated proteins A and B (VAPA and VAPB), which are ER-resident proteins involved in organelle tethering to the ER. In this study, we show that the VAPs serve to tether peroxisomes to the ER through their interaction with the peroxisomal membrane protein acyl-CoA binding domain containing 5 (ACBD5). Furthermore, we present evidence of the importance of this ER–peroxisome tether for both peroxisome maintenance and lipid homeostasis.
Results and discussion
A proximity interaction network for PEX16
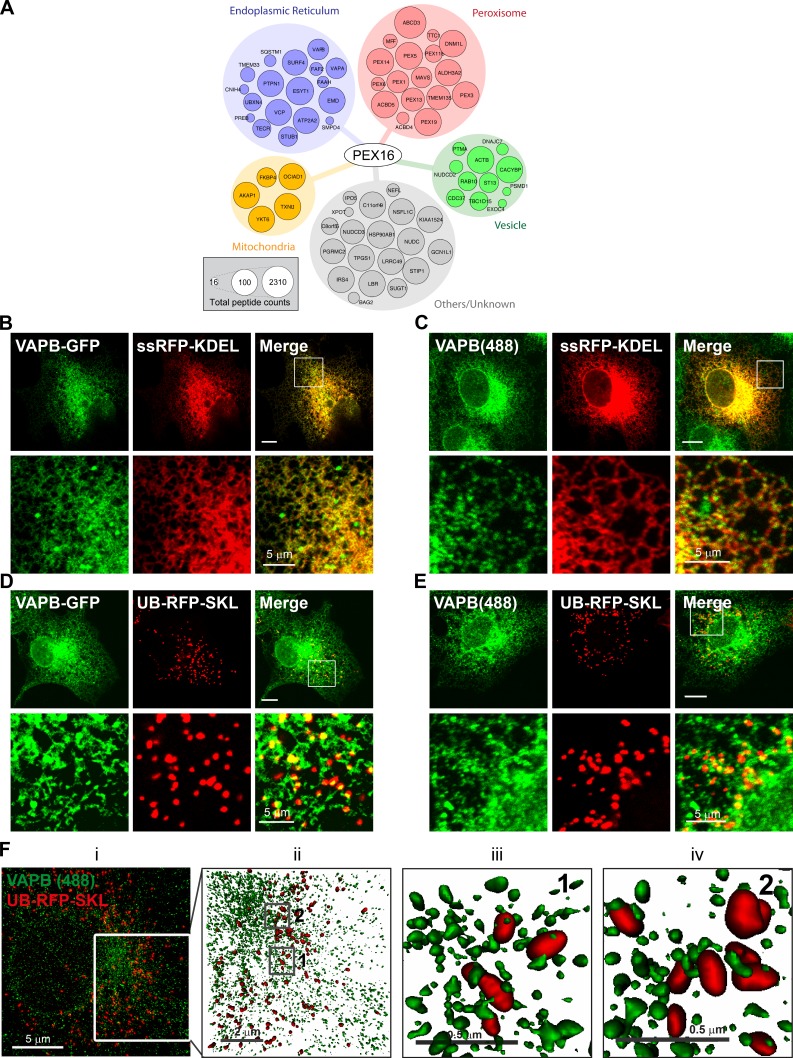
As PEX16 recruits other peroxisomal membrane proteins to the ER and subsequently transports them to peroxisomes (Kim and Mullen, 2013), we reasoned that identifying PEX16-interacting partners could allow us to discover other polypeptides that are important for ER–peroxisome communication. To this end, we conducted BioID (proximity-dependent biotinylation coupled with mass spectrometry [MS]) analysis on PEX16 (Cole et al., 2015; Coyaud et al., 2015; Gupta et al., 2015). We identified 70 high-confidence proximal interactors in this assay, including 17 of the 25 known peroxisomal membrane proteins (Schlüter et al., 2010; Fig. 1 A and Table S1), suggesting that the dataset comprises biologically relevant polypeptides.
Figure 1.
VAPB punctate structures colocalize with peroxisomes. (A) PEX16 interactome. Node size is proportional to peptide counts detected. COS7 cells transiently expressing ssRFP-KDEL (B and C) or UB-RFP-SKL (D and E) and either coexpressing VAPB-GFP (B and D) or immunostained for endogenous VAPB (C and E). The white boxes indicate the magnified area shown below each panel. Bars, 10 µm (or as indicated). (F, left to right) Maximum intensity projection of VAPB–Alexa 488 (green) and UB-RFP-SKL (red) acquired via SIM (i). (ii) Surface projection of the region denoted in panel i of F. Boxes denoted by 1 and 2 in panel ii are magnified in panels iii and iv, respectively. The surface projections demonstrate the apposition of the two organelles in 3D space.
Notably, the interactors also include the organelle-tethering proteins VAPA and VAPB. The VAPs form homodimers or heterodimers on the ER and are found at the membrane contact sites between the ER and many other organelles, including the plasma membrane (PM; Jansen et al., 2011), the Golgi apparatus (Mesmin et al., 2013), mitochondria (De Vos et al., 2012), and endosomes (Rocha et al., 2009; Alpy et al., 2013). To date, it is not known whether VAPs are also required for the formation of an ER–peroxisome tether.
VAPs are juxtaposed to peroxisomes on the ER
To determine whether the VAPs are localized at or near the site of ER–peroxisome contact, we first examined their subcellular localization in mammalian cells. When expressed in COS7 cells, both VAPB-GFP (Fig. 1 B) and VAPA-GFP (Fig. S1 A) displayed ER localization as seen by their colocalization with the ER marker ssRFP-KDEL. Interestingly, despite the strong colocalization, the VAPs appeared more punctate. Examination of endogenous VAPB in COS7 cells showed that most VAPB is localized to punctate structures (Fig. 1 C). To determine whether these VAP puncta colocalized with peroxisomes, we coexpressed VAPB-GFP (Fig. 1 D) or VAPA-GFP (Fig. S1 A) with the peroxisomal marker UB-RFP-SKL and found that some of the VAP puncta colocalized with peroxisomes. Similar colocalization of punctate structures with peroxisomes was also observed when we immunostained for endogenous VAPB (Fig. 1 E). Remarkably, using a structured illumination superresolution approach, we observed that most endogenous VAPB puncta do not perfectly overlap with the peroxisomal marker, but are in juxtaposition to peroxisomes (Fig. 1 F).
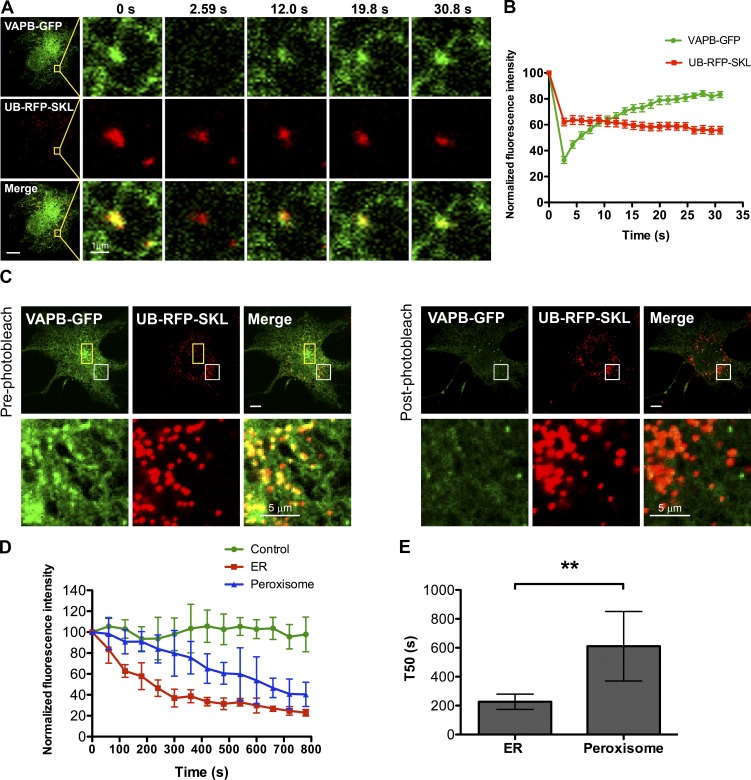
As VAPs are reported to be ER-resident proteins, we reasoned that the VAPs might be concentrating on the ER in juxtaposition to peroxisomes rather than being localized on peroxisomes. To test this, we performed a FRAP assay in COS7 cells coexpressing VAPB-GFP (Fig. 2 A) or VAPA-GFP (Fig. S1 C) with UB-RFP-SKL. We expected that if the peroxisome-localized VAP-GFP punctate structure is on the ER, then its fluorescence should rapidly recover upon photobleaching because of the influx of fluorescent molecules from other portions of the ER. However, those on peroxisomes will not recover during the short recovery time (∼30 s). In this assay, the high 488 laser power used to photobleach the GFP also partially photobleached the UB-RFP-SKL signal (Fig. 2 A and Fig. S1 C). This served as an internal control for a fluorescence signal on peroxisomes that should not recover during the duration of the FRAP assay. Consistent with our hypothesis, the signal for both VAPs on the punctate structures recovered to ∼80% of their initial levels in ∼30 s after photobleaching, whereas the UB-RFP-SKL signal did not recover (Fig. 2 B and Fig. S1 D).
Figure 2.
VAPB localizes in juxtaposition to peroxisomes on the ER. (A) FRAP assay performed in a COS7 cell transiently coexpressing VAPB-GFP and UB-RFP-SKL. Yellow squares indicate the photobleached ROI. (B) FRAP curves. Shown is the normalized fluorescence intensity of VAPB-GFP and UB-RFP-SKL punctate structures within each ROI. Mean ± SD (n = 12). (C) FLIP assay performed in a COS7 cell transiently coexpressing VAPB-GFP and UB-RFP-SKL in a yellow rectangular ROI. Shown is the first frame before photobleaching (pre-photobleach) and the first image after repeated photobleaching (post-photobleach). The white boxes indicate the magnified area shown below each panel. Bars, 10 µm (or as indicated). (D) FLIP curves. Shown is the normalized fluorescence intensity of VAPB-GFP punctate structure juxtaposed to a peroxisome and that of ER-localized VAPB-GFP. The fluorescence intensity of VAPB-GFP in an adjacent cell from the same image serves as a control for imaging induced photobleaching. Mean ± SD (n = 6). (E) Bar graph illustrating the time taken by the ER localized VAPB-GFP and peroxisomal VAPB-GFP in D to drop to 50% of its original level (T50). Mean ± SD (n = 6). **, P < 0.01.
To validate this observation, we performed a fluorescence loss in photobleaching (FLIP) assay in COS7 cells coexpressing either VAPB-GFP (Fig. 2 C) or VAPA-GFP (Fig. S1 B) with UB-RFP-SKL. In this study, we repeatedly photobleached a small region of the ER to deplete the entire ER of VAP-GFP signal. As the ER is interconnected, the photobleaching should result in the loss of VAP-GFP signal on the entire ER, whereas VAP-GFP not localized to the ER will be protected. As seen in Fig. 2 C and Fig. S1 B, depleting the respective VAPB-GFP or VAPA-GFP signal from the ER resulted in the loss of the GFP fluorescence signal from the punctate structures colocalized with peroxisomes. However, the rate of depletion of VAPB-GFP signal associated with peroxisomes was slower than those in other portions of the ER (Fig. 2, D and E), suggesting that the mobility of VAPB-GFP fluorescent molecules juxtaposed to peroxisomes was more constrained.
VAPs interact with ACBD5
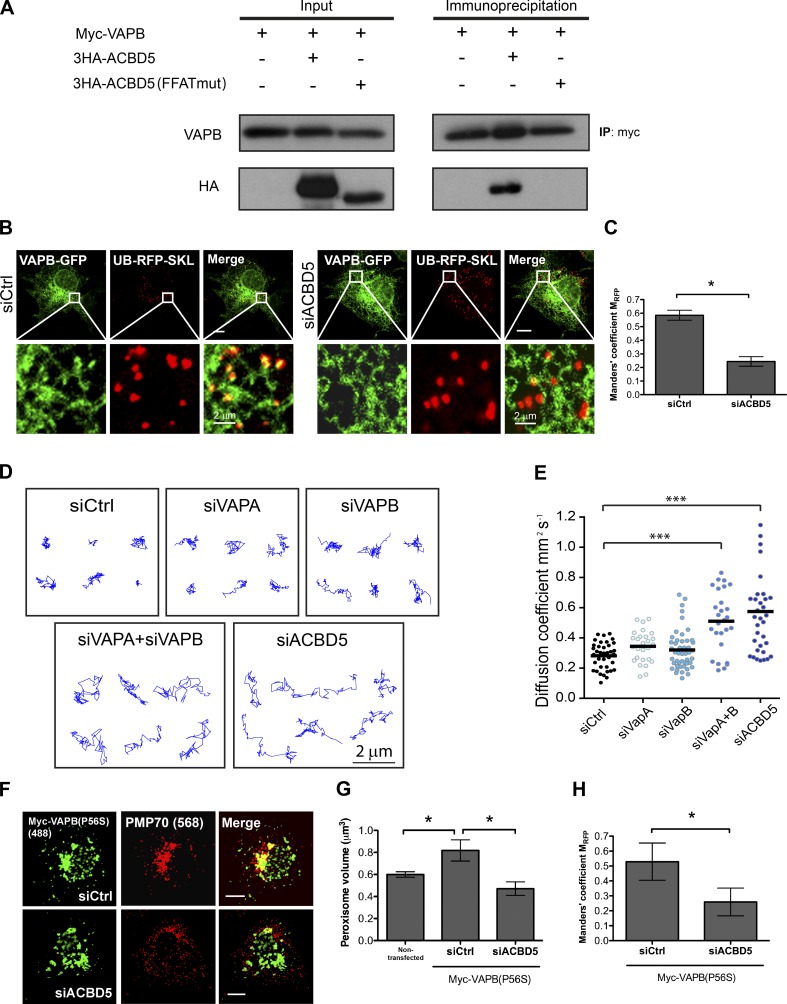
We hypothesized that the constrained VAPs juxtaposed to peroxisomes was because of their interaction with a peroxisomal protein. VAPs are membrane-anchored proteins on the ER that are known to interact with proteins containing a two phenylalanines in an acidic tract (FFAT) motif through their major sperm protein domain (Loewen et al., 2003; Wyles and Ridgway, 2004). As PEX16 does not contain an FFAT domain, we examined the PEX16 BioID dataset (Fig. 1 A) for a peroxisomal protein that possessed an FFAT domain. In the screen, we only found ACBD5 to contain an FFAT domain (Huttlin et al., 2015). Coimmunoprecipitation analysis of VAPs and ACBD5 in HEK293T cells showed that ACBD5 coimmunoprecipitated with both VAPB (Fig. 3 A) and VAPA (Fig. S1 E), but not with the FFAT mutant form of ACBD5, ACDB5(FFATmut) (Fig. 3 A).
Figure 3.
VAPB-ACBD5 tethers peroxisomes to the ER. (A) Coimmunoprecipitation performed in HEK293 cells transiently expressing Myc-VAPB (34 kD) with wild-type or FFAT-motif mutant (mut) ACBD5 (63 kD). (B) COS7 cells treated with indicated siRNAs, and coexpressing VAPB-GFP and UB-RFP-SKL. The white boxes indicate the magnified area shown below each panel. Bars, 10 µm (or as indicated). (C) Bar graph illustrating the Manders’ colocalization coefficient MRFP for UB-RFP-SKL and VAPB-GFP in B. Mean ± SD (n = 3; 20 cells/trial). (D) Representative trajectories of HeLa cells treated with indicated siRNAs and expressing UB-RFP-SKL. Z stacks of single cells were acquired at 40 frames/min, and the center of peroxisomes was tracked over 2 min. (E) The median diffusion coefficient of >27 cells from 3 experiments are graphed (dots) along with the mean (bars). Each video analyzed contained >30 trajectories and each condition >6,000 trajectories. (F) COS7 cells treated with indicated siRNAs, and expressing Myc-VAPB(P56S). Cells were immunostained for Myc tag and endogenous peroxisomal PMP70. Bars, 10 µm. (G and H) Quantification of mean peroxisome volume (G) and the Manders’ colocalization coefficient MRFP for PMP70 and Myc-VAPB(P56S) in F. Mean ± SD (n = 3; 20 cells/trial). *, P < 0.05; ***, P < 0.001. IP, immunoprecipitation.
We next examined whether ACBD5 was responsible for the juxtaposition of VAPB to peroxisomes. In cells depleted of ACBD5, VAPB-GFP puncta juxtaposed to peroxisomes were no longer observed (Fig. 3 B). Quantification of UB-RFP-SKL that colocalized with VAPB-GFP by Manders’ coefficient showed a significant decrease in colocalization between VAPB-positive puncta and peroxisomes in cells depleted of ACBD5 (Fig. 3 C). Interestingly, the expression of endogenous ACBD5 appears to depend on the VAPs, as the depletion of both VAPs resulted in a significant decrease in ACBD5 level, suggesting that the VAPs may stabilize ACBD5 (Fig. S2, A and B).
The VAP–ACBD5 interaction tethers peroxisomes to the ER
We next asked whether the VAP–ACBD5 interaction acts as a tether for peroxisomes to the ER. To test this hypothesis, we reasoned that if VAP–ACBD5 is indeed a tether for peroxisomes to the ER, then disrupting this interaction should result in an increase in peroxisome mobility. To evaluate peroxisome mobility, we determined the diffusion coefficient of peroxisomes in cells depleted of VAPs or ACBD5 in COS7 cells expressing UB-RFP-SKL. Cells depleted with either VAPA or VAPB did not show a visual change in the peroxisome mobility (Fig. 3, D and E; and Fig. S3 A). However, a significant increase in both peroxisome mobility and diffusion coefficient was observed in cells depleted of both VAPs or ACBD5 alone. Together with the localization and immunoprecipitation data, our results suggest that ACBD5 tethers peroxisomes to the ER through its interaction with the VAPs.
An amyotrophic lateral sclerosis (ALS)–associated VAPB mutant requires ACBD5 to induce peroxisome clustering
The proline-to-serine mutation in VAPB at position 56 (P56S) is linked to ALS (Nishimura et al., 2004, 2005). Overexpressing mutant VAPB(P56S) has been shown to induce ER clustering with mitochondria, the Golgi, and endosomes inside cells, and this clustering is thought to cause defects in the functions of these organelles (Navone et al., 2015). Similarly, we found that overexpression of this VAPB mutant resulted in peroxisome aggregation (Fig. 3 F). The peroxisome clustering was validated by the increase in peroxisome volume compared with nontransfected cells (Fig. 3 G). However, the clustering of peroxisomes (Fig. 3, F and G) and colocalization between peroxisome and VAPB(P56S) aggregates (Fig. 3 H) were not observed in cells depleted of ACBD5. These results further indicate that peroxisomes are tethered to the ER through ACBD5 interaction with VAPs.
The VAP–ACBD5 tether is required for peroxisome growth
The multiplication of peroxisomes by growth and division first requires the expansion of the lipid bilayer, leading to elongation followed by fission. Peroxisome fission is mediated by FIS1, MFF, and DLP1/DRP1, and knockdowns of any of these genes have been shown to result in elongated peroxisomes (Farr et al., 2016; Schrader et al., 2016). However, the manner in which peroxisomes obtain membrane lipids for their growth is not known.
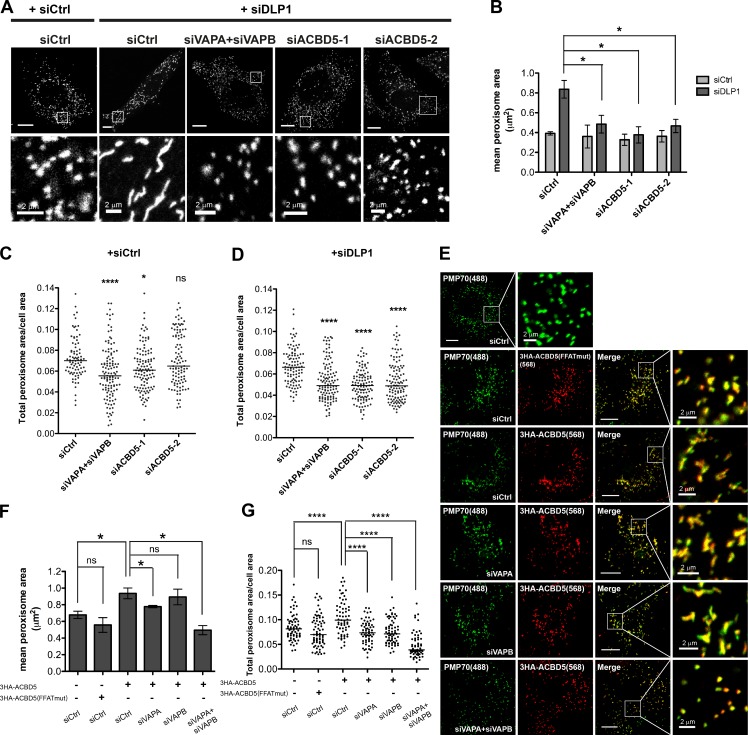
To test whether the VAP–ACBD5 tether is required for lipid transport from the ER to peroxisomes for membrane expansion during peroxisome elongation, we depleted the tethering components in cells in which peroxisomal elongation was induced. We found that depleting VAPs or ACBD5 prevented elongation of peroxisomes in cells predisposed to have elongated peroxisomes because of lack of DLP1 activity (Fig. 4 A and Fig. S2 C). The quantification of mean peroxisome area showed that individual peroxisome was no longer elongated upon DLP1 knockdown (Fig. 4 B). A decrease in the total peroxisome area was also observed (Fig. 4, C and D), suggesting that VAPs and ACBD5 are required for peroxisomal membrane expansion. We also examined the impact of ACBD5 overexpression on peroxisome size. We observed that overexpressing wild-type ACBD5 induced peroxisomal elongation in a VAP-dependent manner, but not in cells expressing the FFAT-motif mutant ACBD5 (Fig. 4, E–G). Together, these results suggest that the VAP–ACBD5 tether is required for peroxisome growth.
Figure 4.
Loss of VAP–ACBD5 tether prevents peroxisomal membrane expansion. (A) HeLa cells treated with indicated siRNAs and immunostained for PMP70. Bars, 10 µm (or as indicated). (B) Quantification of mean peroxisome area in HeLa cells treated with indicated siRNAs. Mean ± SD (n = 3; 30 cells/trial). Quantification of total peroxisome area in HeLa cells either in the absence (C) or presence (D) of DLP1 knockdown and treated with indicated siRNAs. The total peroxisome area of >90 cells from 3 experiments for each siRNA condition are graphed (dots) along with the medium (bars). One-way analysis of variance with Bonferroni correction. (E) COS7 cells treated with indicated siRNAs and expressing wild-type or the FFAT-motif mutant (mut) ACBD5. Cells were immunostained for HA tag and endogenous PMP70. The white boxes indicate the magnified area shown below each panel. Bars, 10 µm (or as indicated). (F) Quantification of mean peroxisome area in COS7 cells in E. Mean ± SD (n = 3; 30 cells/trial). (G) Quantification of total peroxisome area in COS7 cells in E. Quantification similar to C. *, P < 0.05; ****, P < 0.0001.
VAP–ACBD5 interaction plays a role in lipid synthesis
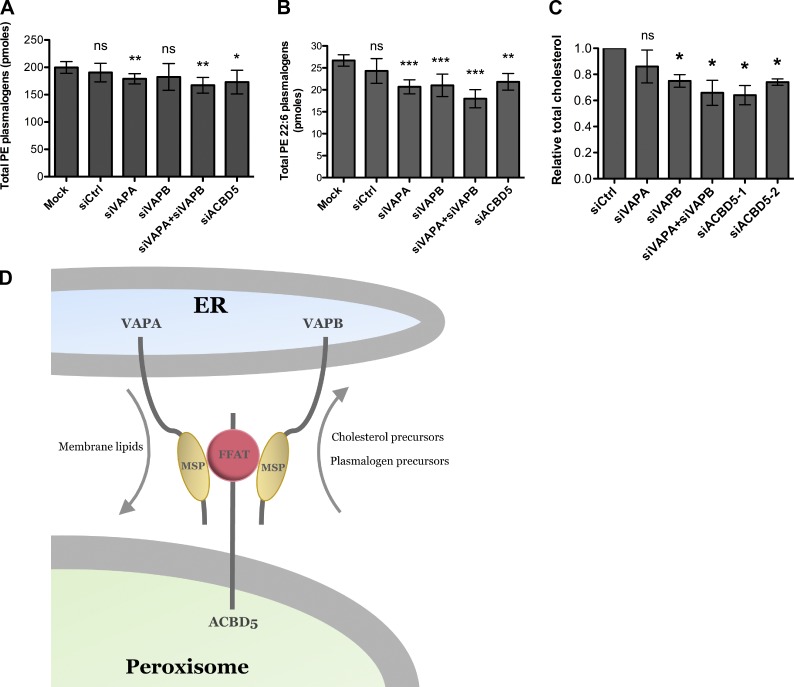
To determine whether lipids are transported from peroxisomes to the ER through the VAP–ACBD5 tether, we examined the cellular levels of two peroxisomal lipids, plasmalogens and cholesterol. The synthesis of plasmalogens is initiated in peroxisomes and subsequently completed in the ER (Braverman and Moser, 2012). Similarly, precursors of cholesterol are synthesized in peroxisomes (Krisans, 1992; Faust and Kovacs, 2014). Moreover, peroxisomes have recently been shown to be involved in trafficking of cholesterol from lysosomes and eventually to the PM (Chu et al., 2015). However, it is not known whether the ER is an intermediate compartment for the trafficking of cholesterol from peroxisomes to the PM. In this study, we tested whether the VAP–ACBD5 tether is required for the maintenance of both plasmalogens and cholesterol levels. We found that depleting both VAPs or ACBD5 alone resulted in a decrease in phosphatidylethanolamine (PE) plasmalogens (Fig. 5, A and B) and total cholesterol levels (Fig. 5 C). However, nonperoxisomal lipids were not affected (Fig. S3, B and C).
Figure 5.
Loss of VAP–ACBD5 tether affects cellular plasmalogen and cholesterol levels. Bar graphs of total PE plasmalogens (A) and total PE 22:6 plasmalogens (B) in HeLa cells treated with indicated siRNAs. Mean ± SD (n = 4). *, P < 0.1; **, P < 0.05; ***, P < 0.01 as compared with mock cells. (C) Quantification of total cholesterol levels in HeLa cells treated with indicated siRNAs using the Amplex Red Cholesterol reagent. The total cholesterol level for each siRNA treatment (n = 3; mean ± SD) was normalized to that in siCtrl-treated cells. *, P < 0.05 as compared with siCtrl-treated cells. (D) Model for ER–peroxisome contact sites. The ER-anchored VAPs bind directly to the FFAT motif containing peroxisomal protein ACBD5 via their major sperm protein (MSP) domains to allow for peroxisome tethering and lipid exchange.
The impact of siRNA treatment on plasmalogen synthesis was modest when compared with prior studies of fibroblast cell lines from patients with constitutional defects in plasmalogen synthesis (i.e., AGPS and GNPAT) in which plasmalogen levels are barely detectable (Itzkovitz et al., 2012). We attribute our observed phenotypes to: (a) the short-term nature of the siRNA experiment; (b) the turnover rate of plasmalogen phospholipid fraction; and (c) alternative pathways of lipid exchange between the ER and peroxisomes (e.g., vesicular intermediates; see Introduction). In summary, we conclude that the VAP–ACBD5 tether is required for the optimal biosynthesis of plasmalogens.
During the preparation of this manuscript, two groups independently reported that patients with mutations in ACBD5 show elevated levels in very long-chained fatty acids (VLCFAs; Ferdinandusse et al., 2016; Yagita et al., 2017). Although we did observe an increasing trend in VLCFA levels in ACBD5 or VAP-depleted cells in our system, the changes are not statistically significant (Fig. S3, B–E). Because much of the VLCFAs are incorporated in phospholipids, the relatively short nature of the transient siRNA knockdown was likely not sufficient for the measurement of a robust change in VLCFAs.
In conclusion, our new insight into the ER–peroxisome tethering advances our understanding of communication between the ER and peroxisomes. We show that peroxisomes are tethered to the ER through the interaction between peroxisomal ACBD5 with the ER-resident VAPs, and this tether is required for the exchange of lipids between them (Fig. 5 D). Finally, as patients with ALS carrying the VAPB(P56S) mutation are reported to have increased cholesterol levels (Marques et al., 2006), it is tempting to speculate that this increase in cholesterol levels may be because of increased ER–peroxisome contact. Further studies on patient cells carrying the VAPB(P56S) mutant will help to illustrate a possible role of the ER–peroxisome tethering in the pathogenesis of ALS.
Materials and methods
Cell culture and transfection
COS7, HEK293T, and HeLa cells were obtained from ATCC. All cells were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Gibco) and 2 mM L-glutamine (Thermo Fisher Scientific) at 37°C in humidified air containing 5% CO2. Plasmids and siRNA were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Prior to live-cell imaging, the medium was changed to CO2-indepent medium (Invitrogen) containing 10% FBS and 2 mM L-glutamine.
Microscopy and analysis
Fluorescence images of mammalian cultured cells were acquired using an LSM 710 laser-scanning confocal microscope with a 63× 1.4 NA oil immersion objective (ZEISS). For peroxisome diffusion analysis, the images were acquired using a DMI6000B inverted fluorescence microscope (Leica Biosystems) with an ImagEM X2 camera (Hamamatsu Photonics).
Live-cell imaging of mammalian cells was performed at 37°C in CO2-independent medium containing 10% FBS and 2 mM L-glutamine. For immunofluorescence, cells were fixed in PFA and permeabilized using 0.1% Triton X-100 in PBS, followed by incubation with appropriate antibodies as specified. FLIP assays were performed using 50 iterations of a 488-nm laser light at full strength in a rectangular region of interest (ROI) repeatedly every 60 s. An image was taken immediately before and after each photobleaching session. FRAP assays were performed using a 488-nm laser light at full strength in a rectangular ROI, and 20 single scan images were collected after the photobleaching section. For measurement of peroxisome volume, Z-stack series were collected.
For peroxisome diffusion analysis, Z stacks of single cells were acquired at 40 frames per min and projected to generate 2D videos. At each frame, individual peroxisomes were fit with multiple Gaussians to determine the center of the organelle with a positional accuracy of <200 nm using the u-track analysis on MatLab software (Jaqaman et al., 2008). Peroxisomes were tracked over 80 frames (2 min) using u-track to determine the median diffusion coefficient of peroxisomes per cell.
The measurements of peroxisome volume, peroxisome area, and Manders’ colocalization coefficient were all performed using Volocity 5.0 software (PerkinElmer). All images were adjusted for brightness and contrast using ImageJ (National Institutes of Health). Figure compositions and merged images were generated using Photoshop CS4 and Illustrator CS2 (Adobe Systems). Unless otherwise indicated, all statistical analysis was performed using a Student’s t test.
Structured illumination microscopy (SIM) imaging
Imaging of samples was performed using structured illumination on the Elyra PS.1 superresolution inverted microscope (ZEISS). Samples were imaged at an effective magnification of 101× (63× objective plus 1.6× optovar tube lens) on an oil immersion objective. Typically, 10–20 slices of 0.110 µm were captured for each field of view for an imaging volume of ∼1.1 to 2.2 µm. 488- and 561-nm laser lines were directed into the microscope optical train via a multimode fiber coupler. The lasers were passed through a diffraction grating, and a series of diffraction orders (−1, 0, and +1) were projected onto the back focal plane of the objective. These wave fronts were collimated in the objective to create a 3D sinusoidal illumination pattern on the sample. The diffraction grating was then rotated and translated throughout the acquisition to create patterned offset images containing higher spatial frequency information as compared with wide-field imaging. Five lateral positions were acquired at each of three diffraction grating rotations (120°) for a total of 15 raw images per slice. SIM imaging with both the 488- and 561-nm laser was performed at 75-ms exposures with laser power varying between 3 and 10% and a gain level of 60–80.
SIM image processing and analysis
Raw SIM image stacks were processed in Zen under the Structured Illumination toolbar. A series of parameters were set to generate an optical transfer function used for 3D reconstruction. The noise filter for Wiener deconvolution was set to a value of 1.0 × 10−4 to maximize the recovery of high spatial frequency information while minimizing illumination pattern artifacts. The maximum isotropy option was left unselected to recover all available frequency information at exactly the 120° rotation angles. Superresolution frequency weighting was set 1.0. Negative values arising as an artifact of the Wiener filter were clipped to zero using the Baseline Cut option. Sectioning filters used to remove the 0-frequencies from the 0th order and nonshifted first order (+1 and −1) were set to default values of 100 and 83, respectively. Processed SIM images were then aligned via an affine transformation matrix of predefined values obtained using 100-nm multicolor Tetraspeck fluorescent microspheres (Thermo Fisher Scientific). SIM data were analyzed for peroxisome–ER association by generating 3D surface projections in Zen.
Coimmunoprecipitation and immunoblot analysis
For coimmunoprecipitation, transfected HEK293T cells were lysed in 1 ml lysis buffer (150 mM NaCl, 50 mM Hepes, 1% Triton X-100, 10% glycerol, 1.5 mM MgCl2, and 1.0 mM ethylene glycol-bis[β-aminoethyl ether]-N,N,N′,N′-tetraacetic acid) supplemented with protease inhibitors. For each immunoprecipitation, a 0.9-ml aliquot of the lysate was incubated with 0.9 µg Myc antibody (EMD Millipore) and 10 µl of a 1:1 slurry of Protein G Sepharose (Bioshop Canada Inc.) overnight. Sepharose beads were washed three times with 1 ml high-salt lysis buffer containing 0.5 M NaCl. The precipitates were analyzed by standard immunoblot procedures.
Biotin-streptavidin affinity purification
BioID (Roux et al., 2012) was performed as previously described (Coyaud et al., 2015). In brief, the full-length human PEX16 coding sequence was amplified by PCR from MGC clone BC004356 and cloned with AscI and NotI restriction sites into our pcDNA5 FRT/TO FLAGBirA* expression vector, and 293 T-REx Flp-In cells stably expressing FLAGBirA*-PEX16 were generated. After selection (DMEM plus 10% FBS plus 200 µg/ml Hygromycin B), 5 × 15-cm diameter plates of subconfluent (60%) cells were incubated for 24 h in complete media supplemented with 1 µg/ml tetracycline (Sigma-Aldrich) and 50 µM biotin (BioShop Canada Inc.). Cells were collected and pelleted (2,000 rpm for 3 min), the pellet was washed twice with PBS, and dried pellets were snap frozen. Cell pellets were resuspended in 10 ml lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.1% SDS, 1:500 protease inhibitor cocktail [Sigma-Aldrich], and 1:1,000 benzonase nuclease [EMD Millipore]), incubated on an end-over-end rotator at 4°C for 1 h, briefly sonicated to disrupt any visible aggregates, and then centrifuged at 16,000 g for 30 min at 4°C. Supernatant was transferred to a fresh 15-ml conical tube. 30 µl packed, pre-equilibrated streptavidin sepharose beads (GE Healthcare) was added and the mixture incubated for 3 h at 4°C with end-over-end rotation. Beads were pelleted by centrifugation at 2,000 rpm for 2 min and transferred with 1 ml lysis buffer to a fresh Eppendorf tube. Beads were washed once with 1 ml lysis buffer and twice with 1 ml of 50 mM ammonium bicarbonate (ammbic; pH 8.3), transferred in ammbic to a fresh Eppendorf tube, and washed two more times with 1 ml ammbic. Washed beads were incubated with 1 µg MS-grade TPCK trypsin (Promega) dissolved in 200 µl of 50 mM ammbic, pH 8.3, overnight at 37°C. The following morning, 0.5 µg MS-grade TPCK trypsin was added, and beads were incubated 2 additional hours at 37°C. Beads were pelleted by centrifugation at 2,000 g for 2 min, and the supernatant was transferred to a fresh Eppendorf tube. Beads were washed twice with 150 µl of 50 mM ammonium bicarbonate, and these washes were pooled with the first eluate. The sample was lyophilized and resuspended in buffer A (0.1% formic acid). One-fifth of the sample was analyzed per MS run.
MS
Liquid chromatography analytical columns (75-µm inner diameter) and precolumns (100-µm inner diameter) were made in-house from fused silica capillary tubing from InnovaQuartz and packed with 100 Å C18-coated silica particles (Magic; Michrom Bioresources). Peptides were subjected to nanoflow liquid chromatography–electrospray ionization–tandem MS (MS/MS), using a 120-min reversed-phase (10–40% acetonitrile and 0.1% formic acid) buffer gradient running at 250 nl/min on a Proxeon EASY-nLC pump in-line with a hybrid linear quadrupole ion trap (Velos LTQ) Orbitrap mass spectrometer (Thermo Fisher Scientific). A parent ion scan was performed in the Orbitrap using a resolving power of 60,000. Simultaneously, up to 20 of the most intense peaks were selected for MS/MS (minimum ion count of 1,000 for activation) using standard collision-induced dissociation fragmentation. Fragment ions were detected in the LTQ. Dynamic exclusion was activated such that MS/MS of the same mass-to-charge ratio (m/z; within a 10-ppm window; exclusion list size 500) detected three times within 45 s were excluded from analysis for 30 s. Data were analyzed using the trans-proteomic pipeline (Deutsch et al., 2010; Pedrioli, 2010) via the ProHits software suite (Liu et al., 2010). For protein identification, Thermo RAW files were converted to the mzXML format using Proteowizard (Kessner et al., 2008) and then searched using X!Tandem (Craig and Beavis, 2004) and Comet (Eng et al., 2013) against Human RefSeq Version 45 (containing 36,113 entries). Search parameters specified a parent ion mass tolerance of 10 ppm and an MS/MS fragment ion tolerance of 0.4 D, with up to two missed cleavages allowed for trypsin. Variable modifications of +16 at M and W, +32 at M and W, +42 at N terminus, and +1 at N and Q were allowed. Proteins identified with a ProteinProphet cutoff of 0.90 (corresponding to ≤1% false discovery rate [FDR]) and with ≥2 unique peptides were analyzed with SAINT Express v.3.3. Each biological replicate was analyzed using two technical replicates. Data were compared with 12 control runs (FlagBirA* alone and no bait control lysates), collapsed to the two highest spectral counts for each prey protein, and the SAINT score cutoff value was set to 0.80 (Bayes FDR <1%) to define high-confidence interactors.
Liquid chromatography MS/MS analysis of plasmalogens and VLCFAs
Cell pellets were homogenized in PBS (Thermo Fisher Scientific). An extraction solution of methanol containing 10 ng each of the internal standards 16:0-d4 Lyso-PAF (15.6 pmol; Cayman Chemical) and 26:0-d4 Lyso PC (Avanti Polar Lipids, Inc.) was added to 50 µg protein cell extract in a glass tube. The samples were incubated on a shaker at RT for 1 h. The samples were transferred to Costar Spin-X centrifuge tube filters (Corning) and centrifuged for 5 min. The filtrates were then transferred to autosampler Verex vials (Phenomenex) for analysis by LC-MS/MS.
A TQD interfaced with an Acquity UPLC system (Waters) was employed for positive-ion electrospray ionization–MS/MS. Plasmalogen species were detected by monitoring multiple-reaction monitoring transitions representing fragmentation of [M+H]+ species to m/z 311, 339, 361, 385, 389, and 390 for compounds with 16:1, 18:1, 20:4. 22:6, 22:4, and 18:0, respectively, at the sn-2 position. Elution of Lyso PCs was detected by monitoring multiple-reaction monitoring transitions representing fragmentation of [M+H]+ to m/z 104. Chromatographic resolution was achieved via the use of a 2.1 × 50-mm, 1.7-µm Acquity UPLC BEH column (Waters). The solvent systems used were mobile phase A, 54.5% water/45% acetonitrile/0.5% formic acid, and mobile phase B, 99.5% acetonitrile/0.5% formic acid, with both solutions containing 2 mM ammonium acetate. Injections of extracts dissolved in methanol were made with initial solvent conditions of 85% mobile phase A/15% mobile phase B. The gradient employed was from 15 to 100% mobile phase B over a period of 2.5 min and held at 100% mobile phase B for 1.5 min before reconditioning the column back to 85% mobile phase A/15% mobile phase B for 1 min at a solvent rate of 0.7 ml/min. A column temperature of 35°C and an injection volume of 5 µl for plasmalogens and 10 µl for Lyso PCs were used.
Plasmids and siRNA
UB-RFP-SKL and ssRFP-KDEL have been described previously (Kim et al., 2006). VAPA-GFP, Myc-VAPA, and VAPB-GFP were gifts from W.S. Trimble (Hospital for Sick Children, Toronto, Ontario, Canada). Myc-VAPB and Myc-VAPB(P56S) were gifts from C.C.J. Miller (King’s College London, London, England, UK; De Vos et al., 2012). The FFAT-motif mutant ACBD5, Myc-VAPB(FFATmut), was generated in which all residues in its FFAT domain (aa 262–271; Murphy and Levine, 2016) were replaced by Ala. Mutagenesis was performed using appropriate forward and reverse mutagenic primers and PCR-based mutagenesis protocols. The ACBD5 ORF was purchased from SPARC BioCentre (SIDNET identification no. 1004942) and cloned into a Myc-N1 vector by standard PCR-based method. A complete description of the construction of all plasmids and primer sequences used in this study is available upon request.
siRNAs were all obtained from Invitrogen. siRNA sequences were: siCtrl, 5′-AAUAAGGCUAUGAAGAGAUAC-3′; siVAPA, 5′-GCGAAAUCCAUCGGAUAGAAA-3′; siVAPB, 5′-GCUCUUGGCUCUGGUGGUUUU-3′; siACBD5-1 (or simply referred to as siACBD5), 5′-GCACAGUGGUUGGUGUAUUUA-3′; siACBD5-2, 5′-CCGUUAAUGGUAAAGCUGAAA-3′; and siDLP1 (sc-43732; Santa Cruz Biotechnology, Inc.).
Reagents
The antibodies used in this study were rabbit monoclonal anti–PMP70 (Epitomics); rabbit polyclonal anti–HA (OriGene); goat anti–rabbit Alexa Fluor 488 secondary antibody (Invitrogen); goat anti–mouse Alexa Fluor 488 secondary antibody (Invitrogen); goat anti–rabbit Alexa Fluor 568 secondary antibody (Invitrogen); rabbit polyclonal anti–VAPB (Sigma-Aldrich); rabbit polyclonal anti–VAPA (Novus Biologicals); mouse anti–DLP1 (BD), rabbit polyclonal anti–ACBD5 (Novus Biologicals); and monoclonal anti–mouse GAPDH (Novus Biologicals). The total cholesterol levels were measured using Amplex Red reagent (Thermo Fisher Scientific). HPLC-grade solvents (methanol, acetonitrile, chloroform, and water) were purchased from Thermo Fisher Scientific. Formic acid was purchased from Sigma-Aldrich.
Online supplemental material
Fig. S1 shows localization and coimmunoprecipitation analysis of VAPA. Fig. S2 shows Western blots for validation of siRNA-mediated knockdowns. Fig. S3 shows sample trajectories for peroxisome mobility analysis and the levels of nonperoxisomal lipids. Table S1 shows raw data and statistical analysis of the spectral counts for PEX16 BioID analysis.
Supplementary Material
Acknowledgments
The work in this study is funded by the Natural Sciences and Engineering Research Council of Canada and a Canadian Institutes of Health Research project grant to P.K. Kim.
The authors declare no competing financial interests.
Author contributions: R. Hua, W.S. Trimble, and P.K. Kim conceived the study. R. Hua, D. Cheng, É. Coyaud, S. Freeman, E. Di Pietro, Y. Wang, A. Vissa, B. Raught, and P.K. Kim performed experiments. R. Hua, É. Coyaud, S. Freeman, E. Di Pietro, C.M. Yip, G.D. Fairn, N. Braverman, J.H. Brumell, W.S. Trimble, B. Raught, and P.K. Kim analyzed the data. R. Hua, and P.K. Kim wrote the manuscript. All of the authors contributed to editing the manuscript.
Footnotes
Abbreviations used:
- ACBD5
- acyl-CoA binding domain containing 5
- ALS
- amyotrophic lateral sclerosis
- ammbic
- ammonium bicarbonate
- FDR
- false discovery rate
- FFAT
- two phenylalanines in an acidic tract
- FLIP
- fluorescence loss in photobleaching
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- m/z
- mass-to-charge ratio
- PE
- phosphatidylethanolamine
- PM
- plasma membrane
- ROI
- region of interest
- SIM
- structured illumination microscopy
- VAP
- VAMP-associated protein
- VLCFA
- very long-chained fatty acid
References
- Agrawal G., and Subramani S.. 2016. De novo peroxisome biogenesis: Evolving concepts and conundrums. Biochim. Biophys. Acta. 1863:892–901. 10.1016/j.bbamcr.2015.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal G., Joshi S., and Subramani S.. 2011. Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 108:9113–9118. 10.1073/pnas.1018749108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpy F., Rousseau A., Schwab Y., Legueux F., Stoll I., Wendling C., Spiegelhalter C., Kessler P., Mathelin C., Rio M.C., et al. 2013. STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J. Cell Sci. 126:5500–5512. 10.1242/jcs.139295 [DOI] [PubMed] [Google Scholar]
- Antonenkov V.D., and Hiltunen J.K.. 2012. Transfer of metabolites across the peroxisomal membrane. Biochim. Biophys. Acta. 1822:1374–1386. 10.1016/j.bbadis.2011.12.011 [DOI] [PubMed] [Google Scholar]
- Aranovich A., Hua R., Rutenberg A.D., and Kim P.K.. 2014. PEX16 contributes to peroxisome maintenance by constantly trafficking PEX3 via the ER. J. Cell Sci. 127:3675–3686. 10.1242/jcs.146282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman N.E., and Moser A.B.. 2012. Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta. 1822:1442–1452. 10.1016/j.bbadis.2012.05.008 [DOI] [PubMed] [Google Scholar]
- Chu B.B., Liao Y.C., Qi W., Xie C., Du X., Wang J., Yang H., Miao H.H., Li B.L., and Song B.L.. 2015. Cholesterol transport through lysosome-peroxisome membrane contacts. Cell. 161:291–306. 10.1016/j.cell.2015.02.019 [DOI] [PubMed] [Google Scholar]
- Cole A., Wang Z., Coyaud E., Voisin V., Gronda M., Jitkova Y., Mattson R., Hurren R., Babovic S., Maclean N., et al. 2015. Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 27:864–876. 10.1016/j.ccell.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyaud E., Mis M., Laurent E.M., Dunham W.H., Couzens A.L., Robitaille M., Gingras A.C., Angers S., and Raught B.. 2015. BioID-based identification of Skp Cullin F-box (SCF)β-TrCP1/2 E3 ligase substrates. Mol. Cell. Proteomics. 14:1781–1795. 10.1074/mcp.M114.045658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R., and Beavis R.C.. 2004. TANDEM: Matching proteins with tandem mass spectra. Bioinformatics. 20:1466–1467. 10.1093/bioinformatics/bth092 [DOI] [PubMed] [Google Scholar]
- David C., Koch J., Oeljeklaus S., Laernsack A., Melchior S., Wiese S., Schummer A., Erdmann R., Warscheid B., and Brocard C.. 2013. A combined approach of quantitative interaction proteomics and live-cell imaging reveals a regulatory role for endoplasmic reticulum (ER) reticulon homology proteins in peroxisome biogenesis. Mol. Cell. Proteomics. 12:2408–2425. 10.1074/mcp.M112.017830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch E.W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., et al. 2010. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 10:1150–1159. 10.1002/pmic.200900375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos K.J., Mórotz G.M., Stoica R., Tudor E.L., Lau K.F., Ackerley S., Warley A., Shaw C.E., and Miller C.C.. 2012. VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum. Mol. Genet. 21:1299–1311. 10.1093/hmg/ddr559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J.K., Jahan T.A., and Hoopmann M.R.. 2013. Comet: An open-source MS/MS sequence database search tool. Proteomics. 13:22–24. 10.1002/pmic.201200439 [DOI] [PubMed] [Google Scholar]
- Farr R.L., Lismont C., Terlecky S.R., and Fransen M.. 2016. Peroxisome biogenesis in mammalian cells: The impact of genes and environment. Biochim. Biophys. Acta. 1863:1049–1060. 10.1016/j.bbamcr.2015.08.011 [DOI] [PubMed] [Google Scholar]
- Faust P.L., and Kovacs W.J.. 2014. Cholesterol biosynthesis and ER stress in peroxisome deficiency. Biochimie. 98:75–85. 10.1016/j.biochi.2013.10.019 [DOI] [PubMed] [Google Scholar]
- Ferdinandusse S., Falkenberg K.D., Koster J., Mooyer P.A., Jones R., van Roermund C.W., Pizzino A., Schrader M., Wanders R.J., Vanderver A., and Waterham H.R.. 2016. ACBD5 deficiency causes a defect in peroxisomal very long-chain fatty acid metabolism. J. Med. Genet. 10.1136/jmedgenet-2016-104132 [DOI] [PubMed] [Google Scholar]
- Gupta G.D., Coyaud É., Gonçalves J., Mojarad B.A., Liu Y., Wu Q., Gheiratmand L., Comartin D., Tkach J.M., Cheung S.W., et al. 2015. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell. 163:1484–1499. 10.1016/j.cell.2015.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., Erdmann R., van der Klei I., and Veenhuis M.. 2014. Evolving models for peroxisome biogenesis. Curr. Opin. Cell Biol. 29:25–30. 10.1016/j.ceb.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua R., and Kim P.K.. 2016. Multiple paths to peroxisomes: Mechanism of peroxisome maintenance in mammals. Biochim. Biophys. Acta. 1863:881–891. 10.1016/j.bbamcr.2015.09.026 [DOI] [PubMed] [Google Scholar]
- Hua R., Gidda S.K., Aranovich A., Mullen R.T., and Kim P.K.. 2015. Multiple domains in PEX16 mediate its trafficking and recruitment of peroxisomal proteins to the ER. Traffic. 16:832–852. 10.1111/tra.12292 [DOI] [PubMed] [Google Scholar]
- Hunt M.C., Tillander V., and Alexson S.E.. 2014. Regulation of peroxisomal lipid metabolism: The role of acyl-CoA and coenzyme A metabolizing enzymes. Biochimie. 98:45–55. 10.1016/j.biochi.2013.12.018 [DOI] [PubMed] [Google Scholar]
- Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., et al. 2015. The BioPlex Network: A systematic exploration of the hHuman interactome. Cell. 162:425–440. 10.1016/j.cell.2015.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz B., Jiralerspong S., Nimmo G., Loscalzo M., Horovitz D.D., Snowden A., Moser A., Steinberg S., and Braverman N.. 2012. Functional characterization of novel mutations in GNPAT and AGPS, causing rhizomelic chondrodysplasia punctata (RCDP) types 2 and 3. Hum. Mutat. 33:189–197. 10.1002/humu.21623 [DOI] [PubMed] [Google Scholar]
- Jansen M., Ohsaki Y., Rega L.R., Bittman R., Olkkonen V.M., and Ikonen E.. 2011. Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic. 12:218–231. 10.1111/j.1600-0854.2010.01142.x [DOI] [PubMed] [Google Scholar]
- Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S.L., and Danuser G.. 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 5:695–702. 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessner D., Chambers M., Burke R., Agus D., and Mallick P.. 2008. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics. 24:2534–2536. 10.1093/bioinformatics/btn323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.K., and Mullen R.T.. 2013. PEX16: A multifaceted regulator of peroxisome biogenesis. Front. Physiol. 4:241 10.3389/fphys.2013.00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.K., Mullen R.T., Schumann U., and Lippincott-Schwartz J.. 2006. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 173:521–532. 10.1083/jcb.200601036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach B., Sun X., Coquelle N., Fagarasanu A., Poirier R.L., and Rachubinski R.A.. 2013. An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 32:2439–2453. 10.1038/emboj.2013.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs W.J., Tape K.N., Shackelford J.E., Duan X., Kasumov T., Kelleher J.K., Brunengraber H., and Krisans S.K.. 2007. Localization of the pre-squalene segment of the isoprenoid biosynthetic pathway in mammalian peroxisomes. Histochem. Cell Biol. 127:273–290. 10.1007/s00418-006-0254-6 [DOI] [PubMed] [Google Scholar]
- Krisans S.K. 1992. The role of peroxisomes in cholesterol metabolism. Am. J. Respir. Cell Mol. Biol. 7:358–364. 10.1165/ajrcmb/7.4.358 [DOI] [PubMed] [Google Scholar]
- Lam S.K., Yoda N., and Schekman R.. 2010. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 107:21523–21528. 10.1073/pnas.1013397107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Zhang J., Larsen B., Stark C., Breitkreutz A., Lin Z.Y., Breitkreutz B.J., Ding Y., Colwill K., Pasculescu A., et al. 2010. ProHits: Integrated software for mass spectrometry-based interaction proteomics. Nat. Biotechnol. 28:1015–1017. 10.1038/nbt1010-1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C.J., Roy A., and Levine T.P.. 2003. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22:2025–2035. 10.1093/emboj/cdg201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques V.D., Barreira A.A., Davis M.B., Abou-Sleiman P.M., Silva W.A. Jr., Zago M.A., Sobreira C., Fazan V., and Marques W. Jr. 2006. Expanding the phenotypes of the Pro56Ser VAPB mutation: Proximal SMA with dysautonomia. Muscle Nerve. 34:731–739. 10.1002/mus.20657 [DOI] [PubMed] [Google Scholar]
- Mesmin B., Bigay J., Moser von Filseck J., Lacas-Gervais S., Drin G., and Antonny B.. 2013. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 155:830–843. 10.1016/j.cell.2013.09.056 [DOI] [PubMed] [Google Scholar]
- Murphy S.E., and Levine T.P.. 2016. VAP, a versatile access point for the endoplasmic reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta. 1861:952–961. 10.1016/j.bbalip.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Navone F., Genevini P., and Borgese N.. 2015. Autophagy and nNeurodegeneration: Insights from a cultured cell model of ALS. Cells. 4:354–386. 10.3390/cells4030354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.K., Liu F., Lai J., Low W., and Jedd G.. 2009. A tether for Woronin body inheritance is associated with evolutionary variation in organelle positioning. PLoS Genet. 5:e1000521 10.1371/journal.pgen.1000521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura A.L., Mitne-Neto M., Silva H.C., Richieri-Costa A., Middleton S., Cascio D., Kok F., Oliveira J.R., Gillingwater T., Webb J., et al. 2004. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 75:822–831. 10.1086/425287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura A.L., Al-Chalabi A., and Zatz M.. 2005. A common founder for amyotrophic lateral sclerosis type 8 (ALS8) in the Brazilian population. Hum. Genet. 118:499–500. 10.1007/s00439-005-0031-y [DOI] [PubMed] [Google Scholar]
- Pedrioli P.G. 2010. Trans-proteomic pipeline: A pipeline for proteomic analysis. Methods Mol. Biol. 604:213–238. 10.1007/978-1-60761-444-9_15 [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S., and Prinz W.A.. 2008. Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 105:15785–15790. 10.1073/pnas.0808321105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., and Neefjes J.. 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell Biol. 185:1209–1225. 10.1083/jcb.200811005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., Raida M., and Burke B.. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196:801–810. 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter A., Real-Chicharro A., Gabaldón T., Sánchez-Jiménez F., and Pujol A.. 2010. PeroxisomeDB 2.0: An integrative view of the global peroxisomal metabolome. Nucleic Acids Res. 38:D800–D805. 10.1093/nar/gkp935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M., Godinho L.F., Costello J.L., and Islinger M.. 2015. The different facets of organelle interplay-an overview of organelle interactions. Front. Cell Dev. Biol. 3:56 10.3389/fcell.2015.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M., Costello J.L., Godinho L.F., Azadi A.S., and Islinger M.. 2016. Proliferation and fission of peroxisomes - An update. Biochim. Biophys. Acta. 1863:971–983. 10.1016/j.bbamcr.2015.09.024 [DOI] [PubMed] [Google Scholar]
- Shai N., Schuldiner M., and Zalckvar E.. 2016. No peroxisome is an island - Peroxisome contact sites. Biochim. Biophys. Acta. 1863:1061–1069. 10.1016/j.bbamcr.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyles J.P., and Ridgway N.D.. 2004. VAMP-associated protein-A regulates partitioning of oxysterol-binding protein-related protein-9 between the endoplasmic reticulum and Golgi apparatus. Exp. Cell Res. 297:533–547. 10.1016/j.yexcr.2004.03.052 [DOI] [PubMed] [Google Scholar]
- Yagita Y., Shinohara K., Abe Y., Nakagawa K., Al-Owain M., Alkuraya F.S., and Fujiki Y.. 2017. Deficiency of a retinal dystrophy protein ACBD5 impairs peroxisomal β-oxidation of very-long-chain fatty acids. J. Biol. Chem. 292:691–705. 10.1074/jbc.M116.760090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Rachubinski D.A., Joshi S., Rachubinski R.A., and Subramani S.. 2008. Dysferlin domain-containing proteins, Pex30p and Pex31p, localized to two compartments, control the number and size of oleate-induced peroxisomes in Pichia pastoris. Mol. Biol. Cell. 19:885–898. 10.1091/mbc.E07-10-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.