Transcytosis plays an important role in establishing cell polarity and in mediating transport of large cargo across epithelial barriers, but its molecular basis is unclear. Nelms et al. present a new dataset of genes involved in receptor-mediated transcytosis and show that the apical and basolateral recycling and transcytotic pathways are genetically separable.
Abstract
Endosome transport by transcytosis is the primary mechanism by which proteins and other large cargo traverse epithelial barriers in normal tissue. Transcytosis is also essential for establishing and maintaining membrane polarity in epithelia and other polarized cells. To identify novel components of this pathway, we conducted a high-throughput RNA interference screen for factors necessary for the bidirectional transcytosis of IgG by the Fcγ receptor FcRn. This screen identified 23 genes whose suppression resulted in a reproducible decrease in FcRn-mediated transcytosis. Pulse-chase kinetic transport assays on four of the top-ranking genes (EXOC2, EXOC7, PARD6B, and LEPROT) revealed distinct effects on the apical and basolateral recycling and transcytotic pathways, demonstrating that these pathways are genetically separable. We also found a strong dependence on PARD6B for apical, but not basolateral, recycling, implicating this cell polarity gene in assembly or maintenance of the apical endosomal system. This dataset yields insights into how vesicular transport is adapted to the specialized functions of differentiated cell types and opens new research avenues into epithelial trafficking.
Introduction
In many tissues throughout the body, a single layer of polarized epithelial cells functions to separate and affect vastly different environments. Large molecules are transported across these epithelial barriers by transcytosis, a process of intracellular endosome traffic that connects one cell surface with the other. Many roles for transcytosis have been documented (Tuma and Hubbard, 2003). Transcytosis of the immunoglobulins, for example, enables immune surveillance of mucosal surfaces lining the gut, lungs, and urogenital tract (Rojas and Apodaca, 2002; Rath et al., 2015). Transcytosis also contributes to the development of cell polarity by enabling the assembly of distinct apical and basolateral plasma membranes, thus fundamentally shaping cell and tissue function (Apodaca et al., 2012). Microbes, viruses, and microbial toxins exploit transcytosis for invasion of the host (Lencer et al., 1995; Bomsel, 1997; Couesnon et al., 2008), and the pathway holds promise for mucosal delivery of therapeutic peptides and proteins (Spiekermann et al., 2002; Li et al., 2011). Here, we report the results of a high-throughput endoribonuclease-prepared short interfering RNA (esiRNA) screen for components of transcytosis. Several of the newly identifed genes are informative of endosome organization in polarized simple epithelia.
The endocytic compartments mediating transcytosis are uniquely adapted to accommodate the polarized cell phenotype (Rodriguez-Boulan et al., 2005; Tzaban et al., 2009; Li et al., 2011). One specialization is the establishment of distinct populations of apical and basolateral early sorting endosomes, which selectively receive cargo from either apical or basolateral surfaces, respectively (Bomsel et al., 1989, 1990; Parton et al., 1989; Sheff et al., 2002). These early sorting endosomes then route internalized cargo to one of several possible destinations, including back to the cell surface where endocytosis originated (the recycling pathway) or to the opposite cell surface via the common/apical recycling endosomes (the transcytotic pathway). The common/apical recycling endosome is another adaptation of the endosomal system of polarized epithelia. It receives cargo from both cell surfaces and serves as a major sorting station for the recycling and transcytotic pathways (Parton et al., 1989; Hughson and Hopkins, 1990; Apodaca et al., 1994; Barroso and Sztul, 1994; Odorizzi et al., 1996; Wang et al., 2000a).
Transcytosis has been best characterized for basolateral-to-apical transport of dimeric IgA (dIgA) by the polymeric immunoglobulin receptor (pIgR; Rojas and Apodaca, 2002). pIgR binds dIgA on the basolateral membrane of mucosal epithelial cells and carries it sequentially into the early basolateral sorting endosome, the common endosome, the apical recycling endosome, and finally to the apical cell surface where the extracellular domain of the receptor is cleaved for release into the lumen as a component of secretory IgA. A complex network of genes direct and regulate pIgR-mediated transcytosis, including several Rab family GTPases (RAB3B, RAB5, RAB11A, RAB17, and RAB25; Hunziker and Peters, 1998; Wang et al., 2000b; van IJzendoorn et al., 2002), the exocyst complex (Oztan et al., 2007), retromer (Vergés et al., 2004), phosphoinositide-3-kinase (Vergés et al., 2007), MYO5B (Lapierre et al., 2001), RAB11FIP5 (Ducharme et al., 2007; Su et al., 2010), the Rab11 GTPase activating protein TBC1D9B (Gallo et al., 2014), the lipid microdomain-associated protein MAL (de Marco et al., 2002), and a signaling cascade mediated through the Src family kinase p62YES (Luton et al., 1999; Su et al., 2010).
Much less is known about apical-to-basolateral transcytosis. One example of transport in this direction is the transcytosis of IgG by the Fcγ receptor FcRn, which physiologically carries its cargo in both directions across epithelial barriers (Roopenian and Akilesh, 2007). FcRn trafficking has been shown to depend on some of the same proteins as pIgR, namely, MYO5B, RAB25 (Tzaban et al., 2009), calmodulin (Dickinson et al., 2008), and components of the actin cytoskeleton (Zhou et al., 2015). However, there are also clear differences, as FcRn transcytosis does not require RAB11A (Tzaban et al., 2009), a central organizing molecule for pIgR transcellular transport. Another model of apical-to-basolateral transcytosis by the Wnt receptor in the Drosophila melanogaster wing imaginal disc, required for transepithelial signaling by Wnt, revealed dependence on the E3 ligase Godzilla (Yamazaki et al., 2016). The involvement of other genes in apical-to-basolateral transcytosis has not been tested, leaving a fragmentary view of this complex multistage and clinically relevant pathway.
To gain new insight into receptor-mediated transcytosis, we conducted an RNAi screen for genes that operate in the trafficking of IgG by FcRn. FcRn provides an especially valuable model for studies on apical-to-basolateral membrane transport because its cargo, IgG, can be readily tagged and manipulated and because FcRn moves in both directions across and through each of the specialized endosomes unique to polarized cells, allowing for direct comparison of the two pathways (Dickinson et al., 1999; Claypool et al., 2004; Tzaban et al., 2009). We tested the effect of knocking down 586 genes: 26 in a pilot screen and 560 in a larger follow-up screen. Our results newly implicate many genes in transcytosis. Seven out of eight top-ranking genes were validated in small-scale transport assays, including genes that have not previously been linked to trafficking in polarized epithelia (LEPROT, VPS13C, CCZ1, and ARMT1), one gene with an established role in other aspects of epithelial cell biology (PARD6B), and genes with previous links to transcytosis (EXOC2 and ACTR3B). Studies on EXOC2 and EXOC7 revealed evidence for genetically separable transcellular and recycling pathways. PARD6B was found to act selectively on the trafficking pathways that intersect with the apical membrane only, as basolateral recycling was unchanged by PARD6B knockdown.
Results
Development of a model for receptor-mediated transcytosis compatible with high-throughput screening
We have previously established a cell culture model of FcRn-mediated transcytosis built upon the MDCK cell line stably expressing the two subunits of human FcRn, human β2m and the human FcRn heavy chain, which was tagged with the HA epitope at the N terminus and GFP at the C terminus (Tzaban et al., 2009). To measure transcytosis, cells are grown on transwell semipermeable filters, which provide independent experimental access to both the apical and basolateral sides of the monolayer. Then, IgG is added to one side of the monolayer (at pH 6.0 to facilitate FcRn-dependent binding and uptake), and the amount of IgG to pass through the filter after a period of time is quantified. In preparation for an RNAi screen, several modifications were made.
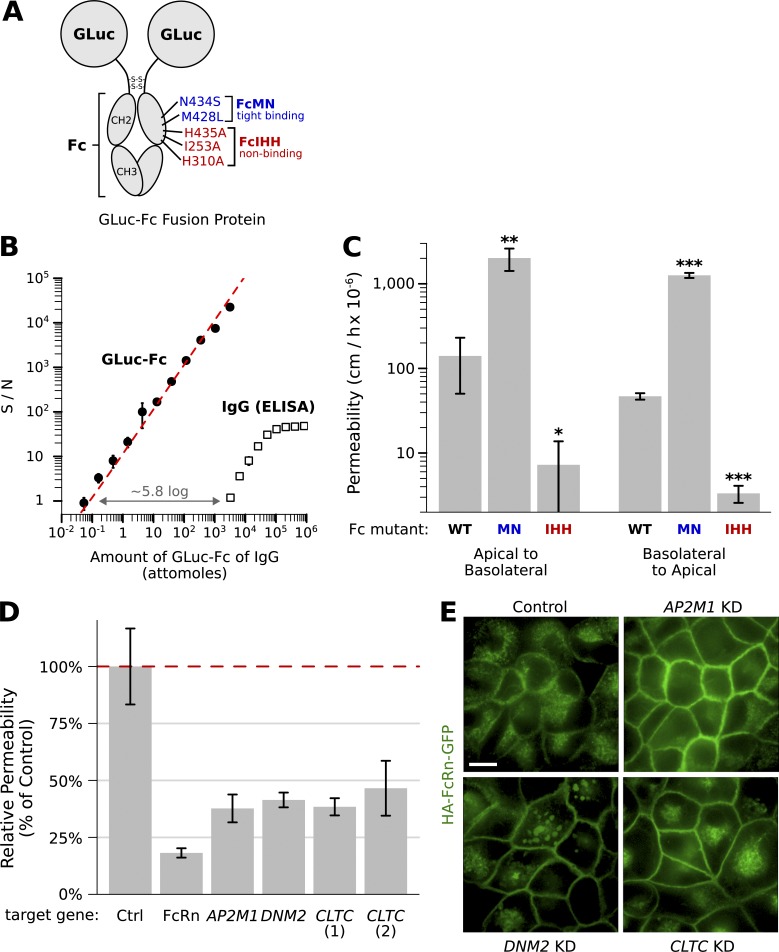
One challenge of the original transcytosis protocol was that the assay used to detect IgG, an ELISA, was not sensitive enough for small-volume (e.g., 96-well) transport studies and furthermore required many binding and washing steps that would be difficult to implement in a high-throughput format. To simplify the assay and increase sensitivity, we constructed a recombinant protein in which the Fc (FcRn-binding) domain of human IgG1 was linked to Gaussia luciferase (Tannous et al., 2005) in place of the variable domains (GLuc-Fc; Fig. 1 A). The Gluc-Fc fusion was produced in CHO cells and purified to >99% homogeneity (Fig. S1 A). The GLuc-Fc fusion can be detected by direct enzymatic assay with approximately five orders of magnitude greater sensitivity and over 2.5 times the dynamic range compared with our original ELISA assay for detecting IgG (Fig. 1 B). To test whether this GLuc-Fc fusion protein crosses MDCK monolayers through an FcRn-mediated pathway, we compared the permeability of GLuc-Fc to that of a similar construct with inactivating mutations in the FcRn-binding domain (Gluc-FcIHH; Fig. 1 A). The FcIHH mutation abolishes binding to FcRn, but not to other immune Fc receptors (Spiekermann et al., 2002; Baker et al., 2011), and thus provides a clean measure of FcRn-independent (i.e., nonspecific) permeability. We found that the wild-type GLuc-FcWT protein was transported in both directions across MDCK monolayers >10 times more efficiently than the nonbinding GLuc-FcIHH mutant (Fig. 1 C), confirming that the luciferase fusion was transported across the monolayer by binding FcRn.
Figure 1.
Validation of a high-throughput model for transcytosis using the IgG trafficking receptor FcRn. (A) Model of the luciferase-Fc fusion protein prepared as ligand for FcRn-dependent trafficking (GLuc-Fc). Point mutations introduced in the Fc-domain that enhanced (blue, GLuc-FcMN) or inhibited (red, GLuc-FcIHH) binding to FcRn are shown. (B) Assay for GLuc-Fc is ∼5.8 logs more sensitive and has a larger dynamic range when compared with our previously established assay for IgG by ELISA (representative titration experiment plotted with n = 2 replicates per concentration). (C) Transcytosis of GLuc-Fc, high-affinity binding GLuc-FcMN, and nonbinding GLuc-FcIHH across MDCK cells expressing HA-tagged FcRn-GFP grown as monolayers on 0.33-cm2 filter supports. Our assay for transcytosis is sensitive enough to measure nonspecific transit of the nonbinding FcRn ligand GLuc-FcIHH. The high-affinity binding GLuc-FcMN crosses the epithelial barrier 20- to 30-fold more efficiently compared with GLuc fused to the wild-type (WT) Fc domain (*, P < 0.05; **, P < 0.01; ***, P < 0.0001; t test adjusted for multiple hypothesis testing with Holm’s method; n = 3). Results were summarized as the mean ± SD of a representative experiment. (D) Results of pilot screen for FcRn-dependent transcytosis. In all cases tested, esiRNA KD of genes required for clathrin-mediated endocytosis inhibited FcRn-dependent transcytosis compared with negative control (cells transfected with nontargeting esiRNA; P < 0.05 for all KDs, t test adjusted for multiple hypothesis testing using Holms method, n = 3). Cells transfected with an esiRNA against FcRn were used as positive controls. (E) Enface confocal images of MDCK FcRn-GFP cell monolayers at steady state after transfection with esiRNA against the indicated genes. In all cases, FcRn-GFP is relocated from the endosomal compartment to the cell surface because of inhibition of clathrin-mediated endocytosis. The morphology of the endosomal compartment is altered in cells lacking dynamin. Bar, 10 μm.
We also took steps to increase the biological signal-to-noise of our transport assays. Several mutations in the FcRn-binding domain of IgG have been identified that bind to FcRn with increased affinity, resulting in enhanced FcRn-dependent trafficking phenotypes in vivo (Zalevsky et al., 2010). We hypothesized that one of these mutations might also lead to more efficient transport in our in vitro cell system. To test this, we introduced two affinity-increasing point mutations into the FcRn-binding region (N434S and M428L, which we call GLuc-FcMN; Fig. 1 A). Indeed, The GLuc-FcMN mutant was transported 20- to 30-fold more efficiently across MDCK monolayers compared with the fusion with wild-type Fc (Fig. 1 C), providing a far greater biological dynamic range. The transcytosis assay using GLuc-FcMN was linear over 3 logs of ligand applied (between 0.01 and 10 µg/ml of GLuc-FcMN; Fig. S1, B and C), indicating that the GLuc-MN fusion does not saturate the receptor under these conditions.
We next established protocols for consistent and efficient gene knockdown (KD) in MDCK cells. We chose to use esiRNAs because they offer a reproducible approach for gene KD in mammalian cell culture (Kittler et al., 2004) and have been shown to produce fewer off-target effects than single, chemically synthesized siRNAs (Kittler et al., 2007). We prepared esiRNAs enzymatically starting from plasmid DNA or a custom MDCK cDNA library (Fig. S1, D and E) and optimized transfection protocols for MDCK cells grown on transwell filters. Using esiRNAs directed against FcRn, we observed an 85% reduction in FcRn transcript levels by quantitative RT-PCR and a corresponding 85% reduction in FcRn-mediated transcytosis (Fig. S1, F and G).
We then adapted our cell culture model to a small-volume 96-well format (0.14-cm2 filter supports). For the 96-well assays, both esiRNA transfections and transport assays were performed with the aid of robotics, allowing tight control over the conditions and timing of each step. In a pilot screen, we observed significant reductions in GLuc-FcMN transepithelial transport by KD of FcRn, as expected, and by KD of genes involved in clathrin-mediated endocytosis (CLTC, DNM2, and AP2M1; Fig. 1 D and Table S1), a pathway previously implicated in FcRn-dependent trafficking (Rodewald and Kraehenbuhl, 1984; Wu and Simister, 2001). When assessed by microscopy, KD for each gene in the clathrin endocytosis pathway redistributed FcRn from its predominantly endosomal location to the cell surface (Fig. 1 E), a phenotype consistent with blockade of endocytosis. These studies validated our experimental system.
Preparing a targeted esiRNA library
We conducted a targeted rather than whole-genome screen for pragmatic reasons, dictated by the cost and feasibility of polarized epithelial monolayer culture in high-throughput formats. To select candidate genes, we adopted a strategy designed to take advantage of known biology without sacrificing the potential to discover unexpected genes and/or gene families. First, we curated a set of 143 genes known to be involved in endosomal trafficking in epithelia (although many of these genes have not yet been tested for a role in transcytosis); we term this the literature-curated gene set (Table S2).
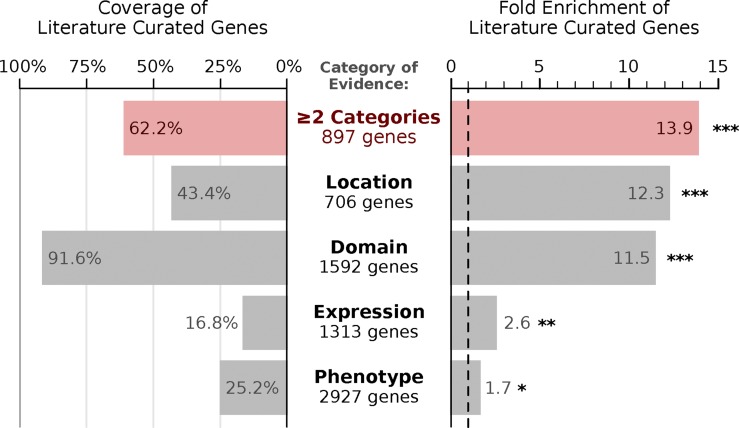
As a second set of candidate genes, we identified genes implicated in epithelial trafficking by one of four indirect lines of evidence: (1) intracellular location, where the gene was localized to the endosomal system by proteomics (Foster et al., 2006; Lapierre et al., 2007; Cao et al., 2008) or by GFP tagging (Huh et al., 2003); (2) protein domain, where the gene contains at least one protein domain implicated in membrane trafficking or endosome dynamics (e.g., Rab GTPase, FYVE-finger, and BAR; Table S2); (3) expression in polarized epithelial cells, where the gene was found to be strongly expressed in simple columnar epithelial cells relative to other cell types (Nelms et al., 2016); and (4) functional phenotype, where the gene was identified in either of two genome-wide RNAi screens for regulators of endocytic trafficking (Balklava et al., 2007; Collinet et al., 2010). Each line of evidence highlighted several established epithelial trafficking genes, and we considered any gene identified in two or more searches to be a potential candidate for our screen (Fig. 2). This uncurated list, termed the high-throughput gene set, was 13.9-fold enriched for literature-defined epithelial trafficking genes (P = 1.5 × 10−84, Fisher’s exact test), containing 89 out of 143 literature-curated genes (62% coverage; Fig. 2). The high-throughput gene set also included many genes that would not have been selected using literature review alone and greatly expanded the capacity for discovery in our targeted screen.
Figure 2.
Selecting candidate genes for an RNAi screen. Evaluation of how effectively different high-throughput categories of evidence recover a literature curated set of 143 epithelial trafficking genes, showing both percent coverage and fold enrichment. *, P < 0.01; **, P < 0.0001; ***, P < 10−10. P-values are estimated using the one-tailed Fisher’s exact test adjusted for multiple hypothesis testing by Holm’s method.
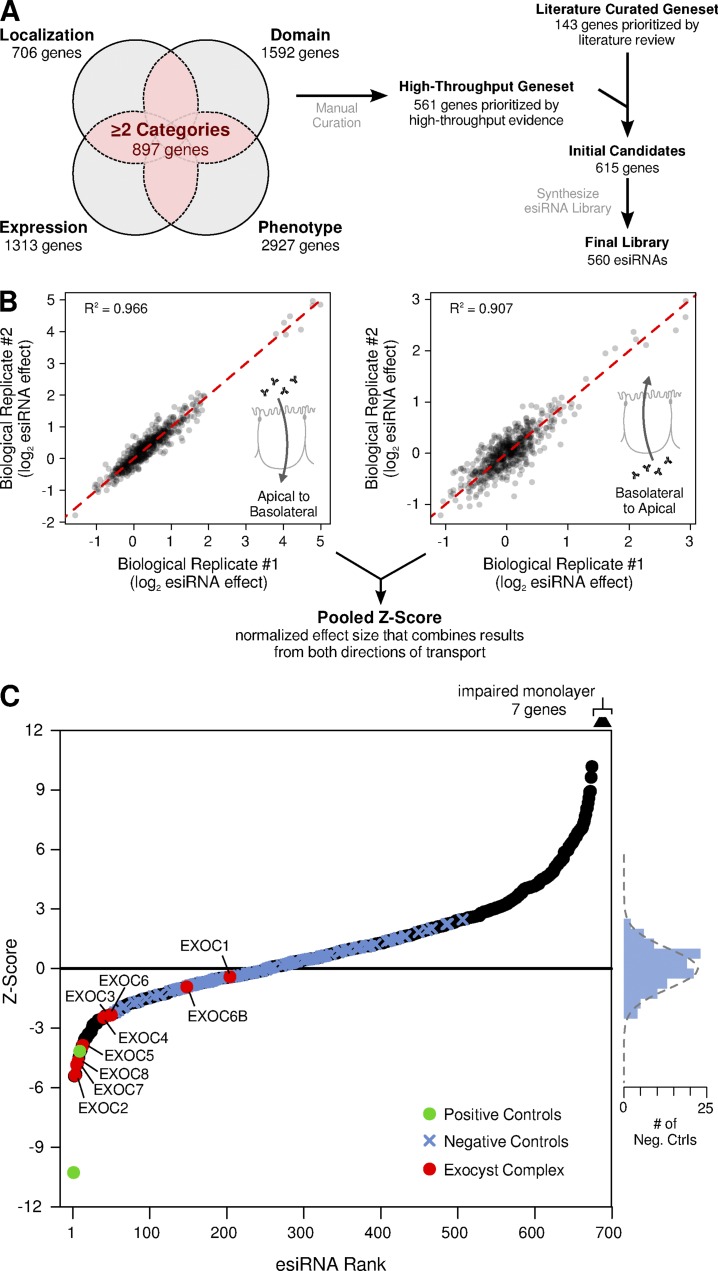
We then manually curated the high-throughput gene set to remove any gene with known function unrelated to regulating membrane trafficking. The two gene sets, high throughput and literature curated, were then combined, resulting in a pooled set of 615 candidate genes (Fig. 3 A and Table S2). An esiRNA library targeting these genes was synthesized using our MDCK cDNA library. In total, synthesis was successful for 560 out of the 615 genes (91%; Fig. 3 A and Table S3); we presume many of the esiRNAs that were not amplified targeted genes not expressed in MDCK cells and were therefore absent from the cDNA library. The final esiRNA library was arrayed on eight 96-well library plates, with 26 controls evenly distributed across each plate (Fig. S2 A).
Figure 3.
Targeted esiRNA screen for genes that regulate FcRn-mediated transcytosis. (A) Schematic of the strategy for candidate gene selection and esiRNA library synthesis. (B) Values of transcytosis (log2 esiRNA effect) for two out of three biological replicates for each esiRNA, plotted for both directions of transcytosis. The biological replicates displayed a high degree of reproducibility, with an R2 of 0.907 for basolateral-to-apical transport and 0.966 for apical-to-basolateral transport. Replicate values of transcytosis for each esiRNA and direction of transport were then pooled together and converted into an overall Z-score. (C) Ordered plot of the final pooled Z-scores. Positive controls (esiRNA against FcRn) are plotted in GREEN. Subunits of the exocyst complex are plotted in red. Negative controls are plotted as blue crosses, and their frequency of distribution is plotted to the right. The dotted gray line on the right shows the expected distribution of negative controls given the standard normal distribution.
A targeted esiRNA screen yields reproducible modifiers of FcRn-mediated transcytosis
The full esiRNA library was screened over a period of three sessions. The raw data displayed substantial systematic variability caused by batch and edge effects of the multiwell format. Several aspects of our experimental design made it possible to control for this systematic variability and identify reproducible hits. First, we included a large number of control wells distributed across each plate (Fig. S2 A); this layout was designed to fit statistician's recommendations for high-throughput screens (Zhang, 2008). Second, we performed the screen with substantial replication (Fig. S2 B): every library plate was screened in triplicate in both transport directions, and GLuc-FcMN concentration was measured in duplicate for every well. The raw data were normalized using a linear mixed-effects model (Yu et al., 2011), resulting in a substantial decrease in variability (Fig. S3). Normalized biological replicates displayed a high degree of reproducibility, with an R2 of 0.907 for basolateral-to-apical transport and 0.966 for apical-to-basolateral transport (Fig. 3 B). Replicate values for each esiRNA were then log-transformed, pooled, and converted into a Z-score, which reflects the number of standard deviations of the phenotype (GLuc-FcMN transport) relative to the negative (mock transfection) controls.
When we compared Z-scores between the two directions of transport, we discovered that the effect of any esiRNA on one direction of transport was always directly proportional to its effect on the other direction (Fig. S4, A and B). This can be explained by the biology of FcRn and the conditions of our transport assays. First, GLuc-FcMN permeability is proportional to the flux of FcRn across the cell; this is caused by several features of FcRn, such as the finding that FcRn trafficking is not dramatically altered by binding to its ligand (Tzaban et al., 2009). Second, because (a) our assays were performed after chronic gene KD and (b) FcRn biosynthesis and degradation are slow relative to recycling and transcytosis, any perturbation in FcRn trafficking will lead to a new steady state where the flux of receptor in both directions across the cell is essentially equal. For example, if an esiRNA KD specifically inhibits basolateral endocytosis, FcRn will accumulate at the basolateral surface until the overall rate of FcRn flux in both directions across the cell is again balanced. Therefore, any perturbation on FcRn trafficking will manifest as an overall increase or decrease in FcRn-mediated transport, regardless of the molecular step affected or direction of Gluc-FcMn transcytosis. We thus pooled together the Z-scores in both directions, resulting in a single pooled Z-score that reflects the chance that an esiRNA affects transcellular FcRn flux.
An ordered plot of pooled Z-scores is shown in Fig. 3 C. The 112 negative control wells closely fit a standard normal distribution (Fig. 3 C, blue crosses and histogram on the right; P = 0.498, Shapiro–Wilk normality test). In contrast, many esiRNAs produced a phenotype far more extreme than the negative controls. For instance, esiRNAs targeting 7 out of 9 genes encoding subunits of the exocyst complex (red circles in Fig. 3 C) resulted in a decrease in transport larger than all negative controls. The exocyst is a large, multimeric protein complex that tethers vesicles to the membrane; it has previously been linked to related membrane trafficking pathways in epithelia (Heider and Munson, 2012), including in basolateral-to-apical transcytosis of pIgR (Oztan et al., 2007), and the identification of the majority of subunits of this complex supports the validity of our results. Furthermore, our top hit, ACTR3B is a component of the ARP2/3 complex recently implicated in IgG trafficking across the intestine in mice (Zhou et al., 2015). All esiRNAs that resulted in a Z-score less than −3 (23 “hits”) are shown in Table 1 and the entire list is shown in Table S4. This Z-score cutoff corresponds to a false discovery rate (FDR) of 0.03, signifying that only approximately one false positive is expected among these 23 genes by chance.
Table 1. Gene KD resulting in decreased transport phenotype.
| Gene Rank | Gene name | Z-score | Reason for inclusion in screen |
|---|---|---|---|
| 1a | ACTR3B | −5.41 | Literature |
| 2a | EXOC2 | −5.38 | Literature |
| 3a | ARL14 | −5.33 | Expression, domain |
| 4 | EXOC7 | −4.85 | Literature |
| 5a | PARD6B | −4.78 | Expression, phenotype |
| 6 | EXOC8 | −4.64 | Literature |
| 7a | VPS13C | −4.47 | Localization, domain, phenotype |
| 8a | CCZ1 | −4.15 | Localization, domain |
| 9 | GNAS | −4.08 | Localization, domain, phenotype |
| 10a | LEPROT | −3.89 | Localization, domain |
| 11 | EXOC5 | −3.87 | Literature |
| 12 | STAM | −3.75 | Localization, domain |
| 13 | DYNC1H1 | −3.58 | Localization, domain, phenotype |
| 14 | DYNC1LI1 | −3.52 | Phenotype, domain |
| 15a | ARMT1 | −3.51 | Expression, phenotype |
| 16 | STX19 | −3.48 | Localization, domain |
| 17 | LRRK2 | −3.37 | Phenotype, domain |
| 18 | RAB10 | −3.30 | Localization, domain, phenotype |
| 19 | VPS26A | −3.29 | Localization, domain |
| 20 | SNX27 | −3.22 | Localization, domain |
| 21 | TRIP11 | −3.19 | Phenotype, domain |
| 22 | TMED2 | −3.16 | Localization, domain |
| 23 | ROCK1 | −3.01 | Phenotype, domain |
Genes selected for secondary validation.
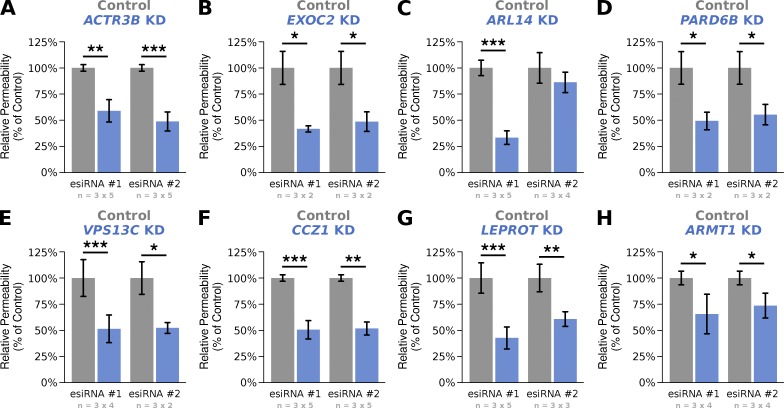
Eight of the top-scoring genes that caused a decrease in GLuc-FcMN transport (Table 1) were selected for validation in transport assays using monolayers grown on larger 0.33-cm2 filter supports. We repeated all validation assays on multiple independent days (to assess day-to-day variability), and with independent esiRNAs that target a distinct region of each gene's transcript (to rule out potential off-target effects). Treatment with all eight original esiRNAs led to a significant reduction in GLuc-FcMN permeability that was reproducible between days (Fig. 4), confirming the phenotype from our screen. In addition, the phenotype was validated for seven out of eight genes using independent esiRNAs, suggesting that these phenotypes are unlikely to be caused by off-target effects. The validated genes include both genes with an established role in epithelial membrane trafficking (e.g., EXOC2 and ACTR3B) and genes that have not been linked to epithelial trafficking before (e.g., LEPROT and ARMT1). Thus, this screen identified several genes previously unknown to be necessary for receptor-mediated transcytosis across polarized epithelia.
Figure 4.
Validation of eight genes selected from the top 15 ranked hits. (A–H) Transcytosis of Gluc-FcMN across MDCK monoloayers transfected with two independent esiRNAs against the indicated genes (blue columns). All experiments were normalized to the value of matched control monolayers transfected with nontargeting esiRNA (gray columns). Each experiment was conducted in triplicate on two to five independent days; the number of replicates and days are indicated below each bar graph in the form of n = (replicates per experiment) × (number of independent experiments). Bar graphs are reported as the weighted mean across experiments (see Materials and methods). Statistical significance was estimated using the Wilcoxon signed-rank test. *, P < 0.05; **, P < 0.001; ***, P < 0.0001.
KD of EXOC2/7, LEPROT, and PARD6B affect diverse steps of the transcytotic and recycling pathways
Trafficking pathways in polarized epithelia involve separate apical and basolateral early endosomal pathways and endosome routes connecting both cell surfaces with each other and with other intracellular organelles. A defect in any of these routes could result in disruption of FcRn-mediated transcytosis. To determine which trafficking steps were disrupted during our screen, we selected four of the top-ranked genes for further characterization. These include two subunits of the exocyst complex (EXOC2 and EXOC7), the polarity regulator PARD6B, and the tetra-spanning membrane protein LEPROT.
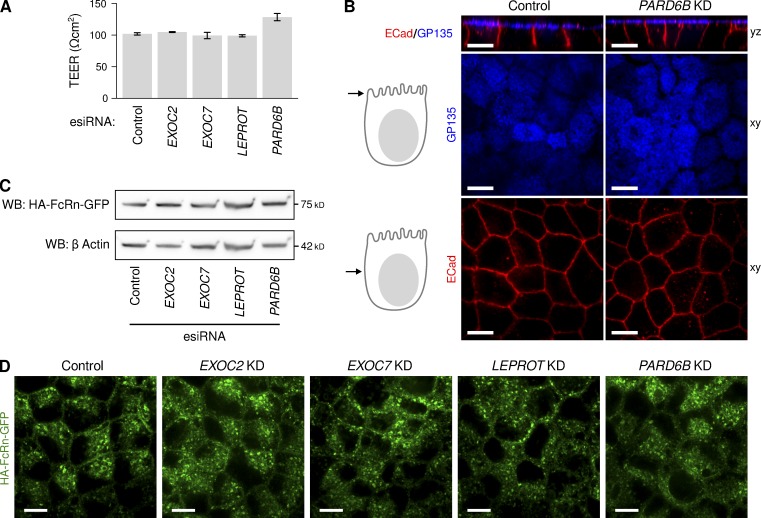
First, we looked for possible indirect effects on membrane polarity or monolayer integrity after knocking down these genes. In each of the four cases tested, the cells assembled into tight epithelial monolayers as assessed by transepithelial electrical resistance (TEER), with PARD6B KD producing a slight increase in TEER and all other genes exhibiting no change (Fig. 5 A). There were no visible defects in monolayer integrity or cell morphology when the cells were imaged by confocal microscopy (Fig. 5 B and Fig. S5), and the membrane polarity markers podocalyxin/GP135 (apical membrane) and E-cadherin (lateral membrane) were normally localized to the appropriate plasma membrane domain (Figs. 5 B and S5 A). We suspect that none of these four genes had a defect in monolayer integrity because they were selected from genes whose KD caused a decrease in GLuc-FcMN permeability. Most gene KDs that disrupt the monolayer in any way are likely to result in an increase in permeability for GLuc-FcMN, and so the genes whose KD led to an increase in permeability during our screen may contain a much higher proportion of genes that affect tight junction integrity.
Figure 5.
Characterization of MDCK-FcRn monolayers after depletion of EXOC2, EXOC7, PARD6B, and LEPROT. (A) Transepithelial electrical resistance, a measure of tight junction function, after esiRNA depletion of the indicated genes (n = 6–10 filters per esiRNA). (B) Immunolocalization of the apical membrane marker GP135 (blue) and lateral membrane marker E-cadherin (red) after esiRNA depletion of the polarity determinant gene PARD6B. Immunolocalization for the same markers are shown for monolayers depleted in EXOC2, EXOC7, and LEPROT in Fig. 4. (C) SDS-PAGE of total cell lysates and immunoblot for the HA-tagged FcRn-EGFP after esiRNA depletion of the indicated genes. Immunoblot for actin on same gel provides control for protein loading. Although the bands for FcRn and corresponding bands for actin have similar morphology, they are distinguished by antibody labeling and running at different positions in same gel. (D) Localization of HA-tagged FcRn-EGFP after esiRNA depletion of the indicated genes by direct confocal microscopy. Additional confocal sections for each gene are shown in Fig. S5. Bars, 10 μm.
We next assessed changes in FcRn expression or localization after gene suppression. Genes that affect the biosynthetic delivery of FcRn could indirectly affect transcytosis, but such phenotypes should be readily distinguished by a reduction in FcRn expression, relocalization to a biosynthetic organelle such as the ER, or both. The total cellular content of FcRn was unaltered by any of the four esiRNAs as determined by Western blot against HA-FcRn-EGFP (Fig. 5 C) and direct epifluorescence microscopy (Figs. 5 D and S5 B). This finding rules out gross biosynthetic defects or mis-sorting of FcRn to the lysosome as causes of the transcytosis phenotype. We also found no change in FcRn localization away from the endosomal compartment or alteration of FcRn-containing endosome morphology (Figs. 5 D and S5 B). This is in contrast to the dramatic shift in localization of FcRn from endosome to plasma membrane after blockade of clathrin-mediated endocytosis (Fig. 1 E), suggesting that EXOC2, EXOC7, LEPROT, and PARD6B all act primarily in steps distinct from endocytosis.
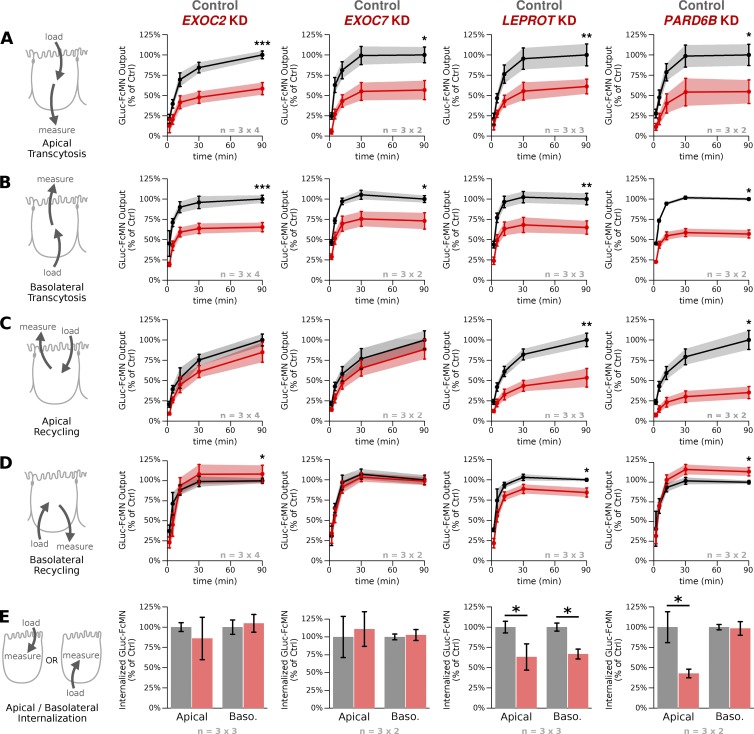
Having found no overt effects on cell monolayer health or FcRn localization, we sought to determine which trafficking pathways were affected by KD of each gene. To this end, we turned to pulse-chase transport assays, where it is possible to measure not only the amount of transepithelial transport (transcytosis), but also the amount of recycling back to the same surface and the amount of cargo internalization. Cell monolayers were loaded with GLuc-FcMN from either the apical or basolateral side for 60 min, washed thoroughly at 4°C, and then either returned to 37°C to measure the time course of apical or basolateral GLuc-FcMN output (recycling or transcytosis) or lysed in detergent to measure GLuc-FcMN internalization.
In the pulse-chase assays, we observed less transcytosis in both directions for all four gene KDs (Fig. 6, A and B), similar to what was observed in the steady-state transport experiments during the screen and subsequent validation (Table 1 and Fig. 4). Unlike transcytosis, however, recycling and endocytic uptake were affected differently by knocking down each gene, revealing three distinct trafficking phenotypes (Fig. 6, C–E). In the first phenotype, shared by both subunits of the exocyst complex (EXOC2 and EXOC7), there was very little change in apical or basolateral recycling or in the amount of GLuc-FcMN internalization from either surface. Thus, exocyst KD appears to specifically affect trafficking into the transcytotic pathway while being dispensable for recycling and internalization. A different phenotype was observed after LEPROT KD, in which recycling and internalization were both inhibited from either surface. This is unlikely to be explained by a biosynthetic or endocytic defect, because FcRn is normally expressed and does not accumulate at the cell surface (Fig. 5 and Fig. S5). Our interpretation is that LEPROT is necessary for a key postendocytic trafficking step (or steps), consistent with the reported effects of LEPROT on leptin receptor trafficking in nonpolarized cells (Séron et al., 2011). Finally, KD of PARD6B had opposing effects on recycling and internalization from each cell surface: whereas apical recycling and cargo uptake were strongly inhibited, basolateral recycling was enhanced and basolateral uptake was unaffected (Fig. 6), suggesting that PARD6B impinges on trafficking pathways particularly relevant to the apical cell surface.
Figure 6.
Pulse-chase transport assays in MDCK monolayers depleted of EXOC2, EXOC7, LEPROT, and PARD6B reveal three distinct phenotypes. FcRn-dependent IgG trafficking in monolayers depleted of the indicated genes (red) is compared with IgG trafficking in control monolayers transfected with nontargeting esiRNA (black). Transcytosis and recycling experiments were normalized to the 90-min time point of matched control monolayers transfected with nontargeting esiRNA, and internalization experiments were normalized to the matched nontargeting control. Each experiment was conducted in triplicate on two to four independent days; the number of replicates and days are indicated below each bar graph in the form of n = (replicates per experiment) × (number of independent experiments). Plots show the weighted mean ± SD across experiments (see Materials and methods). (A and B) Time course of basolaterally and apically directed IgG transcytosis as assessed by 60-min pulse and 90-min chase. (C and D) Time course of apical and basolateral IgG recycling as assessed by 60-min pulse and 90-min chase. For A–D, statistical significance was assessed for the 90-min time point by the Wilcoxon signed-rank test and adjusted for multiple hypothesis testing using Holm’s method. (E) Total cellular uptake of IgG after 60-min pulse. *, P < 0.05; as estimated by the Wilcoxon signed-rank test. **, P < 0.001; ***, P < 0.0001.
Discussion
One major challenge in the field of endosome trafficking is to understand how the general rules of vesicular transport learned from yeast and cultured nonpolarized cells can be adapted to the specialized functions of differentiated cell types. Here, we describe the most comprehensive assessment to date of genes associated with a specialized endosomal transport pathway in polarized epithelial cells. Screens for genes operating in trafficking across polarized mammalian epithelial cells have not been reported before, and several challenges discussed in the Results section were uniquely overcome to make this study successful. Our results identified numerous cases in which the KD of specific genes affects bidirectional transcytosis of IgG by the Fcγ receptor FcRn. We further observed several distinct phenotypic classes, indicating that these genes are likely to operate in diverse stages of the transcytotic pathway and that the apical and basolateral recycling and transcytosis pathways are genetically separable. These results may apply to other constitutively polarized cell types, such as neurons or endothelial cells, which share some molecular mechanisms (Bonifacino, 2014).
An immediate finding from our screen was a clear requirement for the exocyst complex in FcRn-mediated transcytosis. The exocyst is a large, octomeric protein complex that mediates exocytosis of some Golgi- and endosome-derived vesicles at specified regions of the plasma membrane (Heider and Munson, 2012). The complex can act at both apical and basolateral cell surfaces of polarized epithelia and is necessary for apically directed transcytosis of dimeric IgA by pIgR (Oztan et al., 2007). In our pulse-chase studies on EXOC2 and EXOC7, we found that both subunits led to a decrease in bidirectional GLuc-FcMN transcytosis without any significant effect on recycling or uptake from either surface. It is striking that there was very little effect on FcRn recycling after exocyst KD, as many other receptors require the exocyst for this process, including the transferrin receptor (TfR; Prigent et al., 2003) and pIgR (Oztan et al., 2007). One explanation for this difference may be that FcRn follows a different intracellular itinerary. For instance, though there is very strong intracellular colocalization between TfR and FcRn (Ober et al., 2004; Goebl et al., 2008; Tzaban et al., 2009; Jerdeva et al., 2010), recycling by both receptors have a distinct dependence on several genes: TfR recycling in polarized epithelia requires exocysts (Prigent et al., 2003), but not RAB11A (Wang et al., 2000b; Tzaban et al., 2009) and not LEPROT in HeLa cells (Séron et al., 2011). FcRn recycling, on the other hand, does not require exocysts (this study) but requires RAB11A (Ward et al., 2005; Tzaban et al., 2009) and LEPROT (this study). This result emphasizes that the apparent effect of any perturbation of trafficking machinery can be dependent on the specific receptor being studied and provides further evidence that trafficking pathways in epithelial cells are heterogeneous with multiple possible routes back to the cell surface (Goldenring, 2015).
We observed a polarized phenotype for PARD6B KD, with a strong defect on apical uptake and recycling but little effect on, or even an enhancement of, basolateral recycling. PARD6B encodes for a member of a protein family required for the establishment and maintenance of epithelial cell polarity (Chen and Zhang, 2013); it acts as a scaffolding protein in complex with Par3 and atypical protein kinase C to specify the apical membrane domain (Martin-Belmonte and Mostov, 2008). Although the phenotype we observed for PARD6B KD could in principle be an indirect effect caused by a defect in cell polarity, we observed no change in the polarity of two canonical apical and basolateral membrane markers, cell morphology, or tight junction formation. There is mounting evidence that Par6 family members can regulate membrane trafficking independent of their role in cell polarity (Balklava et al., 2007; Shivas et al., 2010), raising the possibility that the PARD6B KD phenotype could be a result of a direct role in endosome function. For instance, expressing a dominant negative Par6b in HeLa cells inhibited TfR internalization and major histocompatibility complex (MHC) class I recycling (Balklava et al., 2007), even though this cell line exhibits no constitutive polarity. Furthermore, Par6c has been shown to directly interact with and regulate trafficking genes, including the exocyst complex (Das et al., 2014).
In mammals, there are three isoforms of Par6 (PARD6A, PARD6B, and PARD6G), and it is unclear to what extent these operate differently in establishing cell polarity or regulating membrane trafficking. PARD6B was the only isoform included in our screen, and it was included because it was predicted to be strongly expressed in polarized epithelia (Nelms et al., 2016). Indeed, antibody staining from the human protein atlas (http://www.proteinatlas.org/; Uhlén et al., 2015) showed that PARD6B was strongly enriched in simple epithelia, whereas the other two Par6 isoforms were not. Moving forward, it will be important to assess any differences in the function of these three isoforms. The epithelial-enriched expression pattern of PARD6B suggests that it may be vital for a function unique to polarized epithelial cells. Given its phenotype in FcRn trafficking, we speculate that PARD6B may contribute to endosome specialization in polarized epithelia, perhaps by helping to establish or regulate an apical membrane-specific endosome compartment.
Other genes identified in this screen have not been studied in the context of polarized epithelial cells before. LEPROT encodes a four-pass transmembrane protein that regulates a postendocytic step of LDL and growth hormone receptor trafficking (Séron et al., 2011). In nonpolarized cells, the requirement for LEPROT is receptor specific, as epidermal growth factor receptor (EGFR) and TfR are unaffected by LEPROT KD (Séron et al., 2011). LEPROT KD resulted in a general slowdown of all FcRn trafficking pathways, suggesting a defect in a rate-limiting step of FcRn transport. Another gene, ARMT1, was recently shown to function in the DNA damage response by negatively regulating PCNA through enzymatic methyltransferase activity (Perry et al., 2015). It is plausible that ARMT1 KD affected FcRn-mediated transcytosis indirectly by altering gene expression, but very little is known about this protein, including its intracellular localization and potential binding partners, and so it may have a direct function in endosomal trafficking distinct from its role in DNA damage.
Our screen implicates many other genes in the process of transcytosis, all with FDRs less than 0.01. Of the validated genes, a few are worthy of further note. ARL14 (Z-score = −5.33) is a small GTPase that recruits MYO1E to MHC class II–containing vesicles to affect movement along the actin cytoskeleton in dendritic cells (Paul et al., 2011). The guanine nucleotide exchange factor for ARL14 is PSD4. PSD4 was not identified in our screen but the PI kinase that regulates its membrane association PIP5K1A scored highly (Z-score = −2.52, FDR < 0.1). The screen also identified the actin-related protein ACTR3B, a component of the ARP2/3 complex (Z-score = −5.41). Both results are consistent with the dependency of endosome trafficking on actin dynamics (Sheff et al., 2002; Guerriero et al., 2006; Salvarezza et al., 2009), and it will be interesting to identify where in the transcytotic pathway ARL14 and ACTR3B operate. Three other genes identified (VPS13C, STAM, and CCZ1) point to sorting steps associated with the transition from early to late endosomes. VPS13C (Z-score = −4.47) encodes a large protein expressed in multiple cell types and of essentially unknown function (Velayos-Baeza et al., 2004). CCZ1 (Z-score = −4.15) assembles with the protein encoded by Mon1 to act as a Rab7 guanine nucleotide exchange factor (Nordmann et al., 2010; Gerondopoulos et al., 2012) and with RABX5/Rab5 to effect the early-to-late endosome transition (Poteryaev et al., 2010). STAM1 (Z-score = −3.75) is a subunit of the ESCRT-0 complex, which also acts at early steps in the early-to-late endosome transition to effect protein sorting (Bache et al., 2003; Ren and Hurley, 2010). One aspect of this transition involves the sorting of selected cargo away from the lysosome, a characteristic feature of trafficking for FcRn (Roopenian and Akilesh, 2007).
We present several resources that will enable research into epithelial trafficking: we identified a large number of genes whose KD affects FcRn-mediated transcytosis and also established more sensitive assays to measure both continuous and pulse-chase FcRn-mediated transport. It should be emphasized that this screen does not provide a complete picture of the genes involved in transcytosis; several may have been missed because of insufficient KD or redundancy with other cellular proteins (e.g., many Rab proteins have isoforms with partially redundant functions). Genes that were not targeted may also play interesting roles in the pathway. In addition, because our cell model was built using the human FcRn in a canine cell line, it is possible that this species difference affected our results; the available data, however, are consistent with the receptor following a conserved intracellular itinerary among epithelial cells from different species where FcRn actively transports cargo.
Of the newly implicated genes we have characterized so far, we find distinct phenotypes on endosomal trafficking, suggesting that these genes operate in diverse stages of the transcytotic pathway. The next steps will be to determine which trafficking events are affected by each gene KD, whether the effects are direct or indirect, and how these genes operate together to maintain the specialized endosomal system of epithelial cells. These results serve as the foundation for future work on the genetic and molecular basis of transcytosis, and advance our understanding of how epithelial cells traffic IgG, sort membrane proteins, and maintain polarity to function at the interface between host and environment.
Materials and methods
Cell culture
An MDCKII cell line stably expressing human β2M and HA-FcRn-EGFP has been previously described (Tzaban et al., 2009). In brief, monomeric EGFP was fused to the FcRn C terminus with a GSSGSS linker between FcRn and EGFP (pcDNA3.1; Invitrogen). The FcRn-EGFP fusion was transfected into MDCKII cells stably expressing human β2M (Claypool et al., 2002). This cell line was sorted by FACS to isolate cells with relatively uniform GFP fluorescence, gating for cells that express detectable but low levels of GFP. The cell line was frozen down after sorting and used within 3 to 15 passages after thawing for all experiments. Cells were maintained in DMEM supplemented with 10% heat-inactivated FBS and penicillin and streptomycin, all obtained from Thermo Fisher Scientific. Cells were routinely confirmed to be negative for mycoplasma using a PCR kit from MD Biosciences.
GLuc-FcMN production and purification
Humanized Gaussia luciferase (Tannous et al., 2005) was cloned into the pcDNA3.1 backbone vector with G418/neomycin resistance (Thermo Fisher Scientific), with an AGGV linker and the Fc domain of human IgG1 on the N terminus and the signal sequence of mouse MHC class I H2-Kb (Platzer and Fiebiger, 2010) on the C terminus. All fusion-protein plasmids were verified by Sanger sequencing. Each fusion protein construct was transfected into CHO cells. These cell lines were placed under selection using G418 and then subcloned to obtain stable cell lines with strong expression of each GLuc-Fc variant. The stable cell lines were grown on Cytodex3 microcarrier beads (GE Healthcare) in Techne spinner flasks for up to 9 d, and media was collected every day for protein purification. Media for protein production was OptiMEM GlutiMAX (Thermo Fisher Scientific) supplemented with 2% ultra-low IgG FBS (Thermo Fisher Scientific), 1 mM CaCl2 (Sigma-Aldrich), nonessential amino acids (Thermo Fisher Scientific), and penicillin and streptomycin (Thermo Fisher Scientific).
Collected media was filtered through a 0.2-µm filter to remove cell debris and incubated with protein A or protein G Sepharose beads (GE) overnight at 4°C. Then the beads were strained and protein was eluted with 100 mM glycine, pH 2.5. Eluted protein was quenched with 5.5% 1 M Tris, pH 9.0, and buffer exchanged for PBS using 30 kD MWCO protein spin concentrators (Thermo Fisher Scientific). Protein concentration was then measured using a NanoDrop (Thermo Fisher Scientific) assuming a default protein mass extinction coefficient at 280 nm of 1 mg ml−1 cm−1. Purified protein was flash frozen and stored at −80°C. After every protein purification, an aliquot was run on an SDS-PAGE Coomassie gel to ensure a single clean band (e.g., Fig. S1 A).
Steady-state transport assays
Cells were transfected at the same time of plating (reverse transfection) using Lipofectomine RNAiMAX (Thermo Fisher Scientific). For the transfections, lipid-esiRNA complexes were prepared by incubating 2 ng/µl esiRNA in OptiMEM (Thermo Fisher Scientific) with 1.5% by volume RNAiMAX for 15 to 30 min. During the incubation period, 34.6 µl lipid-esiRNA complex was added to the apical reservoir of a 6.5 mm polycarbonate transwell plate with 0.4 µm pore size (Corning). Meanwhile, MDCK-FcRn cells were trypsinized and resuspended to a density of 1.2 × 105 cells/ml in DMEM with 10% FBS and without penicillin and streptomycin, and 138.5 µl of this cell solution was added to the apical reservoirs of the transwell plate with lipid-esiRNA complexes. It was important to ensure that the OptiMEM was prewarmed to room temperature and the DMEM was prewarmed to 37°C for a successful transfection. The cells with complexes were then incubated for 16 to 20 h at 37°C without media in the basolateral reservoir. For the follow up experiments (Figs. 4, 5, and 6), a second transfection was subsequently performed to ensure more robust gene KD; in this second transfection, lipid–esiRNA complexes were prepared as for the first transfection and then diluted with DMEM with 10% FBS and added to both the apical and basolateral chambers for 4 to 8 h. After transfection, the media was exchanged for prewarmed DMEM with 10% FBS and penicillin and streptomycin. Media was changed again 3 d after plating, and transport assays were performed the next day.
For the transport assays, media was exchanged for DMEM with 10% ultra-low IgG FBS (Thermo Fisher Scientific), 1 µg/ml GLuc-Fc, and 20 mM MES, pH 6.0, on the input side and DMEM with 10% ultra-low IgG FBS, pH 7.4, on the output side. The filters were incubated for 90 min at 37°C, and then contents on the output side of the filter were gently mixed and 20-µl aliquots withdrawn for protein concentration determination. To measure the concentration of GLuc-Fc, 80 µl of PBS with 12.5 µM coelenterazine (NanoLight Technology) was added to 20 µl media in a white 96-well plate (Thermo Fisher Scientific) and luciferase signal was measured 3 to 10 min later using a Victor X3 plate reader (PerkinElmer).
During the screen, cells were grown on 96-well high-throughput screen transwell plates with polycarbonate membranes and 0.4 µm pore size (Corning). Media volumes were 75 µl and 235 µl for the apical and basolateral chambers, respectively. All assays were performed as described in the previous paragraph with the exception of reduced volumes and the use of automation. Media additions and removal were performed with either a Matrix WellMate (Thermo Fisher Scientific) or an Agilent Bravo Workstation. Luminescence activity was measured with an Envision 2 plate reader (PerkinElmer).
esiRNA synthesis and library construction
esiRNAs are prepared enzymatically as follows (Fig. S1 D). First, primers were designed to amplify a 400- to 600-bp region of a target gene from a cDNA library, appending T7 promoter sites to each end. This target region was optimized for good predicted KD efficiency using the DEQOR algorithm (Henschel et al., 2004). Second, the PCR product was transcribed in vitro to produce long double-stranded RNA. Finally, the double-stranded RNA was spliced into 21-bp fragments by a bacterial RNase, creating a heterogeneous pool of siRNAs. The resulting esiRNA reagent contains a diverse pool of sequences targeting the same gene, which has been experimentally shown to produce more consistent KD and less off target effects than single, chemically synthesized siRNAs (Kittler et al., 2007).
The DNA template for control esiRNAs targeting luciferase (negative control) and FcRn-GFP (positive control) were synthesized from plasmids encoding Gluc-FcWT (this study) and HA-FcRn-GFP (Tzaban et al., 2009), respectively. DNA templates for all other esiRNAs were synthesized from a cDNA library prepared from our MDCK-FcRn cell line (described in the following paragraph). Primers for DNA template synthesis of all esiRNAs, which include a gene-specific region and a T7 promoter site for RNA synthesis, are listed in Table S3. For small-scale experiments (control RNAs in Fig. 1, 4, 5, and 6 and all second esiRNAs in Fig. 4), esiRNAs were synthesized using the New England Biolabs, Inc. T7 HiScribe and SuperScript III kits according to the manufacturer’s instructions. For the pilot and full screen, esiRNAs were synthesized by Eupheria Biotech essentially as described in the previous paragraph except that the MEGAscript kit (Ambion) was used for in vitro transcription and a recombinant Escherichia coli Rnase III tagged with glutathione-S-transferase (Kittler et al., 2007) was used for enzymatic digestion. Primers for DNA template synthesis of all esiRNAs, which include a gene-specific region and a T7 promoter site for RNA synthesis, are listed in Table S3.
The MDCK cDNA library was constructed using a previously described template-switching protocol (Pinto and Lindblad, 2010). In brief, MDCK-FcRn cells were grown on 10-cm-diameter transwell inserts for 4 d with a media change on day 3. Then RNA was purified using the QIAGEN Rneasy mini kit according to the manufacturer’s instructions, and poly(A)+ RNA was enriched with oligo(dT) dynabeads (Thermo Fisher Scientific). Purified RNA was reversed transcribed at 50°C for 30 min, 55°C for 15 min, and 60°C for 15 min using the Superscript III enzyme (Thermo Fisher Scientific) and the oligo(dT) primer 5′-AAGCAGTGGTATCAACGCAGAGTAC(T)30VN. Then, to facilitate the addition of a predetermined sequence at the 3′ end of the cDNA (5′ end of the template RNA) by template switching, MnCl2 was added to a final concentration of 2 mM along with Superscript II reverse transcription and the primer 5′-AAGCAGTGGTATCAACGCAGAGTACGCRGRGRG-3′ (phosphate), and the reaction was incubated at 42°C for 90 min. This protocol leaves an identical priming site on both ends of the cDNA (5′-AAGCAGTGGTATCAACGCAGAGT-3′) that can be used to amplify the library by PCR. The cDNA synthesis resulted in substantial library diversity, with ∼2 × 109 total cDNA molecules as estimated by dilution PCR and 15 out of 20 sequenced clones representing full-length transcripts. Finally, the library was amplified by emulsion PCR using the Miscellula emulsion PCR kit (CHIMERx) and the PCR primer 5′-AAGCAGTGGTATCAACGCAGAGT-3′.
Normalization and hit selection for RNAi screen
To analyze the data from the screen, a strategy was developed to assess esiRNA activity at the same time as normalization, estimating the consistent effect among all esiRNA replicates that cannot be explained by known systematic sources of variation. The method was adapted from (Yu et al., 2011). Let Xeфi be the log-transformed luciferase intensity of well i, treated with esiRNA e, and measured at location ф (ф = {b,p,r,c} for batch b, plate p, row r, and column c). We fit the measured intensities Xeфi to a mixed-effects linear model with fixed esiRNA effects μe, random systematic effects Sф, and residual error εi: Xeфi = μe + Sф + εi, where systematic effects are the sum of batch effects Bb, plate effects Pp, row effects Rpr, and column effects Cpc: Sф = Bb + Pp + Rpr + Cpc. This model was fit separately for each direction of transport using the lme4 R package (Bates et al., 2015). Normalized signal intensities, which have been corrected for systematic errors (Xeфi − Sф), are plotted in Fig. S3.
To assess statistical significance of each esiRNA effect μe, a permutation test was used. A single mock transfection well on the library plate was labeled as a gene named MOCK, and then the mixed linear model fit was repeated, obtaining an estimate of the “esiRNA” effect for a known negative control, μMOCK. This was then repeated for the remaining mock transfection wells (112 in total), providing an empirical null distribution under the mock transfection condition. The null distribution, μMOCK, fits a normal distribution for both directions of transport (P = 0.82, 0.40 for basolateral-to-apical and apical-to-basolateral transport, Shapiro–Wilk test).
A Z-score was then calculated for each gene and direction of transport by dividing the estimated esiRNA effect, μe, by the standard deviation of the empirical null distribution (i.e., the standard deviation of μMOCK). Z-scores for each direction were then pooled using Stouffer's method:
resulting in a single overall Z-score for each esiRNA in the screen.
General cell health characterization
For the general cell health characterization (Fig. 5), MDCK-FcRn cells were subjected to the double-transfection protocol described in the Steady-state transport assays section and then grown for 4 d on transwell filters. For microscopy, cells were fixed using 4% paraformaldehyde (Electron Microscopy Sciences) and then stained and imaged as described previously (Tzaban et al., 2009) using a mouse mAb against GP135 (Ojakian and Schwimmer, 1988) and a rat mAb against E-cadherin (U3254; Sigma-Aldrich). Western blot for HA-FcRn-GFP was performed using a high-affinity rat anti-HA mAb (clone 3F10; Roche) and for β-actin using a mouse mAb (A5316; Sigma-Aldrich). HRP-conjugated secondary antibodies were obtained from Sigma-Aldrich. TEER was measured using an EVOM2 (World Precision Instruments).
Pulse chase assays
For the pulse chase transport assays, MDCK-FcRn cells were subjected to the double-transfection protocol described in the Steady-state transport assays section and grown for 4 d on transwell filters. To load the cells with GLuc-FcMN, media was exchanged for DMEM with 10% ultra-low IgG FBS, 10 µg/ml GLuc-FcMN, and 20 mM MES, pH 6.0, on the input side, and DMEM with 10% ultra-low IgG FBS, pH 7.4 on the output side. The filters were then incubated for 60 min at 37°C and washed three times at 4°C using prechilled DMEM with 10% ultra-low IgG FBS, pH 7.4. At this point, the filters were either returned to 37°C and 20-µl aliquots were withdrawn after gentle mixing for recycling and transcytosis assays or they were lysed using 100 µl PBS with 1% NP-40, 1% FBS, and Complete protease inhibitor cocktail (Thermo Fisher Scientific) for uptake assays. Luciferase activity was measured as described in the Steady-state transport assays section and converted to protein concentration using a linear least squares fit to a standard curve.
Microscope and image acquisition
Confocal images were acquired at 37°C using a spinning disk confocal head (CSU-X1; PerkinElmer) coupled to a fully motorized inverted Axiovert 200-M microscope (ZEISS) equipped with Plan Apochromat; 63×/1.4 oil DIC objective and with a X-Cite 120LED light source (Excelitas) for wide-field illumination. Solid-state lasers (480, 568, and 660 nm) coupled to the spinning head through a fiber optic were used as a light source. An acoustic-optical tunable filter was used to switch between different wavelengths. The imaging system operates under control of SlideBook 6 (Intelligent Imaging Innovations, Inc.) and includes a computer-controlled spherical aberration correction device (SAC; Intelligent Imaging Innovations, Inc.) installed between the objective lens and the charge-coupled device camera (QuantEM:512SC; Photometrics). Postacquisition images were scaled linearly using ImageJ. For immunofluorescence, MDCK monolayers were fixed in 4% paraformaldehyde, washed, and stained for HA-FcRn-EGFP, E-cadherin, or GP135 as described in General cell health characterization. Secondary anti–mouse Fab2 labeled with Alexa Fluor 647 and anti–rat Fab2 labeled with Alexa Fluor 568 antibodies from Thermo Fisher Scientific were used.
Statistics and analysis
For Figs. 1 and 5, results were summarized as the mean ± SD of a representative experiment. Statistical significance was estimated using a Student’s t test adjusted for multiple hypothesis testing with Holm's method. For Figs. 4 and 6, data were pooled from multiple independent experiments. Raw luminescence values from each experiment were normalized by dividing by the mean value of the nontargeting control wells from that experiment (the 90-min time point of the nontargeting control wells for the pulse-chase assay). Results were then summarized as the weighted mean ± the weighted standard deviation between experiments, with weights given by 1 over the variance of the nontargeting control wells in each experiment. The reason for including weights is that a few experiments resulted in a large standard deviation for the normalizing negative control condition, and the weighted mean allowed for a smooth decrease in the value of these noisy experiments without having to assess or remove outliers. No experiments or data points were excluded from the analysis. Statistical significance was estimated using a Wilcoxon signed-rank test and adjusted for multiple-hypothesis testing with Holm’s method.
Online supplemental material
Fig. S1 shows SDS-PAGE of purified recombinant Gluc-Fc fusion proteins, dose dependency of Gluc-FcMN transport, schematic for preparation of esiRNAs and validation of final products, and validation of esiRNA KD for FcRn. Fig. S2 shows the plate layout and replication schema for the high-throughput screen. Fig. S3 shows plots of normalized and unnormalized luciferase measurements from the screen. Fig. S4 shows the comparison of apical to basolateral and basolateral to apical transport. Fig. S5 shows the immunolocalization of apical (GP135) and basolateral (E-cadherin) membrane markers and the localization of HA-tagged FcRn-EGFP after esiRNA depletion of EXOC2, EXOC7, and LEPROT. Table S1 shows the relative permeabilities for all genes used in the pilot screen. Table S2 shows the final list of genes curated for the screen, the list of genes curated in the high-throughput and literature gene sets, and the list of genes curated based on their containing a domain linked to trafficking. Table S3 shows the list of genes and sequences used to prepare esiRNA for the full and pilot screens and the second esiRNAs prepared for the validation studies. Table S4 shows the entire list of ranked Z-scores for each gene studied.
Supplementary Material
Acknowledgments
We thank the staff at the Institute of Chemistry and Cell Biology-Longwood, especially Sean Johnston, for their assistance with establishing and conducting the robotics assays during the screen. We thank Keiko Maeda for helping to reconfirm specificity of Gluc-Fc binding to FcRn and Ramiro Massol, Andrew Weflen, Phi Loung, Marian Neutra, and Daniela Garcia-Castillo for helpful discussions and critical reading of the manuscript.
This project was supported by a National Science Foundation Graduate Research Fellowship to B. Nelms, National Institutes of Health grants R01 DK084424 and R37 DK048106, and a National Institute of Diabetes and Digestive and Kidney Diseases Harvard Digestive Disease Center grant from the National Institutes of Health (P30 DK034854) to W. Lencer.
The authors declare no competing financial interests.
Author contributions: B. Nelms and W. Lencer conceived and designed the study. B. Nelms and N.F. Dalomba acquired data. B. Nelms and W. Lencer analyzed and interpreted data. B. Nelms and W. Lencer drafted and revised the manuscript, with input from N.F. Dalomba.
Footnotes
Abbreviations used:
- dIgA
- dimeric IgA
- esiRNA
- endoribonuclease-prepared short interfering RNA
- FDR
- false discovery rate
- KD
- knockdown
- MHC
- major histocompatibility complex
- pIgR
- polymeric immunoglobulin receptor
- TEER
- transepithelial electrical resistance
- TfR
- transferrin receptor
References
- Apodaca G., Enrich C., and Mostov K.E.. 1994. The calmodulin antagonist, W-13, alters transcytosis, recycling, and the morphology of the endocytic pathway in Madin-Darby canine kidney cells. J. Biol. Chem. 269:19005–19013. [PubMed] [Google Scholar]
- Apodaca G., Gallo L.I., and Bryant D.M.. 2012. Role of membrane traffic in the generation of epithelial cell asymmetry. Nat. Cell Biol. 14:1235–1243. 10.1038/ncb2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache K.G., Raiborg C., Mehlum A., and Stenmark H.. 2003. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278:12513–12521. 10.1074/jbc.M210843200 [DOI] [PubMed] [Google Scholar]
- Baker K., Qiao S.W., Kuo T.T., Aveson V.G., Platzer B., Andersen J.T., Sandlie I., Chen Z., de Haar C., Lencer W.I., et al. 2011. Neonatal Fc receptor for IgG (FcRn) regulates cross-presentation of IgG immune complexes by CD8-CD11b+ dendritic cells. Proc. Natl. Acad. Sci. USA. 108:9927–9932. 10.1073/pnas.1019037108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balklava Z., Pant S., Fares H., and Grant B.D.. 2007. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 9:1066–1073. 10.1038/ncb1627 [DOI] [PubMed] [Google Scholar]
- Barroso M., and Sztul E.S.. 1994. Basolateral to apical transcytosis in polarized cells is indirect and involves BFA and trimeric G protein sensitive passage through the apical endosome. J. Cell Biol. 124:83–100. 10.1083/jcb.124.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Maechler M., Bolker B.M., and Walker S.C.. 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67:1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Bomsel M. 1997. Transcytosis of infectious human immunodeficiency virus across a tight human epithelial cell line barrier. Nat. Med. 3:42–47. 10.1038/nm0197-42 [DOI] [PubMed] [Google Scholar]
- Bomsel M., Prydz K., Parton R.G., Gruenberg J., and Simons K.. 1989. Endocytosis in filter-grown Madin-Darby canine kidney cells. J. Cell Biol. 109:3243–3258. 10.1083/jcb.109.6.3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomsel M., Parton R., Kuznetsov S.A., Schroer T.A., and Gruenberg J.. 1990. Microtubule- and motor-dependent fusion in vitro between apical and basolateral endocytic vesicles from MDCK cells. Cell. 62:719–731. 10.1016/0092-8674(90)90117-W [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S. 2014. Adaptor proteins involved in polarized sorting. J. Cell Biol. 204:7–17. 10.1083/jcb.201310021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Li C., Higginbotham J.N., Franklin J.L., Tabb D.L., Graves-Deal R., Hill S., Cheek K., Jerome W.G., Lapierre L.A., et al. 2008. Use of fluorescence-activated vesicle sorting for isolation of Naked2-associated, basolaterally targeted exocytic vesicles for proteomics analysis. Mol. Cell. Proteomics. 7:1651–1667. 10.1074/mcp.M700155-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., and Zhang M.. 2013. The Par3/Par6/aPKC complex and epithelial cell polarity. Exp. Cell Res. 319:1357–1364. 10.1016/j.yexcr.2013.03.021 [DOI] [PubMed] [Google Scholar]
- Claypool S.M., Dickinson B.L., Yoshida M., Lencer W.I., and Blumberg R.S.. 2002. Functional reconstitution of human FcRn in Madin-Darby canine kidney cells requires co-expressed human beta 2-microglobulin. J. Biol. Chem. 277:28038–28050. 10.1074/jbc.M202367200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool S.M., Dickinson B.L., Wagner J.S., Johansen F.E., Venu N., Borawski J.A., Lencer W.I., and Blumberg R.S.. 2004. Bidirectional transepithelial IgG transport by a strongly polarized basolateral membrane Fcgamma-receptor. Mol. Biol. Cell. 15:1746–1759. 10.1091/mbc.E03-11-0832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinet C., Stöter M., Bradshaw C.R., Samusik N., Rink J.C., Kenski D., Habermann B., Buchholz F., Henschel R., Mueller M.S., et al. 2010. Systems survey of endocytosis by multiparametric image analysis. Nature. 464:243–249. 10.1038/nature08779 [DOI] [PubMed] [Google Scholar]
- Couesnon A., Pereira Y., and Popoff M.R.. 2008. Receptor-mediated transcytosis of botulinum neurotoxin A through intestinal cell monolayers. Cell. Microbiol. 10:375–387. 10.1111/j.1462-5822.2007.01051.x [DOI] [PubMed] [Google Scholar]
- Das A., Gajendra S., Falenta K., Oudin M.J., Peschard P., Feng S., Wu B., Marshall C.J., Doherty P., Guo W., and Lalli G.. 2014. RalA promotes a direct exocyst-Par6 interaction to regulate polarity in neuronal development. J. Cell Sci. 127:686–699. 10.1242/jcs.145037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Marco M.C., Martín-Belmonte F., Kremer L., Albar J.P., Correas I., Vaerman J.P., Marazuela M., Byrne J.A., and Alonso M.A.. 2002. MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells. J. Cell Biol. 159:37–44. 10.1083/jcb.200206033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson B.L., Badizadegan K., Wu Z., Ahouse J.C., Zhu X., Simister N.E., Blumberg R.S., and Lencer W.I.. 1999. Bidirectional FcRn-dependent IgG transport in a polarized human intestinal epithelial cell line. J. Clin. Invest. 104:903–911. 10.1172/JCI6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson B.L., Claypool S.M., D’Angelo J.A., Aiken M.L., Venu N., Yen E.H., Wagner J.S., Borawski J.A., Pierce A.T., Hershberg R., et al. 2008. Ca2+-dependent calmodulin binding to FcRn affects immunoglobulin G transport in the transcytotic pathway. Mol. Biol. Cell. 19:414–423. 10.1091/mbc.E07-07-0658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducharme N.A., Williams J.A., Oztan A., Apodaca G., Lapierre L.A., and Goldenring J.R.. 2007. Rab11-FIP2 regulates differentiable steps in transcytosis. Am. J. Physiol. Cell Physiol. 293:C1059–C1072. 10.1152/ajpcell.00078.2007 [DOI] [PubMed] [Google Scholar]
- Foster L.J., de Hoog C.L., Zhang Y., Zhang Y., Xie X., Mootha V.K., and Mann M.. 2006. A mammalian organelle map by protein correlation profiling. Cell. 125:187–199. 10.1016/j.cell.2006.03.022 [DOI] [PubMed] [Google Scholar]
- Gallo L.I., Liao Y., Ruiz W.G., Clayton D.R., Li M., Liu Y.J., Jiang Y., Fukuda M., Apodaca G., and Yin X.M.. 2014. TBC1D9B functions as a GTPase-activating protein for Rab11a in polarized MDCK cells. Mol. Biol. Cell. 25:3779–3797. 10.1091/mbc.E13-10-0604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondopoulos A., Langemeyer L., Liang J.R., Linford A., and Barr F.A.. 2012. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr. Biol. 22:2135–2139. 10.1016/j.cub.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl N.A., Babbey C.M., Datta-Mannan A., Witcher D.R., Wroblewski V.J., and Dunn K.W.. 2008. Neonatal Fc receptor mediates internalization of Fc in transfected human endothelial cells. Mol. Biol. Cell. 19:5490–5505. 10.1091/mbc.E07-02-0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenring J.R. 2015. Recycling endosomes. Curr. Opin. Cell Biol. 35:117–122. 10.1016/j.ceb.2015.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero C.J., Weixel K.M., Bruns J.R., and Weisz O.A.. 2006. Phosphatidylinositol 5-kinase stimulates apical biosynthetic delivery via an Arp2/3-dependent mechanism. J. Biol. Chem. 281:15376–15384. 10.1074/jbc.M601239200 [DOI] [PubMed] [Google Scholar]
- Heider M.R., and Munson M.. 2012. Exorcising the exocyst complex. Traffic. 13:898–907. 10.1111/j.1600-0854.2012.01353.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henschel A., Buchholz F., and Habermann B.. 2004. DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res. 32:W113–W120. 10.1093/nar/gkh408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughson E.J., and Hopkins C.R.. 1990. Endocytic pathways in polarized Caco-2 cells: identification of an endosomal compartment accessible from both apical and basolateral surfaces. J. Cell Biol. 110:337–348. 10.1083/jcb.110.2.337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., and O’Shea E.K.. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Hunziker W., and Peters P.J.. 1998. Rab17 localizes to recycling endosomes and regulates receptor-mediated transcytosis in epithelial cells. J. Biol. Chem. 273:15734–15741. 10.1074/jbc.273.25.15734 [DOI] [PubMed] [Google Scholar]
- Jerdeva G.V., Tesar D.B., Huey-Tubman K.E., Ladinsky M.S., Fraser S.E., and Bjorkman P.J.. 2010. Comparison of FcRn- and pIgR-mediated transport in MDCK cells by fluorescence confocal microscopy. Traffic. 11:1205–1220. 10.1111/j.1600-0854.2010.01083.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R., Putz G., Pelletier L., Poser I., Heninger A.K., Drechsel D., Fischer S., Konstantinova I., Habermann B., Grabner H., et al. 2004. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 432:1036–1040. 10.1038/nature03159 [DOI] [PubMed] [Google Scholar]
- Kittler R., Surendranath V., Heninger A.K., Slabicki M., Theis M., Putz G., Franke K., Caldarelli A., Grabner H., Kozak K., et al. 2007. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat. Methods. 4:337–344. 10.1038/nmeth1025 [DOI] [PubMed] [Google Scholar]
- Lapierre L.A., Kumar R., Hales C.M., Navarre J., Bhartur S.G., Burnette J.O., Provance D.W. Jr., Mercer J.A., Bähler M., and Goldenring J.R.. 2001. Myosin vb is associated with plasma membrane recycling systems. Mol. Biol. Cell. 12:1843–1857. 10.1091/mbc.12.6.1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre L.A., Avant K.M., Caldwell C.M., Ham A.J., Hill S., Williams J.A., Smolka A.J., and Goldenring J.R.. 2007. Characterization of immunoisolated human gastric parietal cells tubulovesicles: identification of regulators of apical recycling. Am. J. Physiol. Gastrointest. Liver Physiol. 292:G1249–G1262. 10.1152/ajpgi.00505.2006 [DOI] [PubMed] [Google Scholar]
- Lencer W.I., Moe S., Rufo P.A., and Madara J.L.. 1995. Transcytosis of cholera toxin subunits across model human intestinal epithelia. Proc. Natl. Acad. Sci. USA. 92:10094–10098. 10.1073/pnas.92.22.10094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Palaniyandi S., Zeng R., Tuo W., Roopenian D.C., and Zhu X.. 2011. Transfer of IgG in the female genital tract by MHC class I-related neonatal Fc receptor (FcRn) confers protective immunity to vaginal infection. Proc. Natl. Acad. Sci. USA. 108:4388–4393. 10.1073/pnas.1012861108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luton F., Vergés M., Vaerman J.P., Sudol M., and Mostov K.E.. 1999. The SRC family protein tyrosine kinase p62yes controls polymeric IgA transcytosis in vivo. Mol. Cell. 4:627–632. 10.1016/S1097-2765(00)80213-0 [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F., and Mostov K.. 2008. Regulation of cell polarity during epithelial morphogenesis. Curr. Opin. Cell Biol. 20:227–234. 10.1016/j.ceb.2008.01.001 [DOI] [PubMed] [Google Scholar]
- Nelms B.D., Waldron L., Barrera L.A., Weflen A.W., Goettel J.A., Guo G., Montgomery R.K., Neutra M.R., Breault D.T., Snapper S.B., et al. 2016. CellMapper: rapid and accurate inference of gene expression in difficult-to-isolate cell types. Genome Biol. 17:201 10.1186/s13059-016-1062-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., and Ungermann C.. 2010. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20:1654–1659. 10.1016/j.cub.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Ober R.J., Martinez C., Vaccaro C., Zhou J., and Ward E.S.. 2004. Visualizing the site and dynamics of IgG salvage by the MHC class I-related receptor, FcRn. J. Immunol. 172:2021–2029. 10.4049/jimmunol.172.4.2021 [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Pearse A., Domingo D., Trowbridge I.S., and Hopkins C.R.. 1996. Apical and basolateral endosomes of MDCK cells are interconnected and contain a polarized sorting mechanism. J. Cell Biol. 135:139–152. 10.1083/jcb.135.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojakian G.K., and Schwimmer R.. 1988. The polarized distribution of an apical cell surface glycoprotein is maintained by interactions with the cytoskeleton of Madin-Darby canine kidney cells. J. Cell Biol. 107:2377–2387. 10.1083/jcb.107.6.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oztan A., Silvis M., Weisz O.A., Bradbury N.A., Hsu S.C., Goldenring J.R., Yeaman C., and Apodaca G.. 2007. Exocyst requirement for endocytic traffic directed toward the apical and basolateral poles of polarized MDCK cells. Mol. Biol. Cell. 18:3978–3992. 10.1091/mbc.E07-02-0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R.G., Prydz K., Bomsel M., Simons K., and Griffiths G.. 1989. Meeting of the apical and basolateral endocytic pathways of the Madin-Darby canine kidney cell in late endosomes. J. Cell Biol. 109:3259–3272. 10.1083/jcb.109.6.3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P., van den Hoorn T., Jongsma M.L., Bakker M.J., Hengeveld R., Janssen L., Cresswell P., Egan D.A., van Ham M., Ten Brinke A., et al. 2011. A Genome-wide multidimensional RNAi screen reveals pathways controlling MHC class II antigen presentation. Cell. 145:268–283. 10.1016/j.cell.2011.03.023 [DOI] [PubMed] [Google Scholar]
- Perry J.J., Ballard G.D., Albert A.E., Dobrolecki L.E., Malkas L.H., and Hoelz D.J.. 2015. Human C6orf211 encodes Armt1, a protein carboxyl methyltransferase that targets PCNA and is linked to the DNA damage response. Cell Reports. 10:1288–1296. 10.1016/j.celrep.2015.01.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto F.L., and Lindblad P.. 2010. A guide for in-house design of template-switch-based 5′ rapid amplification of cDNA ends systems. Anal. Biochem. 397:227–232. 10.1016/j.ab.2009.10.022 [DOI] [PubMed] [Google Scholar]
- Platzer B., and Fiebiger E.. 2010. The signal peptide of the IgE receptor alpha-chain prevents surface expression of an immunoreceptor tyrosine-based activation motif-free receptor pool. J. Biol. Chem. 285:15314–15323. 10.1074/jbc.M110.104281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D., Datta S., Ackema K., Zerial M., and Spang A.. 2010. Identification of the switch in early-to-late endosome transition. Cell. 141:497–508. 10.1016/j.cell.2010.03.011 [DOI] [PubMed] [Google Scholar]
- Prigent M., Dubois T., Raposo G., Derrien V., Tenza D., Rossé C., Camonis J., and Chavrier P.. 2003. ARF6 controls post-endocytic recycling through its downstream exocyst complex effector. J. Cell Biol. 163:1111–1121. 10.1083/jcb.200305029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath T., Baker K., Pyzik M., and Blumberg R.S.. 2015. Regulation of immune responses by the neonatal fc receptor and its therapeutic implications. Front. Immunol. 5:664 10.3389/fimmu.2014.00664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., and Hurley J.H.. 2010. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 29:1045–1054. 10.1038/emboj.2010.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodewald R., and Kraehenbuhl J.P.. 1984. Receptor-mediated transport of IgG. J. Cell Biol. 99:159s–164s. 10.1083/jcb.99.1.159s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Kreitzer G., and Müsch A.. 2005. Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell Biol. 6:233–247. 10.1038/nrm1593 [DOI] [PubMed] [Google Scholar]
- Rojas R., and Apodaca G.. 2002. Immunoglobulin transport across polarized epithelial cells. Nat. Rev. Mol. Cell Biol. 3:944–955. 10.1038/nrm972 [DOI] [PubMed] [Google Scholar]
- Roopenian D.C., and Akilesh S.. 2007. FcRn: The neonatal Fc receptor comes of age. Nat. Rev. Immunol. 7:715–725. 10.1038/nri2155 [DOI] [PubMed] [Google Scholar]
- Salvarezza S.B., Deborde S., Schreiner R., Campagne F., Kessels M.M., Qualmann B., Caceres A., Kreitzer G., and Rodriguez-Boulan E.. 2009. LIM kinase 1 and cofilin regulate actin filament population required for dynamin-dependent apical carrier fission from the trans-Golgi network. Mol. Biol. Cell. 20:438–451. 10.1091/mbc.E08-08-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séron K., Couturier C., Belouzard S., Bacart J., Monté D., Corset L., Bocquet O., Dam J., Vauthier V., Lecœur C., et al. 2011. Endospanins regulate a postinternalization step of the leptin receptor endocytic pathway. J. Biol. Chem. 286:17968–17981. 10.1074/jbc.M111.224857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff D.R., Kroschewski R., and Mellman I.. 2002. Actin dependence of polarized receptor recycling in Madin-Darby canine kidney cell endosomes. Mol. Biol. Cell. 13:262–275. 10.1091/mbc.01-07-0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivas J.M., Morrison H.A., Bilder D., and Skop A.R.. 2010. Polarity and endocytosis: Reciprocal regulation. Trends Cell Biol. 20:445–452. 10.1016/j.tcb.2010.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiekermann G.M., Finn P.W., Ward E.S., Dumont J., Dickinson B.L., Blumberg R.S., and Lencer W.I.. 2002. Receptor-mediated immunoglobulin G transport across mucosal barriers in adult life: Functional expression of FcRn in the mammalian lung. J. Exp. Med. 196:303–310. (published erratum appears in J. Exp. Med. 2003. 197:1601) 10.1084/jem.20020400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T., Bryant D.M., Luton F., Vergés M., Ulrich S.M., Hansen K.C., Datta A., Eastburn D.J., Burlingame A.L., Shokat K.M., and Mostov K.E.. 2010. A kinase cascade leading to Rab11-FIP5 controls transcytosis of the polymeric immunoglobulin receptor. Nat. Cell Biol. 12:1143–1153. 10.1038/ncb2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous B.A., Kim D.E., Fernandez J.L., Weissleder R., and Breakefield X.O.. 2005. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 11:435–443. 10.1016/j.ymthe.2004.10.016 [DOI] [PubMed] [Google Scholar]
- Tuma P., and Hubbard A.L.. 2003. Transcytosis: Crossing cellular barriers. Physiol. Rev. 83:871–932. 10.1152/physrev.00001.2003 [DOI] [PubMed] [Google Scholar]
- Tzaban S., Massol R.H., Yen E., Hamman W., Frank S.R., Lapierre L.A., Hansen S.H., Goldenring J.R., Blumberg R.S., and Lencer W.I.. 2009. The recycling and transcytotic pathways for IgG transport by FcRn are distinct and display an inherent polarity. J. Cell Biol. 185:673–684. 10.1083/jcb.200809122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. 2015. Proteomics. Tissue-based map of the human proteome. Science. 347:1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- van IJzendoorn S.C., Tuvim M.J., Weimbs T., Dickey B.F., and Mostov K.E.. 2002. Direct interaction between Rab3b and the polymeric immunoglobulin receptor controls ligand-stimulated transcytosis in epithelial cells. Dev. Cell. 2:219–228. 10.1016/S1534-5807(02)00115-6 [DOI] [PubMed] [Google Scholar]
- Velayos-Baeza A., Vettori A., Copley R.R., Dobson-Stone C., and Monaco A.P.. 2004. Analysis of the human VPS13 gene family. Genomics. 84:536–549. 10.1016/j.ygeno.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Vergés M., Luton F., Gruber C., Tiemann F., Reinders L.G., Huang L., Burlingame A.L., Haft C.R., and Mostov K.E.. 2004. The mammalian retromer regulates transcytosis of the polymeric immunoglobulin receptor. Nat. Cell Biol. 6:763–769. 10.1038/ncb1153 [DOI] [PubMed] [Google Scholar]
- Vergés M., Sebastián I., and Mostov K.E.. 2007. Phosphoinositide 3-kinase regulates the role of retromer in transcytosis of the polymeric immunoglobulin receptor. Exp. Cell Res. 313:707–718. 10.1016/j.yexcr.2006.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Brown P.S., Aroeti B., Chapin S.J., Mostov K.E., and Dunn K.W.. 2000a Apical and basolateral endocytic pathways of MDCK cells meet in acidic common endosomes distinct from a nearly-neutral apical recycling endosome. Traffic. 1:480–493. 10.1034/j.1600-0854.2000.010606.x [DOI] [PubMed] [Google Scholar]
- Wang X., Kumar R., Navarre J., Casanova J.E., and Goldenring J.R.. 2000b Regulation of vesicle trafficking in madin-darby canine kidney cells by Rab11a and Rab25. J. Biol. Chem. 275:29138–29146. 10.1074/jbc.M004410200 [DOI] [PubMed] [Google Scholar]
- Ward E.S., Martinez C., Vaccaro C., Zhou J., Tang Q., and Ober R.J.. 2005. From sorting endosomes to exocytosis: Association of Rab4 and Rab11 GTPases with the Fc receptor, FcRn, during recycling. Mol. Biol. Cell. 16:2028–2038. 10.1091/mbc.E04-08-0735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., and Simister N.E.. 2001. Tryptophan- and dileucine-based endocytosis signals in the neonatal Fc receptor. J. Biol. Chem. 276:5240–5247. 10.1074/jbc.M006684200 [DOI] [PubMed] [Google Scholar]
- Yamazaki Y., Palmer L., Alexandre C., Kakugawa S., Beckett K., Gaugue I., Palmer R.H., and Vincent J.P.. 2016. Godzilla-dependent transcytosis promotes Wingless signalling in Drosophila wing imaginal discs. Nat. Cell Biol. 18:451–457. 10.1038/ncb3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Danku J., Baxter I., Kim S., Vatamaniuk O.K., Salt D.E., and Vitek O.. 2011. Noise reduction in genome-wide perturbation screens using linear mixed-effect models. Bioinformatics. 27:2173–2180. 10.1093/bioinformatics/btr359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J., Chamberlain A.K., Horton H.M., Karki S., Leung I.W., Sproule T.J., Lazar G.A., Roopenian D.C., and Desjarlais J.R.. 2010. Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 28:157–159. 10.1038/nbt.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.D. 2008. Novel analytic criteria and effective plate designs for quality control in genome-scale RNAi screens. J. Biomol. Screen. 13:363–377. 10.1177/1087057108317062 [DOI] [PubMed] [Google Scholar]
- Zhou K., Sumigray K.D., and Lechler T.. 2015. The Arp2/3 complex has essential roles in vesicle trafficking and transcytosis in the mammalian small intestine. Mol. Biol. Cell. 26:1995–2004. 10.1091/mbc.E14-10-1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.