van Vugt previews work by Qu et al. linking the metabolic enzyme PGAM1 to the pool of nucleotides needed for proper DNA repair in cancer cells.
Abstract
Phosphoglycerate mutase 1 (PGAM1) functions in glycolysis. In this issue, Qu et al. (2017. J. Cell Biol. https://doi.org/10.1083/jcb.201607008) show that PGAM1 inactivation leads to nucleotide depletion, which causes defective homologous recombination–mediated DNA repair, suggesting that targeting metabolic enzymes increases cancer cell susceptibility to DNA damaging agents.
Deregulated metabolism is a hallmark of cancer. Most normal differentiated cells only display low glycolysis rates, of which the resulting pyruvate is used to produce ATP through aerobic phosphorylation. In contrast, tumor cells have highly increased rates of glycolysis, after which the resulting pyruvate is converted to lactate at the expense of efficient energy production. This rewired metabolism is likely dictated by the elevated requirements of cancer cells and proliferating cells in general for the production of biomass, including fatty acids, amino acids, and nucleotides (Vander Heiden et al., 2009). For biosynthesis of nucleotides, an essential intermediate is NADPH, which is produced predominantly by the pentose phosphate pathway (PPP). In this pathway, glucose-6-phosphate is converted into ribose-5-phosphate, the sugar backbone of nucleotides. In line with the elevated requirement for anabolic metabolism, glycolytic enzymes, including those functioning in the PPP, are found to be up-regulated in various cancers (Durany et al., 2000). Within the glycolytic pathway, the phosphoglycerate mutase enzyme (PGAM, also called PGM), catalyzes the conversion of 3-phosphoglycerate (3PG) to 2PG (Fig. 1 A). Two isoforms of PGAM have been reported: PGAM1 and PGAM2. Both isoforms have similar catalytic activity and exist in hetero- and homodimers (Mikawa et al., 2014). The substrate of PGAM, 3PG, inhibits the enzymatic activity of the PPP component 6-phosphogluconate dehydrogenase. As a consequence, by converting 3PG into 2PG, PGAM functions to stimulate PPP pathway flux and to ensure biosynthesis. This pivotal role in coordinating glycolysis with biosynthesis makes PGAM an attractive cancer therapeutic target, as its inhibition may interfere with essential needs of cancer cells. Indeed, chemical or genetic inhibition of PGAM1 leads to reduced rates of oxidative PPP, declined biosynthesis, and was shown to inhibit tumor growth in a xenograft tumor model (Hitosugi et al., 2012).
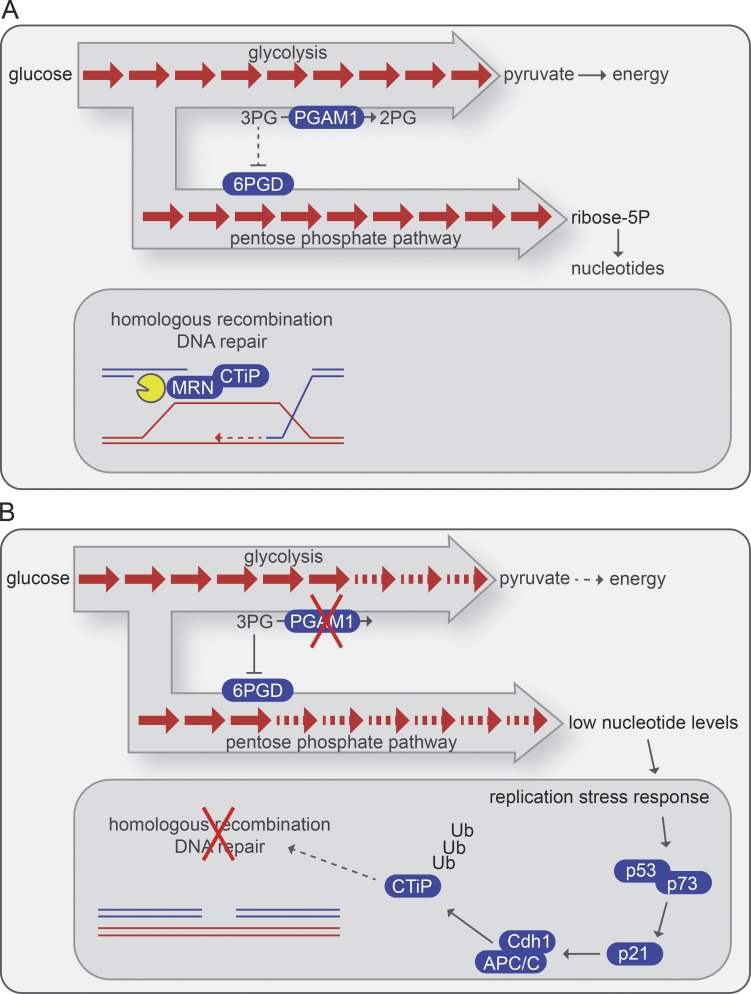
Figure 1.
PGAM1 inhibition blocks repair of DNA breaks through HR. (A) PGAM1 functions in glycolysis by converting 3PG into 2PG. A parallel branch of the glycolysis pathway—the PPP—is used to create biomass, including nucleotides. Under normal circumstances, DNA double-strand breaks are repaired through HR. A critical first step in HR is Mre11–Rad50–Nbs1 (MRN)/CtIP–mediated DNA end resection. (B) When PGAM1 is inactivated, its substrate 3PG accumulates and inhibits 6-phosphogluconate dehydrogenase (6PGD) in the PPP, leading to nucleotide pool depletion. The resulting replication stress response leads to p53/p73-dependent up-regulation of p21 and subsequent activation of the APC/C-Cdh1 E3 ubiquitin ligase. This leads to ubiquitylation and degradation of CtIP, which precludes efficient HR DNA repair.
The nucleotides that are produced as as a result of PPP flux are required for DNA replication in proliferating cells. DNA replication, especially in cancer cells, is not without hazard. Unscheduled firing of replication origins caused by oncogene activation, as well as increased transcriptional activity leading to collisions between the transcription and replication machineries, may stall replication forks and lead to fork collapse (Hills and Diffley, 2014). In addition to sufficient building blocks, replicating cells require proper DNA repair to safeguard genome stability. Homologous recombination (HR) is critical to mend collapsed replication forks. The initial step in HR involves resection of DNA ends to create stretches of single-stranded DNA (ssDNA). In eukaryotic cells, initiation of DNA end resection is governed by the Mre11–Rad50–Nbs1 complex, in conjunction with the DNA double-strand break end resection factor CTBP-interacting protein (CtIP). The resulting ssDNA is ultimately covered with Rad51 filaments, which govern homology search and pairing of the ssDNA with the intact template DNA. Not only does HR facilitate the repair of collapsed replication forks, but non-replication–associated DNA double-strand breaks can also be repaired with high fidelity using HR, at least when cells are in S–G2. Defective HR, for instance, caused by cancer-associated mutations in BRCA1/BRCA2, gives rise to enhanced sensitivity to agents that perturb DNA replication, including poly (ADP-ribose) polymerase (PARP) inhibitors and DNA cross-linking agents such as cisplatin (Evers et al., 2010).
In this issue, Qu et al. describe that inactivation of PGAM1 results in defective DNA repair through HR. Indeed, the researchers observed via SILAC-based proteomics analyses that PGAM1 depletion is associated with changes in protein abundance that underscored metabolic rewiring as well as with perturbations of the levels of proteins involved in the DNA damage response. Notably, PGAM inactivation induced down-regulation of the HR component CtIP. Further characterization of the phenotypes of PGAM1-depleted HeLa cells showed that PGAM1 depletion leads to enhanced sensitivity to DNA damaging agents, including camptothecin, cisplatin, and the PARP inhibitor olaparib (Qu et al., 2017). The requirement for PGAM1 in HR repair involves its enzymatic activity, as Qu et al. (2017) observed that a small molecule inhibitor of PGAM or introduction of a catalytically inactive PGAM mutant effectively interfered with HR repair of DNA double-strand breaks.
In accordance with its role in facilitating DNA end resection, the decreased abundance of CtIP upon PGAM1 depletion prevented DNA end resection as well as subsequent steps in HR, including replication protein A recruitment and Rad51 filament formation at sites of DNA damage. Consequently, Qu et al. (2017) found that genetic or chemical PGAM1 inactivation interfered with functional HR, as judged by repair of I-Sce1–induced DNA breaks in a fluoresence-based HR reporter. Previously, CtIP levels were shown to be under control of the APC/C-Cdh1 E3 ubiquitin ligase (Lafranchi et al., 2014). Activation of the APC/C-Cdh1 normally occurs in a cell cycle–dependent fashion, but can also be triggered in response to DNA damage (Bassermann et al., 2008; Wiebusch and Hagemeier, 2010). In the context of DNA damage, APC/C-Cdh1 activation requires activation of the p53/p21 axis (Wiebusch and Hagemeier, 2010). Also in response to PGAM1 inactivation, APC/C-Cdh1 appeared responsible for CtIP degradation, as Qu et al. (2017) observed that Cdh1 knockdown rescued the decreased CtIP protein levels in PGAM1-depleted cells. Of note, CtIP degradation in response to PGAM1 depletion did not strictly depend on p53. Knockdown of the p53 family member p73 in cells deficient for p53 function, or knock down of p53 in p53 wild-type cancer cells, abolished p21 levels. In these respective cell lines, chromatin immunoprecipitation analyses showed increased p73 or p53 recruitment in the promoter region of the p21 gene in PGAM1-depleted cells, suggesting p73 and p53 drive p21 up-regulation in response to PGAM1 inactivation. In cells in which p53 function is compromised, p73 may functionally compensate for the loss of p53, suggesting that p73 provides cell cycle control in TP53 mutant cancer cells. In this context, it is of interest that expression of PGAM1 itself is under the control of p53 (Kondoh et al., 2005). Wild-type p53 was reported to block PGAM1 expression, whereas expression of a dominant-negative p53 mutant drives PGAM1 expression (Kondoh et al., 2005). These effects could be attributed to the p53 target MDM-2, which directly ubiquitylates and thereby down-regulates PGAM levels (Mikawa et al., 2014). Abberant expression of PGAM1 was shown to facilitate oncogenic transformation and may in part explain the effects of p53 inactivation on tumor cell metabolism (Kondoh et al., 2005).
To assess how PGAM1 inactivation provokes a DNA damage response, Qu et al. (2017) examined nucleotide levels. In good agreement with a role for PGAM1 in promoting PPP flux, PGAM1 inactivation phenocopied the silencing of the PPP enzyme 6-phosphogluconate dehydrogenase and led to lower levels of deoxynucleoside triphosphates. Very likely, it is the ensuing replication stress that resulted in a DNA damage response, with consequent transcriptional activation of p53/p73 and transactivation of their target gene p21.
The APC/C-Cdh1 is negatively regulated by cyclin-dependent kinase (CDK) activity, through inhibitory phosphorylation of Cdh1 by Cdk2 (Lukas et al., 1999). However, up-regulation of the CDK inhibitor p21, as induced by PGAM1 inactivation, will lead to lower CDK activity, and thereby reverses the inhibitory phosphorylation of the APC/C activator Cdh1. Consequently, p21 up-regulation indirectly leads to unscheduled degradation of APC/C-Cdh1 targets. Whether APC/C-Cdh1 activation in response to PGAM1 inactivation leads to selective degradation of APC/C-Cdh1 targets remains unclear. However, the observation that PGAM1 inhibition does not dramatically alter cell cycle profiles, as shown by Qu et al. (2017) using flow cytometry, suggests that the majority of APC/C-Cdh1 substrates (which include many essential cycle cycle regulators) is not significantly affected. If and how the DNA damage-activated APC/C-Cdh1 differentially targets substrates for ubiquitylation remains to be elucidated.
Therapeutic targeting of metabolism pathway components for cancer treatment has been studied for decades. Initially, these efforts were primarily aimed at blocking energy production, to which tumor cells were thought to be addicted. Increasingly, researchers have realized that tumor cells may not per se depend on metabolic flux, but rather may depend on anabolic pathways for the production of biomolecules such as nucleotides, amino acids, and fatty acids (Vander Heiden et al., 2009). The study by Qu et al. (2017) extends this view and shows that modulation of glycolytic pathways also affects secondary pathways that may be essential for tumor cell survival. PGAM1 inhibition interferes with nucleoside biosynthesis and through this mechanism induces HR deficiency. Based on this model, targeting of any enzymatic step involved in nucleoside synthesis may impact HR. Several lines of evidence indeed underscore this model. For instance, the activity of phosphoinositide 3-kinase is required for nucleoside synthesis (Juvekar et al., 2016). Accordingly, inhibition of phosphoinositide 3-kinase interfered with nucleoside synthesis and potently sensitized tumor cells for PARP inhibitor treatment (Juvekar et al., 2012). What remains difficult in these studies, however, is to identify to what extent the synergistic effects can be attributed to defective DNA repair, rather than interference with energy homeostasis or prosurvival signaling.
The observation by Qu et al. (2017) that PGAM1 inhibition blocks HR DNA repair clearly demonstrates that targeting metabolism may come with additional benefits, which can be exploited to further improve cancer treatment. These important insights set the stage for novel combination therapies. Future studies, however, will be required to establish which component of glycolysis (e.g., PGAM1/2) or PPP pathway (e.g., 6-phosphogluconate dehydrogenase) is the most effective therapeutic target to inactivate HR DNA repair, and whether combined targeting of these enzymes may further impair HR and lead to more potent sensitization to DNA damaging agents. In this respect, it is important to realize that ongoing proliferation is required for many genotoxic agents to cause DNA lesions, including PARP inhibitors and cisplatin. Targeting metabolism to sensitize tumors for DNA damaging agents may therefore require a well-balanced approach in which metabolism is affected sufficiently to induce a DNA repair defect, but still allows cancer cells to accumulate the relevant DNA lesions. When such features are established, these novel therapeutic approaches may extend the elegibility for PARP inhibitors beyond tumors with BRCA1/2 mutations. Alternatively, inhibition of PGAM1 or related metabolic enzymes may be used to sensitize tumors for currently used chemotherapeutics, such as cisplatin, that also differentially affect HR-deficient cancers.
Acknowledgments
I apologize to those colleagues whose work could not be cited because of space limitations.
Research in the van Vugt laboratory is funded by the KWF Kankerbestrijding (RUG #2011-5093), the Netherlands Organisation for Scientific Research (NWO-VIDI #91713334), and the H2020 European Research Council (ERC-CoG 682421).
The author declares no competing financial interests.
References
- Bassermann F., Frescas D., Guardavaccaro D., Busino L., Peschiaroli A., and Pagano M.. 2008. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 134:256–267. 10.1016/j.cell.2008.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durany N., Joseph J., Jimenez O.M., Climent F., Fernández P.L., Rivera F., and Carreras J.. 2000. Phosphoglycerate mutase, 2,3-bisphosphoglycerate phosphatase, creatine kinase and enolase activity and isoenzymes in breast carcinoma. Br. J. Cancer. 82:20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers B., Helleday T., and Jonkers J.. 2010. Targeting homologous recombination repair defects in cancer. Trends Pharmacol. Sci. 31:372–380. 10.1016/j.tips.2010.06.001 [DOI] [PubMed] [Google Scholar]
- Hills S.A., and Diffley J.F.X.. 2014. DNA replication and oncogene-induced replicative stress. Curr. Biol. 24:R435–R444. 10.1016/j.cub.2014.04.012 [DOI] [PubMed] [Google Scholar]
- Hitosugi T., Zhou L., Elf S., Fan J., Kang H.-B., Seo J.H., Shan C., Dai Q., Zhang L., Xie J., et al. . 2012. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 22:585–600. 10.1016/j.ccr.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juvekar A., Burga L.N., Hu H., Lunsford E.P., Ibrahim Y.H., Balmañà J., Rajendran A., Papa A., Spencer K., Lyssiotis C.A., et al. . 2012. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2:1048–1063. 10.1158/2159-8290.CD-11-0336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juvekar A., Hu H., Yadegarynia S., Lyssiotis C.A., Ullas S., Lien E.C., Bellinger G., Son J., Hok R.C., Seth P., et al. . 2016. Phosphoinositide 3-kinase inhibitors induce DNA damage through nucleoside depletion. Proc. Natl. Acad. Sci. USA. 113:E4338–E4347. 10.1073/pnas.1522223113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh H., Lleonart M.E., Gil J., Wang J., Degan P., Peters G., Martinez D., Carnero A., and Beach D.. 2005. Glycolytic enzymes can modulate cellular life span. Cancer Res. 65:177–185. [PubMed] [Google Scholar]
- Lafranchi L., de Boer H.R., de Vries E.G.E., Ong S.-E., Sartori A.A., and van Vugt M.A.T.M.. 2014. APC/C(Cdh1) controls CtIP stability during the cell cycle and in response to DNA damage. EMBO J. 33:2860–2879. 10.15252/embj.201489017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C., Sørensen C.S., Kramer E., Santoni-Rugiu E., Lindeneg C., Peters J.M., Bartek J., and Lukas J.. 1999. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature. 401:815–818. 10.1038/44611 [DOI] [PubMed] [Google Scholar]
- Mikawa T., Maruyama T., Okamoto K., Nakagama H., Lleonart M.E., Tsusaka T., Hori K., Murakami I., Izumi T., Takaori-Kondo A., et al. . 2014. Senescence-inducing stress promotes proteolysis of phosphoglycerate mutase via ubiquitin ligase Mdm2. J. Cell Biol. 204:729–745. 10.1083/jcb.201306149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J., Sun W., Zhong J., Lv H., Zhu M., Xu J., Jin N., Xie Z., Tan M., Lin S., et al. . 2017. Phosphoglycerate mutase 1 regulates dNTP pool and promotes homologous recombination repair in cancer cells. J. Cell Biol. 10.1083/jcb.201607008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C., and Thompson C.B.. 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 324:1029–1033. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebusch L., and Hagemeier C.. 2010. p53- and p21-dependent premature APC/C-Cdh1 activation in G2 is part of the long-term response to genotoxic stress. Oncogene. 29:3477–3489. 10.1038/onc.2010.99 [DOI] [PubMed] [Google Scholar]