Abstract
Background
The optimal monitoring schedules and cutoff minimal residual disease (MRD) levels for the accurate prediction of relapse at all time points after allogeneic hematopoietic stem cell transplantation (allo-HSCT) remain unclear in patients with t(8;21) acute myeloid leukemia (AML).
Methods
RUNX1-RUNX1T1 transcript levels were measured in bone marrow samples collected from 208 patients at scheduled time points after transplantation (1530 samples in total).
Results
A total of 92.3% of the requested samples were collected, and 74.0% of patients had complete sample collection. The 1-, 3-, and 6-month RUNX1-RUNX1T1 transcript levels could significantly discriminate between continuous complete remission and a hematologic relapse at 1.5–3, 4–6, and 7–12 months but not at >3, >6, and >12 months, respectively. Over 90% of the 175 patients who were in continuous complete remission had a ≥3-log reduction in RUNX1-RUNX1T1 transcript levels from the time of diagnosis at each time point after transplantation and a ≥4-log reduction at ≥12 months. A <3-log reduction within 12 months and/or a <4-log reduction at ≥12 months was significantly related to a higher 3-year cumulative incidence of relapse (CIR) rate in both the entire cohort and the patients with no intervention after HSCT (58.4 vs. 2.2%, 76.5 vs. 2.0%; all P < 0.0001). Patients who had received a preemptive donor lymphocyte infusion when the increase in RUNX1-RUNX1T1 transcripts was ≤1-log according to the above dual cutoff values had significantly lower 1-year CIR rate after intervention than the patients who had received an infusion when the increase was >1-log (0 vs. 55.0%, P = 0.015).
Conclusions
RUNX1-RUNX1T1 transcripts with a <3-log reduction from diagnosis within 12 months and/or a <4-log reduction at ≥12 months after allo-HSCT could accurately predict relapse and may prompt a timely intervention in patients with t(8;21) AML.
Electronic supplementary material
The online version of this article (doi:10.1186/s13045-017-0414-2) contains supplementary material, which is available to authorized users.
Keywords: RUNX1-RUNX1T1 transcript levels, Acute myeloid leukemia, Allogeneic hematopoietic stem cell transplantation, Relapse, Donor lymphocyte infusion
Background
Although t(8;21) acute myeloid leukemia (AML) is considered to have a good prognosis, relapse occurs in up to 40% of patients treated with chemotherapy [1–6]. Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is a potentially curative approach for patients with acute myeloid leukemia (AML) [7–9]. High-risk t(8;21) AML patients benefited from allo-HSCT in our previously published report [6]. Even after they had received HSCT, 10–20% of patients still experienced relapse, which led to a poor long-term outcome [10, 11].
In patients with t(8;21) AML, the presence of minimal residual disease (MRD), measured by RUNX1-RUNX1T1 transcript levels, is now established as a powerful marker to predict relapse and to direct clinical interventions for patients receiving chemotherapy or transplantation [4–6, 10, 12–16]. Our previous study indicated that a <3-log reduction from diagnosis in RUNX1-RUNX1T1 transcripts at the first 3 months post-HSCT generally identified high relapse risk patients [12].
Additionally, we realized that patients with an early ≥3-log reduction in RUNX1-RUNX1T1 transcripts might experience late relapse post-HSCT. Regarding the prediction of relapse that occurs at all time points after HSCT, only a few case reports and studies with a very small sample size have shown that an increase in the number of RUNX1-RUNX1T1 transcripts occurred prior to hematologic relapse [16–18]. Due to the low incidence of t(8;21) AML [19] and its low proportion of patients undergoing allogeneic transplantation, no large-scale study results have been presented to date. As a result, the exact threshold of MRD levels and monitoring schedules for the accurate prediction of relapse at all time points after allo-HSCT remain unclear.
In the current study, we evaluated the RUNX1-RUNX1T1 transcript levels in 208 patients who were monitored at scheduled time points after HSCT and found that a <3-log reduction from diagnosis within 12 months and/or a <4-log reduction at ≥12 months accurately predicted relapse at all time points after HSCT and led to an effective preemptive donor lymphocyte infusion (DLI).
Methods
Patients, treatment, and samples
A total of 208 t(8;21) AML patients were included in this study. They were in complete remission (CR, first or second) at the time of HSCT and consecutively received allo-HSCT at our institute from March 2006 to May 2016. All patients received induction therapy (one to four courses) to achieve CR, followed by at least 2 cycles of consolidation therapy before receiving a transplant. Indications for allo-HSCT included (1) hematologic relapse, (2) meeting the high-risk criteria that we published (not achieving a ≥3-log reduction after the second consolidation and/or the loss of a ≥3-log reduction during the next six consolidation therapies) [6], (3) c-KIT mutations at the time of diagnosis [20], and (4) the patient’s repeated request. The allo-HSCT conditioning regimen, graft-versus-host disease (GVHD) prophylaxis, modified DLI regimen, and preemptive interferon-a (IFN-a) treatment for MRD were performed as previously described [8, 21, 22]. All patients who received haploidentical, unrelated or cord blood grafts were administered an oral dose of rabbit anti-thymocyte globulin (ATG, 2.5 mg/kg; Sanofi, Gentilly, France) on days five to two before transplantation. A modified DLI or IFN-a were given before hematological relapse as intervention therapy after 3 months post-HSCT following a trial of immunosuppressant withdrawal if patients did not achieve a ≥3-log reduction of RUNX1-RUNX1T1 until 3 months or lost a ≥3-log reduction after HSCT without uncontrolled GVHD or severe infection according to donor availability and willingness.
The monitoring schedule was planned in advance, and the scheduled time points were 0, 1, 2, 3, 4.5, 6, 9, 12, 18, and 24 months post-HSCT. Morphologic evaluations and the quantitative measurement of RUNX1-RUNX1T1 transcripts in bone marrow (BM) samples were performed at the specified time points and when patients showed signs of relapse. The cutoff date for follow-up was August 15, 2016. The median follow-up time after HSCT was 24 (2–89) months in the entire cohort. The study was approved by the Ethics Committee of Peking University People’s Hospital, and all patients or their guardians provided written informed consent to participate in the study in accordance with the Declaration of Helsinki.
Measurement of RUNX1-RUNX1T1 transcript levels
TRIzol reagent (Invitrogen, CA, USA) was used to extract total RNA. A high capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) was used to synthesize complementary DNA (cDNA). TaqMan-based real-time quantitative PCR (RQ-PCR) technology was used as described previously [6, 7, 17]. The primers and probes for ABL and RUNX1-RUNX1T1 were obtained from the report of the Europe Against Cancer Program [23, 24]. Quality control samples were included in each PCR run. All amplifications were performed at least in duplicate. The RUNX1-RUNX1T1 transcript level was calculated as the percentage of RUNX1-RUNX1T1 transcript copies/ABL copies. The pretreatment baseline level of RUNX1-RUNX1T1 transcripts was 400% in our laboratory [6]. The reproducible sensitivity of RQ-PCR is five copies. All of the samples with an undetectable fusion transcript had ≥12,500 copies of ABL to guarantee that at least a 4-log reduction of RUNX1-RUNX1T1 transcript levels (0.04%) could be detected.
Statistical analysis and definitions
The cumulative incidence of relapse (CIR), disease free survival (DFS), and overall survival (OS) were measured from the time of allo-HSCT. Standard definitions of relapse were used. Relapses included BM and/or extra-medullary sites. The events for measuring DFS included death and relapse. The event for measuring OS was death (regardless of the cause), and patients, or their relatives, were queried at the date of last follow-up to determine whether they were still living or censored on the date that they were last known to be alive. Comparisons of the CIR between the groups were calculated by considering the competing risks defined by death and performed with the Gray test. The molecular response (the log reduction of RUNX1-RUNX1T1 transcript levels) was considered a time-dependent covariate, i.e., the start time point of the CIR rate curves according to the transcript levels at a specific time point was that specific time point. Survival functions were estimated using the Kaplan-Meier method and compared using the log-rank test. Comparisons between the two groups were performed using the Mann-Whitney U test for continuous variables and Fisher’s exact test for categorical variables. Receiver operating characteristic (ROC) curves were used to evaluate the effect of RUNX1-RUNX1T1 transcript levels after HSCT on relapse. The level for a statistical significance was set at P ≤ 0.05. R version 2.6.1 (R Foundation for Statistical Computing, Vienna, Austria), SPSS 13.0 (SPSS Inc., Chicago, IL, USA), and GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA, USA) software were used.
Results
Patient characteristics, outcomes, and time of relapse
Patient characteristics at the time of diagnosis and HSCT are shown in Table 1. After receiving allo-HSCT, 33 (15.9%) patients experienced relapse; 25 and 8 patients suffered only hematologic and extra-medullary relapse, respectively. A total of 156 (75.0%) patients were alive at the last follow-up; a total of 22 patients died of relapse, and 30 died due to treatment-related mortality. The median follow-up time after allo-HSCT was 26 (3–89) months for the surviving patients. The 3-year CIR, DFS, and OS rates were 20.4% (95% confidence interval (CI), 9.9–33.6%), 65.4% (95% CI, 57.4–72.2%), and 70.8% (95% CI, 62.9–77.3%), respectively.
Table 1.
Patient characteristics
| Parameters | n = 208 |
|---|---|
| Median age when receiving HSCT, y (range) | 30 (4–57) |
| Gender | |
| Male | 124 (60%) |
| Female | 84 (40%) |
| c-KIT gene at diagnosis | |
| Mutation | 69 (33%) |
| Wild type | 80 (39%) |
| Unknown | 59 (28%) |
| First CR induction courses | |
| 1 | 136 (65%) |
| >1 | 58 (28%) |
| Unknown | 14 (7%) |
| Interval between diagnosis and HSCT | |
| <8 months | 110 (53%) |
| ≥8 months | 98 (47%) |
| Disease status when receiving HSCT | |
| First CR | 179 (86%) |
| Second CR | 29 (14%) |
| Donor source | |
| HLA-matched sibling | 60 (29%) |
| Haploidentical | 135 (65%) |
| Unrelated donor | 10 (5.5%) |
| Cord blood | 1 (0.5%) |
| Conditioning regimen | |
| Chemotherapy based | 204 (98.1%) |
| TBI based | 4 (1.9%) |
The median relapse time point was 6 months (range 1.5–43 months) after HSCT for 33 relapsed patients. A total of 27 (82%) patients experienced relapse within the first year after HSCT (11 at 1.5–3 months, 6 at 4–6 months, and 10 at 7–12 months), and the remaining 6 (18%) patients relapsed after 12 months (at 26–43 months after HSCT).
Sample collection and monitoring implementation
Seventy-four percent (154/208) of patients were collected of all requested samples with qualified ABL copies. Of the 54 patients with incomplete collections, 30 (55.6%) and 14 (25.9%) lacked 1 and 2 qualified requested samples, respectively. In all, RUNX1-RUNX1T1 transcripts were measured in 1530 BM samples in this study, which accounted for 92.3% of the requested number of samples.
A <3-log reduction from diagnosis in RUNX1-RUNX1T1 transcripts within the first 3 months after HSCT predicted relapse
Patients with a <3-log reduction in RUNX1-RUNX1T1 transcripts at 1–3 months after HSCT had a significantly higher 3-year CIR rate than those with a ≥3-log reduction (54.5% [95% CI, 40.3–70.8%]) vs. 11.6% [95% CI, 3.0–31.5%]; P < 0.0001, Fig. 1).
Fig. 1.
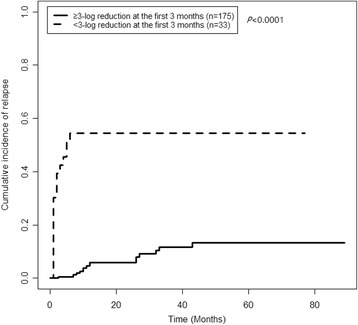
CIR rates of the patients categorized according to their RUNX1-RUNX1T1 transcript levels at the first 3 months after HSCT
A <3-log reduction in RUNX1-RUNX1T1 transcripts within the first 3 months was relevant to early but not to late relapse
Figure 1 shows that some of the patients who had achieved a ≥3-log reduction in RUNX1-RUNX1T1 transcripts at 1–3 months still suffered a relapse. The effect of the early MRD level on relapse was further analyzed in terms of relapse time. The distributions of patients are shown in Fig. 2. A <3-log reduction in RUNX1-RUNX1T1 transcripts at 1–3 months was significantly associated with a higher rate of relapse at 1.5–6 months than a ≥3-log reduction (relapses after 6 months were excluded. 16/31 vs. 1/161, 51.6 vs. 0.6%, P < 0.0001); a <3-log reduction in RUNX1-RUNX1T1 transcripts was not related to relapse after 6 months (relapses at 1.5–6 months were excluded. 2/17 vs. 14/174, 11.8 vs. 8.0%, P = 0.64).
Fig. 2.
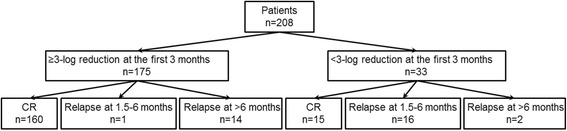
Distribution of the patients according to their reduction in RUNX1-RUNX1T1 transcript levels at 1–3 months and relapse time after allo-HSCT
The RUNX1-RUNX1T1 transcript levels after HSCT only predicted the forthcoming relapse
ROC curve analysis was performed. To evaluate the effect of the 1-month RUNX1-RUNX1T1 transcript levels on relapse, the patients were categorized into three groups: continuous CR, relapse at 1.5–3 months, and relapse after 3 months. The analysis demonstrated that the 1-month RUNX1-RUNX1T1 transcript levels could significantly discriminate continuous complete remission (CR) from relapse at 1.5–3 months (area under the curve [AUC] 0.85, P < 0.0001), but not relapse after 3 months (AUC 0.53, P = 0.62). Similarly, the 3-month RUNX1-RUNX1T1 levels significantly discriminated continuous CR from relapse at 4–6 months (AUC 0.98, P < 0.0001), but not at >6 months (AUC 0.60, P = 0.24). In addition, the 6-month RUNX1-RUNX1T1 levels could discriminate continuous CR from relapse at 7–12 months (AUC 0.75, P = 0.009), but not after 12 months (AUC 0.50, P = 0.98).
The RUNX1-RUNX1T1 transcript levels at 1, 3, and 6 months were individually compared among the patients in continuous CR, the patients who had experienced forthcoming relapse, and the patients who had experienced late relapse. At 1 month, the RUNX1-RUNX1T1 transcript levels in the patients in continuous CR were significantly lower than in the patients who had relapsed at 1.5–3 months, but similar levels to that in the patients who had relapsed after 3 months (Fig. 3a). Likewise, at 3 months, the CR patients had significantly lower RUNX1-RUNX1T1 transcript levels than those who had relapsed at 4–6 months, but similar levels to those patients who had relapsed after 6 months (Fig. 3b). At 6 months, the CR patients had significantly lower RUNX1-RUNX1T1 transcript levels than those who had relapsed at 7–12 months but had levels that were similar to those patients who had relapsed after 12 months (Fig. 3c).
Fig. 3.
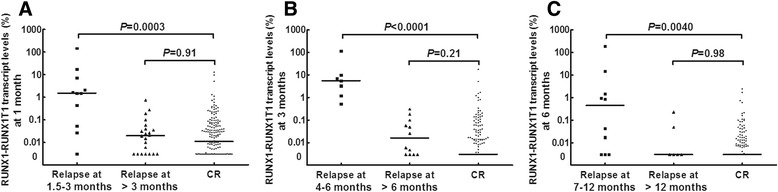
Comparisons of the RUNX1-RUNX1T1 transcript levels at 1, 3, and 6 months among the patients in continuous CR, the patients who had experienced forthcoming relapse and the patients who had experienced late relapse. a RUNX1-RUNX1T1 transcript levels at 1-month post-HSCT. b RUNX1-RUNX1T1 transcript levels at 3 months post-HSCT. c RUNX1-RUNX1T1 transcript levels at 6 months post-HSCT
These results showed that RUNX1-RUNX1T1 transcript levels after HSCT only predicted forthcoming relapse but not late relapse. Therefore, serial monitoring of RUNX1-RUNX1T1 transcripts after HSCT is needed for the accurate prediction of relapse at any time.
Dynamic patterns of RUNX1-RUNX1T1 transcript levels after HSCT in patients in continuous CR
As shown in Table 2, for the patients in continuous CR (n = 175), the RUNX1-RUNX1T1 transcript levels gradually decreased after HSCT, which was demonstrated by the gradual reduction of the median RUNX1-RUNX1T1 transcript levels and the increase of the percentage of patients with a ≥3-log and a ≥4-log reduction in RUNX1-RUNX1T1 transcripts. Over 90% of patients had a ≥3-log reduction from the first month and had a ≥4-log reduction at ≥12 months post-HSCT. In addition, these two frequencies were similar at each time point starting at 12 months (P > 0.05). Therefore, ≥3-log and ≥4-log reductions in RUNX1-RUNX1T1 transcripts were individually chosen as the thresholds for the prediction of relapse within the first year and starting at 12 months post-HSCT, respectively.
Table 2.
Dynamics of RUNX1-RUNX1T1 transcripts post-HSCT in patients in continuous CR
| Month post-HSCT | Number of patients evaluated | Median RUNX1-RUNX1T1 transcript levels (range) | Patients with ≥3-log reduction (%) | Patients with ≥4-log reduction (%) | P value* |
|---|---|---|---|---|---|
| 1 | 174 | 0.011% (0–12.8%) | 163 (93%) | 121 (70%) | <0.0001 |
| 2 | 167 | 0.0062% (0–22.5%) | 160 (96%) | 132 (79%) | <0.0001 |
| 3 | 161 | 0 (0–17.8%) | 153 (95%) | 127 (79%) | <0.0001 |
| 4.5 | 126 | 0 (0–1.5%) | 122 (97%) | 93 (74%) | <0.0001 |
| 6 | 143 | 0 (0–2.4%) | 139 (97%) | 123 (86%) | 0.001 |
| 9 | 113 | 0 (0–4.1%) | 111 (98%) | 99 (88%) | 0.003 |
| 12 | 118 | 0 (0–0.66%) | 116 (98%) | 110 (93%) | 0.10 |
| 18 | 90 | 0 (0–0.30%) | 90 (100%) | 87 (97%) | 0.25 |
| 24 | 72 | 0 (0–0.30%) | 72 (100%) | 68 (94%) | 0.12 |
| 30 | 23 | 0 (0–0.49%) | 22 (96%) | 22 (96%) | 1.0 |
| 36 | 27 | 0 (0–0.005%) | 27 (100%) | 27 (100%) | 1.0 |
*Comparison of the frequency of patients with a ≥3-log reduction with the frequency of patients with a ≥4-log reduction at each time point
A <3-log reduction within 12 months and/or a <4-log reduction at ≥12 months after HSCT predicted relapse at all time points after allo-HSCT
As shown in Fig. 4, both a <3-log reduction in RUNX1-RUNX1T1 transcripts within 12 months and a <4-log reduction in RUNX1-RUNX1T1 transcripts at ≥12 months post-HSCT significantly predicted relapse, respectively (Fig. 4a and b, all P < 0.0001). In addition, patients who had a ≥3-log reduction in RUNX1-RUNX1T1 transcripts within 12 months and a ≥4-log reduction at ≥12 months (defined as low MRD levels after HSCT, n = 151) have significantly lower 3-year CIR rates than patients with a <3-log reduction within 12 months and/or a <4-log reduction at ≥12 months (defined as high MRD levels after HSCT, n = 57) in the entire cohort (2.2% [95% CI, 0–30.6%] vs. 58.4% [95% CI, 45.5–68.7%]; P < 0.0001; Fig. 4c). This result was also observed in the patients with no intervention after HSCT (n = 144 and 21, 2.0% [95% CI, 0–36.5%] vs. 76.5% [95% CI, 74.9–85.5%]; P < 0.0001; Fig. 4d).
Fig. 4.
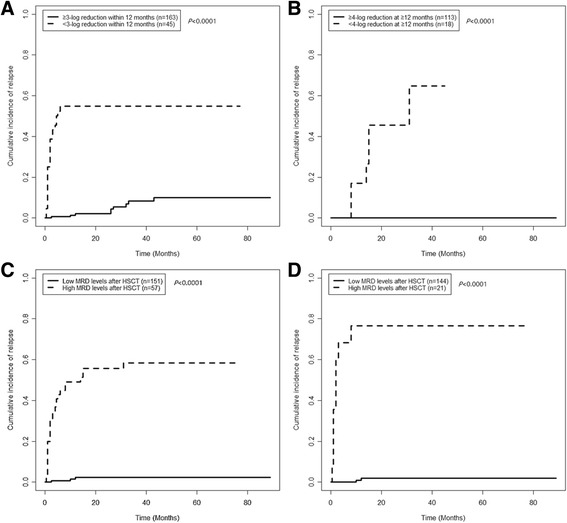
Comparisons of the CIR rates among patients with different MRD levels after HSCT. a The patients were grouped according to whether they achieved a ≥3-log reduction within 12 months. b The patients were grouped according to whether they achieved a ≥4-log reduction at ≥12 months. c All of the patients were grouped according to the dual cutoff values. d The patients with no intervention after HSCT were grouped according to the dual cutoff values
A ≤1-log increase according to the dual thresholds directed effective preemptive DLI
Of the 43 patients who were given intervention therapy, 24 received preemptive DLI (12 also received IFN-a) for the prevention of hematologic relapse. The median time point at which DLI was received was 6 months (range 3–26 months) post-HSCT, and the median RUNX1-RUNX1T1 transcript level was 3.1% (range 0.12–163.0%) at the time of the intervention. The patients were categorized according to their RUNX1-RUNX1T1 transcript levels at the time of DLI. Nine patients received preemptive DLI when their RUNX1-RUNX1T1 transcripts were increased by ≤1-log according to the above dual thresholds (that is, a 2–3-log reduction from diagnosis in RUNX1-RUNX1T1 transcripts within 12 months [n = 8] or a 3–4-log reduction at ≥12 months [n = 1]), and 15 patients received preemptive DLI when their increase was >1-log (<2-log reduction within 12 months [n = 11] or <3-log reduction at ≥12 months [n = 4]). Figure 5 reveals that the patients in the former group had significantly lower 1-year CIR rate after intervention than the patients in the latter group (0 vs. 55.0% [95% CI, 33.9–68.7%]; P = 0.015). This finding indicates that if the cutoff values for prompting preemptive DLI were increased by 1-log, it could no longer effectively prevent relapse.
Fig. 5.
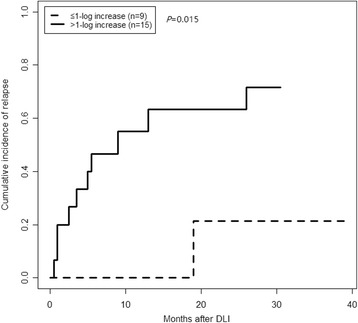
CIR rates of the patients grouped by the increase in RUNX1-RUNX1T1 transcripts compared with the dual thresholds at the time of preemptive DLI
The impact of factors other than the RUNX1-RUNX1T1 transcript levels after HSCT on relapse
As shown in Table 3, mutated c-KIT gene, HLA-matched sibling donor and <3-log reduction of RUNX1-RUNX1T1 transcript levels pre-HSCT were significantly related to a higher 3-year CIR rate (all P < 0.05), and second CR status pre-HSCT tended to be significantly related to a higher 3-year CIR rate (P = 0.071).
Table 3.
The impact of factors other than the RUNX1-RUNX1T1 transcript levels after HSCT on relapse
| Factors | 3-year CIR rate (95% CI) | P value |
|---|---|---|
| WBC count at diagnosis | ||
| ≤ 10 × 109/L | 24.6% (9.1–44.0%) | 0.31 |
| > 10 × 109/L | 17.1% (5.1–35.0%) | |
| c-KIT gene | ||
| Mutation | 38.4% (20.0–56.6%) | 0.003 |
| Wild type | 15.4% (2.4–38.9%) | |
| Karyotype | ||
| Sole t(8;21) | 20.6% (4.3–45.4%) | 0.56 |
| Additional abnormalities | 21.6% (7.6–40.3%) | |
| Course acquired to achieve CR | ||
| 1 | 19.0% (6.1–372%) | 0.20 |
| > 1 | 24.2% (7.9–45.3%) | |
| Time interval from diagnosis to transplant | ||
| < 8 months | 23.5% (9.4–41.2%) | 0.87 |
| ≥ 8 months | 16.4% (4.3–35.5%) | |
| Disease status pre-HSCT | ||
| 1st CR | 18.8% (7.7–33.6%) | 0.071 |
| 2nd CR | 33.3% (9.0–60.4%) | |
| Donor resource | ||
| HLA-matched sibling | 31.4% (13.5–51.2%) | 0.039 |
| Alternative donor | 14.6% (4.2–31.0%) | |
| RUNX1-RUNX1T1 transcript levels pre-HSCT | ||
| ≥ 3-log reduction | 11.1% (0.9–36.4%) | <0.0001 |
| < 3-log reduction | 27.9% (14.6–42.9%) | |
| Acute GVHD | ||
| With | 21.6% (7.5–40.5%) | 0.52 |
| Without | 18.9% (6.0–37.3%) | |
Statistically significant factors are italicized
The impact of acute GVHD on the evolution of the RUNX1-RUNX1T1 transcript levels is shown in the Additional file 1.
Discussion
MRD-directed therapy is a new treatment strategy for patients with AML who receive chemotherapy as well as transplantation. Determination of the cutoff MRD value and the monitoring schedule is the present challenge. In the current study, we demonstrated that the dynamics of RUNX1-RUNX1T1 transcript levels accurately predicted relapse after allo-HSCT in patients with t(8;21) AML. Furthermore, MRD monitoring results directed the preemptive use of DLI, which effectively decreased the occurrence of a hematological relapse.
t(8;21) is a rare disease that accounts for approximately 8% of AML cases [17]. In addition, because t(8;21) is defined as a favorable characteristic, only high-risk patients have been recommended to receive allo-HSCT in recent years [6]. As a result, all of the studies concerning MRD in t(8;21) AML patients who received an allo-HSCT had small sample sizes or were case reports [15–17], except for the study that we published in 2014 [10]. In that paper, we described the effect of changes in RUNX1-RUNX1T1 transcript levels in the first 3 months after allo-HSCT on relapse prediction. To date, no studies have investigated the optimal MRD thresholds and the monitoring schedules for the prediction of relapse at all time points after allo-HSCT in patients with t(8;21) AML.
Several studies have individually shown that a 3-log reduction in RUNX1-RUNX1T1 transcripts is a meaningful threshold at distinct time points during chemotherapy [4–6]. Regarding transplantation, our multicenter data demonstrated that a ≤3-log reduction at the first 3 months after HSCT was an independent factor for CIR, which could be used to rapidly identify those at high risk of relapse [10].
In the present study, we confirmed the predictive value of a 3-log reduction in RUNX1-RUNX1T1 transcripts in the first 3 months for relapse in the entire cohort. However, despite the statistical significance, the majority of late relapse events could not be predicted by the MRD status within the first 3 months. Both the ROC curve analysis and the comparisons of the RUNX1-RUNX1T1 transcripts showed that 1-, 3, and 6-month RUNX1-RUNX1T1 transcript levels could significantly discriminate continuous CR from hematologic relapse that occurred at 1.5–3, 4–6, and 7–12 months but not at >3, >6, or >12 months, respectively. This finding suggested that RUNX1-RUNX1T1 transcript levels after HSCT only predict forthcoming relapse. Therefore, the serial monitoring of RUNX1-RUNX1T1 transcript levels is needed to predict the occurrence of relapse at all time points after HSCT.
Previous studies have demonstrated that a rapid increase in RUNX1-RUNX1T1 transcripts was typically found before relapse occurred [13–18]. This finding suggested that the dynamics of RUNX1-RUNX1T1 after HSCT might predict a relapse. However, the cutoff values and the monitoring time points needed to be determined. In the current study, these determinations were made by the analysis of RUNX1-RUNX1T1 dynamics in patients who were maintained in continuous CR after HSCT. We found that over 90% of patients exhibited a ≥3-log reduction and a ≥4-log reduction in RUNX1-RUNX1T1 transcripts from the first month and at ≥12 months, respectively. Furthermore, the presence of similar frequencies at ≥12 months reflected that these two cutoff values categorized similar amounts of patients into the continuous CR group. Because the cutoff value selection criteria were designed to categorize the highest number of poor-outcome patients into the high-risk group, we developed dual cutoff values: a 3-log reduction for the first year and a 4-log reduction thereafter. As expected, these values accurately predicted relapse in the entire cohort as well as in patients who had received no intervention.
Our previous paper demonstrated that interventions with preemptive DLI decreased the rate of post-HSCT relapse in patients with t(8;21) AML [10]. However, the threshold of RUNX1-RUNX1T1 transcripts for the implementation of an effective intervention remains unclear. In this study, we found that the CIR rate of patients who received DLI when their RUNX1-RUNX1T1 transcripts were within a 1-log increase from the dual thresholds was significantly lower than that of patients receiving DLI at the time of a >1-log increase. Therefore, the effectiveness of preemptive DLI is relevant to RUNX1-RUNX1T1 transcript level at the time of the intervention. A DLI could not prevent hematologic relapse once the leukemic burden was too high. The results demonstrated that the dual cutoff values that we established may direct a timely and effective intervention. However, this result must be confirmed in more cases.
Apart from the RUNX1-RUNX1T1 transcript levels after HSCT, c-KIT mutation status, donor type, and RUNX1-RUNX1T1 transcript levels pre-HSCT were also found to be related to relapse after HSCT in the current cohort. Previous studies have shown the prognostic value of these factors in AML. The c-KIT mutation is a strong poor prognostic factor in t(8;21) AML [6, 10, 20, 25]. MRD levels before allogeneic HSCT have been demonstrated to be associated with adverse outcomes in AML by many studies [26, 27]. We previously reported that high-risk acute leukemia patients receiving haploidentical donor grafts had a significantly lower relapse rate than those receiving HLA-identical sibling donor grafts [28].
The monitoring schedule of this study was planned in advance. In patients who had received allo-HSCT, relapse mostly occurred within the first year, and the relapse rate was highest within the first 3 months; the time interval was therefore set at 1 month within the first 3 months and gradually increased thereafter. A recent report on prospective monitoring in t(8;21) AML patients showed that 71.3% of BM samples that were planned by the protocol were collected [14]. In the present study, 92.3% of the planed samples were collected, and 74.0% of the patients followed the schedule. This finding implied that the results of the current study could represent the effect of this monitoring schedule, and such scheduled monitoring effectively predicted relapse.
Conclusions
The results of the present study indicate that monitoring the dynamics of RUNX1-RUNX1T1 transcripts could predict relapse at all time points after allo-HSCT in patients with t(8;21) AML. A <3-log reduction and a <4-log reduction from diagnosis in RUNX1-RUNX1T1 transcripts within 12 months and at ≥12 months post-HSCT, respectively, were the optimal thresholds to predict relapse. A <1-log increase relative to the above dual thresholds directed the timely and effective use of preemptive DLI. These results should be confirmed in a multi-center study.
Acknowledgements
The authors thank all of the doctors at the institute who participated in this study for providing the follow-up samples and information.
Funding
This work was supported by the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (81621001), the National Nature Science Foundation of China (81570130, 81530046), and the Beijing Municipal Science and Technology Program (Z141100000214011).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Authors’ contributions
XJH designed the research and revised the paper. YZQ and YW analyzed the data and wrote the paper. LPX, XHZ, HC, WH, YHC, FRW, JZW, YC, XDM, XSZ, YJC, and KYL collected and analyzed data. All authors read and approved the final manuscript and submission.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable for individual patient data. This is a pooled analysis.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Peking University People’s Hospital, and all patients or their guardians provided written informed consent to participate in the study in accordance with the Declaration of Helsinki.
Abbreviations
- allo-HSCT
Allogeneic hematopoietic stem cell transplantation
- AML
Acute myeloid leukemia
- AUC
Area under the curve
- CI
Confidence interval
- CIR
Cumulative incidence of relapse
- DFS
Disease-free survival
- DLI
Donor lymphocyte infusion
- GVHD
Graft-versus-host disease
- IFN-a
Interferon-a
- MRD
Minimal residual disease
- OS
Overall survival
- RQ-PCR
Real-time quantitative PCR
Additional file
Impact of acute GVHD on the evolution of RUNX1-RUNX1T1 transcript levels. (DOCX 15 kb)
References
- 1.Schlenk RF, Benner A, Krauter J, Büchner T, Sauerland C, Ehninger G, et al. Individual patient data-based meta-analysis of patients aged 16 to 60 years with core binding factor acute myeloid leukemia: a survey of the German Acute Myeloid Leukemia InterGroup. J Clin Oncol. 2004;22:3741–3750. doi: 10.1200/JCO.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Marcucci G, Mrózek K, Ruppert AS, Maharry K, Kolitz JE, Moore JO, et al. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): a Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:5705–5717. doi: 10.1200/JCO.2005.15.610. [DOI] [PubMed] [Google Scholar]
- 3.Appelbaum FR, Kopecky KJ, Tallman MS, Slovak ML, Gundacker HM, Kim HT, et al. The clinical spectrum of adult acute myeloid leukaemia associated with core binding factor translocations. Br J Haematol. 2006;135:165–173. doi: 10.1111/j.1365-2141.2006.06276.x. [DOI] [PubMed] [Google Scholar]
- 4.Yin JA, O'Brien MA, Hills RK, Daly SB, Wheatley K, Burnett AK. Minimal residual disease monitoring by quantitative RT-PCR in core binding factor AML allows risk stratification and predicts relapse: results of the United Kingdom MRC AML-15 trial. Blood. 2012;120:2826–2835. doi: 10.1182/blood-2012-06-435669. [DOI] [PubMed] [Google Scholar]
- 5.Jourdan E, Boissel N, Chevret S, Delabesse E, Renneville A, Cornillet P, et al. Prospective evaluation of gene mutations and minimal residual disease in patients with core binding factor acute myeloid leukemia. Blood. 2013;121:2213–2223. doi: 10.1182/blood-2012-10-462879. [DOI] [PubMed] [Google Scholar]
- 6.Zhu HH, Zhang XH, Qin YZ, Liu DH, Jiang H, Chen H, et al. MRD-directed risk stratification treatment may improve outcomes of t(8;21) AML in the first complete remission: results from the AML05 multicenter trial. Blood. 2013;121:4056–4062. doi: 10.1182/blood-2012-11-468348. [DOI] [PubMed] [Google Scholar]
- 7.Cornelissen JJ, van Putten WL, Verdonck LF, Theobald M, Jacky E, Daenen SM, et al. Results of a HOVON/SAKK donor versus no-donor analysis of myeloablative HLA-identical sibling stem cell transplantation in first remission acute myeloid leukemia in young and middle-aged adults: benefits for whom? Blood. 2007;109:3658–3666. doi: 10.1182/blood-2006-06-025627. [DOI] [PubMed] [Google Scholar]
- 8.Lu DP, Dong L, Wu T, Huang XJ, Zhang MJ, Han W, et al. Conditioning including antithymocyte globulin followed by unmanipulated HLA-mismatched/haploidentical blood and marrow transplantation can achieve comparable outcomes with HLA-identical sibling transplantation. Blood. 2006;107:3065–3073. doi: 10.1182/blood-2005-05-2146. [DOI] [PubMed] [Google Scholar]
- 9.Shimoni A, Labopin M, Savani B, Volin L, Ehninger G, Kuball J, et al. Long-term survival and late events after allogeneic stem cell transplantation from HLA-matched siblings for acute myeloid leukemia with myeloablative compared to reduced-intensity conditioning: a report on behalf of the acute leukemia working party of European group for blood and marrow transplantation. J Hematol Oncol. 2016;9:118. doi: 10.1186/s13045-016-0347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Wu DP, Liu QF, Qin YZ, Wang JB, Xu LP, et al. In adults with t(8;21)AML, posttransplant RUNX1/RUNX1T1-based MRD monitoring, rather than c-KIT mutations, allows further risk stratification. Blood. 2014;124:1880–1886. doi: 10.1182/blood-2014-03-563403. [DOI] [PubMed] [Google Scholar]
- 11.Yoon JH, Kim HJ, Kim JW, Jeon YW, Shin SH, Lee SE, et al. Identification of molecular and cytogenetic risk factors for unfavorable core-binding factor-positive adult AML with post-remission treatment outcome analysis including transplantation. Bone Marrow Transplant. 2014;49:1466–1474. doi: 10.1038/bmt.2014.180. [DOI] [PubMed] [Google Scholar]
- 12.Weisser M, Haferlach C, Hiddemann W, Schnittger S. The quality of molecular response to chemotherapy is predictive for the outcome of AML1-ETO-positive AML and is independent of pretreatment risk factors. Leukemia. 2007;21:1177–1182. doi: 10.1038/sj.leu.2404659. [DOI] [PubMed] [Google Scholar]
- 13.Leroy H, de Botton S, Grardel-Duflos N, Darre S, Leleu X, Roumier C, et al. Prognostic value of real-time quantitative PCR (RQ-PCR) in AML with t(8;21) Leukemia. 2005;19:367–372. doi: 10.1038/sj.leu.2403627. [DOI] [PubMed] [Google Scholar]
- 14.Willekens C, Blanchet O, Renneville A, Cornillet-Lefebvre P, Pautas C, Guieze R, et al. Prospective long-term minimal residual disease monitoring using RQ-PCR in RUNX1-RUNX1T1-positive acute myeloid leukemia: results of the French CBF-2006 trial. Haematologica. 2016;101:328–335. doi: 10.3324/haematol.2015.131946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lane S, Saal R, Mollee P, Jones M, Grigg A, Taylor K, et al. A > or = 1 log rise in RQ-PCR transcript levels defines molecular relapse in core binding factor acute myeloid leukemia and predicts subsequent morphologic relapse. Leuk Lymphoma. 2008;49:517–523. doi: 10.1080/10428190701817266. [DOI] [PubMed] [Google Scholar]
- 16.Tobal K, Newton J, Macheta M, Chang J, Morgenstern G, Evans PA, et al. Molecular quantitation of minimal residual disease in acute myeloid leukemia with t(8;21) can identify patients in durable remission and predict clinical relapse. Blood. 2000;95:815–819. [PubMed] [Google Scholar]
- 17.Sugimoto T, Das H, Imoto S, Murayama T, Gomyo H, Chakraborty S, et al. Quantitation of minimal residual disease in t(8;21)-positive acute myelogenous leukemia patients using real-time quantitative RT-PCR. Am J Hematol. 2000;64:101–106. doi: 10.1002/(SICI)1096-8652(200006)64:2<101::AID-AJH5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 18.Mitterbauer M, Mitterbauer-Hohendanner G, Sperr WR, Kalhs P, Greinix HT, Fonatsch C, et al. Molecular disease eradication is a prerequisite for long-term remission in patients with t(8;21) positive acute myeloid leukemia: a single center study. Leuk Lymphoma. 2004;45:971–977. doi: 10.1080/10428190310001638913. [DOI] [PubMed] [Google Scholar]
- 19.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- 20.Qin YZ, Zhu HH, Jiang Q, Jiang H, Zhang LP, Xu LP, et al. Prevalence and prognostic significance of c-KIT mutations in core binding factor acute myeloid leukemia: a comprehensive large-scale study from a single Chinese center. Leuk Res. 2014;38:1435–1440. doi: 10.1016/j.leukres.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Yan CH, Liu DH, Liu KY, Xu LP, Liu YR, Chen H, et al. Risk stratification-directed donor lymphocyte infusion could reduce relapse of standard-risk acute leukemia patients after allogeneic hematopoietic stem cell transplantation. Blood. 2012;119:3256–3262. doi: 10.1182/blood-2011-09-380386. [DOI] [PubMed] [Google Scholar]
- 22.Mo XD, Zhang XH, Xu LP, Wang Y, Yan CH, Chen H, et al. Interferon-α: a potentially effective treatment for minimal residual disease in acute leukemia/myelodysplastic syndrome after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2015;21:1939–1947. doi: 10.1016/j.bbmt.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, van der Schoot E, et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR) - a Europe against cancer program. Leukemia. 2003;17:2474–2486. doi: 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]
- 24.Gabert J, Beillard E, van der Velden VH, Bi W, Grimwade D, Pallisgaard N, et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia—a Europe Against Cancer program. Leukemia. 2003;17:2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- 25.Paschka P, Marcucci G, Ruppert AS, Mrózek K, Chen H, Kittles RA, et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study. J Clin Oncol. 2006;24:3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 26.Walter RB, Buckley SA, Pagel JM, Wood BL, Storer BE, Sandmaier BM, et al. Significance of minimal residual disease before myeloablative allogeneic hematopoietic cell transplantation for AML in first and second complete remission. Blood. 2013;122:1813–1821. doi: 10.1182/blood-2013-06-506725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Araki D, Wood BL, Othus M, Radich JP, Halpern AB, Zhou Y, et al. Allogeneic hematopoietic cell transplantation for acute myeloid leukemia: time to move toward a minimal residual disease-based definition of complete remission? J Clin Oncol. 2016;34:329–336. doi: 10.1200/JCO.2015.63.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Liu DH, Xu LP, Liu KY, Chen H, Chen YH, et al. Superior graft-versus-leukemia effect associated with transplantation of haploidentical compared with HLA-identical sibling donor grafts for high-risk acute leukemia: an historic comparison. Biol Blood Marrow Transplant. 2011;17:821–830. doi: 10.1016/j.bbmt.2010.08.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.


