Lang et al. identify E3 ligase TRIM65 as an essential component for the MDA-5 signaling pathway and provide solid evidence showing the importance of ubiquitination in MDA5 oligomerization and activation.
Abstract
MDA5 plays a critical role in antiviral innate immunity by functioning as a cytoplasmic double-stranded RNA sensor that can activate type I interferon signaling pathways, but the mechanism for the activation of MDA5 is poorly understood. Here, we show that TRIM65 specifically interacts with MDA5 and promotes K63-linked ubiquitination of MDA5 at lysine 743, which is critical for MDA5 oligomerization and activation. Trim65 deficiency abolishes MDA5 agonist or encephalomyocarditis virus (EMCV)–induced interferon regulatory factor 3 (IRF3) activation and type I interferon production but has no effect on retinoic acid–inducible I (RIG-I), Toll-like receptor 3 (TLR3), or cyclic GMP-AMP synthase signaling pathways. Importantly, Trim65−/− mice are more susceptible to EMCV infection than controls and cannot produce type I interferon in vivo. Collectively, our results identify TRIM65 as an essential component for the MDA5 signaling pathway and provide physiological evidence showing that ubiquitination is important for MDA5 oligomerization and activation.
Introduction
MDA5 and RIG-I are cytoplasmic viral RNA sensors and can activate type I interferon signaling pathways after virus infection, so they play a critical role in antiviral innate immunity (Takeuchi and Akira, 2008; Loo and Gale, 2011). MDA5 and RIG-I share high sequence similarity and a common signaling adaptor, mitochondrial antiviral signaling (MAVS), but they play nonredundant functions in antiviral immunity by recognizing different viruses or viral RNA (Kato et al., 2006). RIG-I recognizes 5′-triphosphorylated (PPP) blunt-ended double-stranded RNA (dsRNA) or single-stranded RNA hairpins often present in a variety of positive and negative strand viruses (Hornung et al., 2006; Pichlmair et al., 2006; Schlee et al., 2009). MDA5 recognizes relatively long dsRNA in the genome of dsRNA viruses or dsRNA replication intermediates of positive-strand viruses, such as encephalomyocarditis virus (EMCV) and poliovirus (Kato et al., 2006). In recent years, much attention has been paid to the molecular mechanisms that control the activation of RIG-I–mediated antiviral signaling pathways (Goubau et al., 2013; Wu and Hur, 2015; Yoneyama et al., 2015). It is now well established that RIG-I activation is precisely regulated by combinatorial posttranslational modifications (Chan and Gack, 2015; Cao, 2016), but little is known about how MDA5 is activated during RNA virus infection.
Ubiquitination is one of the most versatile posttranslational modifications and plays an important role in signal transduction during the innate antiviral immune response (Zhang et al., 2011; Jiang and Chen, 2012; Li et al., 2016). The role and mechanism of ubiquitination in RIG-I signaling activation have been widely investigated. Ubiquitination of RIG-I mediated by E3 ligase TRIM25 or Riplet is essential for its activation and signal transduction (Gack et al., 2007; Oshiumi et al., 2009, 2010). In addition, unanchored K63-linked polyubiquitin chains can bind to RIG-I CARD domains and then promote its activation and signal transduction (Zeng et al., 2010; Jiang et al., 2012). For MDA5, the role and mechanisms of ubiquitination are poorly understood. Jiang et al. (2012) proposed that unanchored K63-linked polyubiquitin chains can stabilize 2CARD oligomerization of MDA5 and promote its activation, but Wu et al. (2013) reported that MDA5 could be directly activated by a filamentous structure formed by MDA5 oligomerization around dsRNA in a ubiquitin-independent manner. However, because both of these models for MDA5 activation are based on cell-free systems, the physiological role of ubiquitination in MDA5 activation is unclear.
The tripartite motif (TRIM) protein family comprises over 70 members and is involved in various cellular processes, including cell proliferation, differentiation, cell death, and immunity. TRIMs contain an N-terminal RBCC motif composed of a RING domain, one or two B-boxes, and a coiled-coil domain and represent a new class of single RING-finger E3 ubiquitin ligases (Meroni, 2012). Over the past decade, several TRIMs have been reported to regulate signal transduction by ubiquitinating the key components of antiviral innate immunity, including retinoic acid–inducible I (RIG-I), STING, and interferon regulatory factor 3 (IRF3; Ozato et al., 2008; Cai and Wang, 2013; Versteeg et al., 2013). To clarify the mechanisms for MDA5 activation, we searched for previously unknown MDA5 binding partners and identified an MDA5-binding protein, TRIM65, that plays an essential role in MDA5 activation by promoting its ubiquitination and oligomerization.
Results
TRIM65 is an MDA5-binding protein
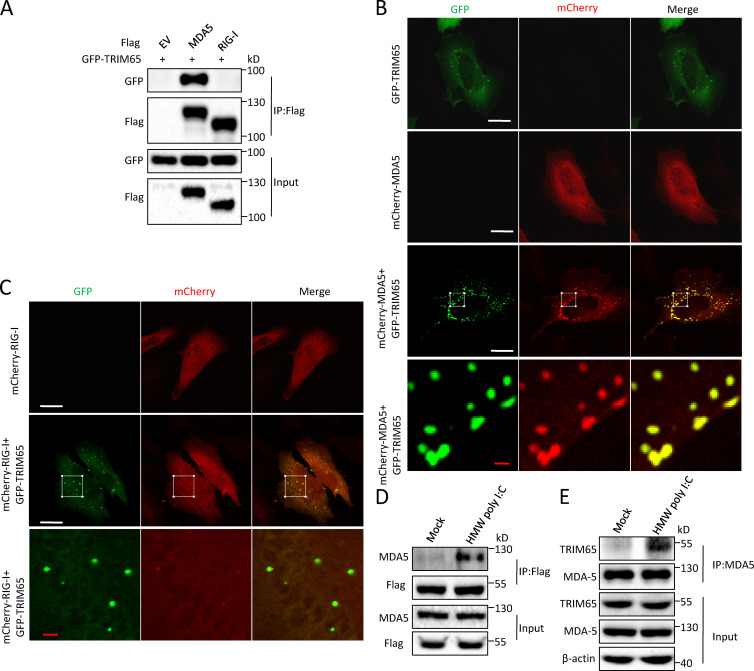
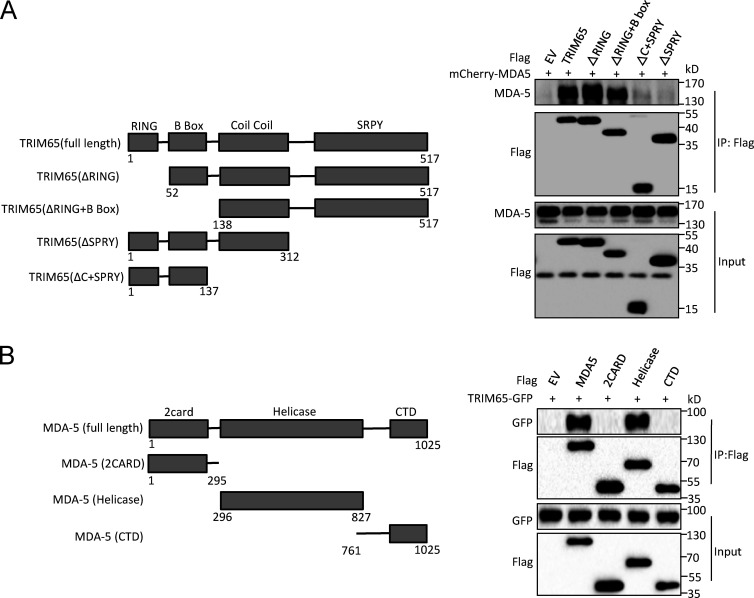
To search for MDA5 binding partners, we used mass spectrometry to identify the proteins associated with MDA5 and found that E3 ligase TRIM65 had the highest number of matched peptides in the precipitates (Li et al., 2014). We confirmed the interaction between TRIM65 and MDA5 in human embryonic kidney T (HEK-293T) cells. Overexpressed TRIM65 strongly bound to full-length MDA5, but not RIG-I, in a coimmunoprecipitation assay (Fig. 1 A), consistent with a recent study (Kamanova et al., 2016). Confocal microscopy showed that overexpressed TRIM65 colocalized with MDA5 and promoted MDA5 to form aggregates in HeLa cells (Fig. 1 B), suggesting that TRIM65 interacts with MDA5 and promotes its oligomerization. In contrast, TRIM65 overexpression could not induce RIG-I to form aggregates (Fig. 1 C). Importantly, high-molecular-weight (HMW) dsRNA polyriboinosinic:polyribocytidylic acid (poly I:C), which is a specific activator for MDA5 (Gitlin et al., 2006; Kato et al., 2008), could promote endogenous MDA5 binding to overexpressed Flag-TRIM65 in HEK-293 cells (Fig. 1 D) or endogenous TRIM65 in THP-1 cells (Fig. 1 E). We further mapped the physical domains of TRIM65 required for this association and found that the SPRY domain was essential for this interaction (Fig. 2 A). Similarly, we also found that TRIM65 associated with helicase domain of MDA5, but not with 2CARD domain or CTD domain (Fig. 2 B). Collectively, these results suggest that TRIM65 is a binding protein of innate immune sensor MDA5.
Figure 1.
TRIM65 interacts with MDA5. (A) Immunoprecipitation (IP) and immunoblot analysis of the interaction of Flag-MDA5 with GFP-TRIM65 in the lysates of HEK-293T cells. EV, empty vector; input, cell extract without immunoprecipitation. (B and C) Confocal microscopy analysis of HeLa cells cotransfected with plasmids of GFP-TRIM65 with mCherry-MDA5 (B) or mCherry–RIG-I (C). The bottom row shows higher-magnification images of white boxes in the row above. Bars: (white) 20 µm; (red) 2 µm. (D) Immunoblot analysis of the interaction of Flag-TRIM65 with endogenous MDA5 in HEK-293 cells stably expressing Flag-TRIM65 stimulated by HMW poly I:C for 1.5 h, followed by IP with anti-Flag antibody. (E) Immunoblot analysis of the endogenous interaction between TRIM65 and MDA5 in THP-1 cells stimulated by HMW poly I:C for 1.5 h, followed by IP with anti-MDA5 antibody. Data are representative of two (A) or three (B–E) independent experiments.
Figure 2.
Mapping interacting domains in TRIM65 and MDA5. (A) Immunoprecipitation (IP) and immunoblot analysis of the interaction of mCherry-MDA5 with Flag-tagged full-length TRIM65 protein or TRIM65 protein without the indicated domains in the lysates of HEK-293T cells. EV, empty vector. (B) IP and immunoblot analysis of the interaction of GFP-TRIM65 with full-length or truncated MDA5 in the lysates of HEK-293T cells. Data are representative of three independent experiments.
TRIM65 is essential for MDA5 activation
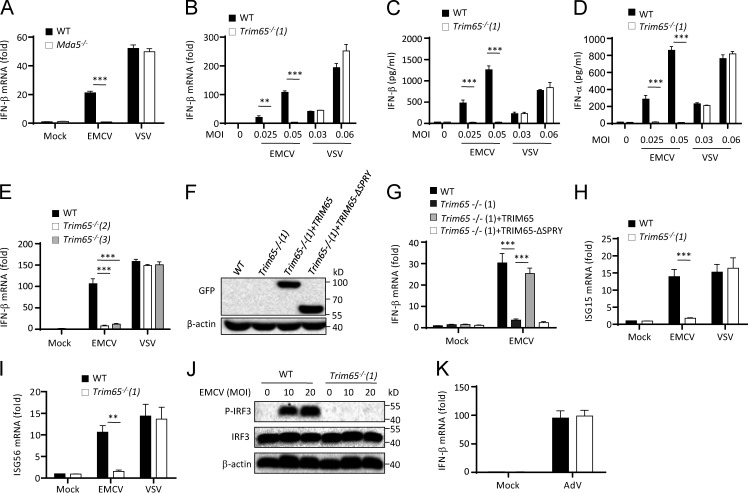
We next investigated whether the observed interaction between TRIM65 and MDA5 is important for MDA5-mediated antiviral innate immune responses. Consistent with a previous study (Kato et al., 2006), deletion of Mda5 blocked EMCV infection–induced IFN-β production but had no effect on IFN-β production induced by vesicular stomatitis virus (VSV; Fig. 3 A), which is known to be specifically recognized by RIG-I (Kato et al., 2006). To examine the role of TRIM65 in EMCV-induced MDA5 activation, we generated Trim65-deficient mice using transcription activator–like effector nuclease (TALEN)–based genome editing technology (Fig. S1). When BMDMs from Trim65−/− (strain 1) mice were infected with viruses, EMCV-induced IFN-β and IFN-α production was completely inhibited, whereas VSV-induced production of interferons was normal (Fig. 3, B–D). The results were confirmed in BMDMs from another two Trim65−/− strains (strains 2 and 3; Fig. 3 E). The deficiency of EMCV-induced IFN-β production was also observed in Trim65−/− MEF cells and could be rescued by overexpression of full-length TRIM65, but not by TRIM65 without SPRY domain, in Trim65−/− cells (Fig. 3, F and G). In addition, the induction of interferon-stimulated genes, such as ISG15 and ISG56, was also inhibited in Trim65−/− BMDMs during EMCV infection (Fig. 3, H and I). Furthermore, Trim65 deficiency blocked EMCV-induced IRF3 phosphorylation, indicating that TRIM65 is essential for MDA5-induced IRF3 activation (Fig. 3 J). In contrast, adenovirus-induced IFN-β production was not changed in Trim65−/− BMDMs (Fig. 3 K), suggesting that TRIM65 is not required for the DNA virus–activated cGAS/STING pathway (Lam et al., 2014).
Figure 3.
Role of TRIM65 in RNA virus-induced activation of MDA5 signaling. (A and B) qPCR analysis of Ifn-β expression in BMDMs from Mda5−/− (A) or Trim65−/− (B) mice infected with indicated MOI of EMCV or VSV for 12 h. (C and D) Secretion of IFN-β (C) or IFN-α (D) by Trim65−/− BMDMs infected with indicated doses of EMCV or VSV for 12 h. (E) qPCR analysis of Ifn-β expression in BMDMs from WT mice or Trim65−/− mice lines infected with EMCV or VSV for 12 h. (F) Immunoblot analysis of indicated proteins in Trim65−/− MEF cells overexpressing GFP-TRIM65 or GFP-TRIM65 without the SPRY domain. (G) qPCR analysis of IFN-β expression in Trim65−/− MEF cells overexpressing GFP-TRIM65 or GFP-TRIM65 without SPRY domain and left stimulated with EMCV for 12 h. (H and I) qPCR analysis of ISG15 (H) or ISG56 (I) expression in WT or Trim65−/− BMDMs infected with EMCV (MOI = 0.05) or VSV (MOI = 0.06) for 12 h. (J) Immunoblot analysis of IRF3 phosphorylation (P-IRF3) in WT or Trim65−/− BMDMs infected with EMCV (MOI = 0.05) for 6 h. (K) qPCR analysis of Ifn-β expression in WT or Trim65−/− BMDMs infected with adenovirus (AdV; MOI = 1,000) for 12 h. Data are from three independent experiments with biological duplicates in each (A–E, G–I, and K; mean and SEM of n = 6) or representative of three independent experiments (F and J). Statistics were analyzed via an unpaired Student’s t test: **, P < 0.01; ***, P < 0.001.
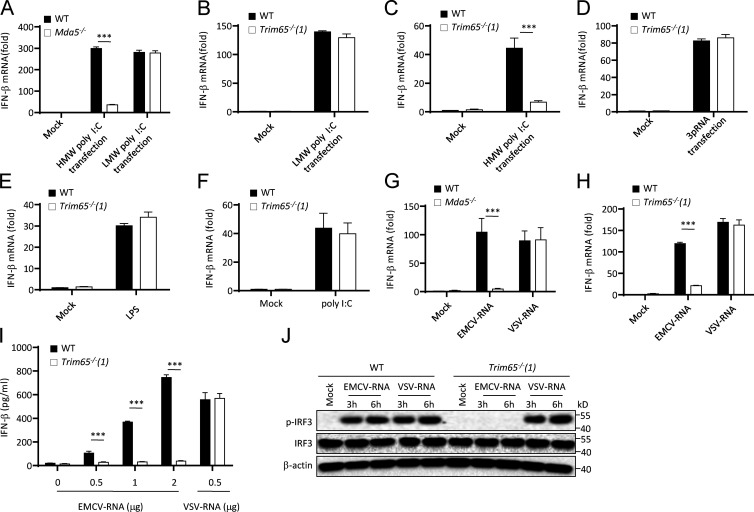
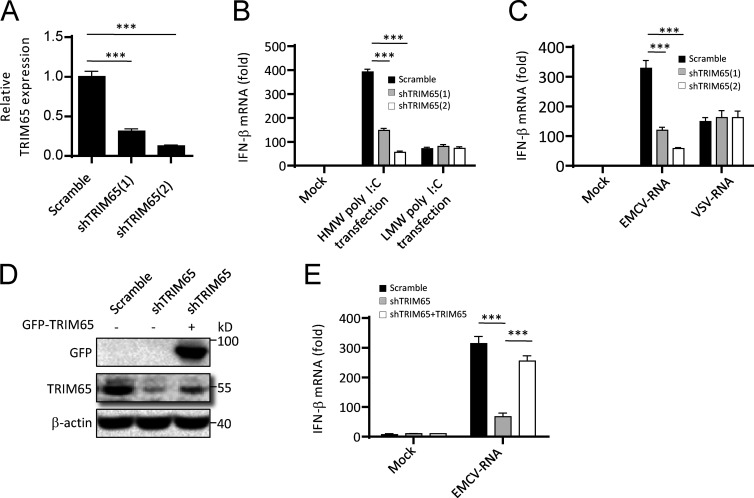
Because MDA5 recognizes long dsRNA or dsRNA replication intermediates to initiate interferon production and antiviral innate immune responses, we asked whether TRIM65 mediated RNA agonist–induced MDA5 activation. Consistent with previous studies (Gitlin et al., 2006; Kato et al., 2008), we confirmed that HMW poly I:C–induced IFN-β production was MDA5 dependent, whereas that of low-molecular-weight (LMW) poly I:C induced was MDA5 independent (Fig. 4 A). When Trim65−/− BMDMs were stimulated with cytosolic HMW poly I:C, LMW poly I:C, 5′-triphosphate RNA (3pRNA, agonist for RIG-I), extracellular LPS (agonist for TLR4), or poly I:C (agonist for TLR3), only HMW poly I:C–induced IFN-β production was blocked as expected (Fig. 4, B–F), confirming that TRIM65 is specifically involved in MDA5 activation. RNA isolated from cells infected with VSV (VSV-RNA) or EMCV (EMCV-RNA) has been reported to activate RIG-I or MDA5, respectively (Kato et al., 2006; Pichlmair et al., 2009). Our results showed that EMCV-RNA–induced IFN-β production depended on not only MDA5, but also TRIM65 (Fig. 4, G–I). Consistent with this, Trim65 deficiency also blocked EMCV-RNA–induced IRF3 phosphorylation (Fig. 4 J). We also tested whether TRIM65 mediates MDA5 activation in human HeLa cells and found that knockdown of TRIM65 expression by shRNA significantly suppressed HMW poly I:C– or EMCV-RNA–induced IFN-β production (Fig. 5, A–C). The impairment of IFN-β production caused by TRIM65 shRNA (which targets the 3′ UTR region of TRIM65 gene)–mediated knockdown could be rescued by TRIM65 overexpression (Fig. 5, D and E). Collectively, these results indicate that TRIM65 plays an essential role in MDA5 signaling activation induced by cytosolic RNA sensing.
Figure 4.
Role of TRIM65 in dsRNA-induced activation of MDA5 signaling. (A–H) qPCR analysis of Ifn-β expression in Mda5−/− (A and G) or Trim65−/− (B–F and H) BMDMs stimulated with cytosolic LMW poly I:C (A and B), HMW poly I:C (A and C), cytosolic 3pRNA (D), extracellular LPS (E), poly I:C (F), and EMCV-RNA or VSV-RNA (G and H) for 6 h. (I) Secretion of IFN-β in Trim65−/− BMDMs stimulated with different doses of EMCV-RNA or VSV-RNA for 18 h. (J) Immunoblot analysis of IRF3 phosphorylation (p-IRF3) in WT or Trim65−/− BMDMs stimulated with cytosolic EMCV-RNA or VSV-RNA for 3 h. Data are from three independent experiments with biological duplicates in each (A–I; mean and SEM of n = 6) or are representative of three independent experiments (J). Statistics were analyzed via an unpaired Student’s t test: ***, P < 0.001.
Figure 5.
Knockdown of TRIM65 suppresses MDA5 activation in human cells. (A) qPCR analysis of TRIM65 expression in HeLa cells stably expressing shRNA against TRIM65. (B and C) qPCR analysis of IFN-β expression in HeLa cells stably expressing shRNA against TRIM65 and left stimulated for 6 h with cytosolic HMW poly I:C (B), LMW poly I:C (B), EMCV-RNA (C), or VSV-RNA (C). (D) Immunoblot analysis of TRIM65 expression in HeLa cells stably expressing shRNA against TRIM65 with or without overexpression of GFP-TRIM65. (E) qPCR analysis of IFN-β expression in HeLa cells stably expressing shRNA against TRIM65 with or without overexpression of GFP-TRIM65 and left stimulated for 6 h with cytosolic EMCV-RNA. Data are from three independent experiments with biological duplicates in each (mean and SEM of n = 6). Statistics were analyzed via an unpaired Student’s t test: ***, P < 0.001.
TRIM65 mediates K63-linked ubiquitination of MDA5 at lysine 743
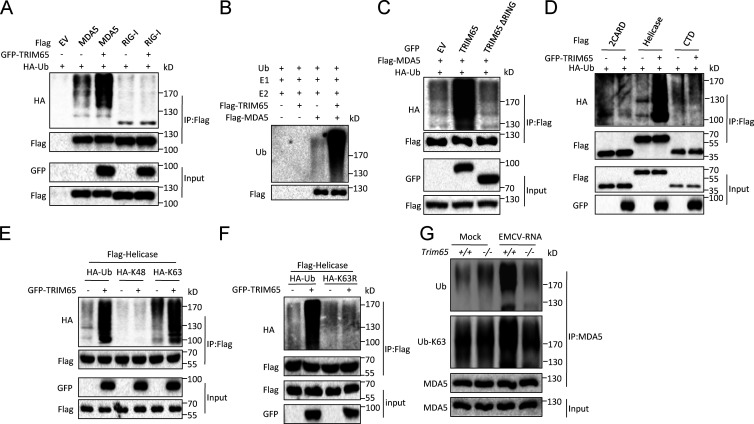
Because TRIM65 can interact with MDA5 and is essential for activation of MDA5 signaling, we investigated how TRIM65 promotes MDA5 activation. Because TRIM65 is an E3 ligase, we then investigated whether TRIM65 mediated MDA5 ubiquitination to promote its activation. First, we found that overexpression of TRIM65 could promote MDA5 ubiquitination in HEK-293T cells but could not induce RIG-I ubiquitination, suggesting the specific role of TRIM65 in MDA5 ubiquitination (Fig. 6 A). TRIM65-mediated MDA5 ubiquitination was also confirmed by in vitro ubiquitination assay (Fig. 6 B). In addition, the RING finger domain was critical for TRIM65-mediated MDA5 ubiquitination (Fig. 6 C). Next, we investigated which domain of MDA5 was ubiquitinated by TRIM65 and found that TRIM65 overexpression in HEK-293T cells could promote helicase domain ubiquitination, but could not ubiquitinate 2CARD or CTD domain of MDA5 (Fig. 6 D). We also investigated which type of ubiquitin linkage was occurring on MDA5 and found that the ubiquitin mutant that contains only one lysine at position 63 (K63) was sufficient for TRIM65-mediated MDA5 ubiquitination, but K48 mutant was not (Fig. 6 E). In addition, TRIM65 could not catalyze the linkage of K63R ubiquitin mutant, which contains a single lysine to arginine mutation at position 63, to MAD5 (Fig. 6 F), suggesting that TRIM65 mediates K63-linked MDA5 ubiquitination. To further confirm the role of TRIM65 in MDA5 ubiquitination, we also examined whether Trim65 deficiency affected MDA5 ubiquitination after recognizing viral RNA. The results showed that EMCV-RNA could induce MDA5 K63-linked ubiquitination in WT BMDMs, but not in Trim65−/− BMDMs (Fig. 6 G). These results indicate that TRIM65 mediates K63-linked ubiquitination of MDA5 at its helicase domain.
Figure 6.
TRIM65 promotes K63-linked polyubiquitination of MDA5. (A) Immunoblot analysis of MDA5 ubiquitination in HEK-293T cells transfected with constructs encoding various combinations (above lanes) of Flag-MDA5, Flag–RIG-I, GFP-TRIM65, and HA-Ub, followed by immunoprecipitation (IP) with anti-Flag antibody. EV, empty vector. (B) In vitro ubiquitination assay for purified MDA5 by TRIM65. (C) Immunoblot analysis of MDA5 ubiquitination in HEK-293T cells transfected with constructs encoding various combinations of Flag-MDA5, GFP-TRIM65, GFP-TRIM65 without RING domain, and HA-Ub, followed by IP with anti-Flag antibody. (D) Immunoblot analysis of ubiquitination of MDA5 domains in HEK-293T cells transfected with constructs encoding various combinations of Flag-tagged MDA5 proteins, GFP-TRIM65, and HA-Ub, followed by IP with anti-Flag antibody. (E and F) Immunoblot analysis of ubiquitination of MDA5 helicase domain in HEK-293T cells transfected with constructs encoding various combinations of Flag-helicase, GFP-TRIM65, HA-Ub, HA-tagged mutant ubiquitin with substitution of alanine for all lysine residues except the residue noted (K48 and K63; E), or HA-tagged mutant ubiquitin containing a single lysine to arginine mutation at position 63 (K63R; F), followed by IP with anti-Flag antibody. (G) Immunoblot analysis of endogenous MDA5 ubiquitination in WT or Trim65−/− BMDMs stimulated with EMCV-RNA for 3 h, followed by IP with anti-MDA5 antibody. Data are representative of three independent experiments.
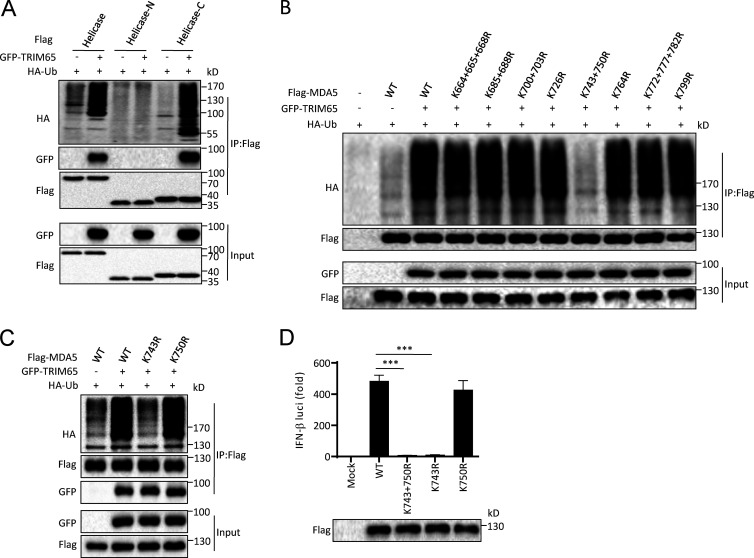
To identify the specific MDA5 lysine site that is ubiquitinated by TRIM65, we first examined which region of MDA5 helicase domain was ubiquitinated by TRIM65. We found that TRIM65 could interact with and ubiquitinate the C terminus of MDA5 helicase domain (helicase-C, aa 541–827) but did not interact with or ubiquitinate the N terminus of helicase domain (helicase-N, aa 295–541; Fig. 7 A), suggesting that the ubiquitinated lysine site is located in the C terminus of helicase domain. There are 15 lysines in this region of human MDA5, so we constructed several mutants in which one to three lysines were changed to arginine. When these mutants were overexpressed in HEK-293T cells together with TRIM65, we found that TRIM65-mediated MDA5 ubiquitination was abrogated when lysines at 743 and 750 were changed to arginines (Fig. 7 B). Further study showed that mutation at lysine 743 of MDA5 (K743R) blocked TRIM65-mediated MDA5 ubiquitination (Fig. 7 C), suggesting that TRIM65 ubiquitinates MDA5 at lysine 743. More importantly, we found that K743R mutation at MDA5 blocked its activity in an IFN-β promoter reporter assay (Fig. 7 D). These results indicate that TRIM65 promotes MDA5 ubiquitination at lysine 743, which is important for MDA5 activation.
Figure 7.
TRIM65 ubiquitinates MDA5 at lysine 743. (A) Immunoblot analysis of ubiquitination of MDA5 helicase domain in HEK-293T cells transfected with constructs encoding various combinations of Flag-helicase proteins, GFP-TRIM65, and HA-Ub, followed by immunoprecipitation (IP) with anti-Flag antibody. Helicase-N, N terminus of helicase domain; Helicase-C, C terminus of helicase domain. (B and C) Immunoblot analysis of MDA5 ubiquitination in HEK-293T cells transfected with constructs encoding various combinations of GFP-TRIM65, HA-Ub, Flag-tagged WT MDA5, or mutants with substitution of arginine for lysine residues at different sites, followed by IP with anti-Flag antibody. (D) Luciferase activity (top) and immunoblot analysis (bottom) of lysates of HEK-293T cells transfected with luciferase reporter constructs driven by promoters of genes encoding IFN-β, plus Flag-tagged WT MDA5 or mutants. Data are representative of three independent experiments (A–C) or from three independent experiments with biological duplicates in each (D, mean and sem of n = 6). Statistics were analyzed via an unpaired Student’s t test: ***, P < 0.001.
TRIM65 mediates MDA5 activation by promoting MDA5 oligomerization
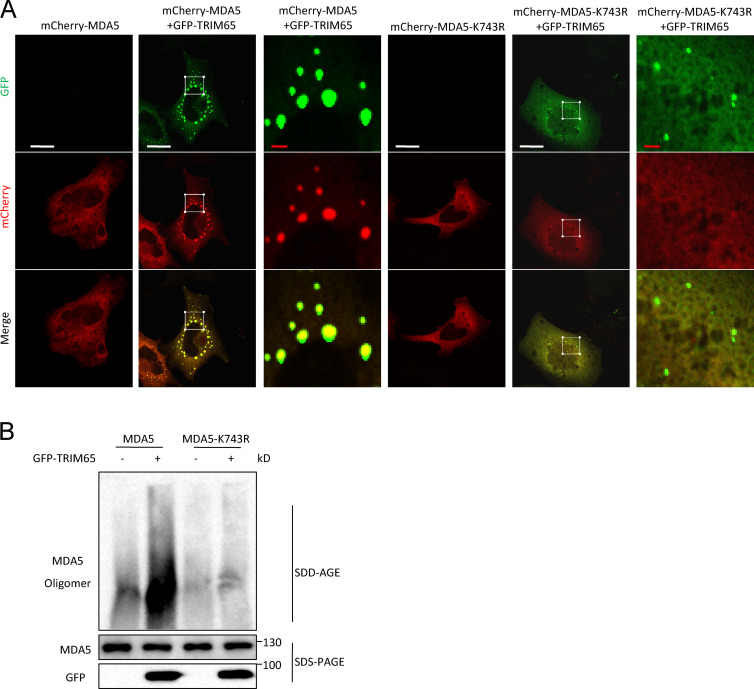
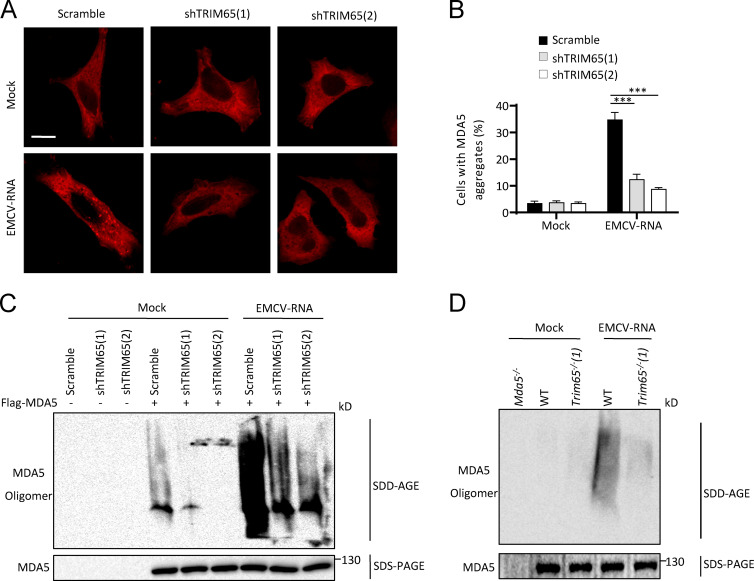
We then investigated how TRIM65-mediated MDA5 ubiquitination at lysine 743 promotes the activation of MDA5. Structural and biochemical data have suggested that the helicase-CTD of MDA5 forms a ring-like structure and cooperatively stacks along dsRNA to assemble a filament, which brings 2CARD into proximity and induces oligomerization of 2CARD (Wu et al., 2013). The 2CARD oligomer then induces MAVS filament formation and downstream signaling activation. In the filament, lysine 743 localizes at the interface between MDA5 monomers, suggesting that the ubiquitination of this site might be critical for the formation and stabilization of MDA5 filament (Wu et al., 2013). Indeed, we found that overexpression of TRIM65 could induce MDA5 to form aggregates in HeLa cells, but K743R mutation at MDA5 blocked this effect (Fig. 8 A). We further confirmed this result using semi-denaturing detergent agarose gel electrophoresis (SDD-AGE; Fig. 8 B), a method for detecting large protein oligomers in studying prions (Alberti et al., 2009; Hou et al., 2011). Consistent with this result, we found that EMCV-RNA treatment could induce MDA5 oligomerization in HeLa cells, but knockdown of TRIM65 expression inhibited this effect (Fig. 9, A–C). In addition, EMCV-RNA–induced oligomerization of endogenous MDA5 was absent in Trim65−/− BMDMs (Fig. 9 D). These results indicate that TRIM65-mediated ubiquitination is essential for MDA5 oligomerization.
Figure 8.
TRIM65-mediated MDA5 oligomerization depends on MDA5 ubiquitination at lysine 743. (A) Confocal microscopy analysis of HeLa cells cotransfected with plasmids for GFP-TRIM65, mCherry-MDA5, or MDA5 mutant (K743R). The third and sixth columns show higher-magnification images of white boxes in the second and fifth columns, respectively. Bars: (white) 20 µm; (red) 2 µm. (B) Immunoblot analysis of MDA5 in HEK-293T cells transfected with constructs encoding various combinations of GFP-TRIM65, MDA5, or MDA5 mutant (K743R) by SDD-AGE or SDS-PAGE assay. Data are representative of three independent experiments.
Figure 9.
EMCV-induced MDA5 oligomerization depends on TRIM65. (A and B) Confocal microscopy analysis (A) or qualification of aggregates (B) in HeLa cells stably expressing shRNA against TRIM65 that were transfected with mCherry-MDA5 for 24 h and then stimulated with EMCV-RNA for 3 h. Bar, 20 µm. (C) Immunoblot analysis of MDA5 by SDD-AGE or SDS-PAGE assay in HeLa cells stably expressing shRNA against TRIM65 that were transfected with Flag-MDA5 for 24 h and then stimulated with EMCV-RNA for 3 h. (D) Immunoblot analysis of MDA5 by SDD-AGE or SDS-PAGE assay in WT or Trim65−/− BMDMs that were primed with LPS for 12 h and then transfected with EMCV-RNA for 3 h. Data are from two independent experiments with biological duplicates in each (mean and SEM of n = 4; B) or are representative of two or three independent experiments (A, C, and D). Statistics were analyzed via an unpaired Student’s t test: ***, P < 0.001.
TRIM65 is essential for host defense against EMCV infection
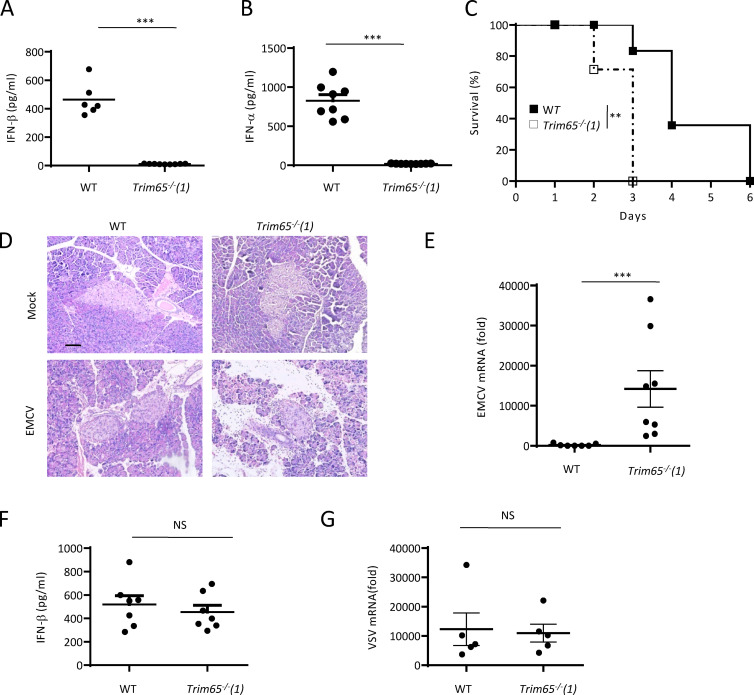
We next examined the in vivo role of TRIM65 in host defense against viral infection. Previous results have shown that the MDA5-mediated antiviral innate immune response is essential for the host to control EMCV infection (Gitlin et al., 2006; Kato et al., 2006), so we then challenged Trim65−/− mice with this virus to see the role of TRIM65 during viral infection. When the mice were infected with EMCV strain D, which primarily replicated in the pancreas, serum IFN-β and IFN-α levels were markedly decreased in Trim65−/− mice compared with control mice (Fig. 10, A and B). Consistent with this finding, Trim65−/− mice were highly susceptible to EMCV infection and showed more severe tissue damage and higher virus titer in the pancreas (Fig. 10, C–E). In contrast, Trim65−/− mice infected with VSV produced normal IFN-β and showed a capacity to control VSV infection similar to that of control mice (Fig. 10, F and G). Thus, these results demonstrate that TRIM65 is important for host defense against viral infection by mediating MDA5 activation in vivo.
Figure 10.
TRIM65 is critical for EMCV-induced MDA5 signaling activation in vivo. (A) Concentration of IFN-β in serum from age- and sex-matched WT (n = 6) or Trim65−/− (n = 9) mice inoculated intravenously with EMCV, assessed 12 h after infection. (B) Concentration of IFN-α in serum from age- and sex-matched WT (n = 8) or Trim65−/− (n = 9) mice inoculated intravenously with EMCV, assessed 12 h after infection. (C) Survival analysis of age- and sex-matched WT (n = 9) or Trim65−/− (n = 7) mice inoculated intravenously with EMCV for 6 d. (D) Histological analysis of pancreas sections from WT or Trim65−/− mice infected with EMCV for 48 h. Bar, 100 µm. (E) qPCR analysis of EMCV RNA in pancreas of WT (n = 8) or Trim65−/− (n = 9) mice infected with EMCV for 48 h. (F) Concentration of IFN-β in serum from age- and sex-matched WT (n = 7) or Trim65−/− (n = 7) mice inoculated intravenously with VSV, assessed 12 h after infection. (G) qPCR analysis of VSV RNA in pancreas of WT (n = 5) or Trim65−/− (n = 5) mice infected with VSV for 24 h. Data are from two or three independent experiments (A, B, and E–G; mean and SEM). Statistics were analyzed via an unpaired Students’ t test (A, B, and E–G) or a generalized Wilcoxon test (C): **, P < 0.01; ***, P < 0.001.
Discussion
MDA5 is an important cytoplasmic viral RNA sensors and can recognize dsRNA replication intermediates to activate the type I interferon signaling pathway after virus infection, so it plays a critical role in antiviral innate immunity (Takeuchi and Akira, 2008; Loo and Gale, 2011), but the mechanisms for MDA5 activation after sensing dsRNA are still unclear. In this study, we demonstrated an essential role of TRIM65-mediated ubiquitination in MDA5 activation and signal transduction. Deletion of Trim65 completely abolished dsRNA agonist or EMCV-induced IRF3 phosphorylation and subsequent interferon production. Mechanically, TRIM65 interacts with MDA5 helicase domain and ubiquitinates this domain at lysine 743, which is important for MDA5 oligomerization. Thus, our results indicate that TRIM65 functions as a critical component of the MDA5 signaling pathway.
Our results demonstrate that TRIM65 has a specific role in MDA5 activation. Depletion of TRIM65 expression blocked MDA5-dependent type I interferon production but had no effect on RIG-I–dependent type I interferon production. In line with this, TRIM65 only interacted with MDA5 but not with RIG-I. This specific interaction of TRIM65 with MDA5 was confirmed by a recent study (Kamanova et al., 2016, in which, they showed that TRIM65 specifically interacted with MDA5 and that its overexpression enhanced MDA5 activation in an IFN-β promoter reporter assay.
Ubiquitination plays a fundamental role in RIG-I activation, and noncovalent ubiquitin chain and TRIM25-mediated covalent ubiquitin conjunction can cooperate to promote the formation of and stabilize the 2CARD tetramer, which then induces MAVS oligomerization and downstream signaling activation (Gack et al., 2007; Zeng et al., 2010; Peisley et al., 2014). However, the role of ubiquitination in MDA5 activation is poorly understood. Jiang et al. (2012) proposed that the 2CARD domain of MDA5 can use the noncovalent ubiquitin chain for oligomerization and subsequent MAVS activation, similarly to the 2CARD domain of RIG-I. Wu et al. (2013) showed that the 2CARD of MDA5 has much lower affinity for K63 ubiquitin chain than RIG-I and that purified 2CARD of MDA5 can spontaneously form oligomers in a ubiquitin-independent manner, suggesting that ubiquitination might not be important for the oligomerization of 2CARD of MDA5. In this study, we demonstrate that TRIM65-mediated ubiquitination of MDA5 is a key step for its activation. In RIG-I activation, TRIM25 ubiquitinates the 2CARD domain of RIG-I and stabilizes its oligomer (Gack et al., 2007). We speculated that TRIM65 might promote MDA5 activation by a similar mechanism. But unexpectedly, the lysine site in MDA5 ubiquitinated by TRIM65 is located in the helicase domain, not in the 2CARD domain. Our results thus provide genetic and physiological evidence showing that ubiquitination plays a critical role in MDA5 activation.
Structural and biochemical data have suggested that purified MDA5 itself can form an oligomer or filament in the presence of dsRNA (Peisley et al., 2011; Berke et al., 2012; Wu et al., 2013), suggesting that binding of dsRNA is sufficient for MDA5 activation in vitro, but the mechanisms for MDA5 oligomerization and activation in vivo are not clear. In this study, we demonstrated that TRIM65-mediated ubiquitination at lysine 743 of the helicase domain is required for MDA5 oligomerization and activation in vivo. Based on structural information and the filament formation model (Wu et al., 2013; del Toro Duany et al., 2015), this lysine site is located on the interface between MDA5 monomers, suggesting that TRIM65-mediated ubiquitination at lysine 743 of MDA5 might be essential for the formation or stabilization of MDA5 filament. The different requirements for MDA5 oligomerization in vitro and in vivo might be caused by the high concentration of MDA5 protein in vitro, but detailed mechanisms need to be further investigated.
Collectively, our study has shown an essential role of TRIM65-mediated ubiquitination in MDA5 oligomerization and activation. This study not only identifies an important checkpoint protein for MDA5-mediated antiviral immunity, but also provides solid evidence showing the importance of ubiquitination in MDA5 oligomerization and activation.
Materials and methods
Mice
Mda5−/− mice have been described (Gitlin et al., 2006). Trim65−/− mice were generated by microinjection of TALEN mRNAs at Cyagen Biosciences (Guangzhou, China). These mice were on a C57BL/6 background; three lines lacking different bases were obtained. All animal experiments were approved by the Ethics Committee of the University of Science and Technology of China.
Reagents
5′-Triphosphate RNA, HMW poly I:C, and LMW poly I:C were from InvivoGen. Lipofectamine 2000 was from Invitrogen. The following antibodies were used for immunoprecipitation and Western blot: anti-GFP (M20004) and anti–β-actin (P30002) antibodies from Abmart; anti-Flag (F1804 and F7425) antibodies from Sigma-Aldrich; anti-MDA5 (21775-1-AP), anti-hemagglutinin (HA; 51064-2-AP), and anti-IRF3 (11312-1-AP) antibodies from Proteintech; anti–p-IRF3 (4947) antibody and mouse anti–rabbit IgG (light-chain specific, #3677) antibody from Cell Signaling Technology; anti-ubiquitin (SC8017) and anti–human TRIM65 (SC138707) antibodies from Santa Cruz Biotechnology, Inc.; and protein G agarose from EMD Millipore. Anti-Flag agarose beads and puromycin were from Sigma-Aldrich (P9620); IFN-β ELISA kit (42400-1) and IFN-α ELISA kit (42120) were from PBL Interferon Source; SYBR premix used for real-time PCR was from Takara Bio Inc. (RR820A); protease inhibitor cocktail was from Sigma-Aldrich (P8340); and PMSF was from Beyotime (ST506). The human recombinant ubiquitin (U-100H), UBE1 (E-305), and His6 UBE2N (Ubc13)/Uev1a complex (E2-664) were from Boston Biochem. HEK-293T, HEK-293, HeLa, L929, and THP-1 cell lines were from ATCC; mycoplasma contamination test results were negative.
Viruses
EMCV was propagated and amplified by infection of a monolayer of HeLa cells. VSV (Indiana strain) was propagated on BHK-21 cells. Adenovirus was from Beijing Five Plus Molecular Medicine Institute. The viruses were stored at −80°C.
Viral infection in vivo
For EMCV infection, 7-wk-old mice were challenged by tail intravenous injection of 5 × 105 pfu EMCV. Serum was prepared after 12 h, and IFN-β or IFN-α production levels were determined by ELISA. The survival of mice infected intraperitoneally with 10 pfu EMCV was monitored every 12 h for 6 d. As for the determination of EMCV virus titers, mice were killed after 48 h, and titers in pancreas were determined by real-time PCR using the following primers: forward, 5′-AATGCCCACTACGCTGGT-3′; and reverse, 5′-GTCGTTCGGCAGTAGGGT-3′. For VSV infection, mice were challenged by tail intravenous injection of 107 pfu virus. After 12 h, serum was collected and IFN-β production levels were determined by ELISA. Titers in livers were determined after 24 h by real-time PCR using the following primers: forward, 5′-TGGGATGACTGGGCTCCATA-3′, and reverse, 5′-CACCATCAGGAAGCTGCGAA-3′.
Preparation of virus RNA
HEK-293 cells were infected with EMCV (MOI = 0.04) or VSV (MOI = 1) for 18 h, followed by total RNA extraction using TRIzol reagent (Takara Bio Inc.), and RNA was stored at −80°C.
Reconstitution of TRIM65 in Trim65−/− MEFs
Full-length cDNA of mouse TRIM65, as well as TRIM65-ΔSPRY, were cloned into GFP-tagged plex-MCS vector and transfected into HEK-293T cells with packaging plasmids to generate lentiviral particles containing TRIM65 or TRIM65-ΔSPRY. The lentiviral particles were then used to infect Trim65−/− MEFs for 48 h, and the cells were used for subsequent experiments.
In vitro ubiquitination assay
Flag-TRIM65 and Flag–MDA-5 were purified through coimmunoprecipitation from overexpressed HEK-293T cells, then eluted by Flag peptide. Ubiquitination reaction was performed with 0.1 µM UBE1, 1 µM Ubc13/Uev1A, 100 µM HA-tagged WT ubiquitin (HA-Ub), and 1 mM DTT in 1× MgATP buffer at 30°C for 1 h. Flag-TRIM65 or Flag–MDA-5 was included where indicated (Jiang et al., 2012).
Luciferase reporter gene assay
HEK-293T cells were seeded on 24-well plates (2 × 105 cells per well) and transfected with reporter vectors for Ifnb–firefly luciferase (100 ng) and Renilla luciferase (50 ng) plus expression vector for WT MDA5 or MDA5 mutants (100 ng). Empty control vector was added so that a total of 250 ng of vector DNA was transfected into each well of cells. 24 h after transfection, cells were collected and the luciferase activity in cell lysates was analyzed with the Dual-Luciferase Reporter Assay System (E1960; Promega).
Generation of HeLa cells expressing shRNA
shRNA clones targeting mRNA of TRIM65 were from Sigma-Aldrich. Sequences of shRNA are as follows: shTRIM65 (1), 5′-GAATTATCGCAATCTGACCTT-3′; and shTRIM65 (2), 5′-CCGTCCTGTCTTGTAGTCTTT-3′. shTRIM65 (1) targets the coding region of TRIM65 gene, and shTRIM65 (2) targets the 3′ UTR region of TRIM65 gene. The protocol for generating HeLa cells stably expressing shRNA has been published (Yan et al., 2013).
Cell preparation and stimulation
BMDMs were derived from tibial and femoral BM cells and cultured in DMEM complemented with 10% FBS, 1 mM sodium pyruvate, and 2 mM l-glutamine (Didierlaurent et al., 2006) in the presence of culture supernatants of L929 mouse fibroblasts. MEFs were generated from 13.5-d-old mouse embryos and cultured in complete DMEM.
For inducing IFN-β, 5 × 105 HeLa cells, 5 × 105 MEFs, or 1.5 × 106 BMDMs were plated in 6-well plates overnight, and the medium was changed to Opti-MEM the next morning. After that, the cells were stimulated with EMCV (MOI = 0.025 or 0.05), VSV (MOI = 0.03 or 0.06), or AdV (MOI = 1,000) for 12 h. For stimulation of TLR3 or TLR4, BMDMs were treated with LMW poly I:C (10 µg/ml) or LPS (50 ng/ml) for 4 h. For cytosol delivery, EMCV-RNA (2 µg/ml), VSV-RNA (500 ng/ml), HMW poly I:C (1 µg/ml), LMW poly I:C (1 µg/ml), or 3pRNA (1 µg/ml) were transfected using Lipofectamine 2000 according to the manufacturer’s protocol (Invitrogen). Cell extracts were analyzed by real-time PCR and immunoblot.
Histological analysis
Mice were infected with 10 pfu EMCV intraperitoneally. After 48 h, pancreases were taken from EMCV-infected mice, fixed with 4% formaldehyde, cut, and stained with hematoxylin and eosin.
ELISA
Mouse IFN-β or IFN-α in supernatants of cell culture or serum was quantified by ELISA according to the manufacturer’s instructions (PBL Interferon Source).
Real-time PCR
BMDM or HeLa cells were dissolved in TRIzol reagent, and total RNA was extracted. cDNA was synthesized from total RNA with an M-MLV Reverse Transcriptase kit according to the manufacturer’s protocol (Invitrogen). SYBR Green premix (Takara Bio Inc.) was used for quantitative PCR (qPCR) with a StepOne Real Time PCR System (Applied Biosystems). GAPDH was used as an internal control. The sequences of primers for qPCR were as follows: mouse Ifn-β forward, 5′-CCAGCTCCAAGAAAGGACGA-3′; and reverse, 5′-GGAGCATCTCTTGGATGGCA-3′; mouse GAPDH forward, 5′-GGTGAAGGTCGGTGTGAACG-3′; and reverse, 5′-CTCGCTCCTGGAAGATGGTG-3′; mouse ISG15 forward, 5′-TGACTGTGAGAGCAAGCAGC-3′; and reverse, 5′-CCCCAGCATCTTCACCTTTA-3′; mouse ISG56 forward, 5′-GAGCCAGAAAACCCTGAGTACA-3′; and reverse, 5′-AGAAATAAAGTTGTCATCTAAATC-3′; human IFN-β forward, 5′-GCACTGGCTGGAATGAGACT-3′; and reverse, 5′-CTGTCCTCCTTGGCCTTCAG-3′; and human GAPDH forward, 5′-GTCAAGGCTGAGAACGGGAA-3′; and reverse, 5′-AAATGAGCCCCAGCCTTCTC-3′.
Transfection and immunoprecipitation
Constructs were transfected into HEK-293T cells through the use of polyethylenimine. After 24 h, cells were collected and resuspended in lysis buffer (50 mM Tris, pH 7.8, 50 mM NaCl, 1% [vol/vol] Nonidet-P40, 5 mM EDTA, and 10% [vol/vol] glycerol). Extracts were immunoprecipitated with anti-Flag beads and then were assessed by immunoblot analysis. BMDMs were transfected with EMCV-RNA for 3 h and then resuspended in lysis buffer, and proteins were immunoprecipitated from extracts with anti-MDA5 antibody and then used for immunoblot analysis.
Confocal microscopy
HeLa cells were plated on coverslips overnight, and then plasmids were transfected using polyethylenimine. After 24 h, cells were transfected with EMCV-RNA (1.5 µg/ml) for 1.5 h, washed three times with PBS, fixed for 15 min with 4% PFA in PBS, and then washed three times with PBST. ZEISS LSM700 was used for confocal microscopy. For quantification of the percentage of cells with mCherry-MDA5 aggregates (>3 aggregates per cell), scanning fields were randomly selected, and at least 100 cells were counted in each slide.
SDD-AGE
The oligomerization of MDA5 was analyzed according to the published protocol (Hou et al., 2011). Cells were lysed with Triton X-100 lysis buffer (0.5% Triton X-100, 50 mM Tris-HCl, 150 mM NaCl, 10% glycerol, 1 mM PMSF, and protease inhibitor cocktail), resuspended in 1× sample buffer (0.5× TBE, 10% glycerol, 2% SDS, and 0.0025% bromophenol blue), and loaded onto a vertical 1.5% agarose gel. (1× TBE contains 89 mM Tris, pH 8.3, 89 mM boric acid, and 2 mM EDTA.) After electrophoresis in the running buffer (1× TBE and 0.1% SDS) for 1 h with a constant voltage of 80 V at 4°C, the proteins were transferred to an Immobilon membrane (EMD Millipore) for immunoblotting.
Statistical analyses
Statistical analyses were performed with an unpaired Student’s t test for two groups (GraphPad Software). Kaplan–Meier plots were constructed, and a generalized Wilcoxon test was used to test for differences in survival between WT and Trim65−/− mice after viral infection. P-values of <0.05 were considered significant.
Online supplemental material
Fig. S1 shows the strategy for the generation of Trim65 mutant mouse lines by TALEN.
Acknowledgments
We thank Dr. Marco Colonna (Washington University, St. Louis, MO) for providing Mda5−/− mice. We thank Dr. Zhengfan Jiang (Peking University, Beijing 100000, China) for providing the VSV strain and Dr. Hong Tang (Wuhan Institute of Virology, Chinese Academy of Sciences, Wu Han 430071, China) for providing the EMCV strain.
This work was supported by the National Basic Research Program of China (grants 2014CB910800 and 2013CB944904), National Natural Science Foundation of China (grants 81273318, 81525013, 31300745, and 81571609), the Young Talent Support Program, and the Fundamental Research Funds for the Central Universities.
The authors declare no competing financial interests.
Author contributions: X. Lang and T. Tang performed experiments; W. Jiang, T. Jin, C. Ding, and R. Zhou designed the research; X. Lang, W. Jiang, and R. Zhou wrote the manuscript; and W. Jiang and R. Zhou supervised the project.
Footnotes
Abbreviations used:
- dsRNA
- double-stranded RNA
- EMCV
- encephalomyocarditis virus
- HA
- hemagglutinin
- HMW
- high molecular weight
- LMW
- low molecular weight
- MAVS
- mitochondrial antiviral signaling
- poly I:C
- polyriboinosinic:polyribocytidylic acid
- qPCR
- quantitative PCR
- RIG
- retinoic acid–inducible
- SDD-AGE
- semi-denaturing detergent agarose gel electrophoresis
- TALEN
- transcription activator–like effector nuclease
- TRIM
- tripartite motif
- VSV
- vesicular stomatitis virus
References
- Alberti, S., Halfmann R., King O., Kapila A., and Lindquist S.. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 137:146–158. 10.1016/j.cell.2009.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke, I.C., Yu X., Modis Y., and Egelman E.H.. 2012. MDA5 assembles into a polar helical filament on dsRNA. Proc. Natl. Acad. Sci. USA. 109:18437–18441. 10.1073/pnas.1212186109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Z., and Wang J.. 2013. TRIM dsDNA sensor to restrict innate immune response. Cell. Mol. Immunol. 10:193–195. 10.1038/cmi.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X. 2016. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 16:35–50. 10.1038/nri.2015.8 [DOI] [PubMed] [Google Scholar]
- Chan, Y.K., and Gack M.U.. 2015. RIG-I-like receptor regulation in virus infection and immunity. Curr. Opin. Virol. 12:7–14. 10.1016/j.coviro.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Toro Duany, Y., Wu B., and Hur S.. 2015. MDA5-filament, dynamics and disease. Curr. Opin. Virol. 12:20–25. 10.1016/j.coviro.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didierlaurent, A., Brissoni B., Velin D., Aebi N., Tardivel A., Käslin E., Sirard J.C., Angelov G., Tschopp J., and Burns K.. 2006. Tollip regulates proinflammatory responses to interleukin-1 and lipopolysaccharide. Mol. Cell. Biol. 26:735–742. 10.1128/MCB.26.3.735-742.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack, M.U., Shin Y.C., Joo C.H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., and Jung J.U.. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 446:916–920. 10.1038/nature05732 [DOI] [PubMed] [Google Scholar]
- Gitlin, L., Barchet W., Gilfillan S., Cella M., Beutler B., Flavell R.A., Diamond M.S., and Colonna M.. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA. 103:8459–8464. 10.1073/pnas.0603082103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau, D., Deddouche S., and Reis e Sousa C.. 2013. Cytosolic sensing of viruses. Immunity. 38:855–869. 10.1016/j.immuni.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung, V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M., et al. . 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 314:994–997. 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- Hou, F., Sun L., Zheng H., Skaug B., Jiang Q.X., and Chen Z.J.. 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 146:448–461. 10.1016/j.cell.2011.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X., and Chen Z.J.. 2012. The role of ubiquitylation in immune defence and pathogen evasion. Nat. Rev. Immunol. 12:35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X., Kinch L.N., Brautigam C.A., Chen X., Du F., Grishin N.V., and Chen Z.J.. 2012. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 36:959–973. 10.1016/j.immuni.2012.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamanova, J., Sun H., Lara-Tejero M., and Galán J.E.. 2016. The Salmonella effector protein SopA modulates innate immune responses by targeting TRIM E3 ligase family members. PLoS Pathog. 12:e1005552. 10.1371/journal.ppat.1005552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J., et al. . 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 441:101–105. 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- Kato, H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., and Akira S.. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601–1610. 10.1084/jem.20080091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E., Stein S., and Falck-Pedersen E.. 2014. Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J. Virol. 88:974–981. 10.1128/JVI.02702-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Chai Q.Y., and Liu C.H.. 2016. The ubiquitin system: a critical regulator of innate immunity and pathogen-host interactions. Cell. Mol. Immunol. 13:560–576. 10.1038/cmi.2016.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Wang L., Fu B., Berman M.A., Diallo A., and Dorf M.E.. 2014. TRIM65 regulates microRNA activity by ubiquitination of TNRC6. Proc. Natl. Acad. Sci. USA. 111:6970–6975. 10.1073/pnas.1322545111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo, Y.M., and Gale M. Jr. 2011. Immune signaling by RIG-I-like receptors. Immunity. 34:680–692. 10.1016/j.immuni.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroni, G. 2012. Genomics and evolution of the TRIM gene family. Adv. Exp. Med. Biol. 770:1–9. 10.1007/978-1-4614-5398-7_1 [DOI] [PubMed] [Google Scholar]
- Oshiumi, H., Matsumoto M., Hatakeyama S., and Seya T.. 2009. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-β induction during the early phase of viral infection. J. Biol. Chem. 284:807–817. 10.1074/jbc.M804259200 [DOI] [PubMed] [Google Scholar]
- Oshiumi, H., Miyashita M., Inoue N., Okabe M., Matsumoto M., and Seya T.. 2010. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe. 8:496–509. 10.1016/j.chom.2010.11.008 [DOI] [PubMed] [Google Scholar]
- Ozato, K., Shin D.M., Chang T.H., and Morse H.C. III. 2008. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8:849–860. 10.1038/nri2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley, A., Lin C., Wu B., Orme-Johnson M., Liu M., Walz T., and Hur S.. 2011. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc. Natl. Acad. Sci. USA. 108:21010–21015. 10.1073/pnas.1113651108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley, A., Wu B., Xu H., Chen Z.J., and Hur S.. 2014. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 509:110–114. 10.1038/nature13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair, A., Schulz O., Tan C.P., Näslund T.I., Liljeström P., Weber F., and Reis e Sousa C.. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 314:997–1001. 10.1126/science.1132998 [DOI] [PubMed] [Google Scholar]
- Pichlmair, A., Schulz O., Tan C.P., Rehwinkel J., Kato H., Takeuchi O., Akira S., Way M., Schiavo G., and Reis e Sousa C.. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 83:10761–10769. 10.1128/JVI.00770-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee, M., Roth A., Hornung V., Hagmann C.A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G., et al. . 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 31:25–34. 10.1016/j.immuni.2009.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, O., and Akira S.. 2008. MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 20:17–22. 10.1016/j.coi.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Versteeg, G.A., Rajsbaum R., Sánchez-Aparicio M.T., Maestre A.M., Valdiviezo J., Shi M., Inn K.S., Fernandez-Sesma A., Jung J., and García-Sastre A.. 2013. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 38:384–398. 10.1016/j.immuni.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, B., and Hur S.. 2015. How RIG-I like receptors activate MAVS. Curr. Opin. Virol. 12:91–98. 10.1016/j.coviro.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, B., Peisley A., Richards C., Yao H., Zeng X., Lin C., Chu F., Walz T., and Hur S.. 2013. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 152:276–289. 10.1016/j.cell.2012.11.048 [DOI] [PubMed] [Google Scholar]
- Yan, Y., Jiang W., Spinetti T., Tardivel A., Castillo R., Bourquin C., Guarda G., Tian Z., Tschopp J., and Zhou R.. 2013. Omega-3 fatty acids prevent inflammation and metabolic disorder through inhibition of NLRP3 inflammasome activation. Immunity. 38:1154–1163. 10.1016/j.immuni.2013.05.015 [DOI] [PubMed] [Google Scholar]
- Yoneyama, M., Onomoto K., Jogi M., Akaboshi T., and Fujita T.. 2015. Viral RNA detection by RIG-I-like receptors. Curr. Opin. Immunol. 32:48–53. 10.1016/j.coi.2014.12.012 [DOI] [PubMed] [Google Scholar]
- Zeng, W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., and Chen Z.J.. 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 141:315–330. 10.1016/j.cell.2010.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Lee A.J., Wu X., and Sun S.C.. 2011. Regulation of antiviral innate immunity by deubiquitinase CYLD. Cell. Mol. Immunol. 8:502–504. 10.1038/cmi.2011.42 [DOI] [PMC free article] [PubMed] [Google Scholar]