Tschurtschenthaler et al. report a Crohn’s disease–like ileitis mediated by IRE1α that develops in mice with intestinal-epithelial Atg16l1 deletion when they age. The authors propose a selective autophagy process involved in the removal of IRE1α clusters during ER stress.
Abstract
ATG16L1T300A, a major risk polymorphism in Crohn’s disease (CD), causes impaired autophagy, but it has remained unclear how this predisposes to CD. In this study, we report that mice with Atg16l1 deletion in intestinal epithelial cells (IECs) spontaneously develop transmural ileitis phenocopying ileal CD in an age-dependent manner, driven by the endoplasmic reticulum (ER) stress sensor IRE1α. IRE1α accumulates in Paneth cells of Atg16l1ΔIEC mice, and humans homozygous for ATG16L1T300A exhibit a corresponding increase of IRE1α in intestinal epithelial crypts. In contrast to a protective role of the IRE1β isoform, hyperactivated IRE1α also drives a similar ileitis developing earlier in life in Atg16l1;Xbp1ΔIEC mice, in which ER stress is induced by deletion of the unfolded protein response transcription factor XBP1. The selective autophagy receptor optineurin interacts with IRE1α, and optineurin deficiency amplifies IRE1α levels during ER stress. Furthermore, although dysbiosis of the ileal microbiota is present in Atg16l1;Xbp1ΔIEC mice as predicted from impaired Paneth cell antimicrobial function, such structural alteration of the microbiota does not trigger ileitis but, rather, aggravates dextran sodium sulfate–induced colitis. Hence, we conclude that defective autophagy in IECs may predispose to CD ileitis via impaired clearance of IRE1α aggregates during ER stress at this site.
Introduction
Crohn’s disease (CD), a remitting-relapsing incurable chronic inflammatory bowel disease (IBD) commonly manifesting in early adulthood, has become a global health issue with rapidly increasing incidence in Asia and other parts of the world, following a steep rise in Europe and North America over the last decades (Molodecky et al., 2012). Although the environmental triggers remain enigmatic, association studies have successfully identified the genomic susceptibility landscape of CD (Jostins et al., 2012; Liu et al., 2015). With ∼200 IBD risk loci scattered across the genome, which are individually mostly associated with minimal incremental risk (Jostins et al., 2012; Liu et al., 2015), it is easily overlooked that a huge proportion of overall explained heritability resides within very few risk genes, which are indeed specific for CD and not shared with other immune-related diseases. Among those, the coding polymorphism ATG16L1T300A is particularly noteworthy (Hampe et al., 2007; Rioux et al., 2007) as it has led to the revelation that impaired autophagy (Murthy et al., 2014), and pathways that impinge upon it (Adolph et al., 2013), may be a main pathogenic disease mechanism in CD (Maloy and Powrie, 2011; Baxt and Xavier, 2015). This view is corroborated by additional risk genes involved in autophagy (e.g., IRGM and LRRK2) and by the discovery that ATG16L1 and NOD2, the latter encoded by the CD-specific risk gene with the largest individual effect size, can interact during autophagosome formation (Cooney et al., 2010; Homer et al., 2010; Travassos et al., 2010). Although CD can arise anywhere in the gastrointestinal tract, both NOD2 and ATG16L1 risk variants are most strongly associated with CD involving the ileum (Prescott et al., 2007).
Abnormalities in the secretory compartment of Paneth cells, specialized intestinal epithelial cells (IECs) located at the base of crypts of the small intestine (Clevers and Bevins, 2013), are a common consequence of hypomorphic ATG16L1 (Cadwell et al., 2008, 2010; Adolph et al., 2013; Deuring et al., 2014), NOD2 (Kobayashi et al., 2005; Wehkamp et al., 2005), IRGM (Liu et al., 2013), and LRRK2 (Zhang et al., 2015). In addition to disordered secretory granules that contain antimicrobial peptides and protein, CD patients and healthy individuals homozygous at the ATG16L1T300A single-nucleotide polymorphism (SNP) characteristically exhibit ER stress in their Paneth cells as indicated by high GRP78 expression (Deuring et al., 2014), which is also observed in mice with IEC-specific deletion of Atg16l1 (Adolph et al., 2013). ER stress in Paneth cells, elicited via deletion of the unfolded protein response (UPR) transcription factor XBP1 in all IECs or Paneth cells specifically, induces a mild, superficial form of enteritis (Kaser et al., 2008; Adolph et al., 2013). In Xbp1ΔIEC mice, ATG16L1-dependent autophagy can partially compensate via yet unknown mechanisms for ER stress. Importantly, in the absence of such compensation, Atg16l1;Xbp1ΔIEC mice develop discontinuous, transmural fissuring ileitis phenocopying CD (Adolph et al., 2013). Here, we investigated the mechanisms engaged by impaired ATG16L1-dependent autophagy that drive ileal inflammation.
Results
Age-dependent manifestation of CD-like ileitis in Atg16l1ΔIEC mice
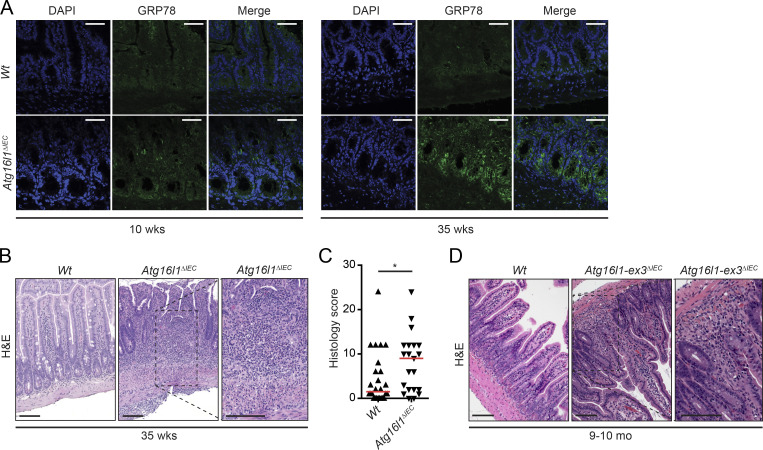
In nematodes, the capacity to resolve ER stress declines rapidly with increasing age (Taylor and Dillin, 2013). In addition, Atg16l1ΔIEC mice (Adolph et al., 2013) and humans homozygous for ATG16L1T300A (Deuring et al., 2014) exhibit increased ER stress in the intestinal epithelium. Therefore, we hypothesized that IECs, which are continuously exposed to a large variety of potential stressors that could have direct and indirect effects on protein folding, would become increasingly dependent on autophagy as a compensatory mechanism to resolve ER stress over time. Indeed, 35-wk-old Atg16l1ΔIEC mice exhibited higher expression of the ER stress marker GRP78 in crypt IECs compared with 10-wk-old Atg16l1ΔIEC mice, whereas Wt mice of either age exhibited minimal GRP78 immunoreactivity (Fig. 1 A). Surprisingly, in light of earlier studies that Atg16l1ΔIEC and Atg16l1HM mice do not develop intestinal inflammation (Cadwell et al., 2008, 2010; Adolph et al., 2013; Conway et al., 2013), 35-wk-old Atg16l1ΔIEC mice developed a transmural ileitis, characterized by patchy infiltration of mononuclear and polymorphonuclear cells that penetrated the mucosa, submucosa, and muscularis propria of the ileum (Fig. 1, B and C). Other parts of the small intestine or the colon were not affected (not depicted). The features and severity of inflammation in 35-wk-old Atg16l1ΔIEC mice were indistinguishable from the CD-like ileitis that develops earlier in life in Atg16l1;Xbp1ΔIEC mice (Adolph et al., 2013), in which the UPR transcription factor XBP1 is co-deleted to induce ER stress (Kaser et al., 2008). Notably, transmural ileitis was also observed in 9–10-mo-old mice with an IEC-specific deletion of a genetically distinct Atg16l1-mutant allele (loxP sites flanking exon 3 rather than exon 1 [Adolph et al., 2013]) and held in a different (Los Angeles rather than Cambridge) animal facility (Fig. 1 D). These studies suggest that environmental and/or microbial stress accumulating over time can serve as a means to uncover the phenotypic consequences of impaired autophagy in the development of CD-like ileitis.
Figure 1.
35-wk-old Atg16l1ΔIEC mice exhibit increased ER stress and transmural CD-like ileitis. (A) Representative images of GRP78 (green) immunoreactivity in 10- and 35-wk-old Atg16l1ΔIEC mice. n = 3. DAPI is shown in blue. Bars, 50 µm. (B and C) Representative H&E images (B) and enteritis histology score of 35-wk-old Atg16l1ΔIEC mice maintained in a mouse norovirus–free facility (Cambridge; C). n = 25/21. The median is shown. *, P < 0.05, Mann–Whitney U test. Note the transmural character of inflammation in Atg16l1ΔIEC mice compared with Wt mice. Bars, 100 µm. (D) Representative H&E images of 9–10-mo-old Atg16l1-ex3ΔIEC mice generated and housed at Cedars-Sinai SPF mouse facility in Los Angeles. The inset shows the transmural character of the inflammation seen in Atg16l1-ex3ΔIEC mice. n = 10. Bars, 100 µm.
IRE1α promotes and IRE1β protects from CD-like ileitis in Atg16l1;Xbp1ΔIEC mice
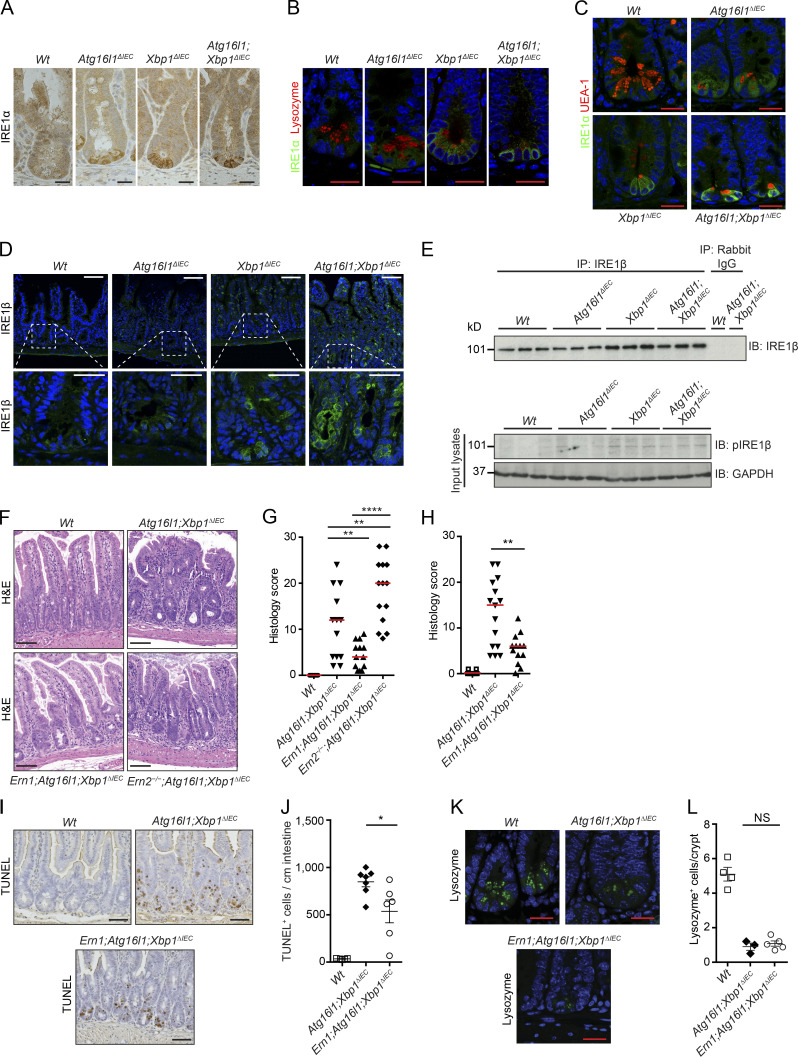
To mechanistically examine the development of ileitis in the setting of impaired autophagy in the intestinal epithelium, we first focused our attention on 10-wk-old Atg16l1;Xbp1ΔIEC mice (Adolph et al., 2013) to allow for a tractable system to study. We had previously noted on immunoprecipitation and immunoblot experiments that total IEC lysates of Atg16l1;Xbp1ΔIEC mice exhibit profoundly increased IRE1α expression and phosphorylation compared with lysates from Xbp1ΔIEC mice (Adolph et al., 2013), but it has remained unclear whether this has a role in transmural ileitis. IRE1α is the ER transmembrane receptor that responds to protein misfolding by activating the XBP1 branch of the UPR, aimed at resolving such ER stress (Tirasophon et al., 2000; Lee et al., 2002; Hetz et al., 2011; Walter and Ron, 2011; Wang and Kaufman, 2016). However, increasing and prolonged ER stress causes IRE1α activation to transition from an adaptive to a maladaptive, pro-death and proinflammatory UPR, associated with IRE1α forming homodimers and large clusters, respectively (Li et al., 2010; Korennykh and Walter, 2012; Ghosh et al., 2014). Whereas IRE1α was barely detectable in Vil-Creneg (Wt) littermate controls, IEC-specific deletion of Atg16l1 or Xbp1 in 10-wk-old Atg16l1ΔIEC and Xbp1ΔIEC mice, respectively, led to increased IRE1α immunoreactivity specifically in small intestinal crypts, with the highest levels detected when both Atg16l1 and Xbp1 were absent as observed in Atg16l1;Xbp1ΔIEC mice (Fig. 2 A). Furthermore, IRE1α immunofluorescence localized most predominantly to Paneth cells, which were identified by antilysozyme and by lectin Ulex europaeus agglutinin 1 (UEA-1) costaining (Fig. 2, B and C). Other highly secretory IECs such as goblet cells, where protein misfolding might be prevalent, did not exhibit increased IRE1α immunoreactivity upon Atg16l1 and/or Xbp1 deletion (Fig. 2, A–C). Whereas IRE1α (encoded by Ern1) at low levels is ubiquitously expressed, the intestinal and bronchial epithelium expresses a paralogue with high sequence similarity, IRE1β (encoded by Ern2; Bertolotti et al., 2001; Iwawaki et al., 2001; Martino et al., 2013), reported to be involved in mucin 2 (MUC2) processing, goblet cell maturation (Tsuru et al., 2013), and enterocyte lipid metabolism (Iqbal et al., 2008). We noted increased immunoreactivity of IRE1β mostly in crypt but also in villus IECs of Xbp1ΔIEC and Atg16l1;Xbp1ΔIEC but not in Atg16l1ΔIEC mice at 10 wk of age when compared with Wt littermates (Fig. 2 D), which was corroborated by immunoprecipitation experiments from purified intestinal crypts (Fig. 2 E).
Figure 2.
IRE1α controls CD-like inflammation. (A) Representative images of IRE1α immunohistochemistry (brown) on ileal sections of 10-wk-old Atg16l1ΔIEC, Xbp1ΔIEC, and Atg16l1;Xbp1ΔIEC mice. n = 3. Hematoxylin is in blue. Bars, 20 µm. (B) Representative confocal images of IRE1α immunofluorescence (green) and Paneth cell–derived lysozyme (red) on ileal sections from the indicated genotypes at 10 wk of age. Note the perinuclear cytoplasmic IRE1α immunoreactivity of Paneth cells from Atg16l1ΔIEC mice that are lysozyme positive on the luminally oriented apex. n = 3. DAPI is in blue. Bars, 20 µm. (C) Representative confocal images of IRE1α immunofluorescence (green) and the Paneth and goblet cell–type–specific lectin UEA-1 (red) on ileal sections from the indicated genotypes at 10 wk of age. n = 3. DAPI is in blue. Bars, 20 µm. (D) Representative confocal images of IRE1β immunofluorescence (green) of the indicated genotypes at 10 wk of age. Note the specific IRE1β immunoreactivity of crypts (insets, bottom) and villus IECs from Atg16l1;Xbp1ΔIEC and Xbp1ΔIEC mice. DAPI is in blue. n = 3. Bars: (top) 50 µm; (bottom) 20 µm. (E) IRE1β immunoprecipitation (IP) and immunoblot (IB) as well as pIRE1β immunoblot in crypt lysates. n = 3. GAPDH was used as the loading control. (F and G) Representative H&E images (F) and their histological score (G) of 10-wk-old mice. n = 28/13/13/9. The median is shown. One-way ANOVA with Bonferroni’s correction was used. Bars, 100 µm. (H) Enteritis histology score of the indicated genotypes housed at the mouse norovirus–positive SPF animal facility (ZVTA) of the Medical University of Innsbruck. n = 12/15/13. The median is shown. Unpaired two-tailed Student’s t test was used. (I and J) Representative images of TUNEL-labeled IECs (brown; I) and quantification (J). n = 5/7/6. The mean ± SEM is shown. Mann–Whitney U test was used. Hematoxylin is in blue. Bars, 50 µm. (K and L) Representative confocal images of lysozyme immunofluorescence in crypt IECs (green; K) and quantification (L). n = 4/3/5. The mean ± SEM is shown. A Mann–Whitney U test was used. DAPI is in blue. Bars, 20 µm. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
To investigate a possible contribution of IRE1α and IRE1β to CD-like ileitis in Atg16l1;Xbp1ΔIEC mice, we generated Ern1;Atg16l1;Xbp1ΔIEC and Ern2−/−;Atg16l1;Xbp1ΔIEC mice, respectively. Fig. 2 (F and G) demonstrates that ileitis is prevented by IEC-specific deletion of Ern1, whereas germline deletion of Ern2 increases the severity of inflammation. The effect of Ern1 deletion was not specific for the particular microbial environment in this specific pathogen–free (SPF) facility (Cambridge) but similarly observed in another SPF facility (Innsbruck), from which the Atg16l1;Xbp1ΔIEC parent strain has been rederived (Fig. 2 H). Protection from ileitis afforded by IEC-specific elimination of IRE1α function in Ern1;Atg16l1;Xbp1ΔIEC mice was also associated with a decrease in the number of dying TdT-mediated dUTP nick-end labeling (TUNEL)+ IECs (Fig. 2, I and J) but, importantly, not with rescue of impaired lysozyme staining in Paneth cells compared with Atg16l1;Xbp1ΔIEC mice (Fig. 2, K and L). Altogether, these experiments identified unexpected divergent roles of the two closely related IRE1 isoforms in the intestinal epithelium, with IRE1α as driver of CD-like ileitis and IRE1β providing a protective function.
IRE1α drives CD-like ileitis in 35-wk-old Atg16l1ΔIEC mice
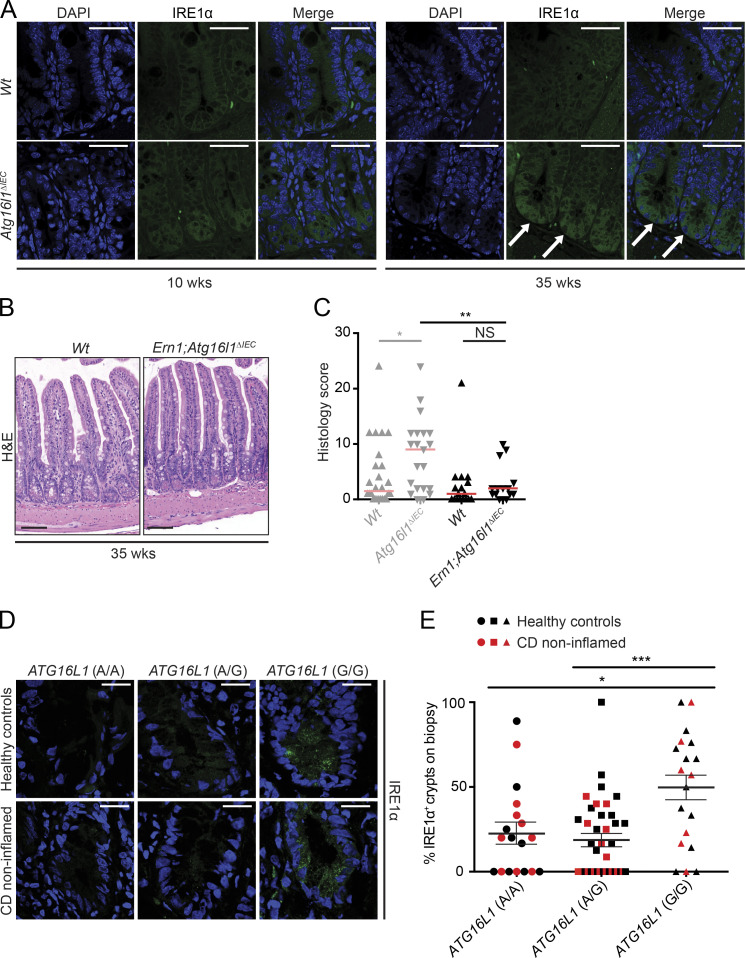
Absence of XBP1 function abolishes cells’ capacity for effective proteostasis causing ER stress, as observed in Xbp1ΔIEC mice, in turn triggering profound IRE1α activation (Kaser et al., 2008). This accounts for the mild, superficial enteritis that develops in Xbp1ΔIEC mice (Adolph et al., 2013). The observation that IRE1α accumulates most profoundly in Atg16l1;Xbp1ΔIEC Paneth cells and drives the transmural CD-like ileitis in Atg16l1;Xbp1ΔIEC mice suggested that autophagy may be directly involved in regulating IRE1α function. However, it remained possible that decreased autophagy function and overactivation of IRE1α caused by Xbp1 deficiency are separate mechanisms and that ileitis is a combinatorial consequence. The spontaneous development of transmural ileitis phenocopying CD in 35-wk-old Atg16l1ΔIEC mice (Fig. 1, B and C) provided us an opportunity to test the hypothesis that IRE1α activity is directly dependent on autophagy function. IRE1α immunofluorescence at the small intestinal crypt bottom was indeed observed to increase from age 10–35 wk in Atg16l1ΔIEC mice, which contrasts with equally minimal staining in Wt littermates of either age (Fig. 3 A). Therefore, we generated Ern1;Atg16l1ΔIEC mice and studied them at 35 wk of age. In marked contrast to Atg16l1ΔIEC mice of the same age, 35-wk-old Ern1;Atg16l1ΔIEC mice housed in the same animal room were indeed protected from developing ileitis (Fig. 3, B and C). These results demonstrate that ATG16L1-dependent autophagy is involved in controlling IRE1α activity and that IRE1α drives inflammation in this mouse model of CD-like ileitis. Thus, decreased ATG16L1 function in IECs without genetic manipulation of UPR effectors such as XBP1 results in IRE1α-dependent spontaneous ileitis.
Figure 3.
IRE1α expression is increased in 35-wk-old Atg16l1ΔIEC mice and ATG16L1T300A patients and drives CD-like ileitis. (A) Representative confocal images of IRE1α immunoreactivity (green; white arrows) in 10- and 35-wk-old Atg16l1ΔIEC mice. n = 3. DAPI is in blue. Bars, 50 µm. (B) Representative H&E images of 35-wk-old Ern1;Atg16l1ΔIEC and Wt mice. Bars, 100 µm. (C) Enteritis histology scores of 35-wk-old Atg16l1ΔIEC mice (the identical scores are depicted in Fig. 1 C) compared with 35-wk-old Ern1;Atg16l1ΔIEC mice and their respective Wt controls. n = 25/21/19/15. The median is shown. A Mann–Whitney U test was used. (D) Representative IRE1α immunoreactivity (green) stratified by the ATG16L1 risk allele (AA/AG/GG; A, healthy allele; G risk allele) in healthy controls (n = 9/22/12) and noninflamed mucosa (n = 9/15/7) of CD patients. DAPI is in blue. Bars, 20 µm. (E) Quantification of IRE1α+ crypts shown in D in healthy controls (n = 9/22/12) and CD patients (n = 9/15/7) according to their ATG16L1 genotype (n = 18/37/19). Mean ± SEM is shown. One-way ANOVA with Bonferroni’s correction was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
IRE1α accumulates in crypts of individuals homozygous for ATG16L1T300A
The ATG16L1T300A polymorphism is common in populations of European ancestry, with a risk allele (G) frequency at rs2241880 of 51 and 57% in healthy individuals and CD patients, respectively (Prescott et al., 2007). Based on the identification of IRE1α as driver of CD-like ileitis in 35-wk-old Atg16l1ΔIEC mice, we assessed IRE1α expression in noninflamed areas of the ileum in CD patients and healthy subjects. Fig. 3 D shows evidence for a granular staining pattern that is localized to the base of intestinal crypts in CD patients and healthy individuals who are homozygous for the risk allele (G/G). Quantitative immunofluorescence in n = 74 healthy individuals and CD patients stratified according to rs2241880 genotype revealed 49.18 ± 7.6% of crypts positive for IRE1α staining in individuals with G/G, compared with 19.4 ± 3.7% and 23.19 ± 6.25% in A/G and A/A genotypes, respectively (Fig. 3 E and Table S1). This suggests that ER stress–inducing noxae impinging on the mucosa of individuals homozygous for ATG16L1T300A may trigger an exaggerated IRE1α response.
Altogether, these data establish a critical role of IRE1α in driving intestinal inflammation consequent to impaired autophagy in the intestinal epithelium and Paneth cells in particular. The experiments that led to this conclusion pose several important questions: first, how do IRE1α and IRE1β relate to each other within the intestinal epithelium and contribute to mucosal homeostasis? Second, how does IRE1α accumulate in IECs of mice and humans with hypomorphic ATG16L1 function? Third, what is the contribution of the microbiota to triggering the spontaneous ileitis observed?
IRE1α and IRE1β are cross-compensatory in the intestinal epithelium
IRE1α and IRE1β are ER transmembrane receptors, which form homodimers in the presence of misfolded proteins, triggering their kinase and endoribonuclease function (Walter and Ron, 2011). IRE1α’s endoribonuclease primarily splices Xbp1 mRNA, causing a frame shift that converts XBP1 into the active transcription factor, whereas IRE1β predominantly engages in regulated IRE1-dependent decay that degrades specific (m)RNAs on the ER membrane (Hetz et al., 2011). Active IRE1α (and presumably IRE1β) forms a scaffold and recruits adapters and signaling molecules (Woehlbier and Hetz, 2011), and upon unabated, unresolvable stress, IRE1α homodimers form large clusters that ultimately trigger cell death (Korennykh and Walter, 2012; Chen and Brandizzi, 2013; Cho et al., 2013; Ghosh et al., 2014; Maurel et al., 2014). The interrelationship between IRE1α and IRE1β has not yet been investigated in vivo, despite their unique coexistence at mucosal surfaces that hints at a particular importance within this locale.
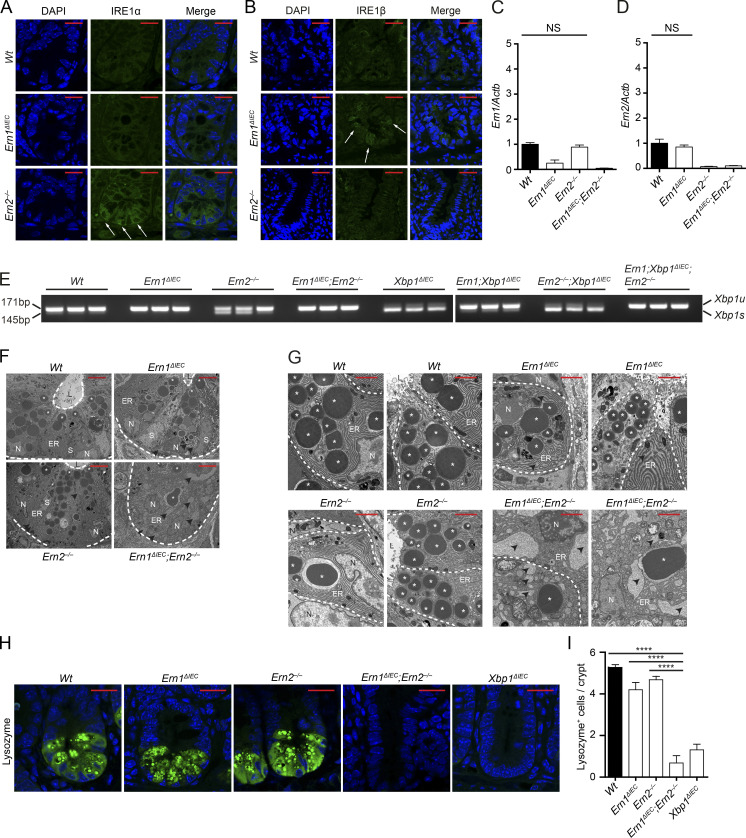
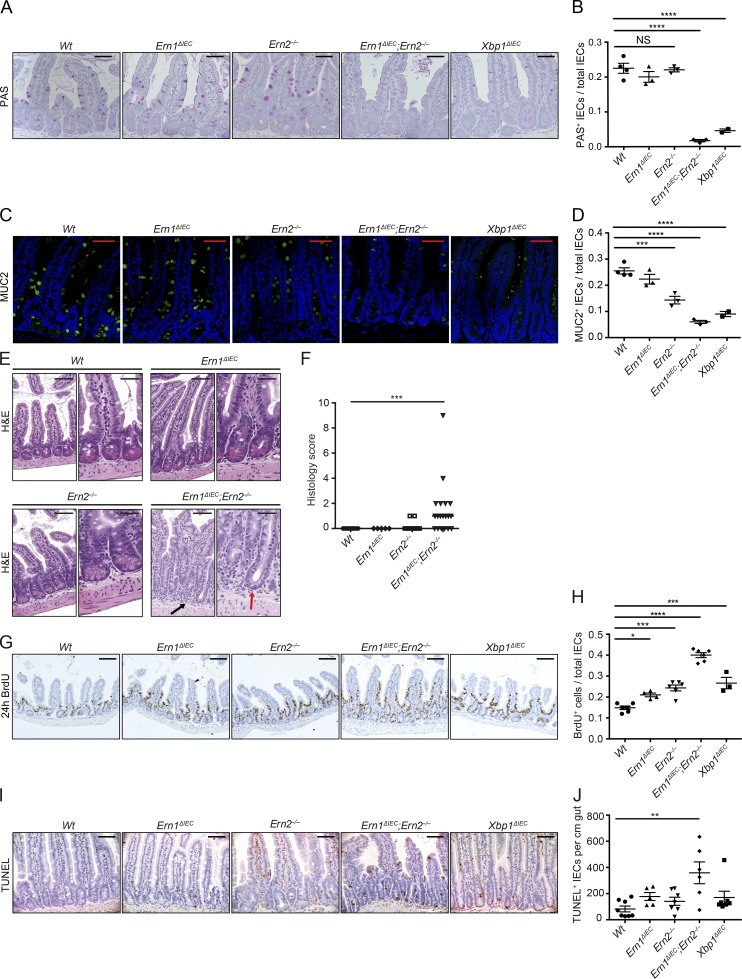
We indeed observed increased staining for IRE1α at the crypt base in Ern2−/− and conversely for IRE1β in Ern1ΔIEC mice (Fig. 4, A and B), whereas Ern1 and Ern2 mRNA expression remained unaltered under these conditions (Fig. 4, C and D), indicating posttranscriptional regulation. As expected, minimal baseline Xbp1 splicing in Wt ileal crypt lysates was absent in Ern1ΔIEC and Ern1ΔIEC;Ern2−/− lysates (Fig. 4 E). Notably, Xbp1 splicing was markedly increased in Ern2−/− ileal crypt lysates (Fig. 4 E), indicating IRE1α overactivation in the absence of IRE1β function. Deletion of the floxed exon 2 of Xbp1 results in IRE1α overactivation as observed through IRE1α accumulation and phosphorylation (Niederreiter et al., 2013) and massive splicing of Xbp1 mRNA (the splicing site encoded by exon 4 is intact in the Xbp1ΔIEC allele; Fig. 4 E), as previously reported (Kaser et al., 2008; Niederreiter et al., 2013). Ern1;Xbp1ΔIEC ileal crypts exhibited some residual Xbp1 splicing, which was completely abrogated in Ern1;Xbp1ΔIEC;Ern2−/− ileal crypts (Fig. 4 E), indicating that IRE1β may contribute to Xbp1 splicing. Consistent with a cross-compensatory role of IRE1α and IRE1β, Ern1ΔIEC;Ern2−/− Paneth cells exhibited a condensed ER and a nearly complete collapse of the secretory compartment on transmission electron microscopy (Fig. 4, F and G), reminiscent of Xbp1ΔIEC mice (Kaser et al., 2008), whereas single-mutant Ern1ΔIEC and Ern2−/− crypts were almost indistinguishable from Wt controls with only minimal distension of the ER (Fig. 4, F and G). Ern1ΔIEC;Ern2−/− mice exhibited an absence of lysozyme immunofluorescence reflecting the massive alterations in the secretory compartment observed in comparison with Ern1ΔIEC and Ern2−/− mice, which exhibited only minimally decreased lysozyme staining compared with Wt controls (Fig. 4, H and I). Both periodic acid–Schiff (PAS; Fig. 5, A and B) and MUC2 (Fig. 5, C and D) staining for mucus-producing goblet cells was nearly completely absent in the small intestines of Ern1ΔIEC;Ern2−/− mice, with decreased MUC2 staining observed in Ern2−/− single-mutant mice (Fig. 5, C and D), consistent with an important role of IRE1β in mucin secretion (Tsuru et al., 2013). Whereas Ern1ΔIEC;Ern2−/− mice developed low levels of spontaneous superficial enteritis (Fig. 5, E and F) accompanied by epithelial hyperproliferation (Fig. 5, G and H) and increased epithelial cell death (Fig. 5, G and H), these changes were not observed in Ern1ΔIEC and Ern2−/− single-mutant mice (Fig. 5, E–J). Together, these data demonstrate that IRE1α and IRE1β complement each other to maintain Paneth cell and intestinal homeostasis, although their roles fundamentally diverge in Atg16l1;Xbp1ΔIEC mice, with IRE1α driving and IRE1β protecting from CD-like ileitis.
Figure 4.
Cross-compensatory up-regulation of IRE1α and IRE1β maintains Paneth cell homeostasis. (A and B) Representative confocal images of IRE1α (A) or IRE1β (B) immunofluorescence (green) of the indicated genotypes at 10 wk of age. Note the cross-compensatory up-regulation of IRE1β and IRE1α in crypts of Ern1ΔIEC and Ern2−/− mice, respectively (white arrows). n = 3. DAPI is in blue. Bars, 20 µm. (C and D) Ern1 (C) and Ern2 (D) mRNA expression measured by qRT-PCR in ileal scrapings of the indicated genotypes normalized to Actb expression. n = 20/10/16/15. The mean ± SEM is shown. One-way ANOVA with Bonferroni’s correction was used. Note that IRE1α and IRE1β (encoded by Ern1 and Ern2) in Ern2−/− and Ern1ΔIEC mice, respectively, are not transcriptionally regulated. (E) Xbp1 splicing in crypt IECs from the indicated genotypes indicative of upstream activation of IRE1α/IRE1β. Note increased Xbp1 splicing in Ern2−/− crypts indicative of upstream activation of IRE1α (also see A), whereas IRE1β appears to have a minor function for Xbp1 splicing in Ern1;Xbp1ΔIEC mice which is absent in Ern1;Xbp1ΔIEC;Ern2−/−. n = 3. (F) High magnification transmission electron microscopy micrographs of ileal crypts depicting Paneth cells from the indicated genotypes. n = 2. Bars, 5 µm. Dashed lines denote crypt unit. (G) High magnification transmission electron microscopy micrographs of ileal crypts depicting Paneth cells from the indicated genotypes. n = 2. Bars, 2 µm. Dashed lines denote single Paneth cells. (F and G) L, lumen; N, nucleus; S, stem cell. White asterisks label Paneth cell granules. Note the absence of intact Paneth cell granules and the dilated ER lumen (black arrowheads). (H and I) Representative confocal images of lysozyme immunoreactivity (green; H) and quantification of lysozyme+ cells per crypt (I) on ileal sections of the indicated genotypes. n = 4/3/4/3/4. The mean ± SEM is shown. One-way ANOVA with Bonferroni’s correction was used. ****, P < 0.0001. DAPI is in blue. Bars, 20 µm.
Figure 5.
Cross-compensation of IRE1α and IRE1β maintains intestinal homeostasis. (A and B) Representative PAS staining on ileal sections from the indicated genotypes at 10 wk of age (A) with quantification of PAS-labeled IECs per total IECs along the crypt villus axis (B). Note the absence of PAS+ goblet cells in Ern1ΔIEC;Ern2−/− mice. n = 4/3/3/3/2. The mean ± SEM is shown. One-way ANOVA with Bonferroni’s correction was used. Hematoxylin counterstaining is in light blue. Bars, 50 µm. (C and D) Representative confocal images of MUC2 immunoreactivity (green) on ileal sections from the indicated genotypes at 10 wk of age (C) with quantification of MUC2+ goblet cells per total IECs along the crypt villus axis (D). Note the absence of MUC2+ goblet cells in Ern1ΔIEC;Ern2−/− mice. n = 4/3/3/3/2. The mean ± SEM is shown. One-way ANOVA with Bonferroni’s correction was used. DAPI is in blue. Bars, 50 µm. (E and F) Representative H&E images (E) of the indicated genotypes at 10 wk of age and their histological score (F). n = 10/5/17/19. The median is shown. A Mann–Whitney U test was used. Note the absence of eosinophilic Paneth cell granule (red arrow) and the surrounding basal infiltration of mononuclear cells in Ern1ΔIEC;Ern2−/− mice (black arrow). Bars: (left) 100 µm; (right) 50 µm. (G and H) Representative images of BrdU-labeled IECs (brown; G) from the indicated genotypes at 10 wk of age after 24 h of BrdU incorporation with quantification of BrdU+ IECs per total IECs along the crypt villus axis (H). n = 6/3/4/6/6. The mean ± SEM is shown. One-way ANOVA with Bonferroni’s correction was used. Hematoxylin counterstaining is in blue. Bars, 100 µm. (I and J) Representative images of TUNEL labeling (brown; I) and quantification of TUNEL+ cells per cm gut on ileal sections (J) of the indicated genotypes at 10 wk of age. n = 8/7/5/7/6. The mean ± SEM is shown. One-way ANOVA with Bonferroni’s correction was used. Hematoxylin counterstaining is in blue. Bars, 50 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
IRE1α accumulates in the absence of selective autophagy receptor optineurin
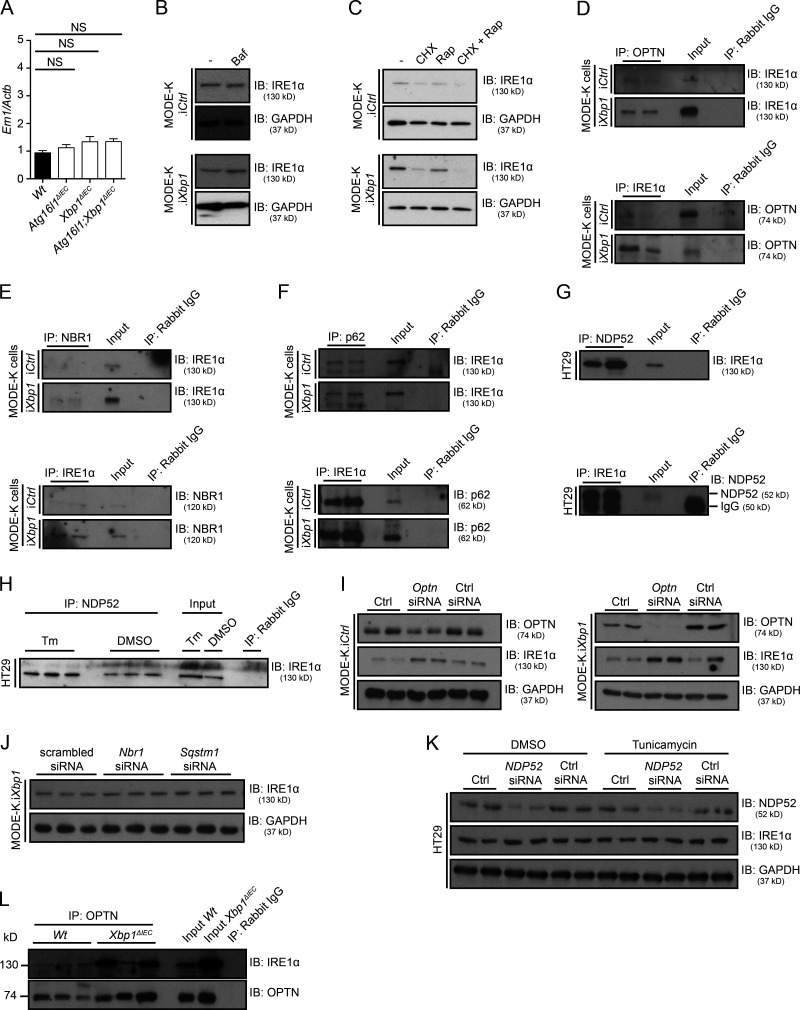
The observed increased IRE1α activity in Atg16l1ΔIEC, Xbp1ΔIEC, and Atg16l1;Xbp1ΔIEC mice could not be explained by transcriptional regulation of Ern1 (Fig. 6 A), suggesting that autophagy may be involved in IRE1α protein turnover. Autophagy engulfs intracellular material, e.g., organelles (mitophagy, pexophagy, and ER-phagy), macromolecular complexes (aggregophagy), or infectious agents (xenophagy), and delivers it to lysosomes for degradation (Levine et al., 2011; Schuck et al., 2014; Khaminets et al., 2015). As reductionist model systems to study the involvement of autophagy in IRE1α protein turnover under baseline and ER stress conditions, we chose the mouse small intestinal epithelial cell line MODE-K expressing control shRNA (MODE-K.iCtrl) and Xbp1-specific shRNA vectors (MODE-K.iXbp1; Kaser et al., 2008), respectively. Bafilomycin A, an inhibitor of vacuolar H+ ATPase that prevents acidification of autophagolysosomes and hence degradation of their cargo, increased IRE1α protein levels in MODE-K.iXbp1 cells, which experience ER stress, but not in MODE-K.iCtrl cells (Fig. 6 B). Conversely, augmentation of autophagy via the mechanistic target of rapamycin inhibitor rapamycin decreased IRE1α protein levels in MODE-K.iXbp1 but not MODE-K.iCtrl cells, whereas inhibition of protein translation via cycloheximide decreased IRE1α levels in both cell lines (Fig. 6 C). This suggested that autophagy may contribute to IRE1α turnover predominantly during ER stress.
Figure 6.
Selective autophagy receptors target IRE1α for degradation. (A) Ern1 mRNA expression measured by qRT-PCR in ileal scrapings from the indicated genotypes and normalized to Actb expression. n = 15/11/10/11. The mean ± SEM is shown. One-way ANOVA with Bonferroni’s correction was used. (B) IRE1α immunoblot (IB) of Xbp1-sufficient (MODE-K.iCtrl) and Xbp1-deficient (MODE-K.iXbp1) MODE-K cells co-cultured for 24 h with bafilomycin (Baf), an inhibitor of vacuolar H+ ATPase that prevents cargo degregation in autophagolysosomes. Data are representative of three independent experiments. (C) IRE1α immunoblot after addition of protein synthesis inhibitor cycloheximide (CHX) and the autophagy inducer rapamycin (Rap) to MODE-K.iCtrl and MODE-K.iXbp1 cells for 24 h. Data are representative of two independent experiments. (D–F) Coimmunoprecipitation (IP) of IRE1α with selective autophagy receptors OPTN (D), NBR1 (neighbor of BRCA1 gene 1 protein; E), and p62 (F) in MODE-K.iCtrl and MODE-K.iXbp1 cells with converse pull-downs also demonstrated. n = 2. Data are representative of three independent experiments. (G) Coimmunoprecipitation of IRE1α with the selective autophagy receptor NDP52 in the human colorectal adenocarcinoma cell line HT29. n = 2. Data are representative of three independent experiments. (H) Coimmunoprecipitation and immunoblot of IRE1α with the selective autophagy receptor NDP52 in HT29 cells at basal conditions (DMSO) and after pharmacological ER stress induction with tunicamycin (Tm). n = 3. Data are representative of two independent experiments. (I) IRE1α and OPTN immunoblots after siRNA silencing of Optn in MODE-K.iCtrl and MODE-K.iXbp1 cells. n = 2. Data are representative of two independent experiments. Ctrl, control. (J) IRE1α immunoblot upon siRNA silencing of Nbr1 or Sqstm1 (p62) in MODE-K.iXbp1 cells. n = 3. Data are representative of two independent experiments. (K) IRE1α and NDP52 immunoblots upon siRNA silencing of NDP52 in HT29 cells with and without tunicamycin treatment for 4 h. n = 2. Data are representative of two independent experiments. (L) Coimmunoprecipitation of OPTN with IRE1α in ileal crypts of Wt and Xbp1ΔIEC mice. n = 3. Data are representative of two independent experiments.
Considering the formation of large clusters of IRE1α during profound and sustained ER stress, we hypothesized that IRE1α might be removed via a selective autophagy process under such conditions. Formerly considered an entirely nonspecific process (bulk [macro-] autophagy), selective autophagy processes have emerged that identify their cargo via specific receptors and adaptors in a ubiquitin-dependent and -independent fashion and deliver them to autophagosomes (Weidberg et al., 2011; Rogov et al., 2014; Stolz et al., 2014; Khaminets et al., 2016). Therefore, we studied the involvement of several known selective autophagy receptors, p62 (sequestosome-1 [Sqstm1]), NBR1 (neighbor of BRCA1 gene 1 [Nbr1]), optineurin (Optn), and NDP52 (CALCOCO2; Wild et al., 2011; Khaminets et al., 2016), with the latter two having previously been implicated in CD (Ellinghaus et al., 2013; Chew et al., 2015; Smith et al., 2015). Fig. 6 (D and E) demonstrates that optineurin and NBR1 reciprocally immunoprecipitated with IRE1α in MODE-K.iXbp1 lysates, whereas biochemical interaction with optineurin was minimal, but detectable, in MODE-K.iCtrl cells. p62 coimmunoprecipitated with IRE1α at equal levels in MODE-K.iCtrl and MODE-K.iXbp1 cells (Fig. 6 F). NDP52 coimmunoprecipitated with IRE1α and vice versa in the human colonic epithelial cell line HT29 under basal conditions (Fig. 6 G) as well as after pharmacological ER stress induction with tunicamycin (Fig. 6 H). Altogether, these data suggest that several selective autophagy receptors biochemically interact with IRE1α. Importantly, silencing of Optn via siRNA resulted in increased IRE1α protein levels in MODE-K.iXbp1 cells and only a minimal increase in MODE-K.iCtrl cells (Fig. 6 I). In contrast to Optn, silencing of Sqstm1 or Nbr1 in MODE-K.iXbp1 did not affect IRE1α protein levels (Fig. 6 J), and co-silencing of these receptors in conjunction with Optn did not have synergistic effects (not depicted). Similarly, silencing of NDP52 in HT29 cells did not increase IRE1α protein levels at baseline or after pharmacological ER stress induction with tunicamycin (Fig. 6 K). Importantly, IRE1α was observed to interact with optineurin in vivo, as demonstrated by detection of IRE1α protein in antioptineurin immunoprecipitates in small intestinal crypt lysates from Xbp1ΔIEC mice, whereas IRE1α was only just above detection limit in immunoprecipitates from Wt crypt lysates (Fig. 6 L). Altogether, these data suggest that during profound or prolonged ER stress, IRE1α may be recruited to autophagosomes via optineurin, followed by IRE1α’s degradation, hence implying selective autophagy as a mechanism for terminating IRE1α signaling during ER stress.
Mice with CD-like ileitis harbor a dysbiotic microbiota
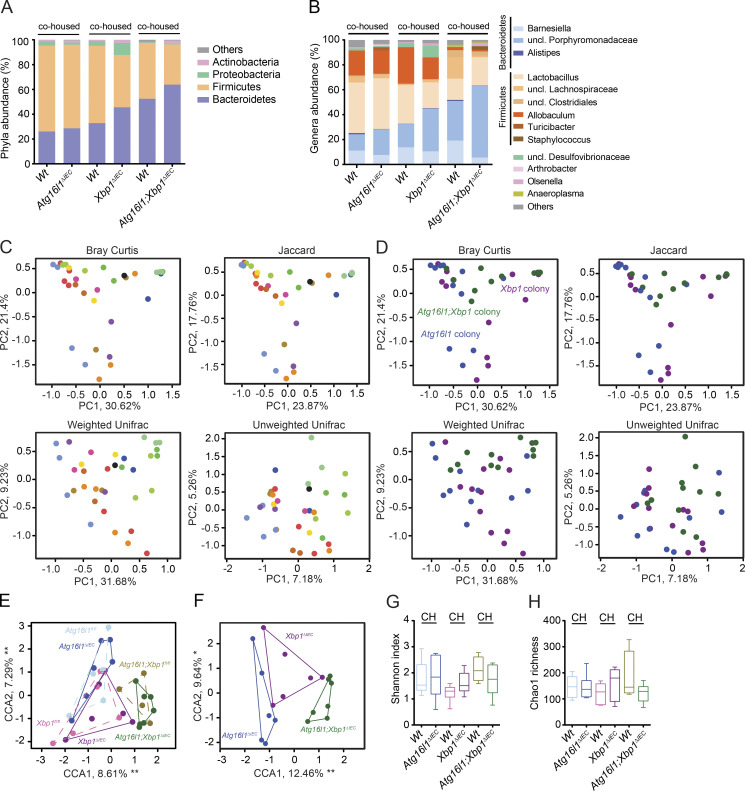
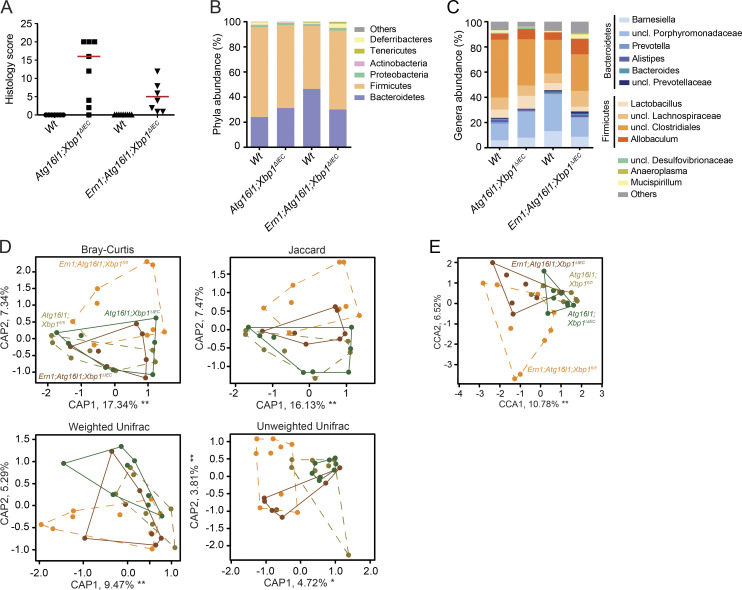
The spontaneous CD-like ileitis observed in Atg16l1;Xbp1ΔIEC mice also provided a unique opportunity to interrogate the contribution of alterations in the microbiota caused by secretory Paneth cell abnormalities to intestinal inflammation. Mice of the Atg16l1;Xbp1 colony (Atg16l1;Xbp1ΔIEC cohoused with their Vil-Creneg Wt [Atg16l1fl/fl;Xbp1fl/fl] littermates) exhibited an overall distinct tissue-adherent ileal microbial composition compared with Atg16l1 (Atg16l1ΔIEC and their Wt [Atg16l1fl/fl] littermates) or Xbp1 (Xbp1ΔIEC and Wt [Xbp1fl/fl] littermates) colonies (adonis; Bray-Curtis: genotype, R2 = 0.14812, P = 0.000999; gender, R2 = 0.07445, P = 0.003996; cage, R2 = 0.38101, P = 0.000999; litter, R2 = 0.02, P = 0.99; Jaccard: genotype, R2 = 0.12052, P = 0.000999; gender, R2 = 0.06340, P = 0.004995; cage, R2 = 0.36132, P = 0.000999; litter, R2 = 0.064, P = 0.99; Fig. 7, A–F), with the maximum microbial shift being observed between Atg16l1;Xbp1ΔIEC and Atg16l1ΔIEC mice (adonis; Bray-Curtis: R2 = 0.22912, P = 0.01598; Jaccard: R2 = 0.18951, P = 0.01199). Specifically, the Atg16l1;Xbp1ΔIEC mice exhibited an increased abundance of Bacteroidetes (Student’s t test with Welch’s correction, P = 0.0295) and decreased Firmicutes (Student’s t test with Welch’s correction, P = 0.0288) compared with Atg16l1ΔIEC mice (Fig. 7, A and B). Differences in bacterial composition were further highlighted by a relatively small overlap of operational taxonomic units between Atg16l1ΔIEC, Xbp1ΔIEC, and Atg16l1;Xbp1ΔIEC mice, respectively (not depicted), whereas species diversity and species richness were comparable between respective colonies (Fig. 7, G and H). Wt littermates of each colony appeared to acquire the respective knockout microbiota, as their microbial compositions were indistinguishable (adonis; Bray-Curtis; Atg16l1 colony, R2 = 0.05133, P = 0.7672; Xbp1 colony, R2 = 0.04037, P = 0.59341; Atg16l1;Xbp1 colony, R2 = 0.10241, P = 0.14885; Fig. 7, E and F).
Figure 7.
Atg16l1;Xbp1ΔIEC mice harbor a dysbiotic and transmissible microbiota. (A and B) Relative abundance of microbial taxa at phylum level (≥1%; A) and genus level (≥1%; B) representing ileal mucosa–adherent microbiota of 18-wk-old cohoused mutant (V-cre+) and Wt littermates (V-cre–). n = 5/6/6/6/4/7. uncl., unclassified. (C and D) Ordination analysis of the constraining factors of the cage effect (each dot represents one mouse; cages are indicated as different colors; C) and genotype effect (each dot represents one mouse; colonies are indicated as different colors; D) in the microbiota analysis of cohoused mice based on principal coordinates analysis (Bray Curtis and Jaccard dissimilarity) and Unifrac (weighted and unweighted). Atg16l1 colony, n = 11; Xbp1 colony, n = 12; Atg16l1;Xbp1 colony, n = 11. (E) CCA plot showing a significant genotype effect (F = 2.2270; P = 0.001) and gender effect (F = 2.2639; P = 0.004) in the indicated groups (n = 5/6/6/6/4/7). Significant genotype effect is shown in D. Cage and litter effects were partialled out in this analysis and are shown in C. (F) CCA plot of knockouts only (n = 6/6/7) constrained for mutation and gender effect (cage and litter effect were partialled out). (G and H) α diversity estimates Shannon’s index analyzing species evenness (G), and Chao 1 determining species richness (H) is shown (n = 5/6/6/6/4/7). Statistical analysis was performed using nonparametric Kolmogorov-Smirnov test. CH, cohoused.
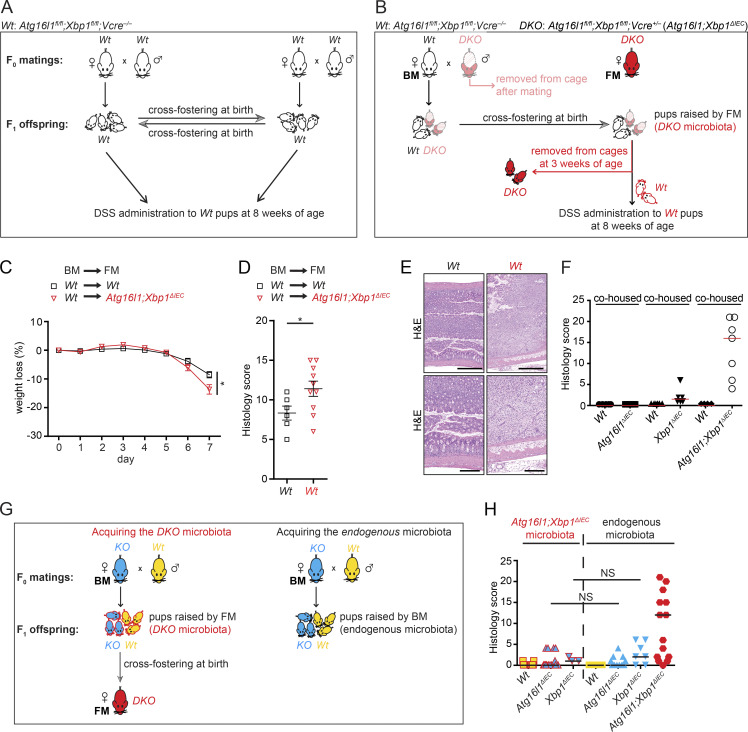
Acquisition of an altered microbiota by Wt mice cohoused with their IEC-mutant littermates afforded an opportunity to dissect functional consequences via cross-fostering studies. Wt litters cross-fostered by Atg16l1;Xbp1ΔIEC mice indeed exhibited increased severity of colitis induced by dextran sodium sulfate (DSS), compared with Wt mice cross-fostered with Wt mice (Fig. 8, A–E). Importantly, all Wt litters and Wt foster dams were V-Creneg;Atg16l1fl/fl;Xbp1fl/fl mice, hence stemming from the Atg16l1;Xbp1ΔIEC colony. Altogether, these data established that the Atg16l1;Xbp1ΔIEC microbiota is indeed dysbiotic and aggravates exogenously induced experimental colitis.
Figure 8.
Microbiota of Atg16l1;Xbp1ΔIEC mice predisposes to colitis but not ileitis. (A) Cross-fostering of Wt (V-cre– Atg16l1fl/fl;Xbp1fl/fl) offspring derived from a Wt (V-cre– Atg16l1fl/fl;Xbp1fl/fl) breeding pair to a Wt (V-cre– Atg16l1fl/fl;Xbp1fl/fl) foster dam and vice versa. Male breeders of the F0 generation were removed from the cage before F1 offspring were born. Cross-fostered offspring were subjected to 3% DSS-induced colitis for 7 d at 8 wk of age (this group of mice is displayed in black in panels C–E), performed in a mouse norovirus–free SPF animal facility (Cambridge). (B) Cross-fostering of Wt (V-cre− Atg16l1fl/fl;Xbp1fl/fl; displayed in black) and Atg16l1;Xbp1ΔIEC (DKO; V-cre+ Atg16l1fl/fl;Xbp1fl/fl; displayed in shaded red) offspring derived from a hemizygous breeding pair (V-cre– Atg16l1fl/fl;Xbp1fl/fl [Wt] × V-cre+ Atg16l1fl/fl;Xbp1fl/fl [DKO]) from a Wt (V-cre− Atg16l1fl/fl;Xbp1fl/fl) dam (biological mother; BM) to a Atg16l1;Xbp1ΔIEC (DKO; V-cre+ Atg16l1fl/fl;Xbp1fl/fl) foster mother (FM; displayed in red). Male breeders of the F0 generation were removed from the cage before F1 offspring was born (displayed in shaded red). Before weaning of cross-fostered offspring, pups were genotyped, and Atg16l1;Xbp1ΔIEC (DKO) littermates were removed from the cage. Remaining Wt offspring that acquired Atg16l1;Xbp1ΔIEC microbiota (framed in red) were subjected to 3% DSS colitis for 7 d at 8 wk of age (this group of mice is displayed in red in panels C–E). (C) Relative weight loss after 3% DSS application for 7 d to 8-wk-old mice after cross-fostering Wt littermates either between Wt dams (displayed in black) or from a Wt dam to an Atg16l1;Xbp1ΔIEC dam after birth (displayed in red). Experimental design is shown in A and B. n = 6/10. The mean ± SEM is shown. Linear mixed-effects model fitted using R (lme package) was used. *, P < 0.05. (D and E) DSS colitis score (D) after 7 d of experiment performed in C with representative H&E images (E). n = 6/10. The mean ± SEM is shown. Unpaired two-tailed Student’s t test was used. *, P < 0.05. Bars: (top) 500 µm; (bottom) 200 µm. (F) Histological score of cohoused mice of the indicated genotypes used for 16S rDNA ribotyping of ileal mucosa–adherent microbiota in Fig. 7 at 18 wk of age. n = 5/6/6/6/4/7. The median is shown. (G, left) Cross-fostering of either Atg16l1ΔIEC offspring (KO; displayed in blue) and their Wt littermates (displayed in yellow) or Xbp1ΔIEC offspring (KO; displayed in blue) and their Wt littermates (displayed in yellow) to an Atg16l1;Xbp1ΔIEC (DKO; displayed in red) foster dam allowing them to acquire the Atg16l1;Xbp1ΔIEC microbiota (framed in red). (Right) Concurrently, non–cross-fostered Atg16l1ΔIEC and Xbp1ΔIEC offspring were raised by their respective biological dams, which allowed them to acquire endogenous microbiota. (H) Enteritis histology score of cross-fostered and Atg16l1;Xbp1ΔIEC microbiota-acquired offspring compared with non–cross-fostered and endogenous microbiota acquired offspring at 8 wk of age. n = 21/18/3/12/13/7/16. The median is shown. Kruskal–Wallis with posthoc Holm’s-corrected Mann–Whitney U test was used.
Ileal dysbiosis does not trigger ileitis
Wt mice cohoused with their Atg16l1;Xbp1ΔIEC littermates did not develop any intestinal inflammation, nor did cohoused Wt littermates of Xbp1ΔIEC mice (Fig. 8 F), despite an almost identical ileal microbial ecology between Wt mice and their respective IEC-mutant littermates (Fig. 7, A and B). Having established the dysbiotic, aggravating effect of the Atg16l1;Xbp1ΔIEC microbiota on DSS colitis (Fig. 8, C–E) and the transferability of the microbiota between IEC-mutant mice, their Wt littermates, and cross-fostered mice (Fig. 7, A–F; and Fig. 8, C–E), we posed the question whether transferable microbial elements might aggravate ileitis in genetically susceptible Atg16l1ΔIEC or Xbp1ΔIEC mice. Cross-fostering of Wt, Atg16l1ΔIEC, and Xbp1ΔIEC litters with Atg16l1;Xbp1ΔIEC foster dams revealed that histology scores and features were indistinguishable from non–cross-fostered mice with the respective genotypes (Fig. 8, G and H). Hence, the genetically susceptible Atg16l1ΔIEC and Xbp1ΔIEC litters cross-fostered with Atg16l1;Xbp1ΔIEC foster dams did not develop CD-like ileitis as observed in Atg16l1;Xbp1ΔIEC mice or 35-wk-old Atg16l1ΔIEC mice. Consistent with ileal dysbiosis not triggering ileal inflammation in this model, Ern1;Atg16l1;Xbp1ΔIEC mice, which were protected from ileitis (Fig. 2, F and G; and Fig. 9 A), exhibited an ileal microbiota similar to Atg16l1;Xbp1ΔIEC mice (Fig. 9, B–E) and Paneth cells that remained profoundly impaired in lysozyme staining (Fig. 2, K and L). Specifically, β diversity did not show any differences between Ern1;Atg16l1;Xbp1 and Atg16l1;Xbp1 colonies (adonis; Bray-Curtis; genotype, R2 = 0.08782, P = 0.08192; gender, R2 = 0.01233, P = 0.90809; litter, R2 = 0.10755, P = 0.74126; Fig. 9 D). Consistently, only marginal significance between both colonies was found in canonical correlation analysis (CCA; genotype, F = 1.7166, P = 0.045; gender, F = 0.6475, P = 0.785; Fig. 9 E). Moreover, no differences were detected in a pairwise comparison between Wt and littermate IEC-mutant mice of both colonies (adonis; Bray-Curtis; Atg16l1;Xbp1 colony, R2 = 0.7456, P = 0.435; Ern1;Atg16l1;Xbp1 colony, R2 = 1.56, P = 0.178; Fig. 9 D). The results demonstrate that IRE1α-dependent development of ileitis is remarkably independent of the dysbiosis present in Atg16l1;Xbp1ΔIEC mice.
Figure 9.
Similar microbiota composition despite amelioration of disease in Ern1;Atg16l1;Xbp1ΔIEC mice. (A) Histological score of mice used for 16S rDNA ribotyping of ileal mucosa–adherent microbiota in B–E at 10 wk of age. n = 7/8/9/7. The median is shown. (B and C) Relative abundance of microbial taxa at phylum level (≥1%; B) and genus level (≥1%; C) representing ileal mucosa–adherent microbiota of 10-wk-old Ern1;Atg16l1;Xbp1ΔIEC mice and their Wt littermates compared with Atg16l1;Xbp1ΔIEC mice. n = 7/8/9/7. uncl., unclassified. (D) Ordination analysis including all constraining factors (genotype, mutation, gender, litter, and cage) based on Bray-Curtis dissimilarity (genotype: R2 = 0.16566, P = 0.1219; gender: R2 = 0.01154, P = 0.9121; litter: R2 = 0.08843, P = 0.8472; and cage: R2 = 0.17390, P = 0.4535), Jaccard index (genotype: R2 = 0.14477, P = 0.1129; gender: R2 = 0.01465, P = 0.9700; litter: R2 = 0.10618, P = 0.8252; and cage: R2 = 0.16193, P = 0.5614), weighted Unifrac distances (genotype: F = 1.0380, P = 0.372; gender: F = 1.1375, P = 0.247; litter: F = 0.9034, P = 0.634; and cage: F = 1.0281, P = 0.411) and unweighted Unifrac distances (genotype: F = 1.0604, P = 0.172; gender: F = 1.0816, P = 0.217; litter: F = 1.0265, P = 0.276; and cage: F = 1.0270, P = 0.304). n = 7/8/9/7. (E) Constrained CCA plot partialled out for cage and litter effect showed a marginal significance for genotype (F = 1.7166 and P = 0.045). n = 7/8/9/7.
Discussion
We report IRE1α as the main driver of spontaneous ileitis phenocopying CD that develops with increasing age in mice mutant for Atg16l1 in the intestinal epithelium. Exaggerated UPR signaling through IRE1α caused by impairment of a selective autophagy process may be an important mechanism in how hypomorphic ATG16L1T300A contributes to disease in the context of yet unknown environmental triggers that cause ER stress. In contrast, alterations in ileal microbiota structure, although present in mice with CD-like ileitis, importantly did not trigger spontaneous ileal but aggravated chemically induced colonic inflammation.
Host genetic alterations can render the microbiota inflammation inducing, e.g., as observed for colitis in Nod2 or Nlrp6 mutants upon exposure to DSS (Elinav et al., 2011; Couturier-Maillard et al., 2013) or for ileitis in the TnfΔARE model (Schaubeck et al., 2016). Because alterations in the secretory compartment, lysozyme, and α-defensin secretion of Paneth cells have emerged as common features observed in carriers of CD-specific risk gene variants (Kobayashi et al., 2005; Wehkamp et al., 2005; Cadwell et al., 2008; VanDussen et al., 2014; Zhang et al., 2015) and microbial community structure differs between CD and healthy individuals (Frank et al., 2007; Gevers et al., 2014; Rehman et al., 2016), it has been conjectured that such host genetically induced dysbiosis might be critical for triggering intestinal inflammation in CD. Our data challenge this hypothesis by demonstrating that host mutant-induced alterations in the ileal microbiota are not sufficient to trigger or promote ileitis, although they might increase severity of established colitis. Nonetheless, the presence of a microbiota is required for developing small intestinal inflammation because Xbp1ΔIEC mice rederived germ free are protected from enteritis (Adolph et al., 2013) and do not accumulate IRE1α in Paneth cells (not depicted). It remains entirely possible that outside of an SPF environment, the altered microbiota might provide a favorable niche for potential pathobionts, e.g., as present in human’s real-life exposome, which may impact on the disease mechanism as environmental factors. Along these lines, host gene mutants have been reported to favor the outgrowth of specific pathobionts that can cause colitis, such as Helicobacter typhlonius in TRUC (Tbx21−/−;Rag2−/− ulcerative colitis) mice (Garrett et al., 2007; Powell et al., 2012), or mutants may be susceptible to opportunistic pathogens, such as Helicobacter hepaticus in Nod2−/− mice (Biswas et al., 2010). Indeed, the pathotype adherent and invasive E. coli, which has been associated with ileal CD (Darfeuille-Michaud et al., 2004) and with granulomatous colitis in boxer dogs (Simpson et al., 2006), is restricted by ATG16L1-dependent autophagy in IEC lines (Lapaquette et al., 2010).
The development of ileitis with increasing lifetime in mice with a null variant of Atg16l1 in IECs has exposed the important role of ATG16L1-dependent autophagy for restraining IRE1α hyperactivation. Although rapid age-dependent decline in the ability to resolve ER stress has been observed in Caenorhabditis elegans (Taylor and Dillin, 2013), aging will likely play only a very subordinate, if any, role as a disease trigger in human ATG16L1T300A carriers; the high risk allele frequency of ATG16L1T300A in itself already argues against such a simple relationship (Jostins et al., 2012). The continual accumulation of stress hits over time that lead to IRE1α engagement may at some point overwhelm the epithelium’s capacity to resolve ER stress, whereby hypomorphic ATG16L1T300A may be an important determinant of that threshold. In fact, the ER serves as a central integrator of the state of the cell (Hotamisligil, 2010), and IECs and Paneth cells at the inner surface are particularly exposed to environmental factors that may impinge on this finely balanced system. Such factors may arise from, for example, infections (Pillich et al., 2012), microbiota constituents (Tashiro et al., 2007), toxins (Sun et al., 2014), or from components of the diet (Nezami et al., 2014; Stancu et al., 2015; Yin et al., 2015). Notably, high fat diet and obesity, the latter associated with markedly increased risk for developing CD (Khalili et al., 2015), have long been known to associate with ER stress in liver and adipose tissue (Hotamisligil, 2010) but also in Paneth cells (Hodin et al., 2011).
The principle mechanism that is activated by the UPR for removal of misfolded proteins, ER-associated degradation, remarkably also controls the turnover of IRE1α via the Sel1L-Hrd1 ER-associated degradation complex under basal conditions (Sun et al., 2015). In contrast, Sel1L-dependent IRE1α degradation is attenuated under conditions of ER stress (Sun et al., 2015), suggesting that optineurin-dependent selective autophagy may then become increasingly important as suggested by our data. As IRE1α dimerization, oligomerization, and higher-order cluster formation are associated with escalating states of IRE1α activation and a transition from homeostatic to pro-death and proinflammatory UPR (Korennykh and Walter, 2012; Ghosh et al., 2014; Maurel et al., 2014), autophagy may provide a more effective process than proteasomal degradation for removing supramolecular clusters of IRE1α. Ubiquitination of IRE1α has been associated with both IRE1α’s activation (Zhu et al., 2014) as well as degradation (Gao et al., 2008; Sun et al., 2015), and it remains to be determined whether this optineurin-dependent autophagic process is dependent on ubiquitination (Wild et al., 2011; Khaminets et al., 2016). Recent studies have identified FAM134b as an ER-resident receptor that regulates ER membrane turnover by selective autophagy (Khaminets et al., 2015). Interestingly, silencing of Fam134b in MODE-K cells did not affect IRE1α levels (not depicted), indicating that autophagy-dependent IRE1α removal may be selective and regulated differently from the bulk process of ER-phagy.
The specific role for the unique expression of both IRE1α and IRE1β in the intestinal epithelium has remained enigmatic. A clue comes from Ern2−/− mice, which are susceptible to DSS-induced colitis (Bertolotti et al., 2001). We now show that they complement each other’s role in protein folding homeostasis within the intestinal epithelium. However, IRE1α and IRE1β have divergent roles in organ inflammation. This feature is conspicuous in the Atg16l1;Xbp1-deficient epithelium and therefore likely reflects divergent effector functions that are independent of XBP1. These effectors appear particularly pertinent to the hyperactive IRE1α observed in the Atg16l1;Xbp1-deficient epithelium (Maurel et al., 2014), and although their precise nature remains to be determined, it seems plausible that they might involve specific assembly of the UPR-osome, regulated IRE1-dependent decay, or cell death mechanisms (Hetz et al., 2011; Walter and Ron, 2011; Korennykh and Walter, 2012; Han et al., 2013).
In conclusion, the mechanism reported herein suggests biological integration of environmental and genetic risk across physiological processes that converge on pathological activation of IRE1α as driver of ileal inflammation. Correspondingly, we show that ATG16L1T300A-related genetic risk associates with IRE1α accumulation in ileal crypts. In this context, it is notable that NOD2, the singly strongest genetic risk factor of CD (Jostins et al., 2012) which, similar to ATG16L1, predisposes for CD involving the ileum (Cuthbert et al., 2002), links IRE1α signaling with inflammation via a noncanonical pathway that does not involve the microbial NOD2 ligand muramyl dipeptide (Keestra-Gounder et al., 2016). It will be intriguing to explore whether IRE1α signaling constitutes a common converging element across this and other CD-specific genetic risk factors that initiate the disease process in CD upon interaction with yet unknown environmental triggers.
Materials and methods
Mice
Villin-Cre;Xbp1fl/fl (Xbp1ΔIEC), Villin-Cre;Atg16l1fl/fl (Atg16l1ΔIEC), Villin-Cre;Atg16l1fl/fl;Xbp1fl/fl (Atg16l1;Xbp1ΔIEC), Villin-Cre;Ern1fl/fl (Ern1ΔIEC), Ern2−/−, and Villin-Cre;Ern1fl/fl;Xbp1fl/fl mice have been described previously (Bertolotti et al., 2001; Kaser et al., 2008; Adolph et al., 2013; Niederreiter et al., 2013). Villin-Cre;Atg16l1fl/fl;Ern1fl/fl (Atg16l1;Ern1ΔIEC), Villin-Cre;Xbp1fl/fl;Ern2−/− (Xbp1ΔIEC;Ern2−/−), Villin-Cre;Atg16l1fl/fl;Xbp1fl/fl;Ern1fl/fl (Atg16l1;Xbp1;Ern1ΔIEC), Villin-Cre;Atg16l1fl/fl;Xbp1fl/fl;Ern2−/− (Atg16l1;Xbp1ΔIEC;Ern2−/−), Villin-Cre;Ern1fl/fl;Ern2−/− (Ern1ΔIEC;Ern2−/−), and Villin-Cre;Ern1fl/fl;Xbp1fl/fl;Ern2−/− (Ern1;Xbp1ΔIEC;Ern2−/−) mice were generated by intercrossing the relevant individual strains. All mice were on a C57BL/6 background except Ern1fl/fl (129/svJ × C57BL/6) and Ern2−/− (F1[DBA/2 × C57BL/6 × 129/svJ]) mice. The Villin-Cre transgene was maintained heterozygously. Mice were housed under SPF conditions at the central biomedical services facility of the University of Cambridge if not stated otherwise. Whereas the Atg16l1fl allele in the aforementioned strains had exon 1 flanked by loxP sites (Adolph et al., 2013), a further strain (on C57BL/6 background) maintained under SPF conditions at the animal facility at Cedars-Sinai Medical Center (CSMC) had exon 3 floxed (Atg16l1-ex3; generated in collaboration with genOway) and was similarly crossed to Villin-Cre transgenic mice (on C57BL/6 background). Experiments were performed with age- and gender-matched mice and included littermate Wt animals as controls where technically possible. Cross-foster experiments were performed according to Fig. 8 (A, B, and G). For cross-foster experiments, breeding pairs from all genotypes were set up simultaneously, and pups of the indicated genotypes and littermate Wt controls were cross-fostered within 24 h upon birth and analyzed at 8–10 wk of age. Mice were handled and all experiments were performed according to institutional guidelines and the approval of relevant authorities (UK Home Office PPL80/2560, Austrian Federal Ministry of Science and Research, and CSMC Animal Care and Use Committee protocol 3723).
Antibodies and reagents
The following antibodies were used for immunoblotting and immunoprecipitation: anti-IRE1β (Bertolotti et al., 2001), anti–phospho-IRE1β (gift from K. Kohno, Nara Institute of Science and Technology, Takayama, Ikoma, Nara, Japan), anti-IRE1α (20790; H-190; polyclonal; Santa Cruz Biotechnology, Inc.), anti-IRE1α (3294; 14C10), anti-NDP52 (9036; polyclonal), anti-NBR1 (9891; D2E6), anti-GAPDH (2118; 14C10), anti–β-actin (4970; 13E5; Cell Signaling Technology), antioptineurin (100000; polyclonal; Cayman), and anti-p62 (PM045; polyclonal; MBL). Immunofluorescence antibodies were anti-IRE1α (20790; H-190), antilysozyme (27958; C19), anti-MUC2 (15334; H-300), anti-IRE1β (Bertolotti et al., 2001; Santa Cruz Biotechnology, Inc.), and anti-GRP78 (ab21685; polyclonal; Abcam). The following secondary antibodies were used: donkey anti–rabbit IgG (DyLight 488; ab96919; polyclonal), donkey anti–goat IgG (DyLight 488; ab96931; polyclonal), donkey anti–rabbit IgG (DyLight 650; ab96894; polyclonal; Abcam), and Clean-Blot (21230; Thermo Fisher Scientific). UEA-1 (RL-1062; Vector Laboratories) was used to stain secretory IECs.
Analysis of microbial composition by 16S rDNA ribotyping
Bacterial DNA from ileal tissue–associated microbes was isolated using the Precellys 24 tissue homogenizer (Bertin Technologies) and FastDNA SPIN kit for Soil (MP Biomedicals) according to the manufacturer’s instructions. The V4 hypervariable region of the bacterial 16S rRNA gene was amplified as described previously (Kozich et al., 2013). The library was sequenced on an Illumina MiSeq system using the MiSeq Reagent kit (v3) with 2 × 250 bp Contig read length configuration and dual indexing. The data output was filtered and trimmed using Mothur package (Schloss et al., 2009), and chimeras were removed using the Uchime (Edgar et al., 2011) algorithm. The trimmed reads were aligned to the SILVA (Quast et al., 2013) reference database for excluding ambiguous sequences and were classified using the Ribosomal Database Project classifier (Wang et al., 2007). The aligned and subsampled datasets were used to compute a distance matrix for categorizing sequences into operational taxonomic units by a 97% similarity cut off. R software was used to analyze α and β diversity (taxonomic classification and principal coordinates analysis plots). Clustering of 16S rDNA V4 ribotyping sequences was done at 3% genetic difference. Bray-Curtis and Jaccard dissimilarities were analyzed using constrained analysis of principle coordinates (capscale function in Vegan for plotting and ANOVA for significance test). The permutational (nonparametric) multivariate ANOVA (NPMANOVA; Adonis; Anderson, 2001; McArdle and Anderson, 2001) based on dissimilarities implemented in Vegan was used to test clustering of β diversity indices for ascertaining differences in community compositions. A p-value of significance was incorporated by comparing the obtained R2 values to that obtained from 1,000 random permutations of the data in each case. In addition, CCA was done to interpret the role of the variables and their multivariate associations among the datasets.
Histology
Hematoxylin and eosin (H&E)–stained sections were assessed for intestinal inflammation using a semiquantitative composite scoring system previously described (Adolph et al., 2013). The total score represents the sum of five histological subscores multiplied by a factor that depends on the extent of the inflammation. The following histological subscores and grading were used (for each parameter: 0, absent; 1, mild; 2, moderate; 3, severe): mononuclear cell infiltrate (0–3), crypt hyperplasia (0–3), epithelial injury/erosion (0–3), polymorphonuclear cell infiltrates (0–3), and transmural inflammation (0, absent; 1, submucosal; 2, one focus extending into muscularis and serosa; 3, up to five foci extending into muscularis and serosa; 4, diffuse). The extent factor represents the proportion of intestine affected by intestinal inflammation: 1, <10%; 2, 10–25%; 3, 25–50%; and 4, >50%. Samples were evaluated in a blinded fashion.
DSS-induced colitis
At 8 wk of age, mice received 3% DSS (36–50 kD; MP Biomedicals) in their drinking water for 7 d. Mouse weight and rectal bleeding was assessed daily. Histological scoring was performed as described previously (Garrett et al., 2007).
Crypt isolation
Crypt isolation protocol was adapted from Sato et al. (2009). In brief, flushed and longitudinally cut intestinal pieces were vortexed in ice-cold PBS for 5 min, transferred to 30 mM EDTA, and vortexed for 5 min and the supernatant was collected, which was repeated for a total of four times. Supernatants were microscopically checked for crypt enrichment and then spun down at 800 g and 4°C followed by RNA isolation (QIAGEN) or protein extraction.
Immunohistochemistry, PAS, TUNEL, and BrdU staining
Immunohistochemistry on formalin-fixed paraffin-embedded sections was performed according to standard protocols as described previously (Adolph et al., 2013). In brief, sections were deparaffinized in Xylol and rehydrated in ethanol. Antigen retrieval was performed in citrate or EDTA buffer for 15 min at subboiling temperature, followed by quenching of endogenous peroxidase activity. Primary antibody was incubated at 4°C overnight, secondary biotinylated antibody for 1 h at room temperature, followed by streptavidin-HRP (Vector Laboratories). Sections were developed using a DAB Peroxidase Substrate kit (Vector Laboratories) and analyzed using a microscope (Axio Observer Z.1) with a camera (AxioCam MRc5) and AxioVision software (release 4.8; ZEISS) or AxioImager Z2 (AxioCam MRc5; ZEISS) with plan-Apochromat lenses and Zen Blue 2012 software. TUNEL using the TUNEL cell death detection kit (Roche) as per the manufacturer’s instructions allowed us to quantify cell death by enumerating TUNEL+ cells adjusted for the intestinal length of the respective sample. IEC proliferation was assessed 24 h after administering 1 mg BrdU (BD) intraperitoneally. Incorporated BrdU was detected by the BrdU in situ detection kit (BD), and BrdU+ IECs were quantified along the crypt villus axis and presented as a ratio to total IECs along that axis. PAS staining was performed according to standard protocols.
Immunofluorescence and confocal microscopy
Formalin-fixed paraffin-embedded sections were deparaffinized in Xylol and rehydrated in ethanol. Sections were antigen retrieved in citrate or EDTA buffer for 15 min at subboiling temperature, blocked (10% donkey serum; Bio-Rad Laboratories), and incubated with primary antibody for 1 h at room temperature. Fluorescently labeled secondary antibodies were incubated for 45 min at room temperature, and slides were mounted with Prolong Gold Antifade reagent with DAPI (Invitrogen). Images were acquired using a confocal microscope (SP5; Leica Biosystems) and LAS AF software.
Transmission electron microscopy
Intestines were harvested and flushed with saline, cut into thin sections, and fixed in 4% glutaraldehyde in 0.1 M Hepes buffer, pH 7.4, for 12 h at 4°C. Sections were rinsed in 0.1 M Hepes buffer for five times and treated with 1% osmium ferricyanide at room temperature for 2 h, followed by rinsing in deionized water for five times and treated with 2% uranyl acetate in 0.05 M maleate buffer, pH 5.5, for 2 h at room temperature. Samples were rinsed in deionized water and dehydrated in 70–100% ethanol, followed by treatment with two changes of dry acetonitrile and infiltration with Quetol epoxy resin. Images were taken with a transmission electron microscope (Tecnai G2; FEI) operated at 120 Kv equipped with a digital camera (AMT XR60B) running Deben software.
Human intestinal biopsies
After obtaining written informed consent, human intestinal biopsies from the terminal ileum of 31 CD patients and 43 healthy controls were collected upon colonoscopy and stored in optimal cutting temperature compound. DNA from EDTA blood samples was extracted (Wizard Genomic DNA purification kit; Promega) according to the manufacturer’s instructions, and genotyping for ATG16L1 (T300A) SNP (rs2241880) was performed as described previously (Deuring et al., 2014). In brief, a 330-bp fragment containing the ATG16L1 (T300A) SNP was amplified using Phusion High-Fidelity PCR (Phusion; Thermo Fisher Scientific) and the following primers: forward, 5′- TTGGAGTCCACAGGTTAG-3′; and reverse, 5′-CAGTAGCTGGTACCCTCACTTC-3′ (Deuring et al., 2014). Subsequently, the DNA fragment was gel extracted from a 1% agarose gel using a Gel Extraction kit (QIAGEN). The forward primer was used for sequencing of the 330-bp ATG16L1 (T300A) amplicon at Source Bioscience Cambridge. All protocols were approved by Cambridgeshire 4 Research Ethics Committee (reference 03/5/012). Patient characteristics are outlined in Table S1.
Cell culture
MODE-K cells cultured in DMEM were transduced with an Xbp1-specific or control shRNA lentiviral vector as described previously (Kaser et al., 2008), and stable clones were established. HT29 cells were cultured in DMEM supplemented with 10% FBS and 5% PenStrep (Gibco). Cells were harvested after seeding in 6-well plates overnight or 72 h after transfection with Nbr1, NDP52, Optn, Sqstm1, or control siRNA (Thermo Fisher Scientific). 50 nM bafilomycin (Sigma-Aldrich), 10 µg/ml cycloheximide (Sigma-Aldrich), 2 µg/ml tunicamycin (Sigma-Aldrich), or 50 nM rapamycin (Sigma-Aldrich) was added to the cell culture for the indicated time.
Silencing via siRNA
siRNA experiments of Nbr1, NDP52, Optn, Sqstm1, or control siRNA (Thermo Fisher Scientific) were conducted according to the manufacturer’s instructions. In brief, MODE-K and HT29 cells were cultured to ∼60% confluence in 6-well plates and transfected with siRNAs suspended in 100 µl Opti-MEM (Gibco) and 6 µl Lipofectamine 2000 (Thermo Fisher Scientific). After 72 h, cultures were lysed in radioimmunoprecipitation assay (RIPA) buffer or Triton X buffer and analyzed by immunoblotting.
RNA extraction and quantitative RT-PCR (qRT-PCR)
RNA was isolated from ileal scrapings or crypt fractions using an RNeasy Mini kit (QIAGEN) and reverse transcribed to cDNA using M-MLV reverse transcriptase (Invitrogen). qRT-PCR was performed with SYBR green mastermix (Eurogentec) on an MX-3005 system (Agilent Technologies). For qRT-PCR, the following pairs of primers were used: mActb (α-actin), 5′-GATGCTCCCCGGGCTGTATT-3′ and 5′-GGGGTACTTCAGGGTCAGGA-3′ (Kaser et al., 2008); mErn1, 5′-GCTCCAGTTTTTCCAGGATG-3′ and 5′-GAGCCACCTTTGTAGGTTCTG-3′ (designed in this study); mErn2, 5′-GATGAGGCAGCAGCAGC-3′ and 5′-GCCCTGCTTCAGGGTCAT-3′ (designed in this study); and mHprt, 5′-GTTAAGCAGTACAGCCCCAAA-3′ and 5′-AGGGCATATCCAACAACAAACTT-3′ (Amthor et al., 2009). Target gene expression was normalized to housekeeping gene expression.
Xbp1 splicing assay
Assessment of Xbp1 splicing status by PCR was described previously (Kaser et al., 2008). In brief, isolated RNA was reverse transcribed (M-MLV reverse transcriptase; Invitrogen), amplified by PCR (HotStarTaq; QIAGEN) using the primers mXbp1 sp. forward 5′-ACACGCTTGGGAATGGACAC-3′ and mXbp1 sp. reverse 5′-CCATGGGAAGATGTTCTGGG-3′ (Iwakoshi et al., 2003), resolved on a 2.5% agarose gel, and spliced Xbp1 (171 bp) and unspliced Xbp1 (145 bp) products were assessed.
Immunoblot analysis
Immunoblot analysis was performed according to standard procedures. In brief, cells, IEC scrapings, or isolated crypts were lysed using RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) or Triton X buffer (10 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 10% glycerol, 0.1% SDS, and 0.5% deoxycholate), supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific). Equal protein amounts eluted in Laemmli buffer were denatured at 95°C and resolved on 8–12.5% SDS-PAGE (Bio-Rad Laboratories), transferred to polyvinylidene fluoride membrane (Bio-Rad Laboratories), blocked with 5% skim milk or 5% BSA in TBS-Tween, and incubated with primary antibody in 5% BSA TBS-Tween overnight. Antibody was detected with HRP-conjugated secondary antibody and visualized with LumiGLO (Cell Signaling Technology).
Immunoprecipitation
Protein A sepharose beads (GE Healthcare) were washed in PBS and in RIPA lysis buffer or Triton X buffer at 4°C overnight. 60 µl of washed sepharose beads resuspended in RIPA or Triton X buffer containing protease and phosphatase inhibitors (Thermo Fisher Scientific) was added to the protein lysates and incubated for 30 min at 4°C. Subsequently, the precleared lysates were incubated with pull-down antibody or with IgG control antibody for 1 h at 4°C, before 60 µl of washed protein A sepharose beads was added and incubated for 1 h at 4°C. After washing the samples in RIPA or Triton X buffer containing protease and phosphatase inhibitors, samples were resuspended in 70 µl of 2× loading buffer before they were boiled at 95°C for 5 min, incubated on ice for 5 min, and run on 10% SDS PAGE.
Statistical analysis
Statistical significance was tested by an unpaired two-tailed Student’s t test or a Mann–Whitney U test as appropriate and considered significant at P < 0.05. In experiments where more than two groups were compared, Kruskal–Wallis followed by Mann–Whitney U and posthoc Bonferroni Holm’s correction or one-way ANOVA with posthoc Bonferroni correction were performed as appropriate. Prism (version 6.0d; GraphPad Software) was used for data analysis.
Online supplemental material
Table S1 is available as an Excel file and shows the characteristics of all patients analyzed for IRE1α immunoreactivity, stratified by ATG16L1 genotype.
Acknowledgments
We thank Drs. James Lee, Andre Franke, and Felix Randow for helpful discussions and Dr. Tim Raine for help with statistical analysis. We are grateful to Elizabeth Andersen and Dr. Jun Inoue, the doctors and nurses at Addenbrooke’s hospital’s endoscopy unit, and participating patients for providing intestinal biopsies. We thank Dr. Jeremy Skepper for assistance with transmission electron microscopy. Expertise and help from the National Institute for Health Research Cambridge Biomedical Research Centre Cell Phenotyping Hub is greatly acknowledged.
This study was supported by the European Research Council under the European Community’s Seventh Framework Program (grant FP7/2007-2013)/ERC, agreement no. 260961 to A. Kaser and grant HORIZON2020/ERC, agreement no. 648889 to A. Kaser), the Wellcome Trust (Investigator Award 106260/Z/14/Z to A. Kaser and Principal Research Fellowship 2008/Z/16/Z to D. Ron), the Cambridge Biomedical Research Centre (A. Kaser), a Medical Research Council PhD for clinicians training fellowship (grant MR/N001893/1 to J. Bhattacharyya), fellowships from the European Crohn’s and Colitis Organization (M. Tschurtschenthaler and T.E. Adolph), the Research Training Group Genes, Environment, and Inflammation supported by the Deutsche Forschungsgemeinschaft (grant RTG 1743/1 to P. Rosenstiel), the SFB877 subproject B9 and CLVIII ExC 306 Inflammation at Interfaces (P. Rosenstiel), and the National Institutes of Health (grants DK044319, DK051362, DK053056, and DK088199 to the Harvard Digestive Diseases Center and grant DK0034854 to R.S. Blumberg).
The authors declare no competing financial interests.
Author contributions: M. Tschurtschenthaler, T.E. Adolph, L. Niederreiter, J.W. Ashcroft, P. Rosenstiel, R.S. Blumberg, and A. Kaser conceived and designed the experiments. M. Tschurtschenthaler, J.W. Ashcroft, T.E. Adolph, and L. Niederreiter together with S. Saveljeva, J. Bhattacharyya, M.B. Flak, and G.M. Fuhler performed and analyzed most experiments. R. Bharti and P. Rosenstiel performed and analyzed 16S rDNA sequencing experiments. M. Parkes supervised collection of human biopsies. K. Kohno, T. Iwawaki, A.M. Smith, H.P. Harding, and D. Ron provided essential reagents and mouse strains. D.Q. Shih and S.R. Targan generated and analyzed new mutant mouse strains. G.M. Fuhler, C.J. van der Woude, and M.P. Peppelenbosch contributed analytic methods. A. Kaser devised and coordinated the project and together with M. Tschurtschenthaler, T.E. Adolph, L. Niederreiter, P. Rosenstiel, and R.S. Blumberg with contributions from all authors, wrote the manuscript.
Footnotes
Abbreviations used:
- CCA
- canonical correlation analysis
- CD
- Crohn’s disease
- DSS
- dextran sodium sulfate
- H&E
- hematoxylin and eosin
- IBD
- inflammatory bowel disease
- IEC
- intestinal epithelial cell
- PAS
- periodic acid–Schiff
- qRT-PCR
- quantitative RT-PCR
- RIPA
- radioimmunoprecipitation assay
- SNP
- single-nucleotide polymorphism
- SPF
- specific pathogen–free
- TUNEL
- TdT-mediated dUTP nick-end labeling
- UPR
- unfolded protein response
References
- Adolph, T.E., Tomczak M.F., Niederreiter L., Ko H.J., Böck J., Martinez-Naves E., Glickman J.N., Tschurtschenthaler M., Hartwig J., Hosomi S., et al. 2013. Paneth cells as a site of origin for intestinal inflammation. Nature. 503:272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amthor, H., Otto A., Vulin A., Rochat A., Dumonceaux J., Garcia L., Mouisel E., Hourdé C., Macharia R., Friedrichs M., et al. 2009. Muscle hypertrophy driven by myostatin blockade does not require stem/precursor-cell activity. Proc. Natl. Acad. Sci. USA. 106:7479–7484. 10.1073/pnas.0811129106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M.J. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26:32–46. [Google Scholar]
- Baxt, L.A., and Xavier R.J.. 2015. Role of autophagy in the maintenance of intestinal homeostasis. Gastroenterology. 149:553–562. 10.1053/j.gastro.2015.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti, A., Wang X., Novoa I., Jungreis R., Schlessinger K., Cho J.H., West A.B., and Ron D.. 2001. Increased sensitivity to dextran sodium sulfate colitis in IRE1β-deficient mice. J. Clin. Invest. 107:585–593. 10.1172/JCI11476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, A., Liu Y.J., Hao L., Mizoguchi A., Salzman N.H., Bevins C.L., and Kobayashi K.S.. 2010. Induction and rescue of Nod2-dependent Th1-driven granulomatous inflammation of the ileum. Proc. Natl. Acad. Sci. USA. 107:14739–14744. 10.1073/pnas.1003363107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell, K., Liu J.Y., Brown S.L., Miyoshi H., Loh J., Lennerz J.K., Kishi C., Kc W., Carrero J.A., Hunt S., et al. 2008. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 456:259–263. 10.1038/nature07416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell, K., Patel K.K., Maloney N.S., Liu T.C., Ng A.C., Storer C.E., Head R.D., Xavier R., Stappenbeck T.S., and Virgin H.W.. 2010. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 141:1135–1145. 10.1016/j.cell.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., and Brandizzi F.. 2013. IRE1: ER stress sensor and cell fate executor. Trends Cell Biol. 23:547–555. 10.1016/j.tcb.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew, T.S., O’Shea N.R., Sewell G.W., Oehlers S.H., Mulvey C.M., Crosier P.S., Godovac-Zimmermann J., Bloom S.L., Smith A.M., and Segal A.W.. 2015. Optineurin deficiency in mice contributes to impaired cytokine secretion and neutrophil recruitment in bacteria-driven colitis. Dis. Model. Mech. 8:817–829. 10.1242/dmm.020362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, J.A., Lee A.H., Platzer B., Cross B.C., Gardner B.M., De Luca H., Luong P., Harding H.P., Glimcher L.H., Walter P., et al. 2013. The unfolded protein response element IRE1α senses bacterial proteins invading the ER to activate RIG-I and innate immune signaling. Cell Host Microbe. 13:558–569. 10.1016/j.chom.2013.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Clevers, H.C., and Bevins C.L.. 2013. Paneth cells: maestros of the small intestinal crypts. Annu. Rev. Physiol. 75:289–311. 10.1146/annurev-physiol-030212-183744 [DOI] [PubMed] [Google Scholar]
- Conway, K.L., Kuballa P., Song J.H., Patel K.K., Castoreno A.B., Yilmaz O.H., Jijon H.B., Zhang M., Aldrich L.N., Villablanca E.J., et al. 2013. Atg16l1 is required for autophagy in intestinal epithelial cells and protection of mice from Salmonella infection. Gastroenterology. 145:1347–1357. 10.1053/j.gastro.2013.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney, R., Baker J., Brain O., Danis B., Pichulik T., Allan P., Ferguson D.J., Campbell B.J., Jewell D., and Simmons A.. 2010. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 16:90–97. 10.1038/nm.2069 [DOI] [PubMed] [Google Scholar]
- Couturier-Maillard, A., Secher T., Rehman A., Normand S., De Arcangelis A., Haesler R., Huot L., Grandjean T., Bressenot A., Delanoye-Crespin A., et al. 2013. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 123:700–711. 10.1172/JCI62236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert, A.P., Fisher S.A., Mirza M.M., King K., Hampe J., Croucher P.J., Mascheretti S., Sanderson J., Forbes A., Mansfield J., et al. 2002. The contribution of NOD2 gene mutations to the risk and site of disease in inflammatory bowel disease. Gastroenterology. 122:867–874. 10.1053/gast.2002.32415 [DOI] [PubMed] [Google Scholar]
- Darfeuille-Michaud, A., Boudeau J., Bulois P., Neut C., Glasser A.L., Barnich N., Bringer M.A., Swidsinski A., Beaugerie L., and Colombel J.F.. 2004. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 127:412–421. 10.1053/j.gastro.2004.04.061 [DOI] [PubMed] [Google Scholar]
- Deuring, J.J., Fuhler G.M., Konstantinov S.R., Peppelenbosch M.P., Kuipers E.J., de Haar C., and van der Woude C.J.. 2014. Genomic ATG16L1 risk allele-restricted Paneth cell ER stress in quiescent Crohn’s disease. Gut. 63:1081–1091. 10.1136/gutjnl-2012-303527 [DOI] [PubMed] [Google Scholar]
- Edgar, R.C., Haas B.J., Clemente J.C., Quince C., and Knight R.. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 27:2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav, E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., Peaper D.R., Bertin J., Eisenbarth S.C., Gordon J.I., and Flavell R.A.. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 145:745–757. 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus, D., Zhang H., Zeissig S., Lipinski S., Till A., Jiang T., Stade B., Bromberg Y., Ellinghaus E., Keller A., et al. 2013. Association between variants of PRDM1 and NDP52 and Crohn’s disease, based on exome sequencing and functional studies. Gastroenterology. 145:339–347. 10.1053/j.gastro.2013.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, D.N., St Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., and Pace N.R.. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA. 104:13780–13785. 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, B., Lee S.M., Chen A., Zhang J., Zhang D.D., Kannan K., Ortmann R.A., and Fang D.. 2008. Synoviolin promotes IRE1 ubiquitination and degradation in synovial fibroblasts from mice with collagen-induced arthritis. EMBO Rep. 9:480–485. 10.1038/embor.2008.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett, W.S., Lord G.M., Punit S., Lugo-Villarino G., Mazmanian S.K., Ito S., Glickman J.N., and Glimcher L.H.. 2007. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 131:33–45. 10.1016/j.cell.2007.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers, D., Kugathasan S., Denson L.A., Vázquez-Baeza Y., Van Treuren W., Ren B., Schwager E., Knights D., Song S.J., Yassour M., et al. 2014. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 15:382–392. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, R., Wang L., Wang E.S., Perera B.G., Igbaria A., Morita S., Prado K., Thamsen M., Caswell D., Macias H., et al. 2014. Allosteric inhibition of the IRE1α RNase preserves cell viability and function during endoplasmic reticulum stress. Cell. 158:534–548. 10.1016/j.cell.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe, J., Franke A., Rosenstiel P., Till A., Teuber M., Huse K., Albrecht M., Mayr G., De La Vega F.M., Briggs J., et al. 2007. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39:207–211. 10.1038/ng1954 [DOI] [PubMed] [Google Scholar]
- Han, J., Back S.H., Hur J., Lin Y.H., Gildersleeve R., Shan J., Yuan C.L., Krokowski D., Wang S., Hatzoglou M., et al. 2013. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15:481–490. 10.1038/ncb2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz, C., Martinon F., Rodriguez D., and Glimcher L.H.. 2011. The unfolded protein response: integrating stress signals through the stress sensor IRE1α. Physiol. Rev. 91:1219–1243. 10.1152/physrev.00001.2011 [DOI] [PubMed] [Google Scholar]
- Hodin, C.M., Verdam F.J., Grootjans J., Rensen S.S., Verheyen F.K., Dejong C.H., Buurman W.A., Greve J.W., and Lenaerts K.. 2011. Reduced Paneth cell antimicrobial protein levels correlate with activation of the unfolded protein response in the gut of obese individuals. J. Pathol. 225:276–284. 10.1002/path.2917 [DOI] [PubMed] [Google Scholar]
- Homer, C.R., Richmond A.L., Rebert N.A., Achkar J.P., and McDonald C.. 2010. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology. 139:1630–1641.e2. 10.1053/j.gastro.2010.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil, G.S. 2010. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 140:900–917. 10.1016/j.cell.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal, J., Dai K., Seimon T., Jungreis R., Oyadomari M., Kuriakose G., Ron D., Tabas I., and Hussain M.M.. 2008. IRE1β inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 7:445–455. 10.1016/j.cmet.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakoshi, N.N., Lee A.H., Vallabhajosyula P., Otipoby K.L., Rajewsky K., and Glimcher L.H.. 2003. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4:321–329. 10.1038/ni907 [DOI] [PubMed] [Google Scholar]
- Iwawaki, T., Hosoda A., Okuda T., Kamigori Y., Nomura-Furuwatari C., Kimata Y., Tsuru A., and Kohno K.. 2001. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat. Cell Biol. 3:158–164. 10.1038/35055065 [DOI] [PubMed] [Google Scholar]
- Jostins, L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., et al. International IBD Genetics Consortium . 2012. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 491:119–124. 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser, A., Lee A.H., Franke A., Glickman J.N., Zeissig S., Tilg H., Nieuwenhuis E.E., Higgins D.E., Schreiber S., Glimcher L.H., and Blumberg R.S.. 2008. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 134:743–756. 10.1016/j.cell.2008.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keestra-Gounder, A.M., Byndloss M.X., Seyffert N., Young B.M., Chávez-Arroyo A., Tsai A.Y., Cevallos S.A., Winter M.G., Pham O.H., Tiffany C.R., et al. 2016. NOD1 and NOD2 signalling links ER stress with inflammation. Nature. 532:394–397. 10.1038/nature17631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili, H., Ananthakrishnan A.N., Konijeti G.G., Higuchi L.M., Fuchs C.S., Richter J.M., and Chan A.T.. 2015. Measures of obesity and risk of Crohn’s disease and ulcerative colitis. Inflamm. Bowel Dis. 21:361–368. 10.1097/MIB.0000000000000283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets, A., Heinrich T., Mari M., Grumati P., Huebner A.K., Akutsu M., Liebmann L., Stolz A., Nietzsche S., Koch N., et al. 2015. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 522:354–358. 10.1038/nature14498 [DOI] [PubMed] [Google Scholar]
- Khaminets, A., Behl C., and Dikic I.. 2016. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 26:6–16. 10.1016/j.tcb.2015.08.010 [DOI] [PubMed] [Google Scholar]
- Kobayashi, K.S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nuñez G., and Flavell R.A.. 2005. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 307:731–734. 10.1126/science.1104911 [DOI] [PubMed] [Google Scholar]
- Korennykh, A., and Walter P.. 2012. Structural basis of the unfolded protein response. Annu. Rev. Cell Dev. Biol. 28:251–277. 10.1146/annurev-cellbio-101011-155826 [DOI] [PubMed] [Google Scholar]
- Kozich, J.J., Westcott S.L., Baxter N.T., Highlander S.K., and Schloss P.D.. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79:5112–5120. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapaquette, P., Glasser A.L., Huett A., Xavier R.J., and Darfeuille-Michaud A.. 2010. Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell. Microbiol. 12:99–113. 10.1111/j.1462-5822.2009.01381.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., and Kaufman R.J.. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16:452–466. 10.1101/gad.964702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, B., Mizushima N., and Virgin H.W.. 2011. Autophagy in immunity and inflammation. Nature. 469:323–335. 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Korennykh A.V., Behrman S.L., and Walter P.. 2010. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc. Natl. Acad. Sci. USA. 107:16113–16118. 10.1073/pnas.1010580107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., Gulati A.S., Cantillana V., Henry S.C., Schmidt E.A., Daniell X., Grossniklaus E., Schoenborn A.A., Sartor R.B., and Taylor G.A.. 2013. Irgm1-deficient mice exhibit Paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 305:G573–G584. 10.1152/ajpgi.00071.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., et al. International IBD Genetics Consortium . 2015. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47:979–986. 10.1038/ng.3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy, K.J., and Powrie F.. 2011. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 474:298–306. 10.1038/nature10208 [DOI] [PubMed] [Google Scholar]
- Martino, M.B., Jones L., Brighton B., Ehre C., Abdulah L., Davis C.W., Ron D., O’Neal W.K., and Ribeiro C.M.. 2013. The ER stress transducer IRE1β is required for airway epithelial mucin production. Mucosal Immunol. 6:639–654. 10.1038/mi.2012.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel, M., Chevet E., Tavernier J., and Gerlo S.. 2014. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 39:245–254. 10.1016/j.tibs.2014.02.008 [DOI] [PubMed] [Google Scholar]
- McArdle, B.H., and Anderson M.J.. 2001. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology. 82:290–297. 10.1890/0012-9658(2001)082[0290:FMMTCD]2.0.CO;2 [DOI] [Google Scholar]
- Molodecky, N.A., Soon I.S., Rabi D.M., Ghali W.A., Ferris M., Chernoff G., Benchimol E.I., Panaccione R., Ghosh S., Barkema H.W., and Kaplan G.G.. 2012. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 142:46–54.e42. 10.1053/j.gastro.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Murthy, A., Li Y., Peng I., Reichelt M., Katakam A.K., Noubade R., Roose-Girma M., DeVoss J., Diehl L., Graham R.R., and van Lookeren Campagne M.. 2014. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 506:456–462. 10.1038/nature13044 [DOI] [PubMed] [Google Scholar]
- Nezami, B.G., Mwangi S.M., Lee J.E., Jeppsson S., Anitha M., Yarandi S.S., Farris A.B. III, and Srinivasan S.. 2014. MicroRNA 375 mediates palmitate-induced enteric neuronal damage and high-fat diet-induced delayed intestinal transit in mice. Gastroenterology. 146:473–83.e3. 10.1053/j.gastro.2013.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreiter, L., Fritz T.M., Adolph T.E., Krismer A.M., Offner F.A., Tschurtschenthaler M., Flak M.B., Hosomi S., Tomczak M.F., Kaneider N.C., et al. 2013. ER stress transcription factor Xbp1 suppresses intestinal tumorigenesis and directs intestinal stem cells. J. Exp. Med. 210:2041–2056. 10.1084/jem.20122341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillich, H., Loose M., Zimmer K.P., and Chakraborty T.. 2012. Activation of the unfolded protein response by Listeria monocytogenes. Cell. Microbiol. 14:949–964. 10.1111/j.1462-5822.2012.01769.x [DOI] [PubMed] [Google Scholar]
- Powell, N., Walker A.W., Stolarczyk E., Canavan J.B., Gökmen M.R., Marks E., Jackson I., Hashim A., Curtis M.A., Jenner R.G., et al. 2012. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity. 37:674–684. 10.1016/j.immuni.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, N.J., Fisher S.A., Franke A., Hampe J., Onnie C.M., Soars D., Bagnall R., Mirza M.M., Sanderson J., Forbes A., et al. 2007. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn’s disease and is independent of CARD15 and IBD5. Gastroenterology. 132:1665–1671. 10.1053/j.gastro.2007.03.034 [DOI] [PubMed] [Google Scholar]
- Quast, C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., and Glöckner F.O.. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman, A., Rausch P., Wang J., Skieceviciene J., Kiudelis G., Bhagalia K., Amarapurkar D., Kupcinskas L., Schreiber S., Rosenstiel P., et al. 2016. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut. 65:238–248. 10.1136/gutjnl-2014-308341 [DOI] [PubMed] [Google Scholar]
- Rioux, J.D., Xavier R.J., Taylor K.D., Silverberg M.S., Goyette P., Huett A., Green T., Kuballa P., Barmada M.M., Datta L.W., et al. 2007. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39:596–604. 10.1038/ng2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov, V., Dötsch V., Johansen T., and Kirkin V.. 2014. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 53:167–178. 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- Sato, T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., and Clevers H.. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459:262–265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Schaubeck, M., Clavel T., Calasan J., Lagkouvardos I., Haange S.B., Jehmlich N., Basic M., Dupont A., Hornef M., von Bergen M., et al. 2016. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut. 65:225–237. 10.1136/gutjnl-2015-309333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss, P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck, S., Gallagher C.M., and Walter P.. 2014. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J. Cell Sci. 127:4078–4088. 10.1242/jcs.154716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, K.W., Dogan B., Rishniw M., Goldstein R.E., Klaessig S., McDonough P.L., German A.J., Yates R.M., Russell D.G., Johnson S.E., et al. 2006. Adherent and invasive Escherichia coli is associated with granulomatous colitis in boxer dogs. Infect. Immun. 74:4778–4792. 10.1128/IAI.00067-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A.M., Sewell G.W., Levine A.P., Chew T.S., Dunne J., O’Shea N.R., Smith P.J., Harrison P.J., Macdonald C.M., Bloom S.L., and Segal A.W.. 2015. Disruption of macrophage pro-inflammatory cytokine release in Crohn’s disease is associated with reduced optineurin expression in a subset of patients. Immunology. 144:45–55. 10.1111/imm.12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancu, C.S., Carnuta M.G., Sanda G.M., Toma L., Deleanu M., Niculescu L.S., Sasson S., Simionescu M., and Sima A.V.. 2015. Hyperlipidemia-induced hepatic and small intestine ER stress and decreased paraoxonase 1 expression and activity is associated with HDL dysfunction in Syrian hamsters. Mol. Nutr. Food Res. 59:2293–2302. 10.1002/mnfr.201500422 [DOI] [PubMed] [Google Scholar]
- Stolz, A., Ernst A., and Dikic I.. 2014. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16:495–501. 10.1038/ncb2979 [DOI] [PubMed] [Google Scholar]
- Sun, C., Wang H., Chen S., Li Z., Li S., and Wang J.. 2014. Recombinant Clostridium difficile toxin B induces endoplasmic reticulum stress in mouse colonal carcinoma cells. Acta Biochim. Biophys. Sin. (Shanghai). 46:973–981. 10.1093/abbs/gmu091 [DOI] [PubMed] [Google Scholar]
- Sun, S., Shi G., Sha H., Ji Y., Han X., Shu X., Ma H., Inoue T., Gao B., Kim H., et al. 2015. IRE1α is an endogenous substrate of endoplasmic-reticulum-associated degradation. Nat. Cell Biol. 17:1546–1555. 10.1038/ncb3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro, E., Hironiwa N., Kitagawa M., Futamura Y., Suzuki S., Nishio M., and Imoto M.. 2007. Trierixin, a novel Inhibitor of ER stress-induced XBP1 activation from Streptomyces sp. 1. Taxonomy, fermentation, isolation, and biological activities. J. Antibiot. 60:547–553. 10.1038/ja.2007.69 [DOI] [PubMed] [Google Scholar]
- Taylor, R.C., and Dillin A.. 2013. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 153:1435–1447. 10.1016/j.cell.2013.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon, W., Lee K., Callaghan B., Welihinda A., and Kaufman R.J.. 2000. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 14:2725–2736. 10.1101/gad.839400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos, L.H., Carneiro L.A., Ramjeet M., Hussey S., Kim Y.G., Magalhães J.G., Yuan L., Soares F., Chea E., Le Bourhis L., et al. 2010. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 11:55–62. 10.1038/ni.1823 [DOI] [PubMed] [Google Scholar]
- Tsuru, A., Fujimoto N., Takahashi S., Saito M., Nakamura D., Iwano M., Iwawaki T., Kadokura H., Ron D., and Kohno K.. 2013. Negative feedback by IRE1β optimizes mucin production in goblet cells. Proc. Natl. Acad. Sci. USA. 110:2864–2869. 10.1073/pnas.1212484110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDussen, K.L., Liu T.C., Li D., Towfic F., Modiano N., Winter R., Haritunians T., Taylor K.D., Dhall D., Targan S.R., et al. 2014. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology. 146:200–209. 10.1053/j.gastro.2013.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, P., and Ron D.. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334:1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Wang, M., and Kaufman R.J.. 2016. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 529:326–335. 10.1038/nature17041 [DOI] [PubMed] [Google Scholar]
- Wang, Q., Garrity G.M., Tiedje J.M., and Cole J.R.. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehkamp, J., Salzman N.H., Porter E., Nuding S., Weichenthal M., Petras R.E., Shen B., Schaeffeler E., Schwab M., Linzmeier R., et al. 2005. Reduced Paneth cell α-defensins in ileal Crohn’s disease. Proc. Natl. Acad. Sci. USA. 102:18129–18134. 10.1073/pnas.0505256102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg, H., Shvets E., and Elazar Z.. 2011. Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80:125–156. 10.1146/annurev-biochem-052709-094552 [DOI] [PubMed] [Google Scholar]
- Wild, P., Farhan H., McEwan D.G., Wagner S., Rogov V.V., Brady N.R., Richter B., Korac J., Waidmann O., Choudhary C., et al. 2011. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 333:228–233. 10.1126/science.1205405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlbier, U., and Hetz C.. 2011. Modulating stress responses by the UPRosome: a matter of life and death. Trends Biochem. Sci. 36:329–337. 10.1016/j.tibs.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Yin, J., Gu L., Wang Y., Fan N., Ma Y., and Peng Y.. 2015. Rapamycin improves palmitate-induced ER stress/NFκB pathways associated with stimulating autophagy in adipocytes. Mediators Inflamm. 2015:272313. 10.1155/2015/272313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Pan Y., Yan R., Zeng B., Wang H., Zhang X., Li W., Wei H., and Liu Z.. 2015. Commensal bacteria direct selective cargo sorting to promote symbiosis. Nat. Immunol. 16:918–926. 10.1038/ni.3233 [DOI] [PubMed] [Google Scholar]
- Zhu, X., Zhang J., Sun H., Jiang C., Dong Y., Shan Q., Su S., Xie Y., Xu N., Lou X., and Liu S.. 2014. Ubiquitination of inositol-requiring enzyme 1 (IRE1) by the E3 ligase CHIP mediates the IRE1/TRAF2/JNK pathway. J. Biol. Chem. 289:30567–30577. 10.1074/jbc.M114.562868 [DOI] [PMC free article] [PubMed] [Google Scholar]