Chen et al. dissect SAP-dependent and SAP-independent SLAM family signaling in the regulation of NKT cell development and follicular T helper cell differentiation using a novel mouse model lacking all seven SLAM family receptors.
Abstract
Signaling lymphocytic activation molecule (SLAM)–associated protein (SAP) mutations in X-linked lymphoproliferative disease (XLP) lead to defective NKT cell development and impaired humoral immunity. Because of the redundancy of SLAM family receptors (SFRs) and the complexity of SAP actions, how SFRs and SAP mediate these processes remains elusive. Here, we examined NKT cell development and humoral immunity in mice completely deficient in SFR. We found that SFR deficiency severely impaired NKT cell development. In contrast to SAP deficiency, SFR deficiency caused no apparent defect in follicular helper T (TFH) cell differentiation. Intriguingly, the deletion of SFRs completely rescued the severe defect in TFH cell generation caused by SAP deficiency, whereas SFR deletion had a minimal effect on the defective NKT cell development in SAP-deficient mice. These findings suggest that SAP-dependent activating SFR signaling is essential for NKT cell selection; however, SFR signaling is inhibitory in SAP-deficient TFH cells. Thus, our current study revises our understanding of the mechanisms underlying T cell defects in patients with XLP.
Introduction
Cell–cell contacts between hematopoietic cells are critical for immune cell selection and differentiation, including the double-positive (DP) selection of natural killer T (NKT) cells in the thymus and the generation of follicular helper T (TFH) cells within germinal centers (GCs; Veillette et al., 2007). NKT cells are specialized T lymphocytes expressing the invariant TCR α chain (mouse Vα14 and human Vα24) and β chain (Vβ8.2, Vβ7, or Vβ2 in mouse and Vβ11 in human; Borg et al., 2007). These cells diverge from the conventional T cell lineage at the DP stage of thymocyte development. In contrast to conventional T cells, which are selected by heterotypic interactions between hematopoietic thymocytes and nonhematopoietic thymic epithelial cells, NKT precursor cells undergo DP selection between two hematopoietic thymocytes (Bendelac, 1995; Wei et al., 2005). Long-term humoral immunity depends on GC responses, in which TFH cells provide help to B cells (Crotty, 2011); consequently, the differentiation of TFH cells also demands contact between hematopoietic cells, that is, T and B cells. The pivotal roles of these two types of contact between immune cells are highlighted by the finding that human signaling lymphocytic activation molecule (SLAM)–associated protein (SAP) mutations resulting in X-linked lymphoproliferative disease (XLP) give rise to the nearly complete loss of NKT cells in the thymus and impaired humoral immunity characterized by reduced TFH cell differentiation (Sayos et al., 1998; Morra et al., 2001; Nichols et al., 2005; Pasquier et al., 2005).
SAP is an adaptor protein with only one SH2 domain, through which SAP can be recruited by SLAM family receptors (SFRs). These hematopoietic cell–specific receptors are composed of seven receptors, namely 2B4, Ly9, CRACC, CD48, SLAM, CD84, and Ly108 (Ma et al., 2007; Cannons et al., 2011). SFRs possess two to four unique immunoreceptor tyrosine-based switch motifs to which SAP and/or its homologue EAT2 (EWS-activated transcript 2) is recruited. Moreover, SFRs are also able to signal through other SH2 domain–containing molecules, particularly in SAP deficiency. Thus, SFRs can mediate SAP-dependent and SAP-independent signaling (Veillette et al., 2009; Dong and Veillette, 2010).
It has been well established that SAP has dual functions in immune regulation (Dong and Veillette, 2010). On one hand, SAP functions as an active signaling molecule. Upon SFR engagement driven by the interaction between two hematopoietic cells, SFR can be phosphorylated by Src family kinases; thus, phosphorylated immunoreceptor tyrosine-based switch motifs have the ability to recruit SAP, which enables recruitment of SH3 domain–containing molecules, such as Fyn kinase (Chan et al., 2003; Latour et al., 2003). Mutation of arginine 78 to alanine (R78A) can abolish Fyn recruitment, which leads to a 50% reduction in NKT cell number (Nunez-Cruz et al., 2008), suggesting that SAP regulates NKT cell development in part via its intrinsic signaling activity. This conclusion was further confirmed by the finding that Fyn deficiency causes impairments in NKT cell development (Nichols et al., 2005; Pasquier et al., 2005). SAPR78A mutant mice also exhibit a defect in Th2 cytokine production (Davidson et al., 2004). However, Fyn-deficient mice and SAPR78A mutant mice have unaltered humoral immunity (Cannons et al., 2006; McCausland et al., 2007). Thus, how SAP-mediated positive signaling plays different role in both types of T cell development and differentiation needs to be examined. On the other hand, SAP acts as natural blocker to prevent other SH2 domain–containing phosphatases from associating with SFRs. These molecules mainly include SH2 domain phosphatase 1 (SHP-1), SHP-2, and SH2 domain inositol phosphatase 1 (SHIP-1; Sayos et al., 1998; Li et al., 2003; Eissmann et al., 2005; Dong et al., 2012; Le Borgne and Shaw, 2012; Zhao et al., 2012; Wu et al., 2016). They are potentially coupled by SFRs, particularly when SAP is absent. It remains to be understood whether SFR-mediated inhibitory signaling initiated by the engagement of hematopoietic cell contact is capable of inhibiting immune cell development and differentiation.
The specificity of SAP binding to SFRs suggests the indispensable role of this family in regulating NKT cell development and TFH cell differentiation. In contrast to SAP-deficient mice, mice lacking a single SFR member exhibit only mild or even no loss of NKT cells. By generating pseudo–double KO chimeras in which SLAM- and Ly108-mediated homotypic interactions are functionally disrupted, Griewank et al. (2007) discovered that NKT cell selection in the thymus was moderately compromised. Recent studies from two groups, in which triply deficient mice lacking SLAM, Ly108, and CD84 were used, also demonstrated the partial requirement of these SFR members in regulating NKT cell development (Hu et al., 2016; Huang et al., 2016). The phenotypic difference in NKT cell numbers between the mice lacking one to three SFR members and SAP-deficient mice strongly implies that other receptors, SFRs or non-SFRs, are required for SAP-mediated NKT cell selection.
The roles of SFRs in TFH cell differentiation are controversial. CD84-deficient mice showed suboptimal GC responses due to impaired TFH cell differentiation after 4-hydroxy-3-nitrophenyl (NP)–OVA immunization (Cannons et al., 2010a). In contrast, CD84 deficiency does not result in significant defects in humoral immunity upon acute viral infection (Kageyama et al., 2012). In mice lacking only SLAM, Ly9, or Ly108, TFH cell differentiation seems to be unperturbed (Graham et al., 2006; McCausland et al., 2007). Unexpectedly, recent studies demonstrated no apparent defects in GC response and TFH cell differentiation in triply deficient mice lacking SLAM, Ly108, and CD84 (Hu et al., 2016; Huang et al., 2016). Therefore, further study is needed to address whether other SFR members are required for TFH cell differentiation and how SAP deficiency causes severe humoral deficiency.
However, comprehensive analysis of the overall role of the SFR family in NKT cell development and TFH cell differentiation remains a challenge because the genes encoding seven SFR members are closely located within same locus on chromosome 1 (Cannons et al., 2011). With the assistance of the clustered regularly interspaced short palindromic repeat (CRISPR) technique, we generated SFR-deficient mice lacking seven SFR members (Chen et al., 2016). We found that SFR-deficient mice show severe defects in NKT cell development but normal GC reactions and TFH cell differentiation, suggesting that SAP-dependent positive SFR signaling is essential for NKT cell selection. Importantly, we reveal that SFR signaling is overall inhibitory in TFH cells and suppresses humoral immunity in the absence of SAP. Thus, our current study revises our understanding of the mechanisms underlying T cell defects in patients with XLP.
Results
SFR-deficient mice display severe defects in NKT cell development
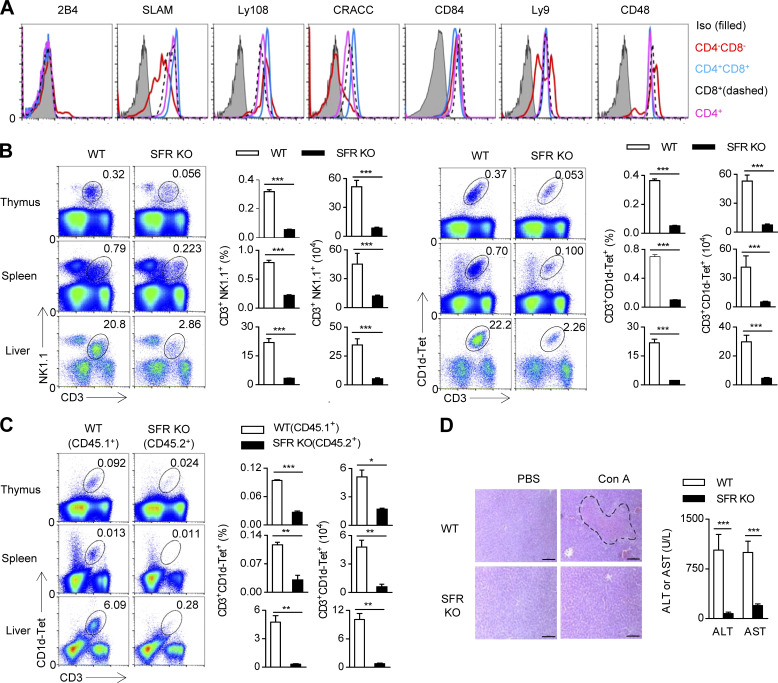
The loss of SAP results in virtually no NKT cells, but deletion of individual SFRs has no apparent effect on NKT cell number. The most plausible explanation for this discrepancy is that SFR redundancy may exist during the thymic selection of NKT cells. First, complete detection of SFRs on mouse thymocytes was performed to comprehensively understand the expression profile of SFRs. Of note, 2B4 is rarely detectable, and high levels of SLAM, Ly108, and CRACC were present in DP thymocytes; CD84 showed increased expression from DP thymocytes, whereas Ly9 and CD48 showed modest changes between DP thymocytes and mature single-positive thymocytes (Fig. 1 A).
Figure 1.
SFR deficiency severely affects NKT cell development. (A) Flow cytometric analysis of SFR expression on CD4−CD8−, CD4+CD8+, CD4+, and CD8+ cell populations in the thymus of WT mice. The corresponding SFR-deficient cells were used as a negative control. The data are representative of three experiments. Iso, isotype. (B) Representative flow cytometry plots and percentages and absolute numbers of the CD3+NK1.1+ NKT cell population (left) or the CD3+CD1d-Tet+ NKT cell population (right) in the thymus, spleen, and liver of the indicated mice. Data represent the mean ± SEM of 10–12 mice per group. ***, P < 0.001. (C) BM chimera assay. Representative flow cytometry plots (left) and percentages and absolute numbers (right) of the CD3+CD1d-Tet+ NKT cell population gated on either the WT (CD45.1+) or the SFR-deficient (CD45.2+) compartment. Data represent the mean ± SEM of three to four mice per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) The indicated mice were treated with Con A for 12 h, and PBS was used as a negative control. H&E staining of liver sections (left) and determination of serum enzyme AST and ALT units (right). The dashed line indicates the necrotic area. Bars, 100 µm. Data represent the mean ± SEM of four to seven mice per group. ***, P < 0.001.
To eliminate the potential redundancy of SFRs, mice lacking all seven SFR members were generated as recently reported (Chen et al., 2016). SFR-deficient mice exhibit normal-sized thymus and peripheral lymphoid organs such as the spleen and LNs (unpublished data), suggesting that SFRs are not required for the development of lymphoid organs. NKT cell numbers were monitored using two different markers, CD3+CD1d-Tet+ and CD3+NK1.1+. Remarkably, the number of CD3+CD1d-Tet+ or CD3+NK1.1+ NKT cells in the thymus was reduced by nearly 90%, whereas the number of NKT cells in peripheral organs such as the spleen and liver was also minimal (Fig. 1 B).
To test whether SFRs are autonomously required for NKT cell development, a mixed chimera assay, in which BM cells isolated from WT and SFR-deficient mice were cotransferred, was performed. As expected, the proportion of NKT cells in SFR-deficient (CD45.2+) compartments was significantly less than that in WT (CD45.1+) compartments (Fig. 1 C). Thus, SFRs intrinsically regulate NKT cell development.
NKT cells are abundant throughout the lives of mice and they are strictly required for Con A–induced fulminate hepatitis. Mice deficient in NKT cells are protected from Con A–induced liver injury (Takeda et al., 2000). To further validate the lack of NKT cells, SFR-deficient mice were administered Con A. As expected, SFR-deficient mice were highly tolerant to Con A challenge and showed less liver injury, as indicated by histological analysis of hepatocyte necrosis and the biochemical determination of two enzymes, ALT and AST (Fig. 1 D); these findings were highly consistent with previous findings in SAP-deficient mice (Furukawa et al., 2009). Thus, SFR-deficient mice indeed have a severe functional defect in NKT cell–mediated autoimmunity in vivo.
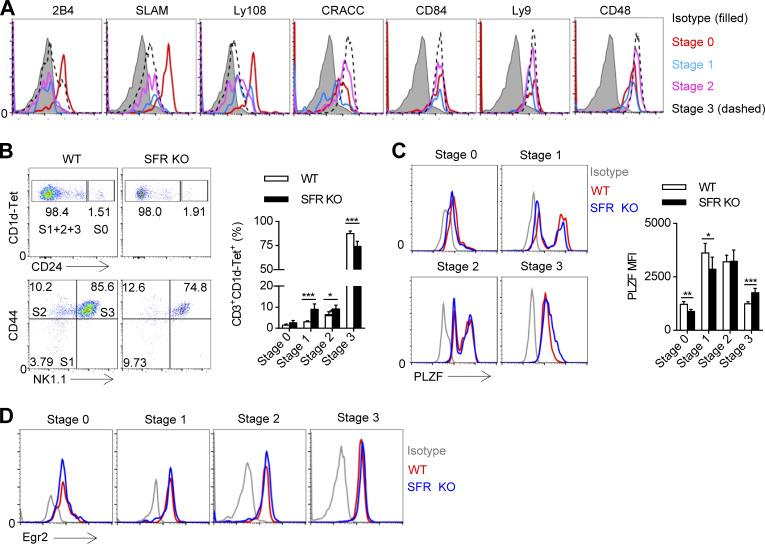
SFR deficiency blocks NKT cell development and promyelocytic leukemia zinc finger (PLZF) induction at an early stage
Based on the surface expression level of CD24, CD44, and NK1.1, developing NKT thymocytes are divided into four stages. 2B4 was only expressed at stage 0 and stage 3 (Fig. 2 A). SLAM and Ly108 was gradually down-regulated starting at stage 0, and this down-regulation continued with NKT cell maturation (Fig. 2 A). In addition, the expression of CRACC and CD48 was gradually up-regulated starting at stage 1, and the expression was maintained at a constant level (Fig. 2 A). Moreover, the expression of CD84 and Ly9 remained nearly unchanged (Fig. 2 A). Thus, multiple SFR members are differentially distributed on developing NKT cells, suggesting the complexity of these families in NKT cell differentiation.
Figure 2.
SFR deficiency blocks NKT cell development and PLZF induction at an early stage. (A) Flow cytometric analysis of SFR expression on developing CD3+CD1d-Tet+ NKT cells in the WT thymus, including CD24hi (stage 0), CD24lowCD44lowNK1.1− (stage 1), CD24lowCD44hiNK1.1− (stage 2), and CD24lowCD44hiNK1.1+ (stage 3) cells. The corresponding SFR-deficient cells were used as a negative control. The data are representative of three experiments. (B) Representative flow cytometry plots (left) and percentages (right) of cells in the four stages of development among gated CD3+CD1d-Tet+ NKT cells in the thymus of the indicated mice, S0: stage 0; S1+2+3: stage 1+2+3. Data represent the mean ± SEM of 4–10 mice per group. *, P < 0.05; ***, P < 0.001. (C) Flow cytometric analysis (left) and mean fluorescence intensity (MFI; right) of PLZF expression on the CD3+CD1d-Tet+ NKT cells at the four development stages in the thymus of the indicated mice. Data represent the mean ± SEM of four to seven mice per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) Flow cytometric analysis of Egr2 expression on the CD3+CD1d-Tet+ NKT cells at the four development stages in the thymus of the indicated mice. The data are representative of three experiments.
To understand which stage requires SFRs for NKT cell differentiation, NKT cells at four developmental stages were compared in SFR-deficient mice. Notably, the proportion of SFR-deficient immature NKT cells, including stage 1 and stage 2 cells, was higher (Fig. 2 B). Therefore, SFR-SAP–mediated signaling is essential for the selection of early developing NKT cells in the thymus.
The transcription factor PLZF is a master regulator of NKT cell development (Savage et al., 2008). To determine whether SFR signaling is necessary for induction of PLZF, its expression in the residual SFR-deficient NKT cells was monitored by intracellular staining. We revealed that the amount of PLZF was markedly reduced around stages 0 and 1, whereas it was unexpectedly up-regulated in the SFR-deficient NKT cells at stage 3 (Fig. 2 C). The altered expression of PLZF in SFR-deficient NKT cells was not due to impaired expression of Egr2 (early growth response gene 2; Fig. 2 D), which is involved in TCR signaling and is essential for NKT cell development (Seiler et al., 2012). Thus, SFR signaling promotes early NKT cell selection, probably by regulating expression of PLZF, but not Egr2.
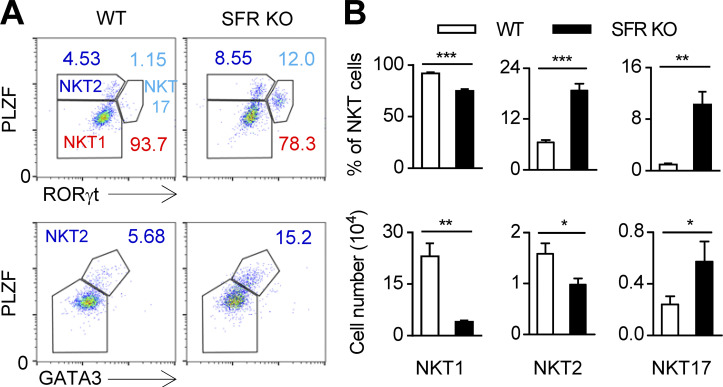
SFR deficiency alters the composition of NKT cell subsets
Three subsets of NKT cells, NKT1, NKT2, and NKT17, have been identified. The fates of these sublineages are determined by the expression of the transcription factors T-bet, GATA3, and retinoid-related orphan receptor γ (RORγt), respectively. To understand whether SFR-SAP signaling controls NKT sublineage specification and differentiation, three NKT subsets among the residual NKT cells in SFR-deficient mice were distinguished by staining with a combination of antibodies against PLZF, RORγt, and GATA3 (O’Hagan et al., 2015). Notably, SFR deficiency profoundly decreased the proportion of NKT1 cells. However, compared with WT populations, the proportions of NKT2 and NKT17 subsets were markedly increased within SFR-deficient NKT cells (Fig. 3, A and B). Accordingly, the total number of thymic NKT1 and NKT2 cells was reduced in SFR-deficient mice. However, the number of thymic NKT17 cells was slightly higher in SFR-deficient mice than in WT mice (Fig. 3 B). Therefore, SFRs are differentially required for the determination of NKT subset fate.
Figure 3.
SFR deficiency alters the composition of NKT cell subsets. (A and B) Representative flow cytometry plots (A) and percentages and absolute numbers (B) of NKT1 (PLZFlowRORγt−), NKT2 (PLZFhiRORγt−), and NKT17 (PLZFintRORγt+) or NKT2 (PLZFhiGATA3hi) of the gated CD3+CD1d-Tet+ NKT cell population in the thymus of the indicated mice. Data represent the mean ± SEM of five mice per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
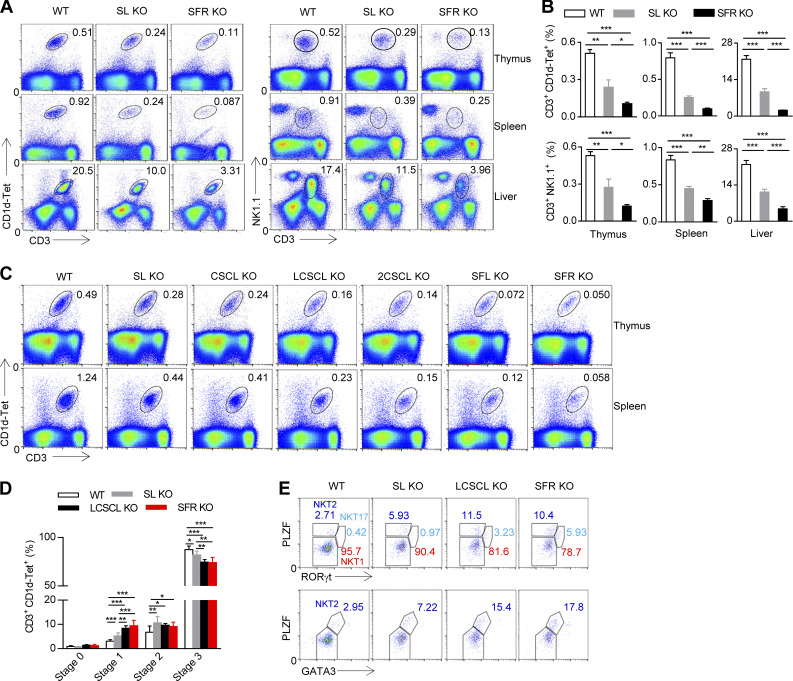
Multiple SFR members, mainly SLAM and Ly108, are involved in NKT cell development
Because a previous study has revealed that SLAM and Ly108 may synergize with each other to regulate NKT cell development in pseudo–double KO mice (Griewank et al., 2007), we first determined whether mice doubly deficient in SLAM and Ly108 had altered NKT cell numbers. We noticed that the mice lacking SLAM and Ly108 only had half the number of thymic NKT cells observed in WT mice (Fig. 4, A and B). Further experiments showed that these mice also had a moderate reduction in the number of NKT cells in peripheral organs, including liver and spleen (Fig. 4, A and B). The residual NKT cells in SLAM and Ly108 double-deficient (SL KO) mice consisted of more immature NKT cells, mainly stage 1 and 2 NKT cells (Fig. 4 D). Although the number of NKT cells was reduced in SL KO mice, the severity of this defect is less pronounced than that in SFR-deficient mice (Fig. 4, A and B), suggesting that other SFRs may be involved in NKT cell development.
Figure 4.
Multiple SFR members participate in the process of NKT cell selection. (A and B) Representative flow cytometry plots (A) and percentages (B) of the CD3+CD1d-Tet+ NKT cell population or the CD3+NK1.1+ NKT cell population in the thymus, spleen, and liver from the indicated mice. The data represent the mean ± SEM of 5–10 mice per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Representative flow cytometry plots of the CD3+CD1d-Tet+ NKT cell population in the thymus of the indicated mice. 2CSCL KO, mice lacking 2B4, CRACC, SLAM, CD84, and Ly108; SL KO, mice lacking SLAM and Ly108; CSCL KO, mice lacking CRACC, SLAM, CD84, and Ly108; LCSCL KO, mice lacking Ly9, CRACC, SLAM, CD84, and Ly108; SFL KO, mice lacking Ly9, CRACC, CD48, SLAM, CD84, and Ly108; SL KO, mice lacking SLAM and Ly108. The data are representative of three experiments. (D) Percentages of NKT cells at four stages of development among the gated CD3+CD1d-Tet+ NKT cells in the thymus of the indicated mice. Data represent the mean ± SEM of three to seven mice per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (E) Representative flow cytometry plots of NKT1, NKT2, and NKT17 cells of the gated CD3+CD1d-Tet+ NKT cell population in the thymus of the indicated mice. The data are representative of three experiments.
Accordingly, we studied several mice that were also missing other SFR members. The additional deletion of CD84 and CRACC did not affect NKT cell number in thymus, as was seen in CSCL mice, whereas further deletion of Ly9 (LCSCL KO) or 2B4 (2CSCL KO) aggravated the defect in NKT cell development compared with CSCL mice, where completely removing Ly9, 2B4 and CD48 further worsened the defective NKT cell development in SFR KO mice (Fig. 4 C). Thus, Ly9 and 2B4 play additional roles in NKT cell development. There are data suggesting that SFR redundancy exists in NKT cell selection. To extensively investigate this redundancy, the proportions of four-stage developing NKT cells among the residual NKT cells in mice differentially lacking SFR members were examined. The additional deletion of Ly9 molecule (LCSCL KO) further blocked NKT cell differentiation at stages 1 and 2 (Fig. 4 D) and also altered the composition of three NKT cell subsets in the mice lacking SLAM and Ly108 (Fig. 4 E). Collectively, multiple SFR pairs, mainly SLAM, Ly108, Ly9, and 2B4/CD48, participate in the hematopoietic cell–driven positive selection of NKT cells and NKT cell fate decisions.
SFR-mediated inhibitory signaling plays a minor role in NKT cell development
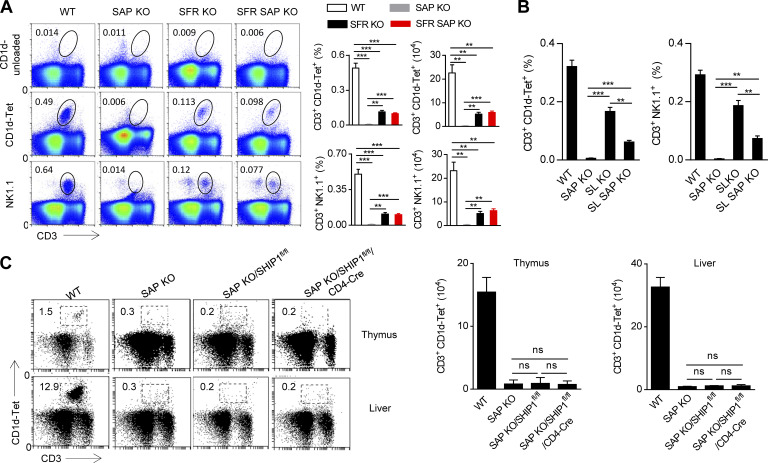
The aforementioned data clearly demonstrated that SFRs are indeed required for NKT cell development. Because SFRs are presumably the only receptors that have the ability to recruit SAP, NKT cell development is orchestrated by SAP-dependent SFR signaling. However, NKT cell development showed a more severe phenotype in SAP-deficient mice than in SFR-deficient mice, suggesting that SAP has additional effects on NKT cell development (Fig. 5 A). We proposed that SFR might also transmit inhibitory signaling to negatively regulate NKT cell development in SAP-deficient mice. To confirm this hypothesis, we sought to test whether the removal of the SFR family could correct the defective NKT cell development in SAP-deficient mice. SAP-deficient mice were then crossed with SFR-deficient mice to yield a mouse lacking SFRs and SAP. Compared with SAP-deficient mice, the NKT cell number in SFR-SAP deficient mice was recovered to ∼10% of WT NKT cells, up to the comparable level with that of SFR-deficient mice (Fig. 5 A). These data suggest that SFRs are general inhibitory receptors for NKT cell selection in SAP-deficient mice; thus, SFR deletion can partially correct the severe defect in NKT cell development in these mice.
Figure 5.
SAP-independent SFR signaling plays a minor role in NKT cell development. (A) Representative flow cytometry plots and percentages and absolute numbers of the CD3+CD1d-Tet+ NKT cell population or the CD3+NK1.1+ NKT cell population in the thymus of the indicated mice. Unloaded CD1d was used as negative control. Data represent the mean ± SEM of three to five mice per group. **, P < 0.01; ***, P < 0.001. (B) Percentages of CD3+CD1d-Tet+ NKT cell population (left) or CD3+NK1.1+ NKT cell population (right) in the thymus of the indicated mice. Data represent the mean ± SEM of three to five mice per group. **, P < 0.01; ***, P < 0.001. (C) Representative flow cytometry plots (left) and absolute numbers (right) of CD3+CD1d-Tet+ NKT cells in the thymus and liver from the indicated mice. Data represent the mean ± SEM of three to four mice per group.
SLAM and Ly108 are two major activating receptors in NKT cell development, and we next examined whether SLAM and Ly108 could be converted to inhibitory receptors in SAP-deficient mice. SLAM–Ly108–SAP triple-deficient mice were bred. Although the additional deletion of these two SFR members largely rescued NKT cell numbers in SAP-deficient mice (Fig. 5 B), the number of NKT cells was much lower in triple-deficient mice than in SLAM-Ly108-deficient mice (Fig. 5 B), suggesting that in addition to Ly108 and SLAM, other SFRs may be inhibitory when SAP is absent.
In summary, SAP-dependent SFR signaling is required for NKT cell development. The defects in NKT cell development caused by SAP deficiency are primarily due to the loss of SFR-mediated positive signaling; however, the gain of SFR-mediated inhibitory signaling very likely aggravates the defect.
The lipid phosphatase SHIP-1 is not responsible for SFR-mediated inhibition of NKT development in SAP-deficient mice
We previously provided biochemical and genetic evidence that the lipid phosphatase SHIP-1 plays a dominant role in the SFR-mediated inhibition of SAP-deficient NK cell function, whereas two other protein phosphatases, SHP1 and SHP2, had a minimal role (Dong et al., 2012). To further examine whether the removal of SHIP1 could rescue impaired NKT cell development in SAP-deficient mice, SAP KO/SHIP1fl/fl/CD4-Cre mice, in which SAP was totally deleted and SHIP1 was only deleted in CD4+ cells, were generated. SAP KO/SHIP1fl/fl/CD4-Cre mice still had a dramatic defect in NKT cell number comparable with that in SAP-deficient mice (Fig. 5 C). These data demonstrate that it is in SAP-deficient mice that SFRs can transmit a low level of SHIP1-independent inhibitory signals, suggesting that the protein phosphatases SHP1 and/or SHP2 may be involved in the process.
SFRs regulate NKT cell development in part by activating the CARMA1–Bcl10–Malt1 (CBM) complex
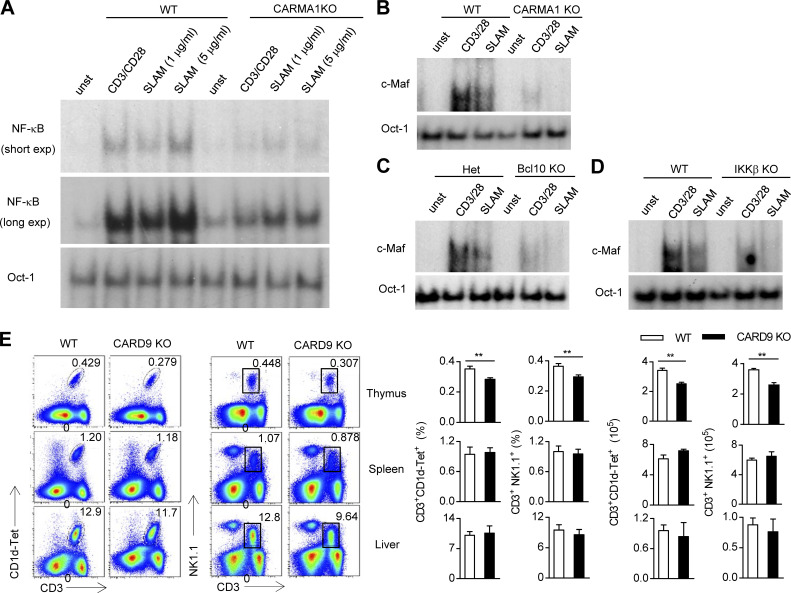
Previous data showed that Fyn kinase is one of most important SH3 domain–containing enzymes that mediate downstream SAP signaling (Chan et al., 2003; Latour et al., 2003). Given that Fyn is required for NKT cell development, we next sought to explore how the SFR/SAP/Fyn cascade triggers downstream signaling, which is necessary for NKT cell development. Previous studies have demonstrated that SLAM regulates Th2 cytokine production by triggering PKC-θ (Cannons et al., 2004) and that the CBM complex is downstream of PKC-θ, which is stimulated by Fyn in NK cells (Rajasekaran et al., 2013). Therefore, we determined whether SLAM engagement could activate the CBM complex in CD4+ T cells as a representative of NKT cells. A series of SLAM antibody concentrations was used to stimulate T cells. Notably, SLAM antibody cross-linking markedly induced the activation of NF-κB and c-avian musculoaponeurotic fibrosarcoma (c-Maf), two major signaling molecules downstream of the CBM complex, in activated WT CD4+ T cells (Fig. 6, A and B). However, the genetic disruption of individual components of the CBM complex, such as CARMA1, could largely abolish the effect, suggesting that the CBM complex is involved in SLAM-mediated activating signaling (Fig. 6, A and B). We also confirmed this result using mice that are deficient in Bcl10, a CBM complex component (Fig. 6 C). Because NF-κB activation is a typical signaling event downstream of the CMB complex, T cells lacking inhibitor of NF-κB kinase (IKK), which can release inhibitor of NF-κB, were resistant to SLAM cross-linking (Fig. 6 D). In light of the fact that Bcl10 has been reported to be essential for NKT cell survival (Schmidt-Supprian et al., 2004), we tried to determine whether the CARD9 (caspase recruitment domain family of adaptor member 9)-containing CBM complex is required for NKT cell development. In contrast to the defect in NKT cells in peripheral organs, CARD9-deficient mice had a lower percentage and absolute number of NKT cells only in the thymus, suggesting that CARD9 is required for NKT cell selection, but not peripheral homeostasis (Fig. 6 E). We also noticed that, compared with SFR-deficient mice, CARD9-deficient mice only showed a moderate reduction in the number of NKT cells in the thymus (Fig. 6 E). Hence, SFRs regulate NKT cell selection partially through the assembly of a CARD9-containing CBM complex.
Figure 6.
SAP-dependent activating SFR signaling regulates NKT cell selection partially by activating the CBM complex. (A) CD4 T cells isolated from WT and CARMA1-deficient (KO) mice were stimulated with anti-CD3 plus anti-CD28 and then restimulated with anti-CD3 plus anti-CD28 or different concentrations of anti-SLAM antibody for 1 h. Nuclear extracts were prepared from these cells and then subjected to electrophoretic mobility shift assays using a 32P-labeled NF-κB probe and Oct-1 as a control probe. Unstimulated (unst) cell extracts were used as a control. (B–D) WT or CARMA1 KO (B), Bcl10 KO (C), or IKKβ KO (D) CD4 T cells activated by IL-6 and anti-CD3 plus anti-CD28 were restimulated with anti-CD3 plus anti-CD28 or an anti-SLAM antibody for 5 h. Nuclear extracts were prepared from these cells and then subjected to electrophoretic mobility shift assays using a 32P-labeled c-Maf probe and an Oct-1 probe as a control. Unstimulated cell extracts were used as a control. (E) Representative flow cytometry plots (left) and percentages and absolute numbers (right) of the CD3+CD1d-Tet+ NKT cell population or the CD3+NK1.1+ NKT cell population in the thymus, spleen, and liver from WT and CARD9-deficient mice. Data represent the mean ± SEM of three to four mice per group. **, P < 0.01.
SFR deficiency does not affect humoral immunity
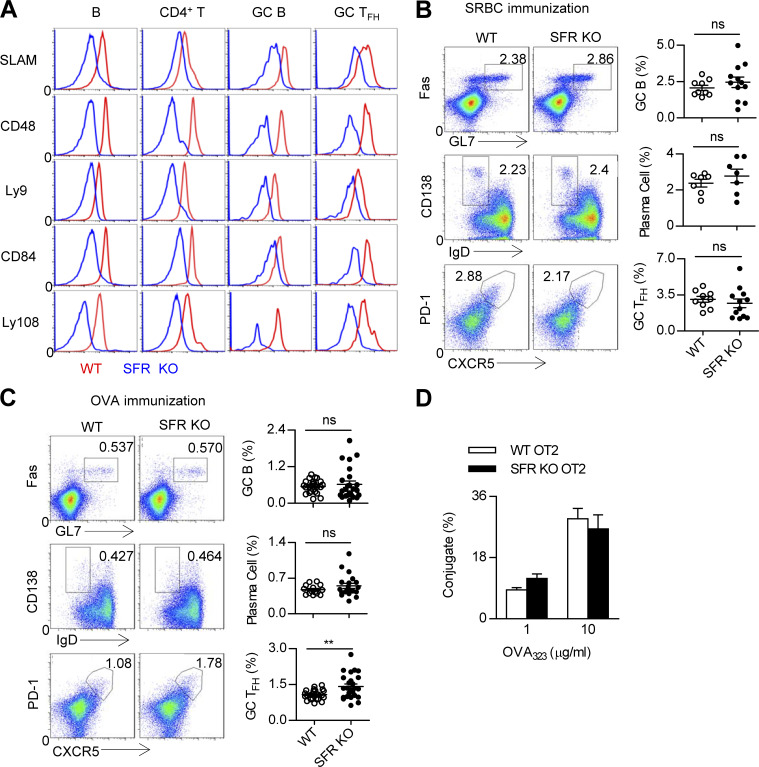
To systematically explore the function of SFRs in T cell contact with B cells, the expression of seven SFRs was detected in splenic CD4+ T cells and B cells. These two types of cells constitutively express five SFR members, namely SLAM, Ly9, CD84, Ly108, and CD48 (Fig. 7 A). Using SFR-deficient cells as a negative control, we detected no expression of 2B4 on either type of cell, whereas a low level of CRACC appeared on splenic B cells but not CD4+ T cells (unpublished data). The expression profiles of SFRs in TFH cells and B cells were further determined after immunization. The results revealed that compared with naive B cells, GC B cells presented an increased amount of SLAM, CD48, Ly9, and Ly108 (Fig. 7 A). Compared with nonimmunized CD4+ T cells or non–TFH cells, the expression of SLAM, CD84, and Ly108 was also up-regulated in GC TFH cells (Fig. 7 A).
Figure 7.
SFR deficiency does not affect humoral immunity. (A) Flow cytometry analysis of SFR expression in resting splenic B cells (CD19+) and CD4 T cells (CD4+CD19−) from nonimmunized WT mice as well as GC B cells (CD19+GL7hiFashi) and GC TFH cells (CD4+CD19−CXCR5hiPD1hi) from SRBC-immunized WT mice. Respective SFR-deficient cells were used as negative controls. The data are representative of three experiments. (B and C) Representative flow cytometry plots (left) and percentages (right) of splenic GC B cells (CD19+FashiGL7hi), plasma cells (CD19+IgDloCD138hi), and GC TFH cells (CD4+CD19−PD1hiCXCR5hi) from WT and SFR KO mice 7 d after SRBC immunization (B) or 9 d after OVA immunization (C). Data represent the mean ± SEM of 9–25 mice per group. **, P < 0.01. (D) Percentage of T–B conjugation (number of CD4+CD19+ cells out of total number of CD4+ cells). OT-II CD4+ T cells from WT and SFR-deficient mice and B cells were pulsed with OVA323–339 peptide at the indicated concentrations. The data are representative of three experiments and show the mean ± SEM of three mice per group.
Next, GC responses were evaluated in SFR-deficient mice immunized with sheep RBCs (SRBCs), which are able to trigger T-dependent and T-independent GC reactions. Unexpectedly, SFR-deficient mice showed normal percentages of GC B cells, plasma cells, and TFH cells, highly comparable with those seen in WT mice (Fig. 7 B). Then, SFR-deficient mice were immunized with OVA, which is a T-dependent antigen. We also failed to find a dramatic effect of SFR deficiency on the generation of GC B cells and plasma cells. However, a slight increase in the number of GC TFH cells was detected in SFR-deficient mice after OVA immunization (Fig. 7 C).
SAP deficiency causes severe defects in humoral immunity largely due to impaired conjugate formation between SAP-deficient T cells and B cells (Qi et al., 2008; Cannons et al., 2010b). To evaluate whether SFRs coordinate the formation of T cell and B cell conjugates, we exploited the flow cytometry–based T cell–B cell conjugation assay in vitro and found that the ability of SFR-deficient OT-II T cells to maintain contact with B cells was nearly comparable to that of WT OT-II T cells (Fig. 7 D). Thus, SFR deficiency in CD4+ T cells does not affect T cell–B cell conjugate formation. Collectively, in the presence of SAP, SFRs are almost dispensable for humoral immunity.
SFRs suppress humoral immunity in SAP-deficient mice
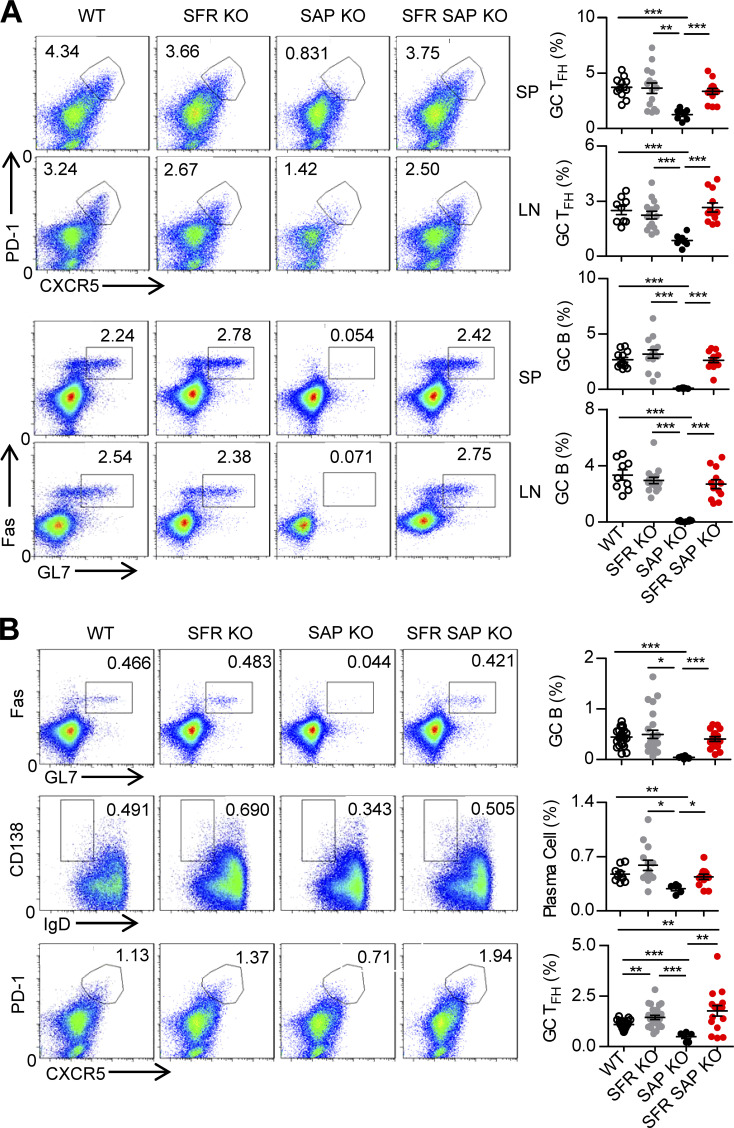
The dispensable role of SFRs in SAP-intact T cell–mediated humoral immunity encouraged us to examine whether the SFRs in SAP-deficient T cells suppress the differentiation of TFH cells and thus provide negative help to GC B cells. To strategically evaluate this, mice lacking both SFRs and SAP were generated. Surprisingly, the additional removal of SFRs from SAP-deficient mice completely rescued the number of GC TFH cells. As a consequence, the defective generation of GC B cells was completely corrected to the same level as WT mice after SRBC immunization (Fig. 8 A). Moreover, similar results were obtained when OVA immunization was used; moreover, the decrease in the number of plasma cells was also reversed (Fig. 8 B). Therefore, these data suggest that in the absence of SAP, SFRs primarily provide a negative signal to dampen TFH cell differentiation, which then impairs subsequent humoral immunity.
Figure 8.
SAP-independent inhibitory SFR signaling leads to severe defects in humoral immunity in SAP-deficient mice. (A) Representative flow cytometry plots and percentages of splenic and LN GC TFH cells (CD4+CD19−PD1hiCXCR5hi) or GC B cells (CD19+FashiGL7hi) from the indicated mice 7 d after SRBC immunization. Data represent the mean ± SEM of 8–15 mice per group. **, P < 0.01; ***, P < 0.001. (B) Representative flow cytometry plots (left) and percentages (right) of splenic GC B cells (CD19+FashiGL7hi), plasma cells (CD19+IgDloCD138hi), and GC TFH cells (CD4+CD19−PD1hiCXCR5hi) from the indicated mice 9 d after OVA immunization. Data represent the mean ± SEM of 5–25 mice per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Discussion
SAP gene mutations cause severe immunodeficiency in humans, including a lack of NKT cells, impairment of TFH cell differentiation, and impaired function of cytotoxic cells such as NK cells and CD8+ T cells (Dong and Veillette, 2010). SAP is considered to be one of the major signaling proteins downstream of the SLAM family, and numerous studies have revealed that the actions of SAP are very likely performed by two types of activities, i.e., active signaling molecule and natural blocker (Dong and Veillette, 2010). However, how SAP deficiency causes those immune deficiencies is still not completely understood, probably because the SFR family is composed of seven members and the potential redundancy prevents immunologists from comprehensively understanding how SFRs transmit SAP-dependent and independent signaling. In the current study, we used novel mice that genetically lack all of the SLAM family members. On the one hand, this model can avoid the redundancy of SFRs in T cells; on the other hand, the genetic removal of all the SFRs is able to synchronously abolish SAP-dependent and SAP-independent signaling. Thus, we found that SFR deficiency causes severe defects in NKT cell development, but not TFH cell differentiation. However, the simultaneous deletion of SFRs can largely correct the TFH cell defects observed in SAP-deficient mice. Therefore, our data provide definitive evidence that can be used to straightforwardly dissect SAP-dependent and SAP-independent SFR signaling in the T cell defects caused by SAP deficiency, and these data also clearly identify new molecular mechanisms underlying T cell defects in patients with XLP.
Our recent study on NK cells also revealed that SFRs are totally inhibitory in SAP-deficient NK cells; therefore, the removal of SFRs from SAP-deficient NK cells completely abolishes the impaired NK cell cytotoxicity (Chen et al., 2016). However, in this study, we revealed the differential effects of SFR deficiency on SAP-deficient T cells. In NKT cells, the removal of SFRs only increases the number of NKT cells by 10%, whereas in TFH cells, it almost completely rescues the severe defect in GC reactions. Therefore, although the presence of SAP family adaptors determines the activity of SFRs in immune cells, variation indeed exists in different immune cells. We conclude that SAP-dependent SFR signaling is critical for NKT cell development and NK cell education (Chen et al., 2016), but it is dispensable for TFH cell differentiation. However, in the context of SAP deficiency, SAP-independent SFR signaling plays a minor role in inhibiting NKT cell development, whereas this signaling can absolutely paralyze NK cell function and TFH cell–mediated humoral immunity. Thus, further studies are needed to address why SAP-dependent and SAP-independent signaling exert differential effects in certain immune cells. Our interpretation is that during NKT cell development, SFRs probably function as essential costimulatory receptors, providing positive signaling in synergy with the invariant TCRs that usually have low to intermediate affinity for lipid antigens. Hence, when the engagement of SFRs is absent, invariant TCRs fail to support NKT cell selection because classical costimulatory receptors are required for NKT cell differentiation, expansion, and homeostasis and are rarely involved in NKT cell lineage commitment and selection (Akbari et al., 2008; Williams et al., 2008). However, several costimulatory molecules, such as ICOS, CD40L, and OX40, were up-regulated in TFH cells after immunization and are essential for TFH cell generation. These receptors probably can compensate for the loss of SFRs.
Through a comprehensive analysis of a collection of mice differentially lacking SFR members, the issue of SFR redundancy in NKT cell development is well resolved in the current study. Several previous studies have revealed that two SFR members, SLAM and Ly108, are critical to NKT cell selection (Griewank et al., 2007). Our study provides further genetic evidence suggesting that two additional SFR members, Ly9 and 2B4, might participate in NKT cell development. Meanwhile, the involvement of CD84 and CRACC in NKT cell development is excluded. The open question is why NKT cells preferentially require SLAM and Ly108, but not CD84 and CRACC, for their selection in thymus. On one hand, we think that these receptors may have a unique thymic SFR expression pattern, that is, SLAM and Ly108 display the highest expression on DP cortical thymocytes with decreased expression on mature single-positive thymocytes (Griewank et al., 2007). On the other hand, SFR members may have the differential ability to activate SAP-dependent activating signaling essential for NKT cell development. For instance, CD84 is generally not activating receptors, at least on NK cells (Dong et al., 2009); studies from other groups suggest that engagement of CRACC can only recruit EAT-2, not SAP, to deliver its downstream signaling (Tassi and Colonna, 2005; Cruz-Munoz et al., 2009). These findings probably help to explain why CRACC and CD84 are dispensable for NKT cell selection although they are constitutively expressed on developing NKT cells.
Our previous study demonstrated dual activities of SAP in the NK cell–mediated killing of hematopoietic cells (Dong et al., 2009). We found that the SAPR78A mutation can lead to a moderate defect in NK cell function compared with SAP-deficient mice, in which SFRs gain inhibitory functions (Dong et al., 2012). SH3-containing Fyn kinase was reported to be downstream of SAP signaling in the regulation of NK cell cytotoxicity and NKT cell development (Eberl et al., 1999; Bloch-Queyrat et al., 2005), as Fyn deficiency also can recapitulate the phenotype observed in SAP-deficient mice. It is largely unknown how the binding of SAP to Fyn kinase transduces active signaling to distal effector molecules. In the current study, we propose a mechanism by which SLAM-SAP signaling is able to activate the CBM complex, which leads to NF-κB activation. The ablation of IKK1, which is downstream of the CBM complex and required for NF-κB activation, prevents the generation of NKT cells (Schmidt-Supprian et al., 2004). Similarly, disruptions in the classical Rel/NF-κB family members, such as NF-κB1/RelA, can intrinsically reduce the number of NKT cells to various degrees (Sivakumar et al., 2003; Stanic et al., 2004). Although NKT cell development needs NF-κB–inducing kinase–dependent RelB activation, this effect is extrinsic and dependent on nonhematopoietic stromal cells (Elewaut et al., 2003). Thus, the generation of NKT cells probably requires classical NF-κB1/RelA signaling. Our study suggests that SLAM-SAP signaling is most likely the upstream receptor that triggers the activation of classical NF-κB via the CBM complex.
Here, we first demonstrated that activation of the CBM complex is critical for NKT cell selection in the thymus. The components of the CBM complex are usually variable. In contrast to the Bcl10-containing CBM complex, which has been revealed to regulate the peripheral homeostasis of NKT cells, we reported here a reduced number of NKT cells in the thymus of CARD9-deficient mice. Therefore, SLAM-SAP signaling might activate NF-κB activation via the CARD9-containing CBM complex, which promotes NKT cell selection but not peripheral homeostasis. Moreover, it was notable that NKT cell numbers are mildly decreased in CARD9-deficient mice, suggesting that a CARD9-independent pathway does exist. Thus, further study is needed to address how distal signaling is elicited by positive signaling involving SLAM–SAP–Fyn.
Here, we clearly show that the defect in humoral immunity in SAP-deficient mice is caused by SFR (in particular Ly108)-mediated inhibitory signaling (Kageyama et al., 2012), but the underlying mechanism through which SAP-independent signaling functions remains elusive. In the context of SAP deficiency, SFRs principally prefer to recruit other SH2 domain–containing molecules. However, SFRs seem to attract different phosphatases to mediate inhibition in different types of immune cells, such as TFH cells, CD8 cells, and NK cells. For example, the SH2 domain–containing inhibitory molecule SHIP1 plays a dominant role in the SFR-mediated inhibition of SAP-deficient NK cell function, whereas SHP1 and SHP2 only have marginal roles in this inhibition (Dong et al., 2012). Although SHIP1 deficiency also hinders invariant NKT cell development (Anderson et al., 2015), removing SHIP1 from SAP-deficient NKT cells cannot rescue the number of NKT cells. In TFH cells, the antibody-mediated cross-linking of SFRs, such as Ly108, induces the phosphorylation of tyrosine residues on SHP1 efficiently; moreover, the biochemical inhibition of SHP1 can alleviate SFR-mediated inhibition in SAP-deficient T cells, suggesting a role for SHP-1 in the SFR-mediated inhibition of TFH cells. However, the chemical inhibitors for these molecules are usually problematic in their specificity, so genetic models lacking these phosphatases are required to precisely dissect the mechanism of SFR-mediated inhibition in various types of SAP-deficient immune cells. Further studies will be helpful in designing strategies to correct immunodeficiency in patients with XLP.
Overall, we have demonstrated that SAP-dependent SFR-mediated positive signaling is essential for NKT cell selection. However, in SAP-deficient TFH cells, SFR-mediated inhibitory signaling can greatly dampen TFH cell differentiation, which is harmful to humoral immunity. These novel findings will be beneficial in elucidating the molecular mechanisms underlying XLP pathogenesis and will also be helpful in designing new strategies to treat immune dysfunction in patients with XLP.
Materials and methods
Mice
OT2 mice and Rag1-γc mice were obtained from The Jackson Laboratory. SAP-deficient mice have been described previously (Dong et al., 2012). Ly108-SAP–deficient, SLAM-Ly108-SAP–deficient, SFR-deficient, and all the other SFR-deficient mice were generated in our laboratory. In all experiments, the littermates were used as controls. All of the mice were bred and maintained in specific pathogen–free animal facilities at Tsinghua University. All of the procedures involving animals were approved by the Animal Ethics Committee of Tsinghua University.
Antibodies
Antibodies against CD3e (145-2C11), CD4 (GK1.5), NK1.1 (PK136), CD19 (eBio1D3), CD24 (30-F1), CD44 (IM7), CD45.1 (A20), CD45.2 (104), CD48 (HM48-1), CXCR5 (SPRCL5), GL7 (GL-7), GATA3 (TWAJ), IgD (11-26C), ICOS (7E.17G9), Ly9 (Ly9ab3), PD1 (J43), PLZF (Mags.21F7), RORγt (AFKJS-9), 2B4 (eBio244F4), 7-AAD, and streptavidin were purchased from eBioscience. Antibodies against CD138 (281–2) and Fas (Jo2) were purchased from BD. Antibodies against SLAM (12F12), CD84 (CD84.7), and Ly-108 (330-AJ) were purchased from BioLegend, whereas CRACC antibody (clone 520914) was provided by R&D Systems. Antibodies against Egr2 were purchased from Abcam, and goat anti–rabbit IgG (H+L) was purchased from Thermo Fisher Scientific. CD1d-tetramer (19156, loaded with PBS57) and empty CD1d control were kindly provided by the National Institutes of Health Tetramer Core Facility.
Cell isolation and cell culture
CD4+ T cells and CD19+ B cells were isolated by positive selection (STEMCELL Technologies) from the spleen. For in vitro T cell activation, CD4+ T cells were stimulated with 3 µg/ml plate-bound anti-CD3 and 1 µg/ml anti-CD28 and were maintained in the presence of 100 U/ml human IL-2. CD19+ B cells were activated in the presence of 1 µg/ml LPS.
Con A–induced acute hepatitis and histological analysis
Mice were intravenously injected with PBS-dissolved Con A (Sigma-Aldrich) at a dose of 10 mg/kg body mass. After 12 h, mice were euthanized for collecting serum and liver. Serum AST and ALT levels were measured by an automatic analyzer. Liver sections were fixated in 4% paraformaldehyde for hematoxylin and eosin (H&E) staining.
BM chimeras
To generate competitive BM chimeras, the equal numbers of BM cells from 4- to 5-wk-old WT B6 (CD45.1+) and SFR-deficient (CD45.2+) mice, which were pretreated with 5-fluorouracil (5-FU) for 5 d, were adoptively transferred into lethally irradiated Rag1-γc recipients (CD45.2+). 6 wk after reconstitution, NKT cell development was analyzed by flow cytometry.
Immunization
Mice were intraperitoneally injected with a mixture of a 100-µl suspension containing 100 µg OVA, 1 µg LPS, and 50% alum. The spleens were removed 9 d after OVA immunization, and 2.5 × 108 fresh SRBCs were diluted in PBS for intraperitoneal and subcutaneous injection. The spleens and LNs were removed 7 d after immunization for FACS analysis.
Conjugate assay
For the T–B conjugate assay, preactivated OT-II CD4+ cells (2.5×105 in 100 µl) were labeled with FITC-conjugated anti-CD4 antibody, and LPS-activated WT B cells were labeled with APC-conjugated anti-CD19 and then pulsed with a series of antigen OVA323–339 concentrations (0, 1, or 10 µg/ml) for 30 min. After washing, 100 µl OT-II CD4+ cells was mixed with an equal volume of WT B cells, gently centrifuged for 5 min at 300 g, and then incubated at 37°C for 30 min. After vortex for 40 s, the frequency of T–B conjugates was enumerated by flow cytometry.
Electrophoretic mobility shift assay
To detect NF-κB and c-Maf activation, T cells isolated from the indicated mice were stimulated with anti-CD3 plus anti-CD28 to increase SLAM expression and then restimulated with anti-CD3 plus anti-CD28 and/or different concentrations of anti-SLAM antibody (1 µg/ml and 5 µg/ml). Nuclear extracts (2–4 µg) were incubated with 1 × 105 cpm of 32P-labeled NF-κB or c-Maf probe probes, Oct-1 probe as a control probe, at room temperature for 15 min. The samples were separated on a native Tris-borate EDTA polyacrylamide gel, which was dried at 80°C for 1 to 2 h, and then exposed to x-ray film.
Statistical analyses
Prism 5 software (GraphPad Software) was used for unpaired Student’s t tests (two tailed).
Acknowledgments
We thank L. Yin for sharing the SAP-deficient mice and Xiaoqian Zhang for help during revision.
Work in the Dong laboratory is supported by the Ministry of Science and Technology of China (grant 2013CB944901) and the Natural Science Foundation of China (grants 81322041, 81273198, 81361128016, and 81471523).
The authors declare no competing financial interests.
Author contributions: S. Chen, Z. Li, and C. Cai conceived the project and designed and performed most experiments. G. Liu, M. Blonska, Y. Wang, D. Li, J. Du, and M. Yang performed experiments and analysis. X. Lin generated critical reagents. Z. Dong led the investigation and wrote the paper.
Footnotes
Abbreviations used:
- CBM
- CARMA1–Bcl10–Malt1
- c-Maf
- c-avian musculoaponeurotic fibrosarcoma
- DP
- double positive
- GC
- germinal center
- IKK
- inhibitor of NF-κB kinase
- PLZF
- promyelocytic leukemia zinc finger
- RORγt
- retinoid-related orphan receptor γ
- SAP
- SLAM-associated protein
- SFR
- SLAM family receptor
- SLAM
- signaling lymphocytic activation molecule
- SRBC
- sheep RBC
- XLP
- X-linked lymphoproliferative disease
References
- Akbari, O., Stock P., Meyer E.H., Freeman G.J., Sharpe A.H., Umetsu D.T., and DeKruyff R.H.. 2008. ICOS/ICOSL interaction is required for CD4+ invariant NKT cell function and homeostatic survival. J. Immunol. 180:5448–5456. 10.4049/jimmunol.180.8.5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, C.K., Salter A.I., Toussaint L.E., Reilly E.C., Fugère C., Srivastava N., Kerr W.G., and Brossay L.. 2015. Role of SHIP1 in invariant NKT cell development and functions. J. Immunol. 195:2149–2156. 10.4049/jimmunol.1500567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac, A. 1995. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med. 182:2091–2096. 10.1084/jem.182.6.2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch-Queyrat, C., Fondanèche M.C., Chen R., Yin L., Relouzat F., Veillette A., Fischer A., and Latour S.. 2005. Regulation of natural cytotoxicity by the adaptor SAP and the Src-related kinase Fyn. J. Exp. Med. 202:181–192. 10.1084/jem.20050449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg, N.A., Wun K.S., Kjer-Nielsen L., Wilce M.C., Pellicci D.G., Koh R., Besra G.S., Bharadwaj M., Godfrey D.I., McCluskey J., and Rossjohn J.. 2007. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 448:44–49. 10.1038/nature05907 [DOI] [PubMed] [Google Scholar]
- Cannons, J.L., Yu L.J., Hill B., Mijares L.A., Dombroski D., Nichols K.E., Antonellis A., Koretzky G.A., Gardner K., and Schwartzberg P.L.. 2004. SAP regulates TH2 differentiation and PKC-θ-mediated activation of NF-κB1. Immunity. 21:693–706. 10.1016/j.immuni.2004.09.012 [DOI] [PubMed] [Google Scholar]
- Cannons, J.L., Yu L.J., Jankovic D., Crotty S., Horai R., Kirby M., Anderson S., Cheever A.W., Sher A., and Schwartzberg P.L.. 2006. SAP regulates T cell-mediated help for humoral immunity by a mechanism distinct from cytokine regulation. J. Exp. Med. 203:1551–1565. 10.1084/jem.20052097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannons, J.L., Qi H., Lu K.T., Dutta M., Gomez-Rodriguez J., Cheng J., Wakeland E.K., Germain R.N., and Schwartzberg P.L.. 2010a. Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity. 32:253–265. 10.1016/j.immuni.2010.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannons, J.L., Wu J.Z., Gomez-Rodriguez J., Zhang J., Dong B., Liu Y., Shaw S., Siminovitch K.A., and Schwartzberg P.L.. 2010b. Biochemical and genetic evidence for a SAP-PKC-θ interaction contributing to IL-4 regulation. J. Immunol. 185:2819–2827. 10.4049/jimmunol.0902182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannons, J.L., Tangye S.G., and Schwartzberg P.L.. 2011. SLAM family receptors and SAP adaptors in immunity. Annu. Rev. Immunol. 29:665–705. 10.1146/annurev-immunol-030409-101302 [DOI] [PubMed] [Google Scholar]
- Chan, B., Lanyi A., Song H.K., Griesbach J., Simarro-Grande M., Poy F., Howie D., Sumegi J., Terhorst C., and Eck M.J.. 2003. SAP couples Fyn to SLAM immune receptors. Nat. Cell Biol. 5:155–160. 10.1038/ncb920 [DOI] [PubMed] [Google Scholar]
- Chen, S., Yang M., Du J., Li D., Li Z., Cai C., Ma Y., Zhang L., Tian Z., and Dong Z.. 2016. The self-specific activation receptor SLAM family is critical for NK cell education. Immunity. 45:292–304. 10.1016/j.immuni.2016.07.013 [DOI] [PubMed] [Google Scholar]
- Crotty, S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Cruz-Munoz, M.E., Dong Z., Shi X., Zhang S., and Veillette A.. 2009. Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function. Nat. Immunol. 10:297–305. 10.1038/ni.1693 [DOI] [PubMed] [Google Scholar]
- Davidson, D., Shi X., Zhang S., Wang H., Nemer M., Ono N., Ohno S., Yanagi Y., and Veillette A.. 2004. Genetic evidence linking SAP, the X-linked lymphoproliferative gene product, to Src-related kinase FynT in TH2 cytokine regulation. Immunity. 21:707–717. 10.1016/j.immuni.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Dong, Z., and Veillette A.. 2010. How do SAP family deficiencies compromise immunity? Trends Immunol. 31:295–302. 10.1016/j.it.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Dong, Z., Cruz-Munoz M.E., Zhong M.C., Chen R., Latour S., and Veillette A.. 2009. Essential function for SAP family adaptors in the surveillance of hematopoietic cells by natural killer cells. Nat. Immunol. 10:973–980. 10.1038/ni.1763 [DOI] [PubMed] [Google Scholar]
- Dong, Z., Davidson D., Pérez-Quintero L.A., Kurosaki T., Swat W., and Veillette A.. 2012. The adaptor SAP controls NK cell activation by regulating the enzymes Vav-1 and SHIP-1 and by enhancing conjugates with target cells. Immunity. 36:974–985. 10.1016/j.immuni.2012.03.023 [DOI] [PubMed] [Google Scholar]
- Eberl, G., Lowin-Kropf B., and MacDonald H.R.. 1999. Cutting edge: NKT cell development is selectively impaired in Fyn-deficient mice. J. Immunol. 163:4091–4094. [PubMed] [Google Scholar]
- Eissmann, P., Beauchamp L., Wooters J., Tilton J.C., Long E.O., and Watzl C.. 2005. Molecular basis for positive and negative signaling by the natural killer cell receptor 2B4 (CD244). Blood. 105:4722–4729. 10.1182/blood-2004-09-3796 [DOI] [PubMed] [Google Scholar]
- Elewaut, D., Shaikh R.B., Hammond K.J., De Winter H., Leishman A.J., Sidobre S., Turovskaya O., Prigozy T.I., Ma L., Banks T.A., et al. 2003. NIK-dependent RelB activation defines a unique signaling pathway for the development of Vα14i NKT cells. J. Exp. Med. 197:1623–1633. 10.1084/jem.20030141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, H., Kitazawa H., Kaneko I., Matsubara M., Nose M., and Ono M.. 2009. Role of 2B4-mediated signals in the pathogenesis of a murine hepatitis model independent of Fas and Vα14 NKT cells. Immunology. 128:e151–e158. 10.1111/j.1365-2567.2008.02936.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, D.B., Bell M.P., McCausland M.M., Huntoon C.J., van Deursen J., Faubion W.A., Crotty S., and McKean D.J.. 2006. Ly9 (CD229)-deficient mice exhibit T cell defects yet do not share several phenotypic characteristics associated with SLAM- and SAP-deficient mice. J. Immunol. 176:291–300. 10.4049/jimmunol.176.1.291 [DOI] [PubMed] [Google Scholar]
- Griewank, K., Borowski C., Rietdijk S., Wang N., Julien A., Wei D.G., Mamchak A.A., Terhorst C., and Bendelac A.. 2007. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 27:751–762. 10.1016/j.immuni.2007.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J.K., Crampton J.C., Locci M., and Crotty S.. 2016. CRISPR-mediated Slamf1Δ/Δ Slamf5Δ/Δ Slamf6Δ/Δ triple gene disruption reveals NKT cell defects but not T follicular helper cell defects. PLoS One. 11:e0156074. 10.1371/journal.pone.0156074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B., Gomez-Rodriguez J., Preite S., Garrett L.J., Harper U.L., and Schwartzberg P.L.. 2016. CRISPR-mediated triple knockout of SLAMF1, SLAMF5 and SLAMF6 supports positive signaling roles in NKT cell development. PLoS One. 11:e0156072. 10.1371/journal.pone.0156072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama, R., Cannons J.L., Zhao F., Yusuf I., Lao C., Locci M., Schwartzberg P.L., and Crotty S.. 2012. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity. 36:986–1002. 10.1016/j.immuni.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latour, S., Roncagalli R., Chen R., Bakinowski M., Shi X., Schwartzberg P.L., Davidson D., and Veillette A.. 2003. Binding of SAP SH2 domain to FynT SH3 domain reveals a novel mechanism of receptor signalling in immune regulation. Nat. Cell Biol. 5:149–154. 10.1038/ncb919 [DOI] [PubMed] [Google Scholar]
- Le Borgne, M., and Shaw A.S.. 2012. SAP signaling: a dual mechanism of action. Immunity. 36:899–901. 10.1016/j.immuni.2012.06.002 [DOI] [PubMed] [Google Scholar]
- Li, C., Iosef C., Jia C.Y., Han V.K., and Li S.S.. 2003. Dual functional roles for the X-linked lymphoproliferative syndrome gene product SAP/SH2D1A in signaling through the signaling lymphocyte activation molecule (SLAM) family of immune receptors. J. Biol. Chem. 278:3852–3859. 10.1074/jbc.M206649200 [DOI] [PubMed] [Google Scholar]
- Ma, C.S., Nichols K.E., and Tangye S.G.. 2007. Regulation of cellular and humoral immune responses by the SLAM and SAP families of molecules. Annu. Rev. Immunol. 25:337–379. 10.1146/annurev.immunol.25.022106.141651 [DOI] [PubMed] [Google Scholar]
- McCausland, M.M., Yusuf I., Tran H., Ono N., Yanagi Y., and Crotty S.. 2007. SAP regulation of follicular helper CD4 T cell development and humoral immunity is independent of SLAM and Fyn kinase. J. Immunol. 178:817–828. 10.4049/jimmunol.178.2.817 [DOI] [PubMed] [Google Scholar]
- Morra, M., Howie D., Grande M.S., Sayos J., Wang N., Wu C., Engel P., and Terhorst C.. 2001. X-linked lymphoproliferative disease: a progressive immunodeficiency. Annu. Rev. Immunol. 19:657–682. 10.1146/annurev.immunol.19.1.657 [DOI] [PubMed] [Google Scholar]
- Nichols, K.E., Hom J., Gong S.Y., Ganguly A., Ma C.S., Cannons J.L., Tangye S.G., Schwartzberg P.L., Koretzky G.A., and Stein P.L.. 2005. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat. Med. 11:340–345. 10.1038/nm1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez-Cruz, S., Yeo W.C., Rothman J., Ojha P., Bassiri H., Juntilla M., Davidson D., Veillette A., Koretzky G.A., and Nichols K.E.. 2008. Differential requirement for the SAP-Fyn interaction during NK T cell development and function. J. Immunol. 181:2311–2320. 10.4049/jimmunol.181.4.2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan, K.L., Zhao J., Pryshchep O., Wang C.R., and Phee H.. 2015. Pak2 controls acquisition of NKT cell fate by regulating expression of the transcription factors PLZF and Egr2. J. Immunol. 195:5272–5284. 10.4049/jimmunol.1501367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier, B., Yin L., Fondanèche M.C., Relouzat F., Bloch-Queyrat C., Lambert N., Fischer A., de Saint-Basile G., and Latour S.. 2005. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J. Exp. Med. 201:695–701. 10.1084/jem.20042432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, H., Cannons J.L., Klauschen F., Schwartzberg P.L., and Germain R.N.. 2008. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 455:764–769. 10.1038/nature07345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekaran, K., Kumar P., Schuldt K.M., Peterson E.J., Vanhaesebroeck B., Dixit V., Thakar M.S., and Malarkannan S.. 2013. Signaling by Fyn-ADAP via the Carma1-Bcl-10-MAP3K7 signalosome exclusively regulates inflammatory cytokine production in NK cells. Nat. Immunol. 14:1127–1136. 10.1038/ni.2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage, A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., and Bendelac A.. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 29:391–403. 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayos, J., Wu C., Morra M., Wang N., Zhang X., Allen D., van Schaik S., Notarangelo L., Geha R., Roncarolo M.G., et al. 1998. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 395:462–469. 10.1038/26683 [DOI] [PubMed] [Google Scholar]
- Schmidt-Supprian, M., Tian J., Grant E.P., Pasparakis M., Maehr R., Ovaa H., Ploegh H.L., Coyle A.J., and Rajewsky K.. 2004. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-κB activation. Proc. Natl. Acad. Sci. USA. 101:4566–4571. 10.1073/pnas.0400885101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, M.P., Mathew R., Liszewski M.K., Spooner C.J., Barr K., Meng F., Singh H., and Bendelac A.. 2012. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol. 13:264–271. (published erratum appears in Nat. Immunol. 2013. 14:413) 10.1038/ni.2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar, V., Hammond K.J., Howells N., Pfeffer K., and Weih F.. 2003. Differential requirement for Rel/nuclear factor κB family members in natural killer T cell development. J. Exp. Med. 197:1613–1621. 10.1084/jem.20022234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanic, A.K., Bezbradica J.S., Park J.J., Matsuki N., Mora A.L., Van Kaer L., Boothby M.R., and Joyce S.. 2004. NF-κB controls cell fate specification, survival, and molecular differentiation of immunoregulatory natural T lymphocytes. J. Immunol. 172:2265–2273. 10.4049/jimmunol.172.4.2265 [DOI] [PubMed] [Google Scholar]
- Takeda, K., Hayakawa Y., Van Kaer L., Matsuda H., Yagita H., and Okumura K.. 2000. Critical contribution of liver natural killer T cells to a murine model of hepatitis. Proc. Natl. Acad. Sci. USA. 97:5498–5503. 10.1073/pnas.040566697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassi, I., and Colonna M.. 2005. The cytotoxicity receptor CRACC (CS-1) recruits EAT-2 and activates the PI3K and phospholipase Cγ signaling pathways in human NK cells. J. Immunol. 175:7996–8002. 10.4049/jimmunol.175.12.7996 [DOI] [PubMed] [Google Scholar]
- Veillette, A., Dong Z., and Latour S.. 2007. Consequence of the SLAM-SAP signaling pathway in innate-like and conventional lymphocytes. Immunity. 27:698–710. 10.1016/j.immuni.2007.11.005 [DOI] [PubMed] [Google Scholar]
- Veillette, A., Dong Z., Pérez-Quintero L.A., Zhong M.C., and Cruz-Munoz M.E.. 2009. Importance and mechanism of ‘switch’ function of SAP family adapters. Immunol. Rev. 232:229–239. 10.1111/j.1600-065X.2009.00824.x [DOI] [PubMed] [Google Scholar]
- Wei, D.G., Lee H., Park S.H., Beaudoin L., Teyton L., Lehuen A., and Bendelac A.. 2005. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J. Exp. Med. 202:239–248. 10.1084/jem.20050413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J.A., Lumsden J.M., Yu X., Feigenbaum L., Zhang J., Steinberg S.M., and Hodes R.J.. 2008. Regulation of thymic NKT cell development by the B7-CD28 costimulatory pathway. J. Immunol. 181:907–917. 10.4049/jimmunol.181.2.907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, N., Zhong M.C., Roncagalli R., Pérez-Quintero L.A., Guo H., Zhang Z., Lenoir C., Dong Z., Latour S., and Veillette A.. 2016. A hematopoietic cell-driven mechanism involving SLAMF6 receptor, SAP adaptors and SHP-1 phosphatase regulates NK cell education. Nat. Immunol. 17:387–396. 10.1038/ni.3369 [DOI] [PubMed] [Google Scholar]
- Zhao, F., Cannons J.L., Dutta M., Griffiths G.M., and Schwartzberg P.L.. 2012. Positive and negative signaling through SLAM receptors regulate synapse organization and thresholds of cytolysis. Immunity. 36:1003–1016. 10.1016/j.immuni.2012.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]