Abstract
Mutations in more than 250 genes are implicated in inherited retinal dystrophy; the encoded proteins are involved in a broad spectrum of pathways. The presence of unsolved families after highly parallel sequencing strategies suggests that further genes remain to be identified. Whole-exome and -genome sequencing studies employed here in large cohorts of affected individuals revealed biallelic mutations in ARHGEF18 in three such individuals. ARHGEF18 encodes ARHGEF18, a guanine nucleotide exchange factor that activates RHOA, a small GTPase protein that is a key component of tight junctions and adherens junctions. This biological pathway is known to be important for retinal development and function, as mutation of CRB1, encoding another component, causes retinal dystrophy. The retinal structure in individuals with ARHGEF18 mutations resembled that seen in subjects with CRB1 mutations. Five mutations were found on six alleles in the three individuals: c.808A>G (p.Thr270Ala), c.1617+5G>A (p.Asp540Glyfs∗63), c.1996C>T (p.Arg666∗), c.2632G>T (p.Glu878∗), and c.2738_2761del (p.Arg913_Glu920del). Functional tests suggest that each disease genotype might retain some ARHGEF18 activity, such that the phenotype described here is not the consequence of nullizygosity. In particular, the p.Thr270Ala missense variant affects a highly conserved residue in the DBL homology domain, which is required for the interaction and activation of RHOA. Previously, knock-out of Arhgef18 in the medaka fish has been shown to cause larval lethality which is preceded by retinal defects that resemble those seen in zebrafish Crumbs complex knock-outs. The findings described here emphasize the peculiar sensitivity of the retina to perturbations of this pathway, which is highlighted as a target for potential therapeutic strategies.
Keywords: retinitis pigmentosa, inherited retinal dystrophy, retinal degeneration, ARHGEF18, p114RhoGEF, apicobasal polarity
Main Text
Inherited retinal dystrophy (IRD) encompasses a clinically and genetically heterogeneous group of disorders characterized by retinal dysfunction or degeneration. Variants in more than 250 genes encoding proteins essential to a wide range of biological pathways including mRNA splicing, posttranslational protein modification, ciliogenesis, cilia protein transport, retinoid recycling in the visual cycle, phototransduction, and retinal development have been found causative of IRD (RetNet).
This report describes mutation of ARHGEF18 (MIM: 616432) as a likely cause of human IRD. The gene encodes ARHGEF18 (also known as p114RhoGEF),1 the Rho/Rac guanine nucleotide exchange factor 18. It has been shown to be involved in the determination of apicobasal (AB) polarity in epithelia and cell-cell junction formation through its action on the small GTPase RHOA.2 The gene is widely expressed, with expressed sequence tags identified in many human tissues including the neurosensory retina (NCBI-UniGene).
The study protocol adhered to the tenets of the Declaration of Helsinki and received approval from the local ethics committee. Written, informed consent was obtained from all participants prior to their inclusion in this study. To gain further insight into the genetic pathology of inherited retinal dystrophy, whole-exome sequencing (WES) has been performed on 230 individuals and whole-genome sequencing (WGS) on a further 599 probands, ascertained from the inherited retinal disease clinics at Moorfields Eye Hospital (MEH), London. The latter cohort forms part of the NIHR-Bioresource Rare Disease consortium in the UK.3
Biallelic mutations in ARHGEF18 were identified in three individuals (Table S1), presenting as simplex cases, each with a retinal dystrophy sharing features with that seen in retinal disease caused by mutation in CRB1 (MIM: 604210).4 For this reason, in all three individuals, Sanger sequencing of CRB1 had been performed but did not identify any potential disease-associated variants. WGS was performed on individuals 1 and 2; the remaining individual (individual 3) underwent WES as previously described.5 In the first instance, resulting coding variant calls were filtered using a list of 236 genes previously implicated in retinal dystrophy.6 No convincing causal variants were identified in these affected individuals (Table S2).
After WGS, individual 1 (GC18203), a 37-year-old female with simplex retinitis pigmentosa (RP [MIM: 268000]), the second of two siblings born to unrelated parents with no family history of eye disease, had 20,863 coding (±8 bp splice region) variants passing standard quality filters. Of these, 360 had a minor allele frequency (MAF) ≤ 0.01 in the publicly available dataset (Exome Aggregation Consortium database [ExAC]). Assuming autosomal-recessive inheritance, five genes contained ≥2 variants (Table S3). Variants were further manually interrogated for variant call quality, MAF in publicly available datasets and our own in-house exome-sequencing dataset (UCL exome project of more than 5,000 individuals), predicted protein impact, and biological plausibility (including protein function, expression profile, and pathway analysis). Of these, two variants were identified in ARHGEF18. The variants were a missense and nonsense absent from ExAC: GRCh37 (hg19) chr19: g.7509101A>G, GenBank: NM_001130955, c.808A>G (p.Thr270Ala) and chr19: g.7527145C>T, GenBank: NM_001130955, c.1996C>T (p.Arg666∗). The missense variant p.Thr270Ala was predicted to be damaging by in silico prediction algorithms (sorting the intolerant from tolerant [SIFT], Polymorphism Phenotyping v2 [PolyPhen-2])7, 8 and affects a highly conserved amino acid residue in the DBL homology (DH) domain (Figure 1).
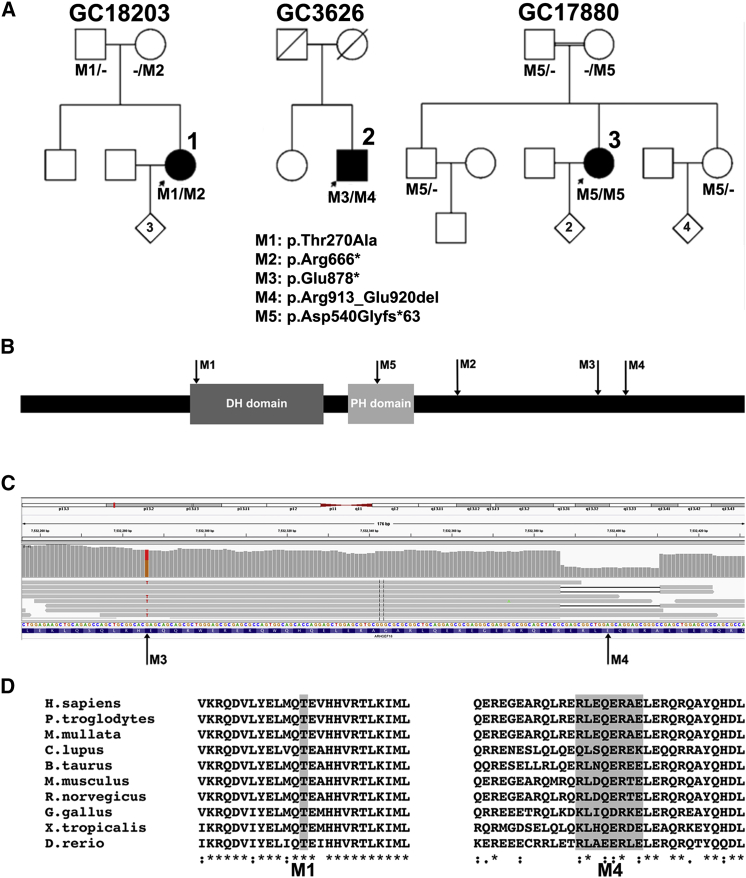
Figure 1.
Variant Analysis of ARHGEF18 in Individuals 1–3
(A) Pedigrees and cosegregation of mutations M1–M5 in families 1–3.
(B) Schematic representation of mutation location in full-length ARHGEF18 including DBL homology (DH) and Plekstrin homology (PH) domains.
(C) IGV visualization of 150 bp paired end reads spanning mutations ARHGEF18, c.2632G>T, and c.2738_2761del in individual 2, showing biallelic state.
(D) Clustal Omega alignment of amino acid residues affected by M1 (missense) and M4 (in-frame deletion) mutations throughout vertebrate orthologues.
Identical analysis was performed on the 21,042 coding variants identified by WGS in individual 2 (GC3626), a 51-year-old male simplex RP-affected individual, the second of two siblings born to unrelated parents with no family history of eye disease. 17 genes with ≥2 variants (MAF ≤ 0.01) were identified (Table S4), among which were two variants comprising a nonsense and in-frame deletion in ARHGEF18: chr19: g.7532286G>T, GenBank: NM_001130955, c.2632G>T (p.Glu878∗) and chr19: g.7532392_7532415del, GenBank: NM_001130955, c.2738_2761del (p.Arg913_Glu920del). The two variants occur within 130 bp in exon 16 of ARHGEF18. Interrogation of the 150 bp paired end reads in this region using the Integrative Genomics Viewer (IGV)9, 10 allowed phasing of the variants on seven reads suggesting they were in trans (Figure 1C). Familial DNA samples were unavailable for segregation analysis. The in-frame deletion of eight amino acid residues removes part of a highly conserved region of the protein (RLEQERAE) (Figure 1D).
Individual 3 (GC17880), an affected individual with simplex RP, the second of three siblings born to first-cousin parents with no family history of eye disease, underwent WES revealing 21,404 coding variants. Of these, 383 were rare (MAF ≤ 0.01 in the publicly available NHLBI GO Exome Sequencing Project dataset [EVS]). Assuming recessive inheritance due to autozygosity, seven genes had homozygous variants affecting the canonical transcript (Table S5), six of which were located within homozygous regions ≥5 Mb identified by prior SNP array autozygosity mapping data (SNP6, Affymetrix). Of these variants, a splice region substitution (chr19: g.7521294G>A, GenBank: NM_001130955, c.1617+5G>A) in ARHGEF18 was the most compelling candidate. This variant is predicted to weaken the canonical splice donor site and lead to out-of-frame skipping of ARHGEF18 exon 8.
The variants in each individual were confirmed to be biallelic by familial segregation analysis in all available relatives (Figure 1A); no unaffected family member available for screening carried two disease alleles. Subsequent direct Sanger sequencing of all coding exons of ARHGEF18 in ten individuals with a similar phenotype and no detectable mutation in CRB14 revealed no further mutations in ARHGEF18. In a cohort of 5,695 individuals who underwent WES (UCL-exome cohort), no rare (MAF ≤ 0.01) loss-of-function (LOF) variants were identified. Four individuals with unrelated phenotypes had predicted biallelic rare missense variants (Table S6).
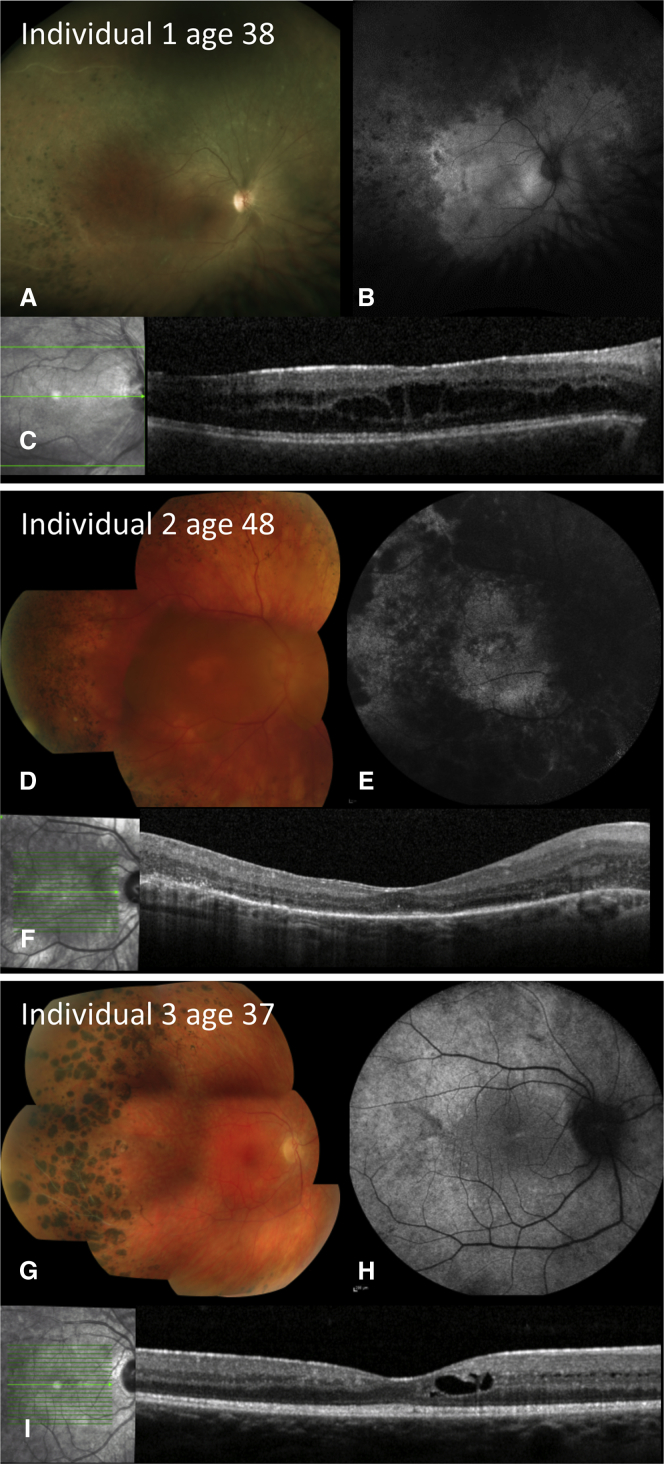
The affected individuals reported here all presented in their third to fourth decades with central visual disturbance, visual field defects, and mild nyctalopia (Table S7). At last review at ages 37, 51, and 38 years for individuals 1, 2, and 3, respectively, visual acuity ranged from 0.18 log MAR (Snellen 20/30) to 1.8 log MAR (Snellen 20/1250); the worst was in the oldest individual. Fundus examination revealed optic disc pallor, attenuated retinal vessels, and irregular mid-peripheral intra-retinal pigment migration. Fundus autofluorescence (FAF) imaging revealed widespread, irregular, peripheral hypo-autofluorescence (Figure 2). Optical coherence tomography (OCT) demonstrated intra-retinal cysts in all affected individuals. Such imaging produces an in vivo cross-section of the retina. A useful landmark to gauge the degree of retinal degeneration is a contiguous line, parallel with the inner retinal surface, that is thought to be formed by reflection from mitochondria in photoreceptor inner segments, the so-called inner-segment ellipsoid line (ISe).11 Individuals 1 and 3 had a preserved ISe throughout the macula; for individual 2 the ISe was retained only in the foveal region. The irregularity of the autofluorescence is distinct from that occurring in primary rod photoreceptor disease and instead resembles that seen in CRB1 retinopathy. Moreover, peripheral nummular pigment was similar to CRB1 retinopathy. In most degenerative dystrophies the retina is thinner than normal, and in these individuals retinal thickness instead resembled that seen in CRB1 retinopathy. Full-field electroretinography (ERG), performed in all individuals at a similar age (29–30 years),12 demonstrated severe generalized retinal dysfunction affecting rod more than cone photoreceptors (Figure S1). The pattern ERG13 was subnormal in individuals 1 and 3, indicating relatively mild macular involvement, but was undetectable in individual 2, consistent with severe macular involvement. There was no clinical evidence of other systemic, neurological, or other epithelial disease in any of the individuals.
Figure 2.
Retinal Abnormalities in ARHGEF18-Related Retinal Dystrophy
Color fundus photographs, 55 degree fundus autofluorescence imaging, and optical coherence tomography (OCT).
(A–C) Individual 1, right eye at age 38 years, showing (A) optic disc pallor, peripheral retinal pigment epithelium (RPE) atrophy, and nummular pigmentation; (B) peripheral patchy reduction of autofluorescence; and (C) reasonably preserved retinal layers on OCT with disruption of the inner segment ellipsoid band and intra-retinal cysts within the inner nuclear layer.
(D–F) Individual 2, right eye at 48 years, showing (D) disc pallor and vessel attenuation with RPE atrophy within the macula and mid-periphery as well as peripheral nummular and dot lesions of hyperpigmentation; (E) extensive loss of autofluorescence in periphery and in a central ring in macula; and (F) loss of outer retina and RPE throughout the macula with small foci of preserved photoreceptors centrally.
(G–I) Individual 3, right eye at 37 years, showing (G) vascular attenuation and occlusion, peripheral RPE atrophy, white dots, and nummular pigmentation; (H) loss of autofluorescence in periphery; and (I) reasonably preserved retinal layers on OCT with disruption of the inner segment ellipsoid band and intra-retinal cysts within the inner nuclear layer.
Cell-cell junctions (tight junctions [TJ] and adherens junctions [AJ]) are important in the establishment of AB polarity. During vertebrate eye morphogenesis, AB polarity of epithelial cells forming the optic vesicle is established and maintained by the migration and accumulation of specific polarity proteins and lipid complexes and the regulation of the actomyosin network in distinct apical and basal membrane domains and the formation of TJ and AJ.14, 15 Interkinetic nuclear migration (IKNM) along the AB polarity axis results in specific positioning of nuclei in the single-cell neuroepithelium and a pseudostratified appearance, and in turn contributes to the cell fate determination during differentiation into the three nuclear layers of the retina.16
Three major classes of protein complexes have been implicated in the establishment and maintenance of AB polarity: the Crumbs, Par, and Scribble complexes that serve as either apical or basolateral determinants. Rho small GTPase family members RHOA, RAC1, and CDC42 are central to the regulation of cell migration, contact adhesion, and the regulation of these apical and basolateral determinants.15 The genes encoding these GTPase family members have not been implicated in retinal disease.15, 17 The activation status of Rho GTPases is determined by the guanine nucleotide bound to them (GTP-active, GDP-inactive), which is, in turn, regulated by guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs).
Regulation of RHOA activation through ARHGEF18 is important for tissue morphogenesis and migration and in the assembly and maintenance of cell-cell junctions, TJ and AJ.14, 15 Cell junctions form intracellular connections essential for control of cell proliferation and morphology and maintenance of tissue integrity. In epithelia, TJ are formed at the apical/lateral border and control the movement of molecules along the paracellular space.18 Molecular mechanisms regulating RHOA activation are crucial components of the pathways that guide TJ assembly and function. ARHGEF18 drives RHOA activation at TJ and thereby regulates actomyosin activity and TJ assembly, epithelial morphology, and dynamics.2, 19, 20
Cell-cell junctions and AB polarity are essential in the function and maintenance of retinal architecture.21, 22 In particular, the outer limiting membrane (OLM) is formed of AJ between Müller glia cells and photoreceptors and the inner/outer segments of photoreceptors are formed from the apical membrane of developing photoreceptors.
All of the individuals harbored genotypes of ARHGEF18 that might conceivably produce some protein function, rather than being definite biallelic nulls. Individuals 1 and 2 had a nonsense mutation in trans with a missense or in-frame deletion, respectively. Reverse transcription-PCR (RT-PCR) and direct sequencing analysis of the ARHGEF18 transcript from lymphocytes of individual 3 (c.1617+5G>A) using PCR primers spanning exons 6–9 identified differently spliced transcripts (Figure S2). Direct sequencing of the PCR-generated products identified a short transcript lacking exon 8 and a weaker band corresponding to the wild-type (WT) transcript (including exon 8). Hence, a low proportion of WT transcript and full-length WT protein is likely to be produced despite this splice-site alteration. Guanine to adenine transitions at position +5 in splice donor sites are recognized pathogenic mutations but have been reported previously to produce some normal mRNA product, for example in the context of cystic fibrosis.23 The downstream consequence of exon 8 skipping would be a termination codon following 62 out-of-frame codons (p.Asp540Glyfs∗63) and a transcript that is likely to undergo nonsense-mediated decay (NMD). The in-frame deletion in individual 2 is predicted to abolish several putative exonic splice enhancer (ESE) motifs.24 However, RT-PCR and direct sequencing of the ARHGEF18 transcript from lymphocytes of individual 2 using PCR primers spanning exons 13–17 identified no alteration in splicing (Figure S2) as a consequence of the deletion.
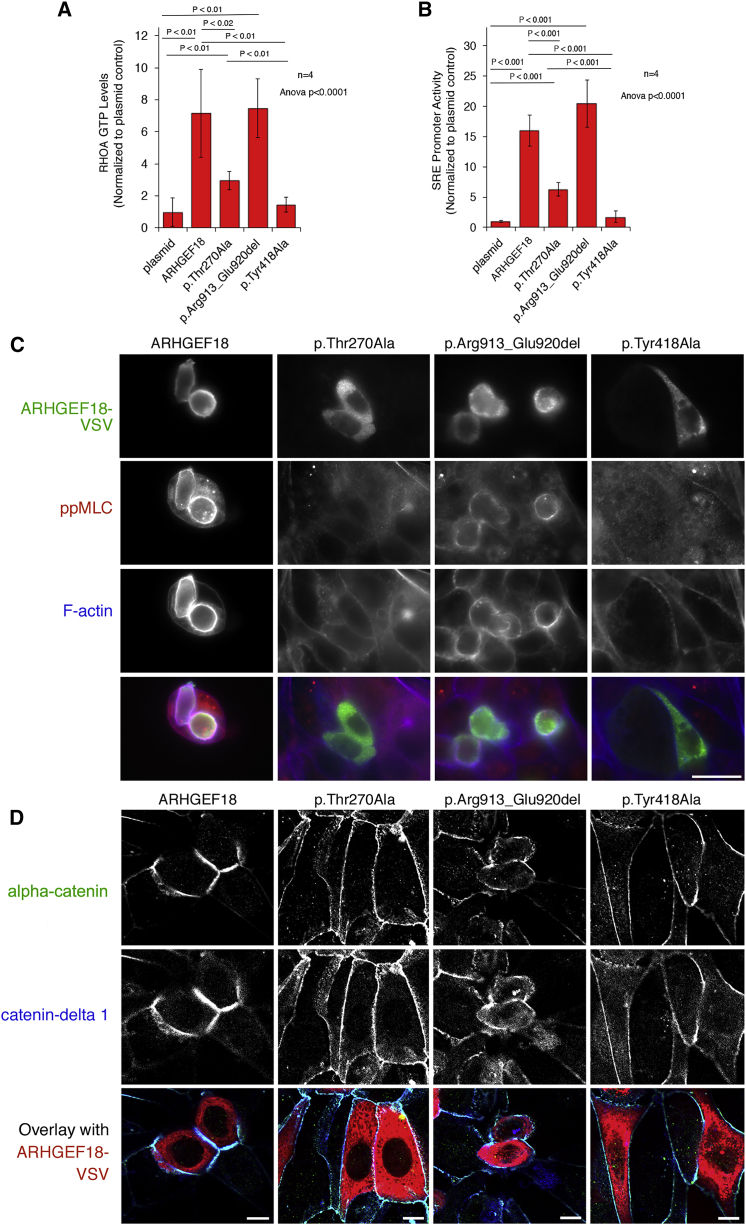
In order to determine the functional consequence and potential pathogenicity of the missense and in-frame deletion variants, HEK293T cells were transfected with expression vectors encoding WT ARHGEF18 (GenBank: NM_001130955)2 or with the p.Thr270Ala substitution or the p.Arg913_Glu920del deletion generated using the Q5 site-directed mutagenesis kit (New England Biolabs) according to manufacturers’ instructions and propagated, purified, and sequenced using standard procedures. A previously characterized catalytically inactive mutant was included as a control (p.Tyr418Ala, previously referred to as p.Tyr260Ala).2 RHOA activation is essential for ARHGEF18 to stimulate TJ assembly. This was tested by measuring RHOA-GTP levels in transfected HEK293T cells using a biochemical 96-well assay that measures binding of RHOA-GTP to the Rho binding domain of Rhotekin (G-LISA, Cytoskeleton, Inc.2). Ectopic expression of the WT protein led to a more than 5-fold stimulation of RHOA-GTP level (Figure 3A). The p.Thr270Ala mutant retained some activity compared to the catalytically inactive p.Tyr418Ala construct as it led to a 3-fold increase in RHOA-GTP level, but was less than 50% of the WT level (Figure 3A). The deletion mutant (p.Arg913_Glu920del) had a similar increase in RHOA-GTP level to the WT protein (Figure 3A).
Figure 3.
Signalling Activity of ARHGEF18 Variants
HEK293T or HCE cells were transfected with cDNAs encoding wild-type or mutant VSV-tagged ARHGEF18.
(A) Lysates of transfected HEK293T cells were assayed for RHOA-GTP levels by G-LISA assay, which measures binding of active RHOA to the GTPase binding domain of Rhotekin.
(B) Serum response element (SRE) element activity was measured using a double luciferase assay with an SRE-containing promoter driving firefly luciferase expression and a CMV promoter expression of renilla luciferase. Firefly to renilla luciferase ratios were calculated and normalized to a plasmid control performed by co-transfecting an empty expression vector. The graphs show averages ± 1 standard deviations, n = 4, indicated are p values from ANOVA and t tests.
(C) Transfected HCE cells were fixed and stained with anti-VSV and anti-phosphorylated myosin regulatory light chain (ppMLC) antibodies along Ato-647-labeled phalloidin to visualize F-actin and imaged by epifluorescence.
(D) Cells transfected as in (C) were stained for the junctional markers alpha-catenin and Catenin delta-1 and imaged by confocal microscopy.
Scale bars represent 20 μm in (C) and 10 μm in (D).
The transcriptional activity of serum response factor (SRF) was measured in transfected cells using a double luciferase reporter assay25 to monitor signaling output of the mutant ARHGEF18 constructs. Similar to the RHOA activation assay, the WT construct led to a strong stimulation of SRF-driven luciferase expression while the missense mutant (p.Thr270Ala) led to a 3-fold reduction in the level of luciferase expression but, as in the RHOA-GTP assay, showed significant activity also if compared to the inactive p.Tyr418Ala mutant; the deletion mutant (p.Arg913_Glu920del) level was unaltered (Figure 3B). Thus, the p.Thr270Ala mutant led to reduced but not abolished RHOA activation and signaling, and the p.Arg913_Glu920del mutant did not affect RHOA activation.
As ARHGEF18 stimulates cortical actomyosin activation leading to TJ assembly and cell rounding in epithelial cells, human corneal epithelial cells (HCEs) were transfected with the WT or mutant expression vectors, including the p.Tyr418Ala catalytically inactive control. Transfection of WT but not the p.Tyr418Ala control led to rounded morphology of the normally flat cells, increased cortical phospho-myosin (pp-MLC), and F-actin staining (Figure 3C). Similar to the GEF inactive mutant (p.Tyr418Ala), the p.Thr270Ala mutant was not recruited to the cell cortex and failed to induce the cortical actomyosin cytoskeleton enrichment (Figure 3C), supporting the conclusion that its activity was strongly reduced. The deletion mutant (p.Arg913_Glu920del) induced some cortical actomyosin reorganization but appeared to do so less efficiently than WT. In addition, its distribution was more patchy and irregular than WT. Both mutations thus affect the normal subcellular localization of ARHGEF18. Only the WT protein induced a strong stimulation of recruitment of junctional proteins when overexpressed (Figure 3D). The two point mutations (p.Thr270Ala and p.Tyr418Ala) failed to induce a response, and the deletion mutation led to cell rounding but only a weak increase in junctional recruitment of TJ markers alpha-catenin and Catenin delta-1, suggesting that the deletion inhibits the normal cellular activity of ARHGEF18 despite showing normal catalytic activity.
The missense variant p.Thr270Ala resides within the DH domain of ARHGEF18, which is the catalytic domain required for guanine nucleotide exchange.26 Thr270 is located within the first alpha-helix of the highly conserved DH domain. This residue is conserved as a threonine or serine in virtually all DH domains throughout nature. In the C. elegans Rac GTPase activating protein, UNC-73, serine or threonine at this residue maintain the catalytic activity whereas mutation to alanine abolishes its activity.26, 27 The hydroxyl group of the serine or threonine at this position in the DH domain is thought to mediate GTPase interaction; hence, substitution of Thr270 of ARHGEF18 may inhibit RHOA activation in this way.
The in-frame deletion occurs in exon 16, downstream of the DH and PH module, and does not directly interfere with the catalytic activity. The STK11 binding domain has been mapped to the C-terminal region of the murine Arhgef18 protein encompassing these deleted amino acid residues, and interaction of STK11 and ARHGEF18 is essential in AJ formation.28 Despite being catalytically active, the in-frame deletion mutant appeared less potent for induction of cortical actomyosin organization than the WT GEF, suggesting that the deletion may indeed affect interactions required for normal cellular ARHGEF18 function, possibly by removing residues required for interactions or for normal folding of the C-terminal domain.
The data indicate that all affected individuals retain some exchange factor activity or native protein. The strong reduction of ARHGEF18 function observed leads to the development of retinal dystrophy in these individuals but heterozygous carriers of LOF mutations are unaffected. The absence of a confirmed biallelic null in the cohort or indeed in the ExAC dataset suggests that complete loss of ARHGEF18 function could be developmentally severe or lethal or may have a more syndromic phenotype. The hypothesis of an embryonic lethal phenotype is supported by the effect of null alleles in medaka fish.29
Perturbation of the AB polarity of epithelial cells is recognized in tumorigenesis and cancer progression17, 30, 31 but to date, only CRB1 of the AB polarity complex encoding genes32 has been implicated in human Mendelian disease. Mutation of CRB1 causes a wide spectrum of retinal disease including Leber congenital amaurosis (LCA), early-onset retinal dystrophy (EORD), RP, and more recently maculopathy and foveal retinoschisis.33, 34, 35, 36 Age of onset and severity are variable, affected individuals often presenting with early-onset severe loss of vision with characteristic sub-retinal white dots, deep nummular pigmentary lesions, and a thickened, disorganized retina with an undetectable ERG in the most severe cases.4, 37 It is of interest that the three individuals reported here resembled clinically those with CRB1 retinopathy although the age of onset was later,4 and this suggests that perturbation of this pathway produces a distinctive human retinal phenotype. The phenotypes of the murine Crb1 knockout, Crb1−/−, and naturally occurring Rd8 truncating mutant are characterized by disruption of AJ between Müller cells and photoreceptors at the OLM, photoreceptor dysplasia, and consequent focal areas of disorganized lamination and degeneration, although the remaining retina provides functional vision.22, 38, 39 The knock-in missense RP mutant CrbC249W has a late-onset degenerative phenotype and can partially rescue the phenotype in Crb1−/− mice.40
Mutation of the apical domain essential Crumbs complex proteins epb41l5 (moe), mpp5a (nok), and crb2a (ome) in the zebrafish (mosaic eyes, nagie oko, and oko meduzy, respectively) all result in AB polarity defects leading to retinal dystrophy characterized by retinal developmental and lamination abnormalities.41, 42, 43, 44, 45 Similarly, the larval lethal medaka fish retinal differentiation mutant (medeka is Japanese for “large eyes”) exhibits disorganization of retinal lamination during embryonic development consequent on a LOF mutation resulting in absence of the ArhGEF18 protein product.29 The phenotype is consequent upon abrogation of ArhGEF18 activity in the developing embryo resulting in disruption of RHOA activation and perturbation of AB polarity characterized by mislocalization of TJ, disorganization of the actin cytoskeleton, and cell proliferation morphology alterations.
The disease mechanism of human ARHGEF18 retinopathy is not yet fully understood and may include developmental and/or degenerative mechanisms. Disruption of ARHGEF18 function in retinal development seems unlikely, as a severe early-onset retinal dystrophy would be a more probable consequence and our three individuals all experienced normal visual function in early life. A more plausible hypothesis is that photoreceptors are peculiarly sensitive to the failure of AJ maintenance than other cells, causing onset of retinal degeneration in adulthood. Similar clinical features and variable age of onset seen in CRB1 retinopathy strengthens the assertion that maintenance of this complex is required for continued photoreceptor viability in humans. The phenotypic similarity of ArhGEF18 and crumbs complex protein knockouts in lower vertebrates reflects the similarity of these in humans. Taken together, these observations suggest that other proteins essential in AB polarity maintenance should be regarded as candidate genes in retinal dystrophies. Furthermore, the pathway provides a target for therapeutic intervention with the potential to ameliorate visual impairment due to this type of retinal dystrophy.
Consortia
UKIRDC (UK Inherited Retinal Dystrophy Consortium) members include Graeme Black, Georgina Hall, Stuart Ingram, Rachel Gillespie, Forbes Manson, Panagiotis Sergouniotis, Chris Inglehearn, Carmel Toomes, Manir Ali, Martin McKibbin, James Poulter, Kamron Khan, Emma Lord, Andrea Nemeth, Susan Downes, Stephanie Halford, Jing Yu, Stefano Lise, Gavin Arno, Alessia Fiorentino, Nikos Ponitkos, Vincent Plagnol, Michel Michaelides, Alison J. Hardcastle, Michael E. Cheetham, Andrew R. Webster, and Veronica van Heyningen.
The members of the NIHR Bioresource – Rare Diseases Consortium are Timothy Aitman, Hana Alachkar, Sonia Ali, Louise Allen, David Allsup, Gautum Ambegaonkar, Julie Anderson, Richard Antrobus, Ruth Armstrong, Gavin Arno, Gururaj Arumugakani, Sofie Ashford, William Astle, Antony Attwood, Steve Austin, Chiara Bacchelli, Tamam Bakchoul, Tadbir K. Bariana, Helen Baxendale, David Bennett, Claire Bethune, Shahnaz Bibi, Maria Bitner-Glindzicz, Marta Bleda, Harm Boggard, Paula Bolton-Maggs, Claire Booth, John R. Bradley, Angie Brady, Matthew Brown, Michael Browning, Christine Bryson, Siobhan Burns, Paul Calleja, Natalie Canham, Jenny Carmichael, Keren Carss, Mark Caulfield, Elizabeth Chalmers, Anita Chandra, Patrick Chinnery, Manali Chitre, Colin Church, Emma Clement, Naomi Clements-Brod, Virginia Clowes, Gerry Coghlan, Peter Collins, Nichola Cooper, Amanda Creaser-Myers, Rosa DaCosta, Louise Daugherty, Sophie Davies, John Davis, Minka De Vries, Patrick Deegan, Sri V.V. Deevi, Charu Deshpande, Lisa Devlin, Eleanor Dewhurst, Rainer Doffinger, Natalie Dormand, Elizabeth Drewe, David Edgar, William Egner, Wendy N. Erber, Marie Erwood, Tamara Everington, Remi Favier, Helen Firth, Debra Fletcher, Frances Flinter, James C. Fox, Amy Frary, Kathleen Freson, Bruce Furie, Abigail Furnell, Daniel Gale, Alice Gardham, Michael Gattens, Neeti Ghali, Pavandeep K. Ghataorhe, Rohit Ghurye, Simon Gibbs, Kimberley Gilmour, Paul Gissen, Sarah Goddard, Keith Gomez, Pavel Gordins, Stefan Gräf, Daniel Greene, Alan Greenhalgh, Andreas Greinacher, Sofia Grigoriadou, Detelina Grozeva, Scott Hackett, Charaka Hadinnapola, Rosie Hague, Matthias Haimel, Csaba Halmagyi, Tracey Hammerton, Daniel Hart, Grant Hayman, Johan W.M. Heemskerk, Robert Henderson, Anke Hensiek, Yvonne Henskens, Archana Herwadkar, Simon Holden, Muriel Holder, Susan Holder, Fengyuan Hu, Aarnoud Huissoon, Marc Humbert, Jane Hurst, Roger James, Stephen Jolles, Dragana Josifova, Rashid Kazmi, David Keeling, Peter Kelleher, Anne M. Kelly, Fiona Kennedy, David Kiely, Nathalie Kingston, Ania Koziell, Deepa Krishnakumar, Taco W. Kuijpers, Dinakantha Kumararatne, Manju Kurian, Michael A. Laffan, Michele P. Lambert, Hana Lango Allen, Allan Lawrie, Sara Lear, Melissa Lees, Claire Lentaigne, Ri Liesner, Rachel Linger, Hilary Longhurst, Lorena Lorenzo, Rajiv Machado, Rob Mackenzie, Robert MacLaren, Eamonn Maher, Jesmeen Maimaris, Sarah Mangles, Ania Manson, Rutendo Mapeta, Hugh S. Markus, Jennifer Martin, Larahmie Masati, Mary Mathias, Vera Matser, Anna Maw, Elizabeth McDermott, Coleen McJannet, Stuart Meacham, Sharon Meehan, Karyn Megy, Sarju Mehta, Michel Michaelides, Carolyn M. Millar, Shahin Moledina, Anthony Moore, Nicholas Morrell, Andrew Mumford, Sai Murng, Elaine Murphy, Sergey Nejentsev, Sadia Noorani, Paquita Nurden, Eric Oksenhendler, Willem H. Ouwehand, Sofia Papadia, Soo-Mi Park, Alasdair Parker, John Pasi, Chris Patch, Joan Paterson, Jeanette Payne, Andrew Peacock, Kathelijne Peerlinck, Christopher J. Penkett, Joanna Pepke-Zaba, David J. Perry, Val Pollock, Gary Polwarth, Mark Ponsford, Waseem Qasim, Isabella Quinti, Stuart Rankin, Julia Rankin, F. Lucy Raymond, Karola Rehnstrom, Evan Reid, Christopher J. Rhodes, Michael Richards, Sylvia Richardson, Alex Richter, Irene Roberts, Matthew Rondina, Elisabeth Rosser, Catherine Roughley, Kevin Rue-Albrecht, Crina Samarghitean, Alba Sanchis-Juan, Richard Sandford, Saikat Santra, Ravishankar Sargur, Sinisa Savic, Sol Schulman, Harald Schulze, Richard Scott, Marie Scully, Suranjith Seneviratne, Carrock Sewell, Olga Shamardina, Debbie Shipley, Ilenia Simeoni, Suthesh Sivapalaratnam, Kenneth Smith, Aman Sohal, Laura Southgate, Simon Staines, Emily Staples, Hans Stauss, Penelope Stein, Jonathan Stephens, Kathleen Stirrups, Sophie Stock, Jay Suntharalingam, R. Campbell Tait, Kate Talks, Yvonne Tan, Jecko Thachil, James Thaventhiran, Ellen Thomas, Moira Thomas, Dorothy Thompson, Adrian Thrasher, Marc Tischkowitz, Catherine Titterton, Cheng-Hock Toh, Mark Toshner, Carmen Treacy, Richard Trembath, Salih Tuna, Wojciech Turek, Ernest Turro, Chris Van Geet, Marijke Veltman, Julie Vogt, Julie von Ziegenweldt, Anton Vonk Noordegraaf, Emma Wakeling, Ivy Wanjiku, Timothy Q. Warner, Evangeline Wassmer, Hugh Watkins, Andrew Webster, Steve Welch, Sarah Westbury, John Wharton, Deborah Whitehorn, Martin Wilkins, Lisa Willcocks, Catherine Williamson, Geoffrey Woods, John Wort, Nigel Yeatman, Patrick Yong, Tim Young, and Ping Yu.
Acknowledgements
This project was supported by The National Institute for Health Research (NIHR) and Biomedical Research Centre (BRC) at Moorfields Eye Hospital and the UCL Institute of Ophthalmology, RP Fighting Blindness, Fight for Sight UK, Moorfields Eye Hospital Special Trustees, and Foundation Fighting Blindness - USA. K.C. was supported by grant funding from the NIHR Bioresource Rare Disease. The UKIRDC was funded by RP Fighting Blindness and Fight for Sight UK. M.E.C. was supported by The Wellcome Trust and F.L.R. was supported by Cambridge Biomedical Research Centre.
Published: January 26, 2017
Footnotes
Supplemental Data include two figures and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.12.014.
Contributor Information
Andrew R. Webster, Email: andrew.webster@ucl.ac.uk.
UK Inherited Retinal Disease Consortium:
Graeme Black, Georgina Hall, Stuart Ingram, Rachel Gillespie, Forbes Manson, Panagiotis Sergouniotis, Chris Inglehearn, Carmel Toomes, Manir Ali, Martin McKibbin, James Poulter, Kamron Khan, Emma Lord, Andrea Nemeth, Susan Downes, Stephanie Halford, Jing Yu, Stefano Lise, Gavin Arno, Alessia Fiorentino, Nikos Ponitkos, Vincent Plagnol, Michel Michaelides, Alison J. Hardcastle, Michael E. Cheetham, Andrew R. Webster, and Veronica van Heyningen
Web Resources
Clustal Omega, http://www.ebi.ac.uk/Tools/msa/clustalo/
ExAC Browser, http://exac.broadinstitute.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
RetNet – Retinal Information Network, https://sph.uth.edu/retnet/home.htm
UniGene, http://www.ncbi.nlm.nih.gov/unigene
Supplemental Data
References
- 1.Niu J., Profirovic J., Pan H., Vaiskunaite R., Voyno-Yasenetskaya T. G Protein betagamma subunits stimulate p114RhoGEF, a guanine nucleotide exchange factor for RhoA and Rac1: regulation of cell shape and reactive oxygen species production. Circ. Res. 2003;93:848–856. doi: 10.1161/01.RES.0000097607.14733.0C. [DOI] [PubMed] [Google Scholar]
- 2.Terry S.J., Zihni C., Elbediwy A., Vitiello E., Leefa Chong San I.V., Balda M.S., Matter K. Spatially restricted activation of RhoA signalling at epithelial junctions by p114RhoGEF drives junction formation and morphogenesis. Nat. Cell Biol. 2011;13:159–166. doi: 10.1038/ncb2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arno G., Hull S., Carss K., Dev-Borman A., Chakarova C., Bujakowska K., van den Born I., Robson A.G., Holder G.E., Michaelides M. Reevaluation of the retinal dystrophy due to recessive alleles of RGR with the discovery of a cis-acting mutation in CDHR1. Invest. Ophthalmol. Vis. Sci. 2016;57:4806–4813. doi: 10.1167/iovs.16-19687. [DOI] [PubMed] [Google Scholar]
- 4.Henderson R.H., Mackay D.S., Li Z., Moradi P., Sergouniotis P., Russell-Eggitt I., Thompson D.A., Robson A.G., Holder G.E., Webster A.R., Moore A.T. Phenotypic variability in patients with retinal dystrophies due to mutations in CRB1. Br. J. Ophthalmol. 2011;95:811–817. doi: 10.1136/bjo.2010.186882. [DOI] [PubMed] [Google Scholar]
- 5.Arno G., Hull S., Robson A.G., Holder G.E., Cheetham M.E., Webster A.R., Plagnol V., Moore A.T. Lack of interphotoreceptor retinoid binding protein, caused by homozygous mutation of RBP3, is associated with high myopia and retinal dystrophy. Invest. Ophthalmol. Vis. Sci. 2015;56:2358–2365. doi: 10.1167/iovs.15-16520. [DOI] [PubMed] [Google Scholar]
- 6.Carss K.J., Arno G., Erwood M., Stephens J., Sanchis-Juan A., Hull S., Megy K., Grozeva D., Dewhurst E., Malka S. Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am. J. Hum. Genet. 2017;100:75–90. doi: 10.1016/j.ajhg.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 8.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spaide R.F., Curcio C.A. Anatomical correlates to the bands seen in the outer retina by optical coherence tomography: literature review and model. Retina. 2011;31:1609–1619. doi: 10.1097/IAE.0b013e3182247535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCulloch D.L., Marmor M.F., Brigell M.G., Hamilton R., Holder G.E., Tzekov R., Bach M. ISCEV Standard for full-field clinical electroretinography (2015 update) Doc. Ophthalmol. 2015;130:1–12. doi: 10.1007/s10633-014-9473-7. [DOI] [PubMed] [Google Scholar]
- 13.Bach M., Brigell M.G., Hawlina M., Holder G.E., Johnson M.A., McCulloch D.L., Meigen T., Viswanathan S. ISCEV standard for clinical pattern electroretinography (PERG): 2012 update. Doc. Ophthalmol. 2013;126:1–7. doi: 10.1007/s10633-012-9353-y. [DOI] [PubMed] [Google Scholar]
- 14.Loosli F. ArhGEF18 regulated Rho signaling in vertebrate retina development. Small GTPases. 2013;4:242–246. doi: 10.4161/sgtp.27061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mack N.A., Georgiou M. The interdependence of the Rho GTPases and apicobasal cell polarity. Small GTPases. 2014;5:10. doi: 10.4161/21541248.2014.973768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norden C., Young S., Link B.A., Harris W.A. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 2009;138:1195–1208. doi: 10.1016/j.cell.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Royer C., Lu X. Epithelial cell polarity: a major gatekeeper against cancer? Cell Death Differ. 2011;18:1470–1477. doi: 10.1038/cdd.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zihni C., Mills C., Matter K., Balda M.S. Tight junctions: from simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 2016;17:564–580. doi: 10.1038/nrm.2016.80. [DOI] [PubMed] [Google Scholar]
- 19.Terry S.J., Elbediwy A., Zihni C., Harris A.R., Bailly M., Charras G.T., Balda M.S., Matter K. Stimulation of cortical myosin phosphorylation by p114RhoGEF drives cell migration and tumor cell invasion. PLoS ONE. 2012;7:e50188. doi: 10.1371/journal.pone.0050188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blomquist A., Schwörer G., Schablowski H., Psoma A., Lehnen M., Jakobs K.H., Rümenapp U. Identification and characterization of a novel Rho-specific guanine nucleotide exchange factor. Biochem. J. 2000;352:319–325. [PMC free article] [PubMed] [Google Scholar]
- 21.van de Pavert S.A., Sanz A.S., Aartsen W.M., Vos R.M., Versteeg I., Beck S.C., Klooster J., Seeliger M.W., Wijnholds J. Crb1 is a determinant of retinal apical Müller glia cell features. Glia. 2007;55:1486–1497. doi: 10.1002/glia.20561. [DOI] [PubMed] [Google Scholar]
- 22.Mehalow A.K., Kameya S., Smith R.S., Hawes N.L., Denegre J.M., Young J.A., Bechtold L., Haider N.B., Tepass U., Heckenlively J.R. CRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum. Mol. Genet. 2003;12:2179–2189. doi: 10.1093/hmg/ddg232. [DOI] [PubMed] [Google Scholar]
- 23.Duguépéroux I., De Braekeleer M. The CFTR 3849+10kbC->T and 2789+5G->A alleles are associated with a mild CF phenotype. Eur. Respir. J. 2005;25:468–473. doi: 10.1183/09031936.05.10100004. [DOI] [PubMed] [Google Scholar]
- 24.Desmet F.-O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsapara A., Luthert P., Greenwood J., Hill C.S., Matter K., Balda M.S. The RhoA activator GEF-H1/Lfc is a transforming growth factor-beta target gene and effector that regulates alpha-smooth muscle actin expression and cell migration. Mol. Biol. Cell. 2010;21:860–870. doi: 10.1091/mbc.E09-07-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aghazadeh B., Zhu K., Kubiseski T.J., Liu G.A., Pawson T., Zheng Y., Rosen M.K. Structure and mutagenesis of the Dbl homology domain. Nat. Struct. Biol. 1998;5:1098–1107. doi: 10.1038/4209. [DOI] [PubMed] [Google Scholar]
- 27.Steven R., Kubiseski T.J., Zheng H., Kulkarni S., Mancillas J., Ruiz Morales A., Hogue C.W., Pawson T., Culotti J. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–795. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- 28.Xu X., Jin D., Durgan J., Hall A. LKB1 controls human bronchial epithelial morphogenesis through p114RhoGEF-dependent RhoA activation. Mol. Cell. Biol. 2013;33:2671–2682. doi: 10.1128/MCB.00154-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herder C., Swiercz J.M., Müller C., Peravali R., Quiring R., Offermanns S., Wittbrodt J., Loosli F. ArhGEF18 regulates RhoA-Rock2 signaling to maintain neuro-epithelial apico-basal polarity and proliferation. Development. 2013;140:2787–2797. doi: 10.1242/dev.096487. [DOI] [PubMed] [Google Scholar]
- 30.Etienne-Manneville S. Polarity proteins in migration and invasion. Oncogene. 2008;27:6970–6980. doi: 10.1038/onc.2008.347. [DOI] [PubMed] [Google Scholar]
- 31.Martin-Belmonte F., Perez-Moreno M. Epithelial cell polarity, stem cells and cancer. Nat. Rev. Cancer. 2011;12:23–38. doi: 10.1038/nrc3169. [DOI] [PubMed] [Google Scholar]
- 32.Assémat E., Bazellières E., Pallesi-Pocachard E., Le Bivic A., Massey-Harroche D. Polarity complex proteins. Biochim. Biophys. Acta. 2008;1778:614–630. doi: 10.1016/j.bbamem.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 33.den Hollander A.I., ten Brink J.B., de Kok Y.J., van Soest S., van den Born L.I., van Driel M.A., van de Pol D.J., Payne A.M., Bhattacharya S.S., Kellner U. Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12) Nat. Genet. 1999;23:217–221. doi: 10.1038/13848. [DOI] [PubMed] [Google Scholar]
- 34.Lotery A.J., Jacobson S.G., Fishman G.A., Weleber R.G., Fulton A.B., Namperumalsamy P., Héon E., Levin A.V., Grover S., Rosenow J.R. Mutations in the CRB1 gene cause Leber congenital amaurosis. Arch. Ophthalmol. 2001;119:415–420. doi: 10.1001/archopht.119.3.415. [DOI] [PubMed] [Google Scholar]
- 35.Tsang S.H., Burke T., Oll M., Yzer S., Lee W., Xie Y.A., Allikmets R. Whole exome sequencing identifies CRB1 defect in an unusual maculopathy phenotype. Ophthalmology. 2014;121:1773–1782. doi: 10.1016/j.ophtha.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent A., Ng J., Gerth-Kahlert C., Tavares E., Maynes J.T., Wright T., Tiwari A., Tumber A., Li S., Hanson J.V.M. Biallelic mutations in CRB1 underlie autosomal recessive familial foveal retinoschisis. Invest. Ophthalmol. Vis. Sci. 2016;57:2637–2646. doi: 10.1167/iovs.15-18281. [DOI] [PubMed] [Google Scholar]
- 37.Jacobson S.G., Cideciyan A.V., Aleman T.S., Pianta M.J., Sumaroka A., Schwartz S.B., Smilko E.E., Milam A.H., Sheffield V.C., Stone E.M. Crumbs homolog 1 (CRB1) mutations result in a thick human retina with abnormal lamination. Hum. Mol. Genet. 2003;12:1073–1078. doi: 10.1093/hmg/ddg117. [DOI] [PubMed] [Google Scholar]
- 38.Aleman T.S., Cideciyan A.V., Aguirre G.K., Huang W.C., Mullins C.L., Roman A.J., Sumaroka A., Olivares M.B., Tsai F.F., Schwartz S.B. Human CRB1-associated retinal degeneration: comparison with the rd8 Crb1-mutant mouse model. Invest. Ophthalmol. Vis. Sci. 2011;52:6898–6910. doi: 10.1167/iovs.11-7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van de Pavert S.A., Kantardzhieva A., Malysheva A., Meuleman J., Versteeg I., Levelt C., Klooster J., Geiger S., Seeliger M.W., Rashbass P. Crumbs homologue 1 is required for maintenance of photoreceptor cell polarization and adhesion during light exposure. J. Cell Sci. 2004;117:4169–4177. doi: 10.1242/jcs.01301. [DOI] [PubMed] [Google Scholar]
- 40.van de Pavert S.A., Meuleman J., Malysheva A., Aartsen W.M., Versteeg I., Tonagel F., Kamphuis W., McCabe C.J., Seeliger M.W., Wijnholds J. A single amino acid substitution (Cys249Trp) in Crb1 causes retinal degeneration and deregulates expression of pituitary tumor transforming gene Pttg1. J. Neurosci. 2007;27:564–573. doi: 10.1523/JNEUROSCI.3496-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei X., Malicki J. nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Nat. Genet. 2002;31:150–157. doi: 10.1038/ng883. [DOI] [PubMed] [Google Scholar]
- 42.Jensen A.M., Walker C., Westerfield M. mosaic eyes: a zebrafish gene required in pigmented epithelium for apical localization of retinal cell division and lamination. Development. 2001;128:95–105. doi: 10.1242/dev.128.1.95. [DOI] [PubMed] [Google Scholar]
- 43.Hsu Y.-C., Willoughby J.J., Christensen A.K., Jensen A.M. Mosaic Eyes is a novel component of the Crumbs complex and negatively regulates photoreceptor apical size. Development. 2006;133:4849–4859. doi: 10.1242/dev.02685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Omori Y., Malicki J. oko meduzy and related crumbs genes are determinants of apical cell features in the vertebrate embryo. Curr. Biol. 2006;16:945–957. doi: 10.1016/j.cub.2006.03.058. [DOI] [PubMed] [Google Scholar]
- 45.Malicki J., Driever W. oko meduzy mutations affect neuronal patterning in the zebrafish retina and reveal cell-cell interactions of the retinal neuroepithelial sheet. Development. 1999;126:1235–1246. doi: 10.1242/dev.126.6.1235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.