Abstract
Genetically determined cell membrane transporters and metabolic enzymes play a crucial role in the transportation of a wide variety of substrates that maintain homeostasis in biological processes. We explored associations between genetic variants in these genes and survival of non-small cell lung cancer (NSCLC) patients by re-analyzing a selected dataset from published genome-wide association studies (GWASs). In the discovery by using the GWAS dataset of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial, we evaluated associations of 1,245 single nucleotide polymorphisms (SNPs) in genes of four transporter families and two metabolic enzyme families with survival of 1,185 NSCLC patients. We then performed a replication analysis in the Harvard University Lung Cancer study (LCS) with 984 NSCLC patients. Multivariate Cox proportional hazards regression and false-discovery rate (FDR) corrections were performed to evaluate the associations. We identified that 21 genotyped SNPs in eight gene regions were significantly associated with survival with FDR ≤ 0.1 in the discovery dataset. Subsequently, we confirmed six SNPs, which were putative functional, in ABCG1 of the ATP-binding cassette transporter family in the replication dataset. In the pooled analysis, two tagging (at r2>0.8 for LD with other replicated SNPs)/functional SNPs were independently associated with survival: rs225388 G>A (adjusted hazards ratio = 1.12, 95% confidence interval = 1.03–1.20, Ptrend = 4.6×10−3) and rs225390 A>G (adjusted hazards ratio = 1.16, 95% confidence interval = 1.07–1.25, Ptrend = 3.8×10−4). Our results indicated that genetic variants of ABCG1 may be predictors of survival of NSCLC patients.
Keywords: lung cancer, drug transporters, overall survival, single nucleotide polymorphism, ABCG1
INTRODUCTION
Globally, lung cancer ranks the most frequent cause of cancer related mortality, with over a million deaths each year.1, 2 Non-small cell lung cancer (NSCLC) is its most common histological type.3 Based on the data from Surveillance, Epidemiology, and End Results (SEER) program, the 5-year survival rate of lung cancer patients was only 16.8% on average between 2004 and 2010.4 Most lung cancer patients remain asymptomatic until they present with advanced disease that is incurable with a 5-year survival rate as low as 4%.4, 5
Outcomes of lung cancer have been improved through screening, more effective systemic therapies, biomarker-defined subpopulations, and multi-modality care.3 Platinum-based chemotherapy remains the mainstay of treatment in either neo-adjuvant and adjuvant therapies for localized disease or systematic therapy for metastatic disease.3, 5 Clinically, body surface area is widely used for drug dose calculation based on the assumption that drug metabolism is the same among different individuals.5, 6 Commonly used clinicopathological variables responsible for prognosis include age, sex, ethnicity, performance status (PS), tumor stage, among others. However, these factors remain insufficient in interpreting the variability in treatment response and clinical outcomes among patients.3 It is estimated that genetic factors account for 20–95% of the variability in drug disposition and pharmacokinetic effects.6 Genetic factors are inherited determinants that could remain unchanged over each individual’s lifetime.6, 7 There is a considerable body of evidence that indicates that single nucleotide polymorphisms (SNPs) could influence short-term response and long-term prognosis in cancer treatment.8, 9 Identifying the role of these genetic factors in carcinogenesis can lead to a better understanding of lung cancer prognosis in humans.
Cell transmembrane transporters play an essential role in regulating substrate disposition, including absorption, distribution and excretion.6 The ATP-binding cassette (ABC) transporters are the largest family of transmembrane proteins, with seven subfamilies that have a predominant role in transporting substrates across the membrane.10 ABC transporters, such as ABCB1, ABCC1, and ABCG2, are well known for their capacity to efflux chemotherapy agents, which are involved in a reduction of intracellular drug concentrations and sensitivity.11, 12 Meanwhile, frequent overexpression of ABC transporter genes reflects their prominent roles in tumor biology.10 Besides ABC transporters, recent reports indicate that copper transporters are also crucial for biological processes.13 Copper is an important cofactor for enzymes in a number of key metabolic activities, such as cytochrome C oxidase in mitochondrion for respiration.13 Both deficiency and excess of copper have a deleterious effect on cellular metabolism.14 Members of the copper transporter family function as uptake transporters, efflux transporters, and copper chaperones, and together they control copper homeostasis in a precise network.14 Recently, copper transporter 1 (CTR1) was proved to disrupt the BRAFV600E signaling and lead to tumorigenesis.15 Solute carriers (SLCs) were noted to transfer diverse compounds with different sizes and structure in intestine, liver and kidney.16, 17 The SLC22 family (organic cation transporters, OCTs) and the SLC47 family (multidrug and toxin extrusion types of transporters, MATEs) mediate organic ion transport and participate in drug transport in cancer treatment.18 In addition, metabolic enzymes, such as metallothionein and glutathione S-transferase, work closely with the previously mentioned transporters in the metabolic mechanism.7 The associations between genetic variants in GST genes and survival have been reported for several malignant diseases.7
In the current study, we hypothesized that the variability in survival of NSCLC patients can be explained by polymorphisms of cell membrane transport-related genes. To this end, we evaluated the association between prognosis in NSCLC patients and SNPs in genes of four transporter families and two metabolic enzyme families.
MATERIALS AND METHODS
Study populations
The discovery phase included 1,185 NSCLC patients obtained from the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial, after application and access approval from National Cancer Institute (NCI). The PLCO is an NCI funded multicenter randomized trial of screening for cancer from ten medical centers in United States between 1993 and 2011.19 The screening trial enrolled 77,500 men and 77,500 women aged 55 to 74. All individuals were randomized to either the intervention arm with screening or the control arm with standard care. The PLCO trial collected blood specimens from the first screening visit and gathered extensive information about each individual, including smoking history, family history of cancer, and demographic information.20 All participants were followed up for at least 13 years after enrollment.20 Genomic DNA extracted from the blood samples was genotyped with Illumina HumanHap240Sv1.0 and HumanHap550v3.0 (dbGaP accession: phs000093.v2.p2 and phs000336.v1.p1).21, 22 In 1,187 Caucasian NSCLC patients from the PLCO, two with the missing follow-up information were excluded. Therefore, the eligible subsets of the PLCO lung cancer dataset for survival analysis included 1,185 NSCLC patients whose clinicopathological variables and genotype data were available. Tumor staging was determined according to the 5th edition American Joint Committee on Cancer (AJCC) staging system. The follow-up time was defined from lung cancer diagnosis to the last follow-up or time of death. Overall survival (OS) was the primary endpoint of the current study, and disease-specific survival (DSS) of lung cancer was also provided. The institutional review boards of each participating institution approved the PLCO trial and the use of biospecimen for further research, and all subjects signed a written informed consent permitting the research represented here.
The validation phase used the GWAS dataset from the Harvard Lung Cancer Susceptibility Study with 984 histology-confirmed Caucasian NSCLC patients. The histological classification of the tumors was done by two staff pulmonary pathologists at the Massachusetts General Hospital. The time of blood collection was within 1–4 weeks of the diagnosis for each patient. DNA was extracted from blood samples by using the Auto Pure Large Sample Nucleic Acid Purification System (QIAGEN Company, Venlo, Limburg, Netherlands). Genotyped data was obtained by using Illumina Humanhap610-Quad arrays, and imputation was performed by using MaCH1.0 based on 1000 Genomes project. Details of the participants in the Harvard study were described previously.23
Gene and SNP selection
In brief, 120 genes from four transporter families and two metabolic enzyme families were selected as candidate genes, including ATP-binding cassette (ABC) transporters, copper transporters, the SLC22 family (organic cation transporters, OCTs), the SLC47 family (multidrug and toxin extrusion types of transporters, MATEs), glutathione S-transferase (GST) and metallothioneins (MT), according to literature reviews10, 14, 16, 17, 24 and online datasets (Reactome, http://www.reactome.org/ and UniProt http://www.uniprot.org/) (Supporting Information Table 1). Genotyped SNPs within these genes and their ± 2 kb flanking regions were selected for association analysis. There were 103 genes from these six families with 1,402 SNPs genotyped in PLCO. SNPs were selected by using the following criteria: SNPs located on autosomal chromosomes; minor allelic frequency (MAF) ≥ 5%; genotyping rate ≥ 95% and Hardy-Weinberg equilibrium (HWE) ≥ 1×10−6. As a result, 1,245 genotyped SNPs from 100 genes were extracted from the PLCO genotyping data (dbGaP accession: phs000093.v2.p2 and phs000336.v1.p1).21, 22, 25 Twenty-one genotyped SNPs in eight genes showed associations with NSCLC overall survival and also passed multiple testing corrections by the false discovery rate (FDR) method. These eight gene regions were further imputed, for which we filtered the imputed SNPs with the criteria of MAF ≥ 5%, genotyping rate ≥ 95% and HWE ≥ 1×10−6. In the end, 2,327 qualified imputed SNPs were identified and used to test their associations with survival of NSCLC patients. We also combined two approaches to choose the imputed SNPs. First, SNPs passed the threshold of P-value 0.001 were chosen for further analysis. Second, potentially functional SNPs, predicted by SNPinfo and RegulomeDB, with a P-value less than 0.05 were also retained.26, 27 SNPinfo incorporates functional predictions of protein structure, gene regulation, splicing, and microRNA binding.26 We used RegulomeDB to identify the SNPs with previously reported links to expression quantitative trait loci (eQTL).27
Statistical analysis
Cox proportional hazards regression models were used to estimate the hazards ratio (HR) and 95% confidence interval (CI) for the associations of demographic and clinical characteristics with OS. Associations between SNPs and OS (in additive models) were obtained by both univariate and multivariable Cox regression analyses performed with GenABEL package of R software with adjustment for age, sex, smoking status, histology, tumor stage, chemotherapy, radiotherapy and surgery.28 For multiple testing corrections, the FDR approach was used with a cut-off value of 0.1 to lower the probability of false positive findings.29 Imputation was performed with IMPUTE2 according to 1000 Genomes CEU data (phase 1 release V3). SNPs with info value ≥ 0.8 were used for further analysis. Inverse variance weighted meta-analysis was performed to combine the results of discovery and validation studies. Cochran’s Q statistics and I2 were carried out to access an inter-study heterogeneity. Fixed effect models were used when no heterogeneity was found between two studies (Q > 0.10 and I2 < 25.0%); otherwise, random effects models were used. The meta-analysis of the two studies was performed by PLINK 1.07.
Pairwise linkage disequilibrium (LD) was estimated by using the data from 1000 Genomes Project of 373 European individuals. The number of risk genotypes was summarized to evaluate the combined effects of all the tagging SNPs. Kaplan-Meier curve and log-rank test were used to evaluate the effects of risk genotypes on the cumulative probability of OS. The heterogeneity of associations between subgroups in stratified analyses was assessed by using the Chi-square-based Q-test.
In analyzing associations between SNPs and corresponding gene expression, we performed linear regression analysis by using the R software. Gene expression levels were obtained from Geuvadis RNA sequencing project of 1000 Genomes samples in 373 European descendants [91 Northern Europeans from Utah (CEU), 95 Finnish in Finland (FIN), 94 Great Britain (GBR) and 95 Toscani in Italia (TSI)].30 Methylation quantitative trait loci (meQTL) associations were assessed by Genevar in the Multiple Tissue Human Expression Resource (MuTHER) project.31 Differences of ABCG1 mRNA expression between paired tumor tissues and adjacent normal tissues were examined by t-test in the Cancer Genome Atlas (TCGA) lung cancer data (http://cancergenome.nih.gov/) (RNASeqV2.Level_3.1.8.0).32, 33 There were 107 lung cancer cases with paired tumor tissues and adjacent normal tissues, including 50 cases of squamous cell carcinoma and 57 cases of adenocarcinoma. Associations of ABCG1 expression levels and lung cancer overall survival were accessed by Cox-regression analysis and log-rank test in the TCGA dataset, and all 625 lung cancer patients with follow-up information were of European descents. All statistical analyses were carried out by SAS software (version 9.1.3; SAS Institute, Cary, NC, USA), unless otherwise specified.
RESULTS
Basic characteristics of study populations
The overall workflow is shown in Supporting Information Fig. 1. Basic characteristics of 1,185 NSCLC patients from the PLCO are described in Table 1. The median age of the patients was 71 years, and the median survival time of all patients was 23.77 months. Of these 1,185 patients, 798 (67.3%) died at the last follow-up (Table 1). In multivariate analyses, seven of the nine selected variables were found to be significantly associated with NSCLC OS. These variables were age at diagnosis (HR=1.26, >71 vs. ≤71), sex (HR=0.80, female vs. male), smoking status (HR=1.79, current vs. never; HR=1.69, former vs. never), histology (HR=1.33, other vs. adenocarcinoma), stage (HR=2.70, IIIB–IV vs. I–IIIA), chemotherapy (HR=0.42, Yes vs. No), surgery (HR=0.26, Yes vs. No). Additionally, we also evaluated associations of all these variables with DSS (Supporting Information Table 2). The median DSS is 27.43 months. Similar to the analyses of OS, seven variables significantly associated with NSCLC DSS in multivariate analyses : age at diagnosis (HR=1.20, >71 vs. ≤71), sex (HR=0.85, female vs. male), smoking status (HR=1.68, current vs. never; HR=1.60, former vs. never), histology (HR=1.25, other vs. adenocarcinoma), stage (HR=2.96, IIIB–IV vs. I–IIIA), chemotherapy (HR=0.41, Yes vs. No), surgery (HR=0.27, Yes vs. No). We compared the demographics and clinical characteristics between the PLCO study and the Harvard study, including age, sex, smoking status, and histology and stage of lung cancer (Supporting Information Table 3). Both studies included a Caucasian population, but the two study populations had some differences in the distribution of age, sex, tumor histology and stage. However, all these factors were adjusted for in the multivariate Cox models for survival analyses.
Table 1.
Associations of demographics and clinical characteristics with overall survival in NSCLC patients from the PLCO study.
| Characteristics | No. | Median Survival Timea |
Univariate Analysis | Multivariate Analysisb | ||||
|---|---|---|---|---|---|---|---|---|
| Patients | Deaths (%) | HR (95%) | P | HR (95%) | P | |||
| Total | 1185 | 798 (67.3) | 23.77 | |||||
| Agec | ≤71 | 636 | 400 (62.9) | 36.53 | 1.00 | 1.00 | ||
| >71 | 549 | 398 (72.5) | 14.60 | 1.74 (1.51–2.00) | <0.0001 | 1.26 (1.09–1.46) | 0.002 | |
| Sex | Male | 698 | 507 (72.6) | 19.33 | 1.00 | 1.00 | ||
| Female | 487 | 291 (59.8) | 31.77 | 0.73 (0.63–0.84) | <0.0001 | 0.80 (0.69–0.94) | 0.005 | |
| Smoking status |
Never | 115 | 63 (54.8) | 43.50 | 1.00 | 1.00 | ||
| Current | 423 | 272 (64.3) | 23.70 | 1.34 (1.02–1.76) | 0.038 | 1.79 (1.32–2.44) | 0.0002 | |
| Former | 647 | 463 (71.6) | 21.30 | 1.51 (1.16–1.96) | 0.002 | 1.69 (1.26–2.27) | 0.0005 | |
| Pack years | ≤49 | 603 | 399 (66.2) | 23.77 | 1.00 | 1.00 | ||
| >49 | 581 | 398 (68.5) | 23.97 | 1.05 (0.92–1.21) | 0.458 | 1.01 (0.87–1.17) | 0.936 | |
| Missing | 1 | |||||||
| Histology | Adenocarcinoma | 577 | 348 (60.3) | 36.37 | 1.00 | 1.00 | ||
| Squamous cell carcinoma | 285 | 192 (67.4) | 29.83 | 1.19 (1.00–1.42) | 0.057 | 1.14 (0.94–1.37) | 0.180 | |
| Others | 323 | 258 (79.9) | 10.87 | 1.89 (1.60–2.22) | <0.0001 | 1.33 (1.12–1.57) | 0.003 | |
| Stage | I – IIIA | 655 | 315 (48.1) | 80.13 | 1.00 | 1.00 | ||
| IIIB – IV | 528 | 482 (91.3) | 6.87 | 5.08 (4.37–5.91) | <0.0001 | 2.70 (2.23–3.27) | <0.0001 | |
| Missing | 2 | |||||||
| Chemotherapy | No | 639 | 367 (57.4) | 53.03 | 1.00 | 1.00 | ||
| Yes | 538 | 423 (78.6) | 14.83 | 1.85 (1.60–2.14) | <0.0001 | 0.42 (0.35–0.50) | <0.0001 | |
| Missing | 8 | |||||||
| Radiotherapy | No | 762 | 450 (59.1) | 40.50 | 1.00 | 1.00 | ||
| Yes | 415 | 340 (81.9) | 12.33 | 1.94 (1.68–2.24) | <0.0001 | 0.90 (0.77–1.05) | 0.160 | |
| Missing | 8 | |||||||
| Surgery | No | 637 | 566 (88.9) | 8.27 | 1.00 | 1.00 | ||
| Yes | 540 | 224 (41.5) | 103.33 | 0.15 (0.13–0.18) | <0.0001 | 0.26 (0.21–0.33) | <0.0001 | |
| Missing | 8 | |||||||
Median survival time for overall survival;
Multivariate cox regression analyses were adjusted for all factors listed in this table;
The median age is 71 years. The median pack year is 49.
Multivariate analyses of associations between SNPs and NSCLC OS in the PLCO study
Multivariate Cox models were used to assess the associations of 1,245 SNPs with OS in the presence of age, sex, smoking status, histology, tumor stage, chemotherapy, radiotherapy and surgery (Supporting Information Fig. 2, as summarized in the Manhattan plot). Of these 1,245 SNPs, 104 SNPs were individually significantly associated with OS at P < 0.05 in an additive genetic model. After multiple test adjustment, 21 SNPs in eight genes (ABCA2, ABCA4, ABCA12, ABCC1, ABCC4, ABCC6, ABCG1 and SLC22A5) remained significant with FDR ≤ 0.1 (Table 2).
Table 2.
Associations between the 21 genotyped SNPs and overall survival of NSCLC patients.
| SNP | Chr. | Position (hg19) | Gene | Location | Allelea | EAFb | Overall Survival | ||
|---|---|---|---|---|---|---|---|---|---|
| HR (95%CI)C | PC | FDRC | |||||||
| rs1931575 | 1 | 94533014 | ABCA4 | Intron | A/G | 0.25 | 1.23 (1.09–1.38) | 8.44E-04 | 0.07 |
| rs2970968 | 2 | 216001990 | ABCA12 | Intron | G/A | 0.21 | 1.21 (1.07–1.36) | 1.75E-03 | 0.10 |
| rs274554 | 5 | 131724950 | SLC22A5 | Intron | G/A | 0.17 | 0.77 (0.67–0.88) | 1.65E-04 | 0.04 |
| rs274550 | 5 | 131728712 | SLC22A5 | Intron | A/C | 0.18 | 0.77 (0.68–0.88) | 1.81E-04 | 0.04 |
| rs7705826 | 5 | 131732456 | SLC22A5 | 3' downstream | G/T | 0.18 | 0.77 (0.68–0.88) | 1.81E-04 | 0.04 |
| rs7048567 | 9 | 139904037 | ABCA2 | Exon | G/A | 0.31 | 1.24 (1.11–1.38) | 8.54E-05 | 0.04 |
| rs4773843 | 13 | 95839495 | ABCC4 | Intron | C/T | 0.17 | 1.25 (1.10–1.43) | 8.42E-04 | 0.07 |
| rs4148486 | 13 | 95843150 | ABCC4 | Intron | T/C | 0.41 | 0.84 (0.76–0.93) | 7.48E-04 | 0.07 |
| rs1678384 | 13 | 95848393 | ABCC4 | Intron | A/G | 0.50 | 0.83 (0.76–0.92) | 4.03E-04 | 0.06 |
| rs2274410 | 13 | 95860288 | ABCC4 | Intron | A/C | 0.35 | 0.84 (0.76–0.93) | 1.08E-03 | 0.08 |
| rs7331142 | 13 | 95860574 | ABCC4 | Intron | T/C | 0.35 | 0.84 (0.76–0.93) | 7.60E-04 | 0.07 |
| rs3782964 | 13 | 95863465 | ABCC4 | Intron | C/T | 0.16 | 1.28 (1.12–1.47) | 2.60E-04 | 0.05 |
| rs7330519 | 13 | 95875418 | ABCC4 | Intron | T/C | 0.30 | 1.22 (1.09–1.36) | 3.90E-04 | 0.06 |
| rs4780585 | 16 | 16095445 | ABCC1 | Intron | G/A | 0.42 | 1.18 (1.07–1.31) | 1.56E-03 | 0.10 |
| rs246221 | 16 | 16138322 | ABCC1 | Exon | T/C | 0.30 | 1.20 (1.08–1.34) | 8.27E-04 | 0.07 |
| rs35596 | 16 | 16152940 | ABCC1 | Intron | T/C | 0.23 | 1.22 (1.08–1.37) | 1.05E-03 | 0.08 |
| rs2299670 | 16 | 16220858 | ABCC1 | Intron | A/G | 0.34 | 0.84 (0.76–0.93) | 1.21E-03 | 0.08 |
| rs3213473 | 16 | 16256767 | ABCC6 | Intron | G/T | 0.17 | 0.79 (0.70–0.91) | 6.64E-04 | 0.07 |
| rs6498606 | 16 | 16260342 | ABCC6 | Intron | T/C | 0.19 | 0.79 (0.69–0.89) | 1.82E-04 | 0.04 |
| rs8057824 | 16 | 16260443 | ABCC6 | Intron | G/A | 0.19 | 0.78 (0.69–0.89) | 1.32E-04 | 0.04 |
| rs225406 | 21 | 43699230 | ABCG1 | Intron | C/T | 0.10 | 1.31 (1.11–1.54) | 1.39E-03 | 0.09 |
HR: Hazard ratio. FDR: False discovery rate.
Reference/effect allele;
Effect allele frequency;
Multivariate cox regression analyses were adjusted for age, sex, smoking status, histology, tumor stage, chemotherapy, radiotherapy and surgery.
After imputation and quality controls for the SNP inclusion as described earlier, 2,327 imputed SNPs in these eight genes remained (Supporting Information Table 4), of which 64 SNPs with P < 0.001 or 22 SNPs with P < 0.05 and potential functions predicted by SNPinfo and eQTL annotation of RegulomeDB were selected for further analysis. After removal of five duplicated SNPs, 81 SNPs (69 in the ABC transporter family and 12 in SLC22 transporter family) were found to be associated with OS in the PLCO study (Supporting Information Table 5). Similarly, 80 SNPs of these 81 SNPs remained significantly associated with DSS of lung cancer, except for one SNP with marginal significance (Supporting Information Table 5).
Validation analysis with Harvard dataset and meta-analysis of two studies
To substantiate the findings from the PLCO dataset, we re-analyzed the 81 SNPs in an independent patient population from the Harvard study. Of these 81 SNPs, six putative functional SNPs in the intronic region of ABCG1 were found to be significantly associated with NSCLC OS in the replication dataset (Table 3). In the subsequent meta-analysis of these two studies (Supporting Information Table 6–7), poorer overall survival of NSCLC was associated with rs225388 A allele, rs74757 A allele, rs225390 G allele, rs225396 T allele, rs170438 T allele and rs170439 T allele. In further LD analysis of these six replicated SNPs, except for rs225390, other five SNPs were in high LD with each other (all r2 > 0.8) (Supporting Information Fig. 3–4). Compared to the other four LD SNPs, rs225388 showed a higher level of histone H3 lysine 4 monomethylation (H3K4me1) enrichment relevant to the ENCODE project on UCSC gene regulation information (http://genome.ucsc.edu/).Therefore, these two tagging SNPs, rs225388 and rs225390, were chosen from the replicated SNPs for additional analyses, including assessments of their combined effect and potentially functional relevance.
Table 3.
Meta-analyses of the six replicated SNPs in ABCG1 from the two studies.
| SNP | Chr. | Position | Gene | Allele a | PLCO study b | Harvard study c | Meta-analysis d | |||
|---|---|---|---|---|---|---|---|---|---|---|
| HR (95%CI) | P | HR (95%CI) | P | HR (95%CI) | P | |||||
| rs225388 | 21 | 43684456 | ABCG1 | G/A | 1.11 (1.00–1.23) | 4.28E-02 | 1.13 (1.00–1.26) | 4.66E-02 | 1.12 (1.03–1.20) | 4.60E-03 |
| rs74757 | 21 | 43684549 | ABCG1 | G/A | 1.11 (1.01–1.23) | 3.89E-02 | 1.13 (1.00–1.26) | 4.63E-02 | 1.12 (1.04–1.21) | 4.16E-03 |
| rs225390 | 21 | 43684585 | ABCG1 | A/G | 1.17 (1.05–1.31) | 3.90E-03 | 1.14 (1.01–1.28) | 3.56E-02 | 1.16 (1.07–1.25) | 3.83E-04 |
| rs225396 | 21 | 43687354 | ABCG1 | C/T | 1.11 (1.00–1.23) | 4.28E-02 | 1.13 (1.00–1.27) | 4.21E-02 | 1.12 (1.04–1.21) | 4.21E-03 |
| rs170438 | 21 | 43687567 | ABCG1 | G/A | 1.11 (1.00–1.23) | 4.26E-02 | 1.13 (1.01–1.27) | 4.00E-02 | 1.12 (1.04–1.21) | 4.01E-03 |
| rs170439 | 21 | 43687647 | ABCG1 | C/T | 1.11 (1.00–1.23) | 4.48E-02 | 1.13 (1.01–1.28) | 3.93E-02 | 1.12 (1.04–1.21) | 4.15E-03 |
Chr.: Chromosome; HR: Hazard ratio.
Reference/effect allele;
In the PLCO study, multivariate analyses were adjusted for age, sex, smoking status, histology, tumor stage, chemotherapy, radiotherapy and surgery;
In the Harvard study, multivariate analyses were adjusted for age, sex, smoking status, histology, tumor stage, chemotherapy, radiotherapy, surgery and principal components;
Fixed effect models were used when no heterogeneity was found between two studies (Q > 0.10 and I2 < 25.0%); otherwise, random effects models were used.
Combined analysis and stratified analysis of the two tagging SNPs in ABCG1
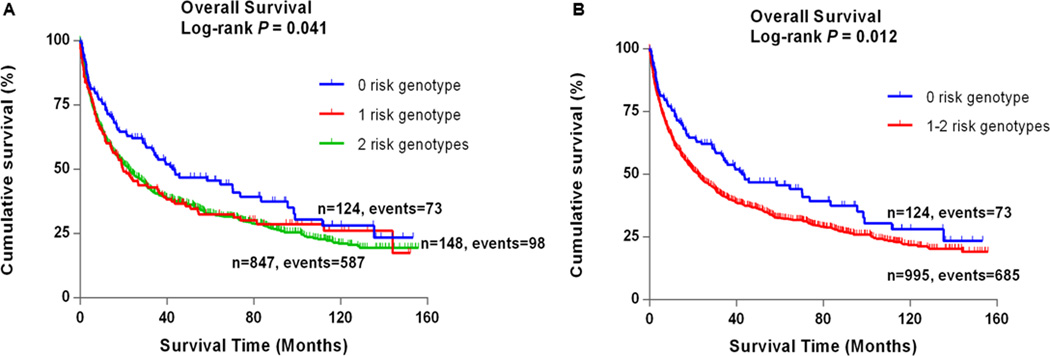
In the PLCO study, rs225388 and rs225390 in ABCG1 were associated with NSCLC OS, with a variant-allele attributed HR of 1.11 (95% CI: 1.00–1.23, P = 0.043) and 1.17 (95% CI: 1.05–1.31, P = 0.004), respectively (Table 4). Compared with their corresponding reference genotypes in a dominant genetic model, patients with rs225390 GA and rs225390 GG genotypes had an increased risk of death (HR = 1.29, 95% CI = 1.00–1.67 and P = 0.050 for GA; HR = 1.45, 95% CI = 1.12–1.87 and P = 0.004; and HR = 1.37, 95% CI = 1.07–1.75 and P = 0.012 for GA+GG, Table 4) in multivariate analyses in 1,185 NSCLC patients of the PLCO study. Meanwhile, rs225388 was associated with a marginally increased risk effect (HR = 1.23, 95% CI = 1.01–1.51 and P = 0.044 for AA, and HR = 1.17, 95% CI = 0.99–1.39 and P = 0.061 for GA+AA, Table 4) on survival in a dominant model. To provide better-estimated hazards of survival, we combined rs225388 GA+AA and rs225390 GA+GG into a genetic score to define the combined risk genotypes. All patients were allocated into three groups with zero, one and two genetic risk scores. Per-unit increased genetic risk score was associated with an increased risk of death after adjustment for other covariates (HR = 1.14, 95% CI = 1.02–1.27, P = 0.024) (Table 4). We next dichotomized all patients into a low-risk group (patients with 0 risk score) and a high-risk group (patients with 1–2 risk scores). We observed that the high-risk group notably had 1.38 fold increase risk of death (95% CI = 1.08–1.76, P = 0.010) associated with the risk genotypes. The analysis of lung cancer DSS showed the results similar to that of OS and that the high-risk group had significantly poorer prognosis (HR = 1.43, 95% CI = 1.11–1.86 and P = 0.007) (Supporting Information Table 8). To further visualize the HR effects, we present Kaplan-Meier survival curves of the associations between OS and risk genotypes in Fig. 1. In stratified analyses, patients with 1–2 risk scores exhibited significantly poor survival in subgroups of older age, current smokers, patients with squamous cell carcinoma, IIIB–IV stage patients, patients with chemotherapy and patients without surgery (Supporting Information Table 9 and Supporting Information Fig. 3). Heterogeneity between subgroups was observed by tumor histology, tumor stage and chemotherapy (P for heterogeneity = 0.013, 0.046 and 0.049, respectively). Therefore, we further conducted interaction analyses, but no significant multiplicative interactions were among the low or high-risk genotype groups (P > 0.05 for all) and tumor histology, tumor stage and chemotherapy (Supporting Information Table 10).
Table 4.
Associations between the two tagging SNPs on ABCG1 and overall survival of NSCLC patients.
| Genotype | Frequency | Univariate analysis | Multivariate analysis c | |||
|---|---|---|---|---|---|---|
| All | Death (%) | HR (95%CI) | P | HR (95%CI) | P | |
| ABCG1 | ||||||
| rs225388 a | ||||||
| GG | 294 | 183 (62.2) | 1.00 | 1.00 | ||
| GA | 597 | 408 (68.3) | 1.13 (0.95–1.35) | 0.170 | 1.15 (0.96–1.37) | 0.131 |
| AA | 272 | 195 (71.7) | 1.25 (1.02–1.53) | 0.031 | 1.23 (1.01–1.51) | 0.044 |
| GA+AA | 869 | 603 (69.4) | 1.17 (0.99–1.38) | 0.069 | 1.17 (0.99–1.39) | 0.061 |
| Trend | 1.12 (1.01–1.24) | 0.031 | 1.11 (1.00–1.23) | 0.043 | ||
| GG+GA | 891 | 591 (66.3) | 1.00 | 1.00 | ||
| AA | 272 | 195 (71.7) | 1.15 (0.98–1.35) | 0.092 | 1.13 (0.96–1.33) | 0.157 |
| rs225390 a | ||||||
| AA | 124 | 73 (58.9) | 1.00 | 1.00 | ||
| GA | 491 | 332 (67.6) | 1.29 (1.00–1.66) | 0.051 | 1.29 (1.00–1.67) | 0.050 |
| GG | 509 | 358 (70.3) | 1.45 (1.13–1.86) | 0.004 | 1.45 (1.12–1.87) | 0.004 |
| GA+GG | 1000 | 690 (69.0) | 1.37 (1.07–1.74) | 0.011 | 1.37 (1.07–1.75) | 0.012 |
| Trend | 1.17 (1.05–1.31) | 0.004 | 1.17 (1.05–1.31) | 0.004 | ||
| GA+AA | 615 | 405 (65.9) | 1.00 | 1.00 | ||
| GG | 509 | 358 (70.3) | 1.18 (1.03–1.36) | 0.021 | 1.81 (1.02–1.36) | 0.024 |
| Number of risk genotypes b | ||||||
| 0 | 124 | 73 (58.9) | 1.00 | 1.00 | ||
| 1 | 148 | 98 (66.2) | 1.35 (0.99–1.82) | 0.055 | 1.39 (1.02–1.88) | 0.037 |
| 2 | 847 | 587 (69.3) | 1.37 (1.07–1.74) | 0.012 | 1.38 (1.08–1.76) | 0.011 |
| Trend | 1.14 (1.02–1.27) | 0.022 | 1.14 (1.02–1.27) | 0.024 | ||
| 0 | 124 | 73 (58.9) | 1.00 | 1.00 | ||
| 1–2 | 995 | 685 (68.8) | 1.36 (1.07–1.73) | 0.012 | 1.38 (1.08–1.76) | 0.010 |
The genotype rates of rs225388 and rs225390 were 98% and 95%, respectively;
Risk genotypes were rs225388 GA, rs225388 AA, rs225390 GA and rs225390 GG;
Adjusted by age, sex, smoking status, histology, tumor stage, chemotherapy, radiotherapy and surgery.
Figure 1.
Kaplan-Meier analysis for patients with NSCLC by the combined risk genotypes (a) by 0, 1 and 2 risk genotypes (log-rank test: p) and (b) by 0 and 1–2 risk genotypes (log-rank test: p) in the PLCO study.
Functional analysis of tagging SNPs in ABCG1
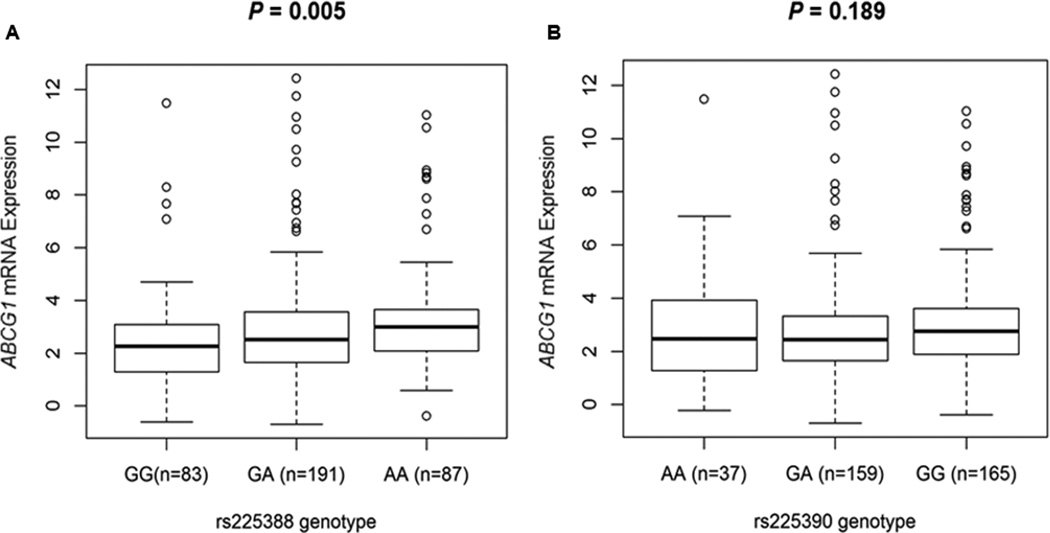
Both rs225388 and rs225390 are predicted to be located at transcription factor binding sites by the SNPinfo online tool. To provide biologically plausible support for the observed associations and prediction, we evaluated the correlations between the two SNPs and ABCG1 mRNA expression levels by their genotypes, using RNA sequencing data of the 373 European descendants in 1000 Genomes Project. Relative expression levels of higher than five time’s interquartile range from the mean value were defined as outliers (Supporting Information Fig. 6).34 After outlier samples were eliminated, 361 samples remained in the analysis. Consistent with the observed associations, the risk A allele of rs225388 was associated with significantly higher levels of ABCG1 mRNA expression (P = 0.005) (Fig. 2A). Although a significant correlation between rs225390 and ABCG1 mRNA expression was not observed, the mean value of expression levels was higher in individuals with the risk G allele of rs225390 (P = 0.189) (Fig. 2B). Moreover, we noted a significant association between rs225388 genotypes and cis-meQTL effects of ABCG1 (P = 1.84× 10−3 for probe: cg01881899 and P = 1.80× 10−2 for probe: cg00222799) (Supporting Information Fig. 7). These findings suggested that rs225388, but not rs225390, could modulate the gene expression levels to influence the function of ABCG1.
Figure 2.
Correlation of ABCG1 relative mRNA expression with genotypes of (a) rs225388 and (b) rs225390 in 373 lymphoblastoid cell lines by using data from 1,000 Genomes Project European decedents.
ABCG1 mRNA expression in TCGA lung cancer dataset
By using the TCGA datasets, we evaluated mRNA expressions levels of ABCG1 in 107 paired tumor and adjacent normal tissue samples in NSCLC (Supporting Information Fig. 8) (39, 40). Lung cancer tissues had a marginally higher expression, compared with that in the normal tissues (mean value of relative expression level, 1,155.5 in tumor vs. 983.7 in normal tissue, P = 0.066). Moreover, patients with higher mRNA expression levels of ABCG1 in tumor tissue showed a poorer overall survival (adjusted HR = 1.44, 95%CI = 1.06–1.95, P = 0.018, Supporting Information Fig. 9).
DISCUSSION
In the current study, we investigated the associations between genetic variants in 100 genes of six gene families (four transporter families and two metabolic enzyme families) and survival of NSCLC in a two-phase analysis of previously published independent GWAS datasets. We first identified ABCG1 rs225388 G>A and rs225390 A>G as predictors of overall survival. Specifically, the risk alleles, rs225388 A and rs225390 G, were associated with poorer survival in patients, and the effect was more pronounced in those patients with combined risk genotypes of these two SNPs. We further confirmed functional relevance of these two SNPs by assessing their correlations with their mRNA expression levels in publicly available datasets. These are some preliminary findings that warrant further investigation for biological relevance.
The ABC transporter family translocate a wide variety of substrates across extra- and intracellular membranes.10 There is some well-established evidence that members of the ABC family contribute to chemoresistance through the efflux of anticancer agents, of which ABCB1, also known as MDR1 or P-glycoprotein, is the most extensively characterized member of the ABC family.11, 12 Besides, ABC transporters are also involved in the transporting some substrates that are relevant to carcinogenesis. These substrates include cyclic nucleotides, prostaglandins, leukotrienes, and cholesterol metabolites.35, 36
In the subgroup of the ABCG family, ABCG2 is known to be involved in resistance to anthracyclines in breast cancer, and it also refers as breast cancer resistance protein (BCRP).35, 37 However, unlike ABCG2, other members of the ABCG subfamily promote cholesterol efflux from cells and regulate intracellular cholesterol homeostasis.37 Cholesterol is an essential molecule for both biophysical structure and metabolism of the cell.38 Dysregulation of cholesterol could be involved in the pathogenesis of cardiovascular and malignant diseases.38 ABCG1 effluxes excess cholesterol to high-density lipoprotein (HDL) particles for reverse cholesterol transport, which is an essential path to elimination of intercellular cholesterol.39 For example, Abcg1 knockout mice exhibited massive accumulation of neutral lipids and phospholipids in hepatocytes and in macrophages.40 It is also known that the macrophage plays a fundamental part in cancer-related inflammation, which is one of the biological hallmarks of cancer.41, 42 Recently, some evidence suggested that ABCG1 deficiency could transform macrophages from a M2 tumor-promoting phenotype into a M1 tumor-fighting phenotype.39 Deficiency of ABCG1 in vivo reduced tumor growth and enhanced tumor cellular apoptosis.39 In the lung cancer TCGA dataset, we also found that ABCG1 mRNA expression levels were higher in tumor tissues than in normal counterparts. Moreover, higher expression of ABCG1 was associated with poorer survival in lung cancer patients in the TCGA dataset. It should be noted that statins, which could block the pathway of cholesterol synthesis, was also reported to decrease ABCG1 gene expression in macrophages and associated with reduced rate of cancer specific mortality in lung cancer patients.43, 44 Those results implied that statin and ABCG1 might share a similar molecular mechanism for their effects on lung cancer survival, which warrants further functional studies. Collectively, ABCG1 may play a part in carcinogenesis and tumor progression.
In the present study, we found two potentially functional SNPs, rs225388 and rs225390, in the intron region of ABCG1 to be associated with survival of NSCLC. Both these SNPs are predicted to have a transcription factor binding ability by the SNPinfo online tool, so are the other four replicated SNPs that were in high LD with rs225388 (rs74757, rs225396, rs170438, and rs170439). Despite the relatively known function of protein-coding regions, a vast majority of regulatory elements in the non-coding regions are being identified, which expands our knowledge of the human genome.45 According to the ENCODE project data from UCSC, rs225388 and rs225390 showed considerable levels of H3K4me1 enrichment, which may be associated with enhancers (Supporting Information Fig. 4).46 Histone modifications modify the accessibility of chromatin during transcription and influence gene expression.46 Moreover, rs225388 causes a change in ABCG1 methylation status in the meQTL analysis. However, we were unable to examine rs225390, because there was no related information in the MuTHER study. Furthermore, the observed association between the increasing number of risk allele of rs225388 A and ABCG1 mRNA expression levels was in a linear manner. All these findings could provide possible biological insights into the mechanism underlying the observed association with survival in NSCLC patients.
There are some limitations in the current study. First, we only used available GWAS datasets from Caucasian populations. Genetic variants show some diversity by different ethnic background, and our findings may not be generalizable to other ethnic populations. Second, some top SNPs from the PLCO study were not validated in the Harvard study. Different distributions of the basic characteristics between two studies could partially explain the reason of non-validated SNPs. Additional validation might provide more evidences for these findings. Third, although we did perform interaction analyses between SNPs and some clinical factors, the sample size of the subgroups was limited for us to detect such an interaction among gene-gene and gene-environment, which widely exist in diseases. Last, only a few clinicopathological factors were considered for analyses, and the information about nutrition status, socioeconomic status and details in cancer treatment were not available for us.
In conclusion, we conducted a two-phase association analysis of 100 cell membrane transport-related genes and NSCLC survival in two independent GWAS datasets. Two SNPs, rs225388 and rs225390, were identified as the prognostic predictors for NSCLC. Further investigation with larger populations and biological assessments of ABCG1 will be needed to validate these findings.
Supplementary Material
Brief description.
Cell transmembrane transporters play an essential role in regulating substrate disposition, including absorption, distribution and excretion. In the present study of re-analyzing published genome-wide association study (GWAS) datasets, we found that two genetic variants, rs225388 G>A and rs225390 A>G, in the ABCG1 of ATP-binding cassette family could modulate survival in over 2000 NSCLC patients in an allele-dose response manner. The identified genetic variants could translate into clinical use for prognostic assessment and personalized management of lung cancer patients.
Acknowledgments
The authors thank the National Cancer Institute for access to NCI’s data collected by the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial. The statements contained herein are solely those of the authors and do not represent or imply concurrence or endorsement by NCI. The author would also like to acknowledge dbGaP repository for providing the cancer genotyping datasets. The accession numbers for the datasets of lung cancer are phs000336.v1.p1 and phs000093.v2.p2. Qingyi Wei was supported by a start-up funds from Duke Cancer Institute, Duke University Medical Center and support from the Duke Cancer Institute as part of the P30 Cancer Center Support Grant (Grant ID: NIH CA014236). Yanru Wang was sponsored by Collaborative Innovation Center for Cancer Personalized Medicine, Nanjing Medical University. The Harvard Lung Cancer Susceptibility Study was supported by NIH grants CA092824, CA074386, and CA090578 to David C. Christiani. We thank all individuals who participated in this project.
Abbreviation
- ABC
ATP-binding cassette
- CI
confidence interval
- DSS
disease-specific survival
- FDR
false discovery rate
- GWAS
genome-wide association study
- HR
hazards ratio
- LD
linkage disequilibrium
- NSCLC
non-small cell lung cancer
- OS
overall survival
- PLCO
Prostate, Lung, Colorectal and Ovarian
- SNP
single nucleotide polymorphism
- SEER
Surveillance, Epidemiology, and End Results program
- TCGA
The Cancer Genome Atlas
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, Shepherd FA. Non-small-cell lung cancer. Lancet. 2011;378:1727–1740. doi: 10.1016/S0140-6736(10)62101-0. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 5.National Comprehensive Cancer Network. Non-small cell lung cancer (Version 6.2015) 2015 doi: 10.6004/jnccn.2015.0071. [DOI] [PubMed] [Google Scholar]
- 6.Evans WE, McLeod HL. Pharmacogenomics--drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 7.Ekhart C, Rodenhuis S, Smits PH, Beijnen JH, Huitema AD. An overview of the relations between polymorphisms in drug metabolising enzymes and drug transporters and survival after cancer drug treatment. Cancer Treat Rev. 2009;35:18–31. doi: 10.1016/j.ctrv.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Coate L, Cuffe S, Horgan A, Hung RJ, Christiani D, Liu G. Germline genetic variation, cancer outcome, and pharmacogenetics. J Clin Oncol. 2010;28:4029–4037. doi: 10.1200/JCO.2009.27.2336. [DOI] [PubMed] [Google Scholar]
- 9.Hu L, Wu C, Zhao X, Heist R, Su L, Zhao Y, Han B, Cao S, Chu M, Dai J, Dong J, Shu Y, et al. Genome-wide association study of prognosis in advanced non-small cell lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res. 2012;18:5507–5514. doi: 10.1158/1078-0432.CCR-12-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 12.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 13.Puig S, Thiele DJ. Molecular mechanisms of copper uptake and distribution. Curr Opin Chem Biol. 2002;6:171–180. doi: 10.1016/s1367-5931(02)00298-3. [DOI] [PubMed] [Google Scholar]
- 14.Gupta A, Lutsenko S. Human copper transporters: mechanism, role in human diseases and therapeutic potential. Future Med Chem. 2009;1:1125–1142. doi: 10.4155/fmc.09.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brady DC, Crowe MS, Turski ML, Hobbs GA, Yao X, Chaikuad A, Knapp S, Xiao K, Campbell SL, Thiele DJ, Counter CM. Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature. 2014;509:492–496. doi: 10.1038/nature13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koepsell H, Endou H. The SLC22 drug transporter family. Pflugers Arch. 2004;447:666–676. doi: 10.1007/s00424-003-1089-9. [DOI] [PubMed] [Google Scholar]
- 17.Motohashi H, Inui K. Multidrug and toxin extrusion family SLC47: physiological, pharmacokinetic and toxicokinetic importance of MATE1 and MATE2-K. Mol Aspects Med. 2013;34:661–668. doi: 10.1016/j.mam.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Burger H, Loos WJ, Eechoute K, Verweij J, Mathijssen RH, Wiemer EA. Drug transporters of platinum-based anticancer agents and their clinical significance. Drug Resist Updat. 2011;14:22–34. doi: 10.1016/j.drup.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Hocking WG, Hu P, Oken MM, Winslow SD, Kvale PA, Prorok PC, Ragard LR, Commins J, Lynch DA, Andriole GL, Buys SS, Fouad MN, et al. Lung cancer screening in the randomized Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. J Natl Cancer Inst. 2010;102:722–731. doi: 10.1093/jnci/djq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oken MM, Marcus PM, Hu P, Beck TM, Hocking W, Kvale PA, Cordes J, Riley TL, Winslow SD, Peace S, Levin DL, Prorok PC, et al. Baseline chest radiograph for lung cancer detection in the randomized Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial. J Natl Cancer Inst. 2005;97:1832–1839. doi: 10.1093/jnci/dji430. [DOI] [PubMed] [Google Scholar]
- 21.Tryka KA, Hao L, Sturcke A, Jin Y, Wang ZY, Ziyabari L, Lee M, Popova N, Sharopova N, Kimura M, Feolo M. NCBI's Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–D979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhai R, Yu X, Wei Y, Su L, Christiani DC. Smoking and smoking cessation in relation to the development of co-existing non-small cell lung cancer with chronic obstructive pulmonary disease. Int J Cancer. 2014;134:961–970. doi: 10.1002/ijc.28414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 25.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager M, Bergen AW, Li Q, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. American journal of human genetics. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 30.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grundberg E, Meduri E, Sandling JK, Hedman AK, Keildson S, Buil A, Busche S, Yuan W, Nisbet J, Sekowska M, Wilk A, Barrett A, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. American journal of human genetics. 2013;93:876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 35.Fletcher JI, Haber M, Henderson MJ, Norris MD. ABC transporters in cancer: more than just drug efflux pumps. Nat Rev Cancer. 2010;10:147–156. doi: 10.1038/nrc2789. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi K, Kimura Y, Nagata K, Yamamoto A, Matsuo M, Ueda K. ABC proteins: key molecules for lipid homeostasis. Med Mol Morphol. 2005;38:2–12. doi: 10.1007/s00795-004-0278-8. [DOI] [PubMed] [Google Scholar]
- 37.Kusuhara H, Sugiyama Y. ATP-binding cassette, subfamily G (ABCG family) Pflugers Arch. 2007;453:735–744. doi: 10.1007/s00424-006-0134-x. [DOI] [PubMed] [Google Scholar]
- 38.Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- 39.Sag D, Cekic C, Wu R, Linden J, Hedrick CC. The cholesterol transporter ABCG1 links cholesterol homeostasis and tumour immunity. Nat Commun. 2015;6:6354. doi: 10.1038/ncomms7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy MA, Barrera GC, Nakamura K, Baldan A, Tarr P, Fishbein MC, Frank J, Francone OL, Edwards PA. ABCG1 has a critical role in mediating cholesterol efflux to HDL and preventing cellular lipid accumulation. Cell Metab. 2005;1:121–131. doi: 10.1016/j.cmet.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Wong J, Quinn CM, Gelissen IC, Jessup W, Brown AJ. The effect of statins on ABCA1 and ABCG1 expression in human macrophages is influenced by cellular cholesterol levels and extent of differentiation. Atherosclerosis. 2008;196:180–189. doi: 10.1016/j.atherosclerosis.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 44.Cardwell CR, Mc Menamin U, Hughes CM, Murray LJ. Statin use and survival from lung cancer: a population-based cohort study. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2015;24:833–841. doi: 10.1158/1055-9965.EPI-15-0052. [DOI] [PubMed] [Google Scholar]
- 45.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.