Abstract
Hypomorphic mutations in the zinc finger domain of NF-κB essential modulator (NEMO) cause X-linked hyper-IgM syndrome with ectodermal dysplasia (XHM-ED). Here we report that patient B cells are characterized by an absence of Ig somatic hypermutation (SHM) and defective class switch recombination (CSR) despite normal induction of activation-induced cytidine deaminase (AID) and Iε-Cε transcripts. This indicates that AID expression alone is insufficient to support neutralizing antibody responses. Furthermore, we show that patient B cells stimulated with CD40 ligand are impaired in both p65 and c-Rel activation, and whereas addition of IL-4 can enhance p65 activity, c-Rel activity remains deficient. This suggests that these NF-κB components have different activation requirements and that IL-4 can augment some but not all NEMO-dependent NF-κB signaling. Finally, using microarray analysis of patient B cells we identified downstream effects of impaired NF-κB activation and candidate factors that may be necessary for CSR and SHM in B cells.
Introduction
Higher vertebrates rely on the diversity of the antibody repertoire to combat infectious pathogens. The cognate interaction between CD40 ligand (CD40L) expressed on activated CD4+ T cells and CD40 expressed on B cells leads to B cell proliferation and Ig class switch recombination (CSR) from IgM toward the production of IgG, IgA, and IgE (1). Humans and mice with mutations in the genes encoding CD40L or CD40 have skewed IgM antibody responses and a markedly diminished or absent IgG response to protein antigens. Whereas the role of CD40L/CD40 interaction is established, the molecular mechanisms associated with Ig CSR and somatic hypermutation (SHM) have not been precisely delineated.
Activation-induced cytidine deaminase (AID) is a putative RNA or DNA editing enzyme that is specifically expressed in B cells in response to combined CD40L and IL-4 signaling. B cells from humans and mice lacking AID develop normally but fail to undergo CSR or SHM in response to antigen challenge (2, 3). AID overexpression in non–B cells can induce somatic mutation and CSR in plasmid DNA substrates, suggesting that AID functions alone, or that factors necessary for its function are ubiquitously expressed (4). However, the means by which AID regulates B cell terminal differentiation remains undefined.
NF-κB essential modulator (NEMO, also known as IKKγ) is a scaffolding protein that binds to 2 proteins with intrinsic kinase activity, IKKα and IKKβ (5). Upon cell stimulation, IKKα and IKKβ become activated, leading to the phosphorylation and subsequent degradation of the inhibitors of NF-κB (IκBs). This frees NF-κB to translocate to the nucleus and activate transcription of target genes. As a result of its central position in NF-κB signaling, large genomic rearrangements in NEMO cause incontinentia pigmenti, a lethal disease in males that causes abnormalities of the skin, hair, nails, teeth, and CNS in carrier females. In contrast, hypomorphic mutations in NEMO cause anhidrotic ectodermal dysplasia with immunodeficiency in affected males, a clinical condition characterized by the absence of sweat glands, a paucity of hair follicles, and heterogeneous immunodeficiency states (6–8).
We have shown previously that patients with missense mutations in the zinc finger domain of NEMO have X-linked hyper-IgM syndrome with ectodermal dysplasia (XHM-ED) (6). B cells from these patients are of the naive phenotype, invariably coexpress surface IgM and IgD, and fail to undergo Ig CSR in vitro when stimulated with CD40 agonists in vitro. Interestingly, NF-κB activation by other members of the TNF or the toll-like receptor superfamily are relatively preserved in XHM-ED, suggesting that mutations in the zinc finger domain primarily impair CD40 signaling in hematopoietic cells (6).
In this report, we show that the Ig variable region in B cells from XHM-ED patients is devoid of SHM. Furthermore, B cells fail to activate c-Rel in response to CD40 stimulation, even in the presence of IL-4 (which partly restores p65 activation). Importantly, in vitro activation of XHM-ED B cells with soluble CD40L and IL-4 induced normal levels of Iε-Cε germline transcripts and expression of the AID gene. This suggests that the expression of additional genes, perhaps regulated by CD40-mediated c-Rel activation, are necessary for Ig CSR. To identify such genes, we used oligonucleotide microarrays to show that B cells in XHM-ED have specific impairments in the expression of RAD50, LYL1, DNA ligase IV (LIG4), and other factors relating to nonhomologous recombination that previously have not been linked to B cell differentiation.
Results
XHM-ED patients show defects in CSR and SHM.
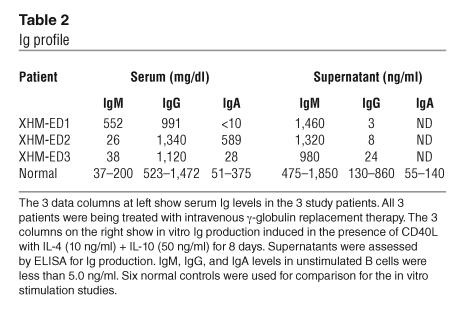
Three unrelated male patients (designated XHM-ED1, XHM-ED2, and XHM-ED3) were diagnosed with XHM-ED, defined by markedly diminished serum levels of IgG and IgE and normal or elevated levels of serum IgM (Tables 1 and 2). Two of these patients have been previously described (6, 9). Mutation analysis revealed that the 3 patients had missense mutations in the zinc finger domain of NEMO affecting cysteine at position 417. All patients presented in childhood with upper respiratory or severe bacterial infections. From the time of diagnosis, all patients were treated with intravenous γ-globulin replacement therapy.
Table 1.
Patient B cell characteristics
Table 2.
Ig profile
Germline homozygous mutations in the AID gene cause an autosomal recessive form of hyper-IgM syndrome (2). Recently it has been shown that the ectopic expression of AID and sterile germline transcripts are sufficient to activate SHM and CSR in hybridomas and non–B cells (4). We therefore examined the expression of Iε-Cε germline transcripts and AID in B cells of XHM-ED patients and normal controls by real-time PCR. Stimulation of B cells with CD40L and IL-4 induced a pronounced increase in Iε-Cε and AID mRNA in XHM-ED at levels comparable to those observed in normal controls (Table 1). Remarkably, despite expression of normal levels of AID and Iε-Cε, XHM-ED B cells fail to undergo CSR in vitro as evidenced by very low to undetectable IgE after culture with CD40L and IL-4. In contrast, normal control B cells produced robust IgE responses under these conditions (Table 1). Interestingly, 1 of the 3 XHM-ED patients had high levels of serum IgA despite the failure to synthesize IgA in response to CD40L stimulation in vitro (Table 2). The fact that humans and mice with CD40L deficiency have also been reported to have IgA suggests a CD40-independent mechanism for IgA synthesis in this patient.
Patients with C417R NEMO mutation have a virtual absence of IgD–CD27+ B cells, which are typical memory B cells. To confirm failure of memory B cell differentiation in these patients, somatic mutations in the heavy chain V3-23 Ig V gene a member of the VH3 family expressed in B cells, were analyzed (10). The frequency of mutation of heavy chain VDJ rearrangements obtained from CD19+ blood B cells from 3 XHM-ED patients was examined and compared with healthy controls (Table 1). There were significant differences (P < 0.001, 95% confidence level) in the mean mutation frequencies between the healthy and the XHM-ED patients (Table 1). The calculated mutation frequency in 3 patients with XHM-ED ranged from 0.09% to 0.19% per bp and was comparable to that obtained by amplification with Taq polymerase alone (0.1% per bp). These results demonstrate that the hypermutation process is impaired in XHM-ED patients and imply that while AID and germline transcripts are necessary for mutational targeting, additional factors regulated by NF-κB are also necessary.
CD40-mediated NF-κB activation in XHM-ED.
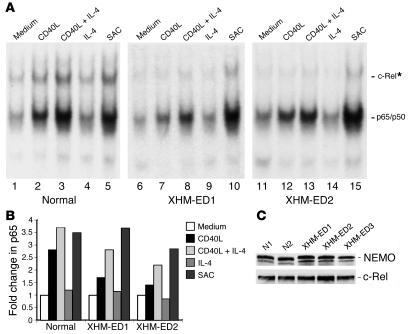
In initial studies, we more precisely defined the effect of the C417R mutation on NF-κB activation in B cells. In particular, we measured p65 and c-Rel activation by electrophoretic mobility shift assay in cellular lysates prepared from B cells from normal controls and 2 unrelated patients (XHM-ED1 and XHM-ED2) after stimulation with CD40L, CD40L plus IL-4, IL-4 alone, or Staphylococcus aureus Cowan’s strain I (Figure 1A). In contrast to the normal control, p65 and c-Rel activity in B cells from XHM-ED patients was impaired after stimulation with CD40L, whereas p65 and c-Rel activation mediated by Staphylococcus aureus Cowan’s strain I was less affected (Figure 1B). This is consistent with our previous finding that mutations in the zinc finger domain of NEMO lead to a specific impairment in CD40 signaling in B cells and monocytes (6). Among other observations noted in patients with hypomorphic mutations in the C-terminal region of NEMO are low NEMO protein levels (7, 11). However, as shown in Figure 1C, both NEMO and c-Rel protein levels are relatively well-preserved in XHM-ED patients, suggesting that the impairments in NF-κB signaling are not a result of low levels of these signaling pathway constituents. Because it has recently been reported that crossregulation exists between the JAK2 and the NF-κB signaling pathways (12), we also measured the effect of IL-4 stimulation on NF-κB signaling. IL-4 stimulation of B cells by itself did not activate NF-κB. However, when B cells were costimulated with IL-4 plus CD40L, enhanced p65 DNA binding was observed in XHM-ED patients and normal controls compared with those receiving CD40L stimulation alone, suggesting that IL-4 can enhance NF-κB signaling. In contrast, CD40-mediated c-Rel activation was unaffected in normal controls with IL-4 costimulation, and remained markedly diminished in patients with XHM-ED. These data highlight the role of c-Rel activation for B cell differentiation and suggest that p65 and c-Rel have different activation requirements.
Figure 1.
IL-4 synergizes with CD40L in activating p65 binding but not c-Rel binding in XHM-ED B cells. (A) DNA binding activity of the NF-κB complexes. CD19+ B cells were isolated from 2 unrelated patients with the C417R mutation and a normal control. Cells were stimulated in the presence of cycloheximide with CD40L, CD40L + IL-4, IL-4 alone, or Staphylococcus aureus Cowan’s strain I (SAC) for 2 hours. Cellular extracts were prepared and analyzed by electrophoretic mobility shift assay for NF-κB binding activity. *For supershift, all samples were incubated with anti_c-Rel antibody prior to incubation with the labeled probe. One representative experiment of 3 is shown. (B) Quantification of p65 activity. The band shifts for p65 shown in A were quantified and are represented as fold change in comparison to unstimulated cells (set at 1.0). In contrast to the normal control, both XHM-ED1 and XHM-ED2 showed impaired p65 activation in response to CD40 stimulation alone and were enhanced with IL-4 costimulation. (C) NEMO and c-Rel protein levels in B cells. Cytoplasmic extracts were prepared from purified B cells from 2 normal controls and 3 patients with XHM-ED and subjected to immunoblotting. Blots were probed with anti-NEMO or anti_c-Rel antibody.
Microarray analysis of stimulated B cells from XHM and XHM-ED patients.
In order to delineate CD40L-dependent NF-κB target genes that modulate Ig CSR and SHM, we used DNA microarrays to compare gene expression profiles of B lymphocytes from patients with XHM and XHM-ED. We reasoned that the inherent defect in the B cells of XHM-ED patients is a specific CD40-triggered event, and would be accompanied by differences in gene expression. In contrast, B cells in XHM are intrinsically normal and can be activated to undergo terminal differentiation with CD40 agonists. We isolated CD19+IgM+IgD+ B cells from patients with XHM and from patients with XHM-ED. We then stimulated these cells with CD40L or with CD40L plus IL-4, and compared gene expression patterns at 0, 6, and 24 hours. The change in gene expression for each gene was computed as a ratio of its expression in control unstimulated cells to that in cells stimulated with CD40L alone or CD40L plus IL-4 (Figure 2).
Figure 2.
IL-4 “rescues” the transcriptional regulation of many genes in B cells of XHM-ED patients. B cells from XHM and XHM-ED patients were stimulated with CD40L alone or CD40L + IL-4 for 6 hours or 24 hours. Each row represents the ratio of expression in stimulated vs. unstimulated cells for each gene. Each column represents the data from an independent stimulation experiment. A color bar shows the magnitude of gene expression changes as a ratio of expression: significantly induced (red), unchanged (black), or repressed (green). (A) Genes regulated by CD40L stimulation in XHM. Inclusion in the analysis required at least a 3-fold change (6 hours vs. 0 hours, or 24 hours vs. 0 hours) with a P value < 0.01 for at least 3 comparisons in 2 different XHM patients in whom CD40 signaling is normal; 1,270 probe sets met this criteria. (B) Genes not regulated by CD40L stimulation in XHM-ED. Of the 1,270 probe sets that showed regulation in XHM B cells, 275 were identified that showed no significant regulation in the B cells from 2 independent XHM-ED patients in response to CD40 stimulation. (C) Genes induced with CD40L + IL-4 stimulation. The expression of the 275 probe sets that have impaired expression in XHM-ED B cells in response to CD40L stimulation alone were reclustered with data from XHM and XHM-ED B cells stimulated with CD40L and IL-4. Note the restored expression of many genes in XHM-ED patients in response to CD40L + IL-4 stimulation compared with CD40L alone. Gene identities can be seen in Supplemental Table 1.
In order to find genes that were abnormally regulated in the XHM-ED B cells, the following analysis was performed. The 4 ratios for the XHM samples (0 hours versus 6 hours and 0 hours versus 24 hours for XHM-1 and XHM-2) were filtered for probe sets with a greater than 3-fold change in expression level and with a P value less than 0.01 for 3 of the 4 comparisons. This analysis was designed to stringently select for genes that showed consistent and significant expression changes, and yielded 1,270 probe sets (Figure 2A and Supplemental Table 1; supplemental material available at http://www.jci.org/cgi/content/full/114/11/1593/DC1). Further analysis of the expression of these 1,270 probe sets identified 275 that showed no significant regulation in the B cells from the 2 XHM-ED patients at either 6 hours or 24 hours (Figure 2B and Supplemental Table 1). This includes genes such as CD69, CD95, and CD86 that have been previously shown at the protein level not to be regulated in XHM-ED B cells by CD40L (6).
Next we assessed the effect of adding IL-4 to CD40L on the expression of these 275 probe sets. Interestingly, addition of IL-4 “rescued” the regulation of many of the genes (Figure 2C and Supplemental Table 1), including the cell surface markers CD86, CD95, and CD69. FACS analysis of B cells from these patients stimulated with CD40L plus IL-4 confirmed this observation at the protein level (Supplemental Figure 1B).
XHM-ED B cells display defects in the gene expression program induced by CD40L plus IL-4 that correlate with defects in CSR.
As indicated above, XHM-ED B cells fail to undergo CSR in the presence of CD40L and IL-4 despite apparently normal expression of AID and Iε-Cε, suggesting the requirement for additional cofactors. To identify CD40-regulated factors required for CSR, we compared the expression profiles of XHM and XHM-ED B cells stimulated with CD40L and IL-4. A common signature was defined in both XHM profiles that represents genes with a significant change in expression (P < 0.01) when comparing 6 hours to 0 hours and 24 hours to 0 hours. This identified 2,359 and 2,741 probe sets, respectively, that were significantly regulated in the 2 XHM patient B cell profiles at 6 hours or 24 hours (Figure 3). These common signatures represent the “normal” expression programs (6 hours and 24 hours) induced by CD40L plus IL-4 (Figure 3A). The 6-hour and 24-hour common signatures were then compared with the individual signatures (significant regulation, P < 0.01) at the relevant time points for the XHM-ED patient B cells (XHM-ED1 and XHM-ED2, duplicate time course). Any gene represented in the common signature but absent from the 4 individual signatures for that time point was selected for further analysis. This yielded 112 probe sets at the 6-hour time point and 166 probe sets at the 24-hour time point. Combining these 2 gene sets gave 295 unique probe sets, and reclustering allowed definition of different patterns of abnormal gene regulation in comparing the XHM and XHM-ED B cells (Figure 3B and Supplemental Table 1). These patterns include genes whose expression does not change in the mutant cells, irrespective of the time point, as well as those that show either delayed regulation or only transient regulation. Microarray findings were validated by independent assay of mRNA expression via real-time PCR (6 genes, Supplemental Figure 1A) and by flow cytometric analysis of protein expression (3 proteins, Supplemental Figure 1B).
Figure 3.
Transcriptional program of B cells stimulated with CD40L + IL-4. B cells from XHM and XHM-ED patients were stimulated with CD40L + IL-4 for 6 hours or 24 hours. Each row represents the ratio of expression in stimulated vs. unstimulated cells for each gene. Each column represents the data from an independent stimulation experiment. (A) Genes induced with CD40L + IL-4 stimulation. Probe sets were selected for inclusion in the cluster using the following inclusion criteria: induction greater than 3-fold (6 hours vs. 0 hours, or 24 hours vs. 0 hours) with a P value < 0.01 for at least 3 comparisons in 2 independent XHM patients. The 1,418 probe sets that met these criteria were hierarchically clustered. (B) Genes with impaired expression after CD40L + IL-4 stimulation in XHM-ED. Probe sets were included if they showed significant regulation (P < 0.01) in both XHM patients at a particular time point but showed either no regulation or reciprocal regulation for that time point in the 4 XHM-ED ratios. The 295 probe sets that met these criteria were hierarchically clustered. (C) Probe sets were selected (see B) that showed significant upregulation (P < 0.01) in the 2 XHM ratios (either 6 hours or 24 hours) but showed no upregulation at any time point in XHM-ED ratios. The 80 probe sets that met these criteria were hierarchically clustered. Note that ratio values with P > 0.01 were set to log (ratio) = 0. Green indicates genes downregulated with respect to t = 0 hours for each patient and red indicates upregulated genes. Gene identities can be accessed in Supplemental Table 1.
Figure 3C shows a subset of 78 genes from Figure 3B that are upregulated in XHM B cells from both patients in response to CD40L plus IL-4 but are not upregulated at any time point in either of the duplicate stimulations of the XHM-ED patient B cells. A complete listing of the 78 genes is given in Table 3. At least 10 of the genes in Table 3 may have roles in Ig CSR and SHM based on their molecular characteristics. These include LYL1 and TFAP4, candidates for switch region binding factors; MRE11 and RAD50, which may actively participate in the recombination process; and LIG4, a ligase previously implicated in CSR. The correlation between the failure to activate c-Rel in response to CD40 stimulation in B cells from XHM-ED patients and defective expression of known c-Rel genomic targets such as c-myc in B cells and IL-12 in monocytes suggests that many of the differentially expressed genes identified in this study may be c-Rel genomic targets (6, 13).
Table 3.
Genes from XHM-ED B cells that show abnormal regulation in response to CD40L plus IL-4
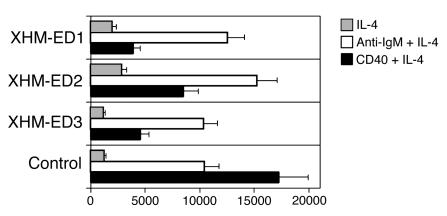
Some of the genes that fail to show normal upregulation in B cells from XHM-ED patients stimulated with CD40L and IL-4 are genes that can be functionally categorized as regulating cell cycle or proliferative responses. Because of the impairment in the expression of genes related to cell cycle progression, the proliferation of XHM-ED B cells was measured and compared with normal controls. B cells from patients with XHM-ED showed a partial defect in proliferation in response to CD40L and IL-4 stimulation but proliferated normally in response to anti-IgM and IL-4 stimulation (Figure 4). In vitro activation of PBMCs by anti-IgM, CD40L, and IL-4 did not lead to IgE production in vitro (A. Jain, unpublished observation). The fact that the defect in proliferation in the B cells of XHM-ED patients can be corrected with stimulation of the antigen receptor suggests that their failure to undergo Ig CSR and SHM is not simply a result of diminished proliferative capacity.
Figure 4.
XHM-ED B cells have a specific CD40 pathway proliferation defect. CD19+ B cells from XHM-ED patients and normal controls were cultured in triplicate for 72 hours with IL-4 (gray bars), CD40L + IL-4 (black bars), or goat anti-human IgM Fab + IL-4 (white bars). Data are the mean ± SD proliferative response and are representative of 3 experiments.
Discussion
In this study, we examined 3 unrelated patients with XHM-ED with a substitution of a highly conserved amino acid at position 417 of NEMO that likely disrupts the putative zinc finger domain. The phenotype observed in these patients is relatively homogeneous and comprises 3 consistent abnormal features: defective CSR, impaired mutational targeting in the Ig variable region, and the persistence of IgM+IgD+ naive B cells in the peripheral blood. Interestingly, 1 of the 3 XHM-ED patients had high serum levels of IgA despite the failure to synthesize IgA in response to stimulation with CD40L and IL-4 plus IL-10 in vitro. Although the precise mechanism for the presence of IgA in the serum of this patient with XHM-ED awaits further study, it may relate to toll-like receptor signaling, which remains largely intact in XHM-ED patients (6). In support of this hypothesis are previous reports of IgA synthesis in vertebrates by commensal bacteria in the absence of T cells, presumably through stimulation of B cells through toll-like receptors (14). Furthermore, serum IgA has also been observed in humans with CD40L deficiency and can be induced in CD40L–/– mice in the absence of CD40 stimulation (15, 16). Taken together these observations suggest a CD40-independent mechanism for serum IgA in some patients with XHM-ED.
Hypomorphic mutations outside the zinc finger domain of NEMO have been associated with osteopetrosis, a marked incidence of infection with Mycobacterium avium complex (MAC), and lymphedema (7, 8). Levels of γ-globulin are often normal in these other patients, and the finding of ectodermal dysplasia is not consistently preserved. Genetic or environmental factors may generate some of this variability. However, one can also suggest that other hypomorphic mutations outside the zinc finger domain of NEMO impair NF-κB activation by Rank, toll-like receptors, or VEGF, while still being functional in the CD40 and ectodysplasin receptor signaling pathways, thus implying that there may exist stimulus-specific cofactors to activate the IKK complex.
Remarkably, a large portion of the gene expression program induced by CD40L in B cells is intact in XHM-ED patients, despite the presence of demonstrable biochemical and phenotypic defects. This implies either that pathways other than NF-κB, such as ERK or JNK, regulate many of these genes, or that NF-κB signaling is not completely blocked by NEMO C417R mutations. Support for the latter hypothesis comes from the observation that there is some, although reduced, activation of p65/NF-κB binding activity in XHM-ED B cells stimulated with CD40L. IL-4 costimulation leads to an enhancement of p65/NF-κB activation and is associated with the restored regulation of a number of CD40 target genes in XHM-ED patients. JAKs have been shown to phosphorylate IκBα under certain stimulation conditions (12). However, we did not observe enhanced phosphorylation of IκBα in response to CD40L and IL-4 stimulation in normal controls (unpublished observation), and future studies will be needed to precisely define the mechanism by which IL-4 costimulation enhances p65 activation.
The mechanism by which the zinc finger domain of NEMO regulates NF-κB activation is not fully understood. Recently, it has been shown that among the nonproteolytic functions of ubiquitin is to positively regulate IKK complex activity (17–19). The ubiquitination of K399 in the zinc finger of NEMO was found to be necessary for NF-κB activation in T cells after stimulation with PMA and ionomycin (19). In addition, the C417R NEMO mutation was specifically shown to have impaired ubiquitination in response to genotoxic stress, and this was also associated with reduced IKK complex activity (18). Our studies of B cells from patients with the C417R NEMO mutation demonstrate a specific impairment in c-Rel activation in response to CD40 and IL-4 stimulation. Further experiments will be needed to address whether differences in NEMO ubiquitination can explain differences in the activation of various NF-κB dimers. The hypothesis that c-Rel may transmit specific signals important for CSR is supported by our finding that c-Rel binding activity is absent in B cells from XHM-ED patients after stimulation with CD40L and IL-4, and by the phenotype of NF-κB1/c-Rel knockout mice (20, 21). These mutants show normal lymphopoiesis, but have severe impairment in humoral immunity characterized by low to absent serum Ig’s and a failure to mount antigen-specific antibody responses.
Previous experiments have shown that AID expression in fibroblasts leads to somatic mutation and nonhomologous recombination in artificial plasmid substrates (4, 22). This is in contrast to our finding that XHM-ED patient B cells can upregulate AID and germline transcripts normally in response to stimulation with CD40L plus IL-4, but fail to undergo CSR. The high levels of AID expression in non–B cells achieved by ectopic expression may account for some of these differences. However, another plausible hypothesis is that AID at physiological levels requires additional cofactors that are not ubiquitous in expression and are regulated by NF-κB in B cells. These putative CD40-responsive cofactors may be necessary to efficiently target AID to the relevant site and/or facilitate its function. In this context it is interesting to note the failure of upregulation of the E47-related proteins LYL1 and TFAP4 in XHM-ED B cells. SNAP, a binding activity at the Sγ3 site, has been partially characterized and shown to have epitopes in common with the E47 family (23). Such binding activities have been suggested to be necessary for recruitment of the putative recombinase (24, 25). Notably, LYL1 has been shown to bind to E12 and E47 as well as the p105 precursor of NF-κB1/p50 (26, 27), a protein known to be essential for CSR. Promoter mapping studies show that p50 homodimers bind to the Sγ3 promoter at a so-called SNIP site and presumably configure the DNA in a structure that facilitates μ-γ3 switch recombination. A recently demonstrated function of AID is in the formation of NBS1-containing nuclear foci at the constant heavy chain region within the Ig locus in cells undergoing CSR (28). NBS1, in combination with MRE11 and RAD50, recognizes hairpins or cruciforms, structures that have been associated with switch region function (29). Our data showing failure to upregulate MRE11 and RAD50 associated with absence of CSR provides strong support for these proteins as cofactors in AID-dependent switch recombination. The final step in CSR likely involves the re-ligation or repair of juxtaposed switch regions following cleavage by the putative recombinase. LIG4 is a strong candidate for involvement in this, based on data presented here and previous observations that show that genetic variants in LIG4 may modulate a predisposition to multiple myeloma, a tumor characterized by aberrant Ig CSR (30).
Comparison of genes regulated in ex vivo B cells stimulated with CD40L with those identified in previous studies looking at different B cell developmental stages suggests some similar features (31, 32). A proliferation signature consisting of upregulation of PCNA and cyclin B1 and downregulation of p21/WAF1 is observed in B cells stimulated with CD40L compared with untreated B cells, and in tonsillar centroblasts compared with naive B cells (32). In addition, genes involved in apoptosis (BIK, TOSO), DNA repair (MLH1, MSH6, RAD2), and adhesion (CD62L) show similar patterns of regulation in various studies. Furthermore, a number of these genes were also identified by previous microarray analysis of germinal center B cells (31). Taken together, these studies validate the gene signatures obtained by ex vivo stimulation as reflecting changes that occur in the in vivo setting.
IL-4 stimulation of XHM-ED patient B cells can induce modest AID mRNA expression (unpublished observation). This finding is consistent with recent observations in normal human and murine B cells and suggests that CD40 stimulation synergizes with signals delivered by IL-4 to optimally induce AID expression (33, 34). While our data suggest that c-Rel activation is not essential for AID expression in B cells, the precise intracellular mechanism by which CD40 contributes to AID regulation awaits further investigation. XHM-ED patients lack B cells in the peripheral blood that exhibit SHM in the Ig variable region, despite normal induction of AID in response to CD40L and IL-4 stimulation. These findings suggest that AID does not function alone for SHM and requires other factors regulated by NF-κB signaling for mutational targeting in B cells. The requirement for AID cofactors for SHM is also supported by recent reports showing that B cells from humans with mutations in the C-terminal domain of AID have normal SHM activity but are devoid of CSR, and that the DNA cleavage activity of AID requires de novo protein synthesis (35, 36). While it is not known if AID forms such complexes, these results suggest that the C-terminal domain of AID may interact with CSR-restricted cofactors, while other regions of AID are necessary for SHM. One can also suggest from our finding of the near absence of SHM and CSR, despite normal induction of AID in the B cells from XHM-ED patients, that AID-specific cofactors necessary for both SHM and CSR are regulated by NF-κB in B cells.
CD40 stimulation also leads to the activation of the nonclassical (NEMO-independent) NF-κB pathway that involves the activation of IKKα through NIK kinase and leads to the phosphorylation of p100 and its subsequent processing to p52 (37). The NIK/IKKα/NF-κB2 signaling pathway is required for proper lymphoid organ development and B cell survival, and is also activated by the B cell survival factor BlyS/BAFF (38). While the precise role of this pathway in B cell terminal differentiation awaits further study, mice transgenic for BAFF have an increased number of B cells, elevated serum Ig levels, and anti-DNA autoantibodies (39). Furthermore, treatment with anti-CD40L or a soluble decoy receptor for BAFF has been shown to ameliorate murine lupus and is characterized by a sharp decline in mortality, autoantibody development, and renal damage (40). Taken together, these findings suggest that the NIK/IKKα/NF-κB2 signaling pathway has a role in the development of autoimmunity. Further definition of this NEMO-independent signaling pathway may lead to the identification of additional factors that influence mutational activity.
In summary, the zinc finger domain of NEMO exerts a regulatory role in B cell maturation. Hypomorphic mutations in this region obstruct normal CD40-mediated activation of NF-κB and impair the expression of target genes that are necessary for B cell CSR and SHM. The expression of AID can be induced normally in XHM-ED, and we have identified putative cofactors that may be necessary for its function. Future studies will be needed to address how these cofactors work coordinately with AID to produce efficient neutralizing antibody responses.
Methods
Patients and protocols.
Patients were studied at the Clinical Center, NIAID, NIH (protocol 89-1-0158). The diagnosis of XHM-ED and XHM was established in the patients by medical and family history and Ig profile. For molecular conformation, DNA and total RNA were extracted from lymphocytes using standard methods. PCR of genomic DNA was performed with primers flanking each exon using primers and PCR cycling conditions previously described (6). cDNA was obtained from RNA by RT-PCR using NEMO-specific primers. The PCR products were purified and cycle sequenced at both the 5′ and 3′ ends using dye terminator dideoxynucleotides. Unaffected family members of patients, other normal volunteers, and immunologically normal individuals in other NIH studies served as controls. The Institutional Review Board of NIAID approved the open protocol and informed consent was obtained from all patients or their parents before enrollment in the study.
Quantitative real-time RT-PCR.
Total RNA was prepared from CD19-positive B cells using TRIzol reagent (Invitrogen Corp.) and treated with RQ1 RNase-free DNase (Promega Corp.). cDNA was synthesized using random hexamers and SuperScript II or ThermoScript reverse transcriptase (Invitrogen Corp.) following the manufacturer’s protocol. Quantitative real-time PCR was performed in a 25-μl reaction in an ABI Prism 7700 Sequence Detector (Applied Biosystems) using TaqMan Universal PCR Master Mix (Applied Biosystems). Specific oligonucleotide primers were used to amplify 100–200 bp fragments from each genes along with an oligonucleotide probe dual-labeled with 5′-6FAM and 3′-TAMRA. The primers and probe combinations were determined by Primer Express software (Applied Biosystems). Normalization was done by including a sample with primers and probe for 18S rRNA (Applied Biosystems). A threshold was set in the linear range of the amplification curve, and the number of cycles needed to reach it was calculated for every gene.
Mutation analysis.
B cells were isolated using Dynal anti-CD19–coated magnetic beads according to the manufacturer’s instructions. Total RNA extraction was performed using TRIzol reagent (Invitrogen Corp.), and cDNA was synthesized using methods previously described (41). PCR was then performed using Taq DNA polymerase (Roche Diagnostics Corp.) and the following primers: the V3-23 leader exon (5′-GGCTGAGCTGGCTTTTTCTTGTGG-3′) and Cμ (5′-TCACAGGAGACGAGGGGGAA-3′). The PCR products were purified and cloned using the TA cloning kit (Invitrogen Corp.) and sequenced using M13 universal primers.
Electrophoretic mobility shift assays.
Cellular lysates were prepared by resuspending cell pellets in 100 mM NaCl, 50 mM Tris-Cl at pH 8.0, 0.5% NP-40, 50 mM NaF, 30 mM sodium pyrophosphate, 1 mM Na3VO4, 0.6% diisopropyl fluorophosphate (Sigma-Aldrich), and 1 Complete protease inhibitor mixture (Roche Diagnostics Corp.). Binding reactions for c-Rel and RelA (p65) were performed using the c-Rel Nushift Plus kit (Active Motif Inc.) following the manufacturer’s recommendations. The AGCTTGGGGTATTTCCAGCCG oligonucleotide used as a probe binds to c-Rel and p65, and the supershift was performed using a c-Rel–specific antibody. A Typhoon fluorescent imager (Amersham Biosciences) using ImageQuant software was used for quantification.
To determine NEMO and c-Rel protein levels, supernatants were collected from cell lysates and the protein concentration was determined by the Bio-Rad protein assay method with BSA as a standard. Samples were then applied to SDS-PAGE followed by transfer onto nitrocellulose membranes. The membranes were probed with either anti-NEMO (BD) or anti–c-Rel antibody (Active Motif Inc.). This was followed by HRP-conjugated donkey anti-mouse IgG (Amersham Biosciences) and developed by the chemiluminescence method.
Flow cytometry.
The specimens were stained by flow cytometry with the whole-blood lysis technique and analyzed using a FACScan (BD Biosciences – Immunocytometry Systems) with CellQuest software. The monoclonal antibodies used included anti-CD40 (5C3), anti-IgM (G20-127), anti-IgD (IA62), anti-CD19 (HIB19), anti-CD3 (Leu-4), and anti-CD27 (Becton, Dickinson and Co.). Irrelevant antibodies of the IgG1, IgG2a, and IgG2b subclasses were used to ascertain background staining. To calculate absolute numbers of each lymphocyte subgroup, the percentage of cells staining positive was multiplied by the absolute count of peripheral-blood lymphocytes as determined by Coulter counter (Celdyne Corp.), followed by a differential leukocyte count in a blood sample obtained simultaneously. For CD69, CD95, and CD86 expression, B cells were isolated from PBMCs using anti-CD19 magnetic beads according to the manufacturer’s instructions (Dynal Inc.). Cells (1 × 106) were cultured in 1 ml RPMI with 10% FCS, and CD40L trimer was used at 2 μg/ml.
In vitro Ig production.
PBMCs were isolated by Ficoll-Hypaque density centrifugation. B cells were isolated with Dynal anti-CD19–coated magnetic beads according to the manufacturer’s instructions. Cells (2 × 105 per ml) were stimulated in RPMI with 10% FCS, CD40L (2 μg/ml), and IL-4 (10 ng/ml; PeproTech Inc.). Ig levels were determined 14 days after stimulation by ELISA.
Proliferation assay.
CD19+ B cells (1 × 105) from patients or normal controls were stimulated in RPMI 1640, 10% FCS with CD40L (2 μg/ml) plus IL-4 (10 ng/ml), or CD40L plus IL-4 and anti-IgM Fab (100 ng/ml; Southern Biotechnology Associates Inc.) for 72 hours. Subsequently, the cells were pulsed with 1 μCi of tritiated thymidine for 8 hours. Cells were then harvested and thymidine incorporation was measured using a 1450 microbeta liquid scintillation counter (Wallac).
Gene array analysis.
RNA was prepared from PBMCs by standard protocols. Expression profiles were established by hybridization of labeled cRNA to U95A chips (Affymetrix Inc.), consisting of approximately 12,000 human genes from UniGene (Build 95). Total RNA (5 μg) from individual samples was labeled according to standard protocols (Affymetrix Inc.) and hybridized to individual arrays. Hybridized chips were processed as recommended and scanned images were analyzed using Resolver software (Rosetta Inpharmatics LLC).
Supplementary Material
Acknowledgments
We thank the families studied for their invaluable contribution to this project; D. Prosser, K. Kerkof, M. Timour, and S. Dorje for DNA microarray support; D. Silber and J. Neimela for DNA sequencing; W. Strober and P. Lipsky for helpful discussions; and U. Siebenlist, P. Schwartzberg, and R. Geha for critical review of the manuscript. This work was supported by the Immune Deficiency Foundation and the National Foundation of Ectodermal Dysplasia.
Ashish Jain and Chi A. Ma contributed equally to this work.
Footnotes
Nonstandard abbreviations used: AID, activation-induced cytidine deaminase; CD40L, CD40 ligand; CSR, class switch recombination; IκB, inhibitor of NF-κB; LIG4, DNA ligase IV; NEMO, NF-κB essential modulator; SHM, somatic hypermutation; XHM-ED, X-linked hyper-IgM syndrome with ectodermal dysplasia.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 2.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 4.Yoshikawa K, et al. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 2002;296:2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- 5.Karin M, Ben-Neriah Y. Phos-phorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 6.Jain A, et al. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat. Immunol. 2001;2:223–228. doi: 10.1038/85277. [DOI] [PubMed] [Google Scholar]
- 7.Doffinger R, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat. Genet. 2001;27:277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 8.Zonana J, et al. A novel X-linked disorder of immune deficiency and hypohidrotic ectodermal dysplasia is allelic to incontinentia pigmenti and due to mutations in IKK-gamma (NEMO) Am. J. Hum. Genet. 2000;67:1555–1562. doi: 10.1086/316914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orange JS, et al. Deficient natural killer cell cytotoxicity in patients with IKK-/NEMO mutations. J. Clin. Invest. 2002;109:1501–1509. doi:10.1172/JCI200214858. doi: 10.1172/JCI14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart AK, Huang C, Stollar BD, Schwartz RS. High-frequency representation of a single VH gene in the expressed human B cell repertoire. J. Exp. Med. 1993;177:409–418. doi: 10.1084/jem.177.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makris C, Roberts JL, Karin M. The carboxyl-terminal region of IkappaB kinase gamma (IKKgamma) is required for full IKK activation. Mol. Cell. Biol. 2002;22:6573–6581. doi: 10.1128/MCB.22.18.6573-6581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Digicaylioglu M, Lipton SA. Erythropoietin-mediated neuroprotection involves cross-talk between Jak2 and NF-kappaB signalling cascades. Nature. 2001;412:641–647. doi: 10.1038/35088074. [DOI] [PubMed] [Google Scholar]
- 13.Sanjabi S, Hoffmann A, Liou HC, Baltimore D, Smale ST. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc. Natl. Acad. Sci. U. S. A. 2000;97:12705–12710. doi: 10.1073/pnas.230436397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macpherson AJ, et al. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 15.Chirmule N, Tazelaar J, Wilson JM. Th2-dependent B cell responses in the absence of CD40-CD40 ligand interactions. J. Immunol. 2000;164:248–255. doi: 10.4049/jimmunol.164.1.248. [DOI] [PubMed] [Google Scholar]
- 16.Atkinson TP, et al. Leukocyte transfusion-associated granulocyte responses in a patient with X-linked hyper-IgM syndrome. J. Clin. Immunol. 1998;18:430–439. doi: 10.1023/a:1023286807853. [DOI] [PubMed] [Google Scholar]
- 17.Tang ED, Wang CY, Xiong Y, Guan KL. A role for NF-kappaB essential modifier/IkappaB kinase-gamma (NEMO/IKKgamma) ubiquitination in the activation of the IkappaB kinase complex by tumor necrosis factor-alpha. J. Biol. Chem. 2003;278:37297–37305. doi: 10.1074/jbc.M303389200. [DOI] [PubMed] [Google Scholar]
- 18.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115:565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 19.Zhou H, et al. Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature. 2004;427:167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- 20.Pohl T, et al. The combined absence of NF-kappa B1 and c-Rel reveals that overlapping roles for these transcription factors in the B cell lineage are restricted to the activation and function of mature cells. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4514–4519. doi: 10.1073/pnas.072071599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zelazowski P, et al. B cells genetically deficient in the c-Rel transactivation domain have selective defects in germline CH transcription and Ig class switching. J. Immunol. 1997;159:3133–3139. [PubMed] [Google Scholar]
- 22.Okazaki IM, Kinoshita K, Muramatsu M, Yoshikawa K, Honjo T. The AID enzyme induces class switch recombination in fibroblasts. Nature. 2002;416:340–345. doi: 10.1038/nature727. [DOI] [PubMed] [Google Scholar]
- 23.Ma L, Hu B, Kenter AL. Ig S gamma-specific DNA binding protein SNAP is related to the helix-loop-helix transcription factor E47. Int. Immunol. 1997;9:1021–1029. doi: 10.1093/intimm/9.7.1021. [DOI] [PubMed] [Google Scholar]
- 24.Wuerffel R, et al. Switch recombination breakpoints are strictly correlated with DNA recognition motifs for immunoglobulin S gamma 3 DNA-binding proteins. J. Exp. Med. 1992;176:339–349. doi: 10.1084/jem.176.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenter AL, Wuerffel R, Sen R, Jamieson CE, Merkulov GV. Switch recombination breakpoints occur at nonrandom positions in the S gamma tandem repeat. J. Immunol. 1993;151:4718–4731. [PubMed] [Google Scholar]
- 26.Miyamoto A, Cui X, Naumovski L, Cleary ML. Helix-loop-helix proteins LYL1 and E2a form heterodimeric complexes with distinctive DNA-binding properties in hematolymphoid cells. Mol. Cell. Biol. 1996;16:2394–2401. doi: 10.1128/mcb.16.5.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrier R, et al. Physical interaction of the bHLH LYL1 protein and NF-kappaB1 p105. Oncogene. 1999;18:995–1005. doi: 10.1038/sj.onc.1202374. [DOI] [PubMed] [Google Scholar]
- 28.Petersen S, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tashiro J, Kinoshita K, Honjo T. Palindromic but not G-rich sequences are targets of class switch recombination. Int. Immunol. 2001;13:495–505. doi: 10.1093/intimm/13.4.495. [DOI] [PubMed] [Google Scholar]
- 30.Roddam PL, et al. Genetic variants of NHEJ DNA ligase IV can affect the risk of developing multiple myeloma, a tumour characterised by aberrant class switch recombination. J. Med. Genet. 2002;39:900–905. doi: 10.1136/jmg.39.12.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaffer AL, et al. Signatures of the immune response. Immunity. 2001;15:375–385. doi: 10.1016/s1074-7613(01)00194-7. [DOI] [PubMed] [Google Scholar]
- 32.Klein U, et al. Transcriptional analysis of the B cell germinal center reaction. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dedeoglu F, Horwitz B, Chaudhuri J, Alt FW, Geha RS. Induction of activation-induced cytidine deaminase gene expression by IL-4 and CD40 ligation is dependent on STAT6 and NFkappaB. Int. Immunol. 2004;16:395–404. doi: 10.1093/intimm/dxh042. [DOI] [PubMed] [Google Scholar]
- 34.Zhou C, Saxon A, Zhang K. Human activation-induced cytidine deaminase is induced by IL-4 and negatively regulated by CD45: implication of CD45 as a Janus kinase phosphatase in antibody diversification. J. Immunol. 2003;170:1887–1893. doi: 10.4049/jimmunol.170.4.1887. [DOI] [PubMed] [Google Scholar]
- 35.Ta VT, et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat. Immunol. 2003;4:843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 36.Doi T, Kinoshita K, Ikegawa M, Muramatsu M, Honjo T. De novo protein synthesis is required for the activation-induced cytidine deaminase function in class-switch recombination. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2634–2638. doi: 10.1073/pnas.0437710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coope HJ, et al. CD40 regulates the processing of NF-kappaB2 p100 to p52. EMBO J. 2002;21:5375–5385. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dixit V, Mak TW. NF-kappaB signaling. Many roads lead to madrid. Cell. 2002;111:615–619. doi: 10.1016/s0092-8674(02)01166-2. [DOI] [PubMed] [Google Scholar]
- 39.Mackay F, Schneider P, Rennert P, Browning J. BAFF AND APRIL: a tutorial on B cell survival. Annu. Rev. Immunol. 2003;21:231–264. doi: 10.1146/annurev.immunol.21.120601.141152. [DOI] [PubMed] [Google Scholar]
- 40.Early GS, Zhao W, Burns CM. Anti-CD40 ligand antibody treatment prevents the development of lupus-like nephritis in a subset of New Zealand black x New Zealand white mice. Response correlates with the absence of an anti-antibody response. J. Immunol. 1996;157:3159–3164. [PubMed] [Google Scholar]
- 41.Levy Y, et al. Defect in IgV gene somatic hypermutation in common variable immuno-deficiency syndrome. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13135–13140. doi: 10.1073/pnas.95.22.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.