Abstract
Mitral valve prolapse (MVP) is a common human phenotype, yet little is known about the pathogenesis of this condition. MVP can occur in the context of genetic syndromes, including Marfan syndrome (MFS), an autosomal-dominant connective tissue disorder caused by mutations in fibrillin-1. Fibrillin-1 contributes to the regulated activation of the cytokine TGF-β, and enhanced signaling is a consequence of fibrillin-1 deficiency. We thus hypothesized that increased TGF-β signaling may contribute to the multisystem pathogenesis of MFS, including the development of myxomatous changes of the atrioventricular valves. Mitral valves from fibrillin-1–deficient mice exhibited postnatally acquired alterations in architecture that correlated both temporally and spatially with increased cell proliferation, decreased apoptosis, and excess TGF-β activation and signaling. In addition, TGF-β antagonism in vivo rescued the valve phenotype, suggesting a cause and effect relationship. Expression analyses identified increased expression of numerous TGF-β–related genes that regulate cell proliferation and survival and plausibly contribute to myxomatous valve disease. These studies validate a novel, genetically engineered murine model of myxomatous changes of the mitral valve and provide critical insight into the pathogenetic mechanism of such changes in MFS and perhaps more common nonsyndromic variants of mitral valve disease.
Introduction
Marfan syndrome (MFS) is a common, autosomal-dominant, systemic disorder of connective tissue, with an estimated prevalence of 1 in 5,000–10,000 individuals (1). It is caused by mutations in FBN1, the gene encoding fibrillin-1 (2), the principal component of extracellular matrix microfibrils. Clinical manifestations of MFS include skeletal deformities, ocular lens dislocation, lung pathology, and cardiac complications, such as aortic dissection and mitral valve prolapse (MVP) and dysfunction. Mitral valve disease is the leading indication for surgery and cause of death in young children with MFS, and there are currently no known or proposed medical therapies for prophylactic treatment of valve disease. While infants presenting with the most severe and rapidly progressive form of MFS can show mitral valve dysfunction at birth, manifestations are highly variable in the classic form of MFS, and myxomatous changes are not easily quantified using noninvasive imaging modalities, precluding a precise understanding of the natural history of disease. To our knowledge, neither recapitulation nor mechanistic exploration of mitral valve pathology in mouse models of MFS has previously been documented.
Little is known about the precise pathogenetic sequence that culminates in either primary or Marfan-related myxomatous mitral valve disease. Increased production of both collagen, particularly type III collagen, and glycosaminoglycans has been documented in myxomatous valves (3), and some postulate that this may be the result of a leaflet response to repeated mechanical stress (4). The prevailing hypothesis is that impact-induced damage to valvular endothelium leads to the release of vasoactive mediators that affect the subendothelial tissue. One such mediator is endothelin, which can cause cell proliferation and collagen synthesis in fibroblasts and smooth muscle cells (5, 6). Indeed, a correlation has been found between increased endothelin-receptor density and disease severity in a canine model of mitral valve prolapse (7). Increased activity of NADPH-diaphorase, which reflects nitric oxide synthase expression, has additionally been documented in canine myxomatous mitral valve leaflets (8). Thus, nitric oxide, which can antagonize the actions of endothelin (9), has also been implicated in the pathogenesis of myxomatous mitral valve changes. The precise contribution of these and other potential mediators of disease pathogenesis remains unclear. Furthermore, in cases where MVP is associated with a known genetic defect, such as in MFS, mitral valve disease may not simply manifest repeated injury but rather may reflect altered morphogenetic events during development and/or postnatal remodeling of the atrioventricular (AV) valves.
The contribution of fibrillin-1, which is traditionally viewed as a structural matrix protein, to the process of valvulogenesis is not an obvious one. Indeed, some manifestations of MFS, such as lens dislocation, could plausibly be caused by a structural deficiency of fibrillin-1. However, others, including AV valve disease, are less easily reconciled by simple loss of mechanical tissue integrity and more likely manifest perturbation of cell number and performance. In addition to its role as a structural component of the extracellular matrix microfibril, fibrillin-1 regulates TGF-β activation, and it targets and concentrates TGF-β at specific locations through its interaction with latent TGF-β–binding proteins (LTBPs) (10, 11). LTBPs form a bridge between matrix microfibrils and latency-associated peptide (LAP), the TGF-β propeptide that remains noncovalently linked to TGF-β and aids in matrix sequestration. These data suggest that the inactive large latent complex (TGF-β, LAP, and LTBP) is stabilized and/or less prone to activation by virtue of interaction with fibrillin-1 and perhaps other components of the extracellular matrix. Prior use of fibrillin-1–deficient mice revealed that fibrillin-1 regulates the activation of TGF-β in the developing lung and that enhanced signaling in the fibrillin-1–deficient state contributes directly to failed distal alveolar septation (12). In this study we demonstrate the contribution of TGF-β–induced cellular events to both morphogenic and functional perturbations of the AV valves in a murine model of MFS.
Results
Morphometric analyses of fibrillin-1–deficient mitral valves.
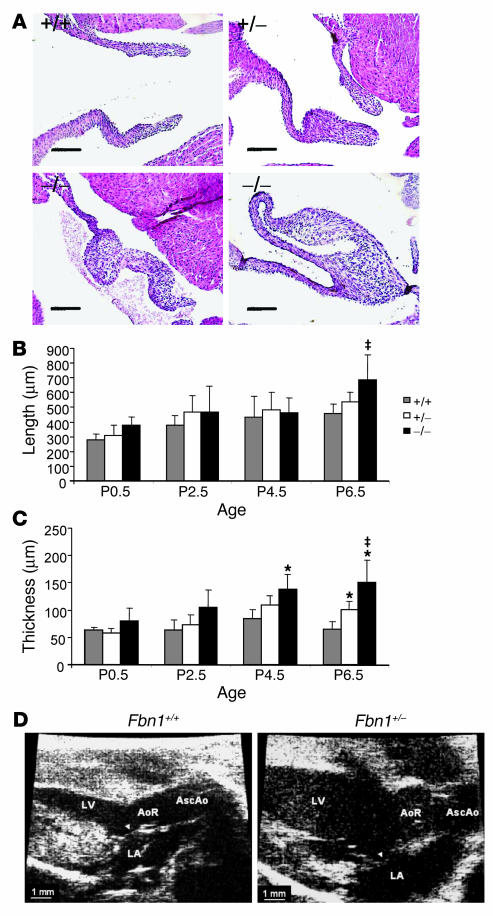
We performed a dedicated assessment of mitral valve pathology in a murine model of MFS at multiple time points in early postnatal life (P0.5, P2.5, P4.5, and P6.5). The model used in our studies contains a cysteine substitution (C1039G) in an epidermal growth factor–like domain in fibrillin-1 (13), representative of the most common class of mutation observed in MFS. Although heterozygous (Fbn1C1039G/+) mice do not die prematurely, they show the progressive deterioration of aortic wall architecture characteristic of patients with MFS. Mice homozygous with respect to this knock-in mutation (Fbn1C1039G/C1039G) fully recapitulate the vascular phenotype of MFS, as evidenced by death at P7–P10 from aortic dissection (13). Both Fbn1C1039G/+ and Fbn1C1039G/C1039G mice showed dramatic alterations in mitral valve architecture, including an increase in leaflet length and thickness (Figure 1A). Fbn1C1039G/C1039G mutant valves uniquely showed bizarre configurations in which leaflet tips folded back and fused to more proximal segments. Morphometric analyses confirmed these visual observations. A progressive increase in leaflet length was noted during postnatal development (Figure 1B), with a significant difference between Fbn1+/+ and Fbn1C1039G/C1039G mice appearing by P6.5. Mutant valves also showed a progressive increase in thickness during postnatal development (Figure 1C), with Fbn1C1039G/+ mice demonstrating intermediate values between the Fbn1+/+ and Fbn1C1039G/C1039G animals. Differences achieved significance in Fbn1C1039G/C1039G valves by P4.5, and by P6.5, differences between all genotypes were statistically significant. Echocardiograms performed at 9 months of age documented functional consequences of altered AV valve morphology including prolapse of the mitral valve with left atrial and ventricular enlargement (Figure 1D) and mitral valve regurgitation (data not shown). These data validated this fibrillin-1–deficient mouse as a bona fide model of myxomatous mitral valve disease and documented a predominant contribution of postnatal events.
Figure 1.
Histologic and morphometric assessment of mitral valve architecture in P6.5 mice. Fbn1 genotypes are indicated as follows: +/+ (Fbn1+/+), +/− (Fbn1C1039G/+), and −/− (Fbn1C1039G/C1039G). (A) Representative mitral valve sections from each genotype at P6.5 showing increased length and thickness in mutant valves as compared to wild-type littermates. Magnification, ×20. Scale bars: 100 μm. (B) Morphometric analysis of mitral valve length during the first week of postnatal life. Fbn1C1039G/C1039G valves were significantly longer by P6.5 when compared with those of Fbn1+/+ animals (–;P –; 0.05). (C) Morphometric analysis of mitral valve thickness during the first week of postnatal life. Increased valve thickness in Fbn1C1039G/C1039G versus Fbn1+/+ mice was significant by P4.5 (*P –; 0.005 vs. Fbn1+/+), and by P6.5, differences between all genotypes were statistically significant (*P –; 0.005 vs. Fbn1+/+; –;P –; 0.05 vs. Fbn1C1039G/+). Error bars indicate 95% confidence intervals. (D) Echocardiographic parasternal long axis views of 9-month Fbn1C1039G/+ and Fbn1+/+ mouse hearts. The anterior mitral valve leaflet (arrowhead) shows increased length and thickening, as well as systolic prolapse into the LA in fibrillin-1–;deficient mice. AoR, aortic root; AscAo, ascending aorta; LA, left atrium.
TGF-β activity and signaling in fibrillin-1–deficient mitral valves.
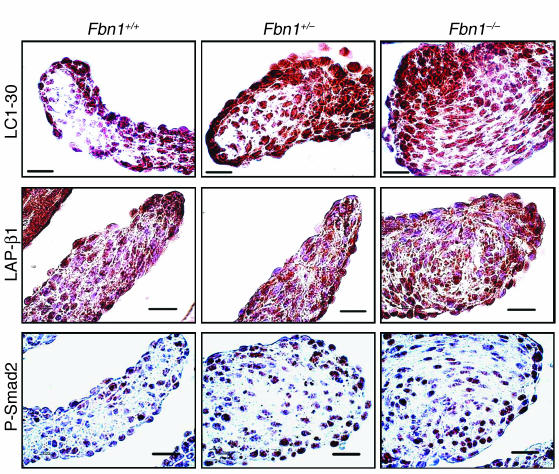
We assessed TGF-β activity in mutant valves using a polyclonal antibody (LC1-30) specific for the free and active form of TGF-β (14) (Figure 2). A graded increase in immunoactivity was observed in the valve leaflets in Fbn1C1039G/+ and Fbn1C1039G/C1039G animals. If this simply reflected an increase in production and secretion of TGF-β, we would expect to see a corresponding increase in the amount of LAP-β1, as the abundance of this peptide reflects that of stored, latent TGF-β. In contrast, we observed no apparent difference in the level of LAP-β1 between genotypes (Figure 2). Use of an antibody directed against the phosphorylated form of Smad2 (P-Smad2), which reflects receptor-mediated activation of the intracellular TGF-β response pathway, allowed us to assess TGF-β signaling (Figure 2). An increase in P-Smad2 in the mutant valves was observed. Taken together, these data are consistent with increased cytokine activation in the mutant mitral valves and demonstrate that the increased free TGF-β can support increased signaling in this developmental context.
Figure 2.
TGF-β activity and signaling in mitral valves of fibrillin-1–;deficient mice. Immunohistochemical analysis using antibodies specific to free and active TGF-β (LC1-30), LAP-β1, and P-Smad2 in wild-type (Fbn1+/+) mice and littermates either heterozygous (Fbn1+/–;) or homozygous (Fbn1–;/–;) for mutation C1039G. Mutant valves show increased TGF-β activity and signaling but not increased expression of LAP-β1, indicative of increased TGF-β activation rather than cytokine production. Magnification, ×100. Scale bars: 20 μm.
Phenotypic rescue of mitral valve phenotype after TGF-β antagonism in vivo.
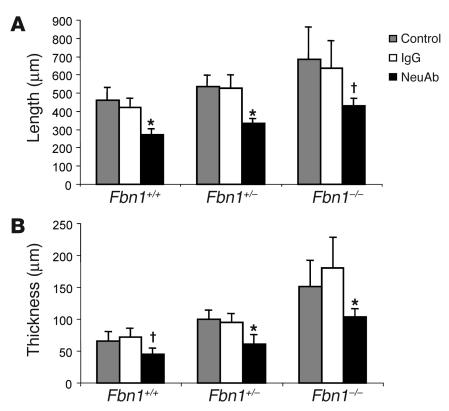
Further investigation of a cause-and-effect relationship between excess TGF-β activation and signaling and altered mitral valve morphogenesis was accomplished using TGF-β antagonism in vivo. Pregnant mice received intraperitoneal injection of TGF-β–neutralizing antibodies at days 14.5 and 17.5 postcoitum, and mitral valves were examined in pups at P6.5. Both TGF-β isoforms 1 and 2 are neutralized in vivo and in vitro by this antibody (15, 16). As shown in Figure 3, valve leaflet length and thickness were significantly decreased in all 3 genotypes after TGF-β antagonism. There was clearly an effect of TGF-β antagonism on valve length and thickness in Fbn1+/+ animals, but there was an increased effect in mutant animals with full normalization in Fbn1C1039G/+ mice. These data demonstrate a role for TGF-β as a physiologic regulator of mitral valve morphology and support a causal relationship between TGF-β dysregulation and mitral valve disease in MFS.
Figure 3.
Phenotypic rescue after TGF-β antagonism. (A) Morphometric analysis of mitral valve length at P6.5 in untreated (control) mice, in mice receiving irrelevant IgG (IgG), and in mice receiving TGF-β–;neutralizing Ab (NeuAb). Leaflet length was significantly shorter in neuAb-treated mice. (B) Morphometric analysis of mitral valve thickness after TGF-β antagonism. Valves from NeuAb-treated mice were significantly less thick than those from IgG-treated counterparts for all genotypes Error bars indicated 95% confidence intervals. *P –; 0.005 vs. IgG; P –; 0.01 vs. IgG.
Elucidation of TGF-β–related effectors contributing to myxomatous mitral valve disease using expression analyses.
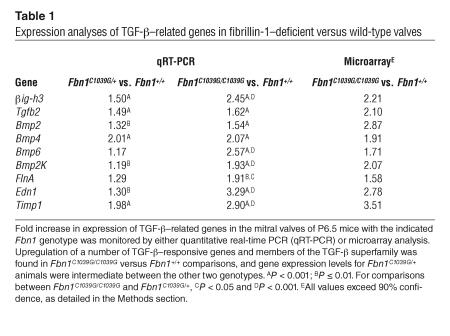
Our next initiative was to identify effector molecules downstream of TGF-β signaling that contribute to morphogenesis of the AV valves. A number of candidate genes that are either regulated by TGF-β or are members of the TGF-β superfamily showed increased expression in valve leaflets derived from fibrillin-1–deficient mice at P6.5, as assessed by quantitative real-time PCR. Informatively, these initially selected candidates, as well as other TGF-β–responsive genes, also exhibited increased expression on microarray analysis (Table 1). A comparison of Fbn1C1039G/+ with Fbn1+/+ animals showed an intermediate gene expression difference for most of the genes that were tested. The results of these expression analyses further support the hypothesis that increased TGF-β activation and signaling contributes to the pathogenesis of mitral valve disease in the fibrillin-1–deficient state.
Table 1.
Expression analyses of TGF-β–;related genes in fibrillin-1–;deficient versus wild-type valves
Exploration ofβIGH3, cellular proliferation, apoptosis, and bone morphogenetic proteins in the mitral valve.
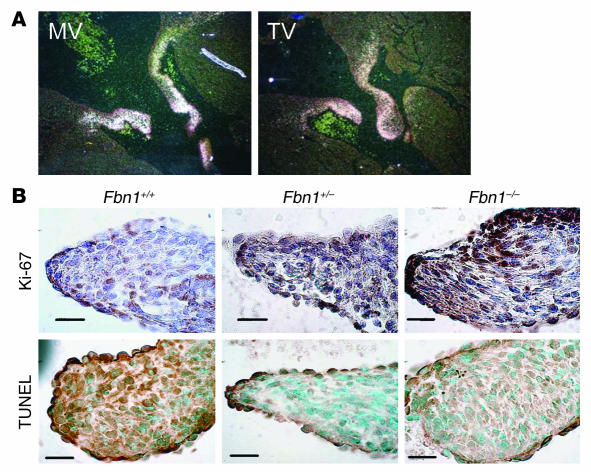
One gene that we found to be upregulated in our murine expression analyses was TGF-β–inducible gene H3 (βig-h3). Due to technical limitations using fetal mouse hearts, the prenatal expression of βIGH3 was assessed using in situ hybridization in fetal bovine hearts. This revealed substantial and specific expression of βIGH3 in the mitral and tricuspid valves of late-gestation embryos (Figure 4A), providing initial evidence of a role for βIGH3 in AV valve development.
Figure 4.
Potential downstream effectors of the myxomatous phenotype. (A) βIGH3 expression in late embryonic bovine mitral (MV) and tricuspid (TV) valves, as assessed by in situ hybridization. (B) Assessment of cell proliferation and apoptosis in P6.5 murine mitral valves using Ki-67 and TUNEL staining, respectively. Mutant valves showed both increased cell proliferation and decreased apoptosis. Magnification, ×100. Scale bars: 20 μm.
In order to determine what processes might contribute to myxomatous changes of the mitral valve in MFS, we stained valves with Ki-67 and TUNEL, markers of cellular proliferation and apoptosis, respectively. We found that fibrillin-1–deficient valves exhibited both increased cell proliferation as well as decreased apoptosis as compared to valves from wild-type littermates (Figure 4B).
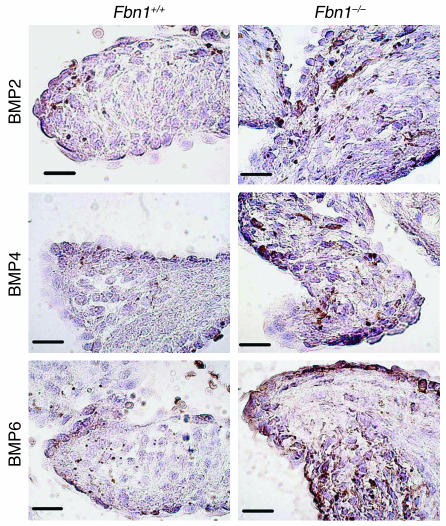
Finally, we examined the expression of bone morphogenetic proteins (BMPs) 2, 4, and 6, all of which were found to be upregulated in mutant valves by expression analyses (Figure 5). Valves from mice homozygous for the fibrillin-1 mutation showed increased expression of BMPs 2, 4, and 6, consistent with our expression analysis data.
Figure 5.
Expression of BMPs 2, 4, and 6 in fibrillin-1–;deficient mitral valves. Fbn1C1039G/C1039G (Fbn1–;/–;) valves exhibited increased expression of BMPs 2, 4, and 6, as compared to Fbn1+/+ valves. Magnification, ×100. Scale bars: 20 μm.
Discussion
Myxomatous degeneration of the mitral valve with consequent valve prolapse and dysfunction is an extremely common phenotype, affecting approximately 5% of individuals in the general population. Given this frequency, there has been a surprising lack of mechanistic insight and genetically defined animal models available for investigating the pathogenesis of this phenotype. Through our studies, we have verified that the fibrillin-1–deficient mouse is a valid model of acquired myxomatous mitral valve disease. To our knowledge, this is the first genetically defined model for this common human phenotype. We have also documented increased TGF-β activity and signaling that correlates temporally and spatially with mitral valve pathology and have furthered the hypothesis of cause and effect between increased TGF-β signaling and mitral valve disease through TGF-β antagonism in vivo. As discussed, the pathogenesis of myxomatous mitral valve disease, both primary and Marfan-related, remains unclear, but current hypotheses have not invoked TGF-β as a potential mediator.
The results of our expression analyses have allowed us to begin to explore pathogenetic mechanisms and candidate effectors involved in mitral valve disease in the context of excess TGF-β signaling. One set of genes of interest are those that are regulated by TGF-β, including βIGH3, endothelin-1 (EDN1), and tissue inhibitor of metalloproteinase 1 (TIMP1). βIGH3 is an extracellular matrix protein whose expression is induced by TGF-β in several cell lines. Mutations in βIGH3 underlie a number of corneal dystrophies involving amyloid deposition in the cornea (17). The physiologic function of βIGH3 has not been well characterized, but in certain contexts, it has been shown to inhibit cell attachment (18) and support cell proliferation (19). βIGH3 is expressed in the murine endocardial cushions and heart during early embryonic valvulogenesis (20). We have shown that it is also robustly and specifically expressed in late embryonic bovine AV valve development. As previously discussed, EDN1 is postulated to play a role in the development of primary myxomatous mitral valve disease. Its expression is induced by TGF-β (21), and it is able to both support cell proliferation and suppress apoptosis (22). In addition to increased TGF-β signaling, myxomatous, fibrillin-1–deficient valves may be subject to increased mechanical stress, providing a second mechanism by which EDN1 expression could be upregulated. A third TGF-β–induced gene, TIMP1 (23), is involved in processes requiring extracellular matrix remodeling, including cellular proliferation and migration. We therefore hypothesize that myxomatous changes of the mitral valve in MFS may manifest decreased cell adhesion and increased cell proliferation and survival due to excess βIGH3, EDN1, and/or TIMP1 activity imposed by a deficiency of fibrillin-1. In keeping with this hypothesis, we documented increased cell proliferation and decreased apoptosis in mutant valves.
Mutant valve leaflets also showed upregulated expression of filamin α (FLNA), a cytoskeletal actin-binding protein that coordinates localization and activation of TGF-β receptor-activated Smads, particularly Smad2 (24). FLNA thus serves as a positive regulator of TGF-β signaling. Gain-of-function mutations in FLNA are responsible for X-linked inheritance of Melnick-Needles syndrome and frontometaphyseal dysplasia (25), both of which include mitral valve prolapse. Hence, increased expression of FLNA may be both a consequence of and a contributor to excess TGF-β signaling in fibrillin-1–deficient valve leaflets.
TGF-β isoforms are known to be significantly involved in AV valve formation in both mouse and chick (26), and mice deficient in Smad6, an inhibitor of TGF-β and BMP signaling, exhibit valvular hyperplasia (27). BMP2 is required for development of endocardial cushion mesenchyme in mouse (28) and acts synergistically with TGF-β3 to initiate epithelial-mesenchymal transition in chick (29). BMP4 helps to regulate mesenchymal cell proliferation in AV endocardial cushions in mouse (30), and BMP6:BMP7 double mutants display underdeveloped and misplaced valves (31). The overt AV valvular hyperplasia seen in mice that are deficient in heparin-binding EGF is associated with a dramatic increase in activated Smad1/4/5, indicative of dysregulation of BMPs and excessive BMP signaling (32). Taken together, these results suggest that precise regulation of multiple members of the TGF-β superfamily is required to achieve proper valve morphogenesis (33). Interestingly, a number of members of the TGF-β superfamily, including TGF-β2 and BMPs 2, 4, and 6, were found to be upregulated in our expression analyses. One explanation is that TGF-β is capable of positive feedback via both autoinduction and cross-regulation of other family members (34). With regard to the upregulation of BMPs, there are data that suggest that fibrillins can interact directly with and perhaps concentrate BMPs at sites of intended action (35, 36). Thus, it is difficult to predict whether the increase in transcript expression for selected BMPs would translate to increased activity or simply reflects feedback induction due to failure of cytokine concentration. Our observation of increased immunostaining for BMPs 2, 4, and 6 in mutant valve leaflets and upregulation of the BMP-responsive gene Bmp2k favors the former hypothesis. There is likely a complex network of altered signaling cascades involving multiple members of the TGF-β superfamily and their downstream effectors that contributes to altered mitral valve architecture and function in MFS. As illustrated in the case of endothelin-1, mechanisms previously thought to account for myxomatous changes in primary MVP may also be relevant to mitral valve disease in MFS. This work provides a novel context within which to further explore the pathogenesis of acquired myxomatous degeneration of the mitral valve in both Marfan syndrome and more common, non-syndromic variants of disease.
Methods
Mice.
All mouse protocols were approved by the Animal Care and Use Committee of Johns Hopkins University School of Medicine. Creation of the mouse line harboring Fbn1 mutations (C1039G) has been previously described (13). All analyses were performed after back-crossing this mutation into the C57BL/6J background, allowing valid comparisons between litters. For morphometric studies, 3–10 mice of each relevant genotype at each designated time point between P0.5–P6.5 were examined (3–5 for P0.5–P4.5 and 7–10 for P6.5). Ten mice per genotype at P6.5 were used to generate RNA for quantitative real-time PCR. For microarray analyses, mitral valves from 8 mice for each genotype were harvested at P6.5. Genotyping was performed as previously described (13).
Microscopic examination.
Mice were sacrificed with an inhalation overdose of halothane (Sigma-Aldrich). For morphometric analysis and immunohistochemistry, hearts were fixed in 10% buffered formalin overnight, arranged in 1.5% agar prior to paraffin embedding, and slides were prepared using 7-μm sections. Sections of paraffin-embedded tissue were prepared and stained with hematoxylin and eosin. Overnight incubations of P6.5 sections with primary antibodies recognizing active TGF-β (LC1-30, provided by K. Flanders, National Cancer Institute, Bethesda, Maryland, USA), LAP-β1 (R&D Systems), P-Smad2 (Cell Signaling Technology), and BMPs 2, 4, and 6, or Ki-67 (all from Santa Cruz Biotechnology Inc.) were performed. In each set of stains, negative controls which omitted the primary antibody were included. P0.5 sections were TUNEL-labeled using the TdT-FragEL DNA Fragmentation Detection Kit (Oncogene Research Products). Immunohistochemical and TUNEL stains were examined at ×100 magnification using an Eclipse E400 microscope (Nikon Inc.) and were compared to an internal 10-μm calibration standard included in each image.
Valve morphology.
Tissue sections were prepared as above and stained with hematoxylin and eosin. Mitral valves were examined at ×20 magnification and were compared to a 50-μm calibration standard included in each image. Mitral valve length was determined by examination of serial sections and by measuring the distance along the center of the leaflet from the valve origin to the leaflet tip. Mitral valve thickness was measured at the thickest point perpendicular to the length axis of the leaflet. Only sections in which the full leaflet tip could be viewed were included in analyses. Morphometric data from mice of the same genotype at a given age were combined and used to generate 95% confidence intervals. A Student’s t test was used to assess statistical significance. Echocardiography was performed using the Visualsonics Vevo 660 V1.3.6 imaging system and a 35-MHz transducer (Visualsonics model RMV603). Animals were sedated with inhalation of 1% isoflurane (Sigma-Aldrich), and heart rates were maintained at 500–550 bpm.
TGF-β–neutralizing antibody treatment.
Female heterozygotes who were pregnant from timed matings with male heterozygotes received intraperitoneal injection of TGF-β–neutralizing antibody (R&D Systems) on E14.5 and E17.5. The antibody was diluted in PBS (pH 7.4) to a concentration of 1 mg/ml and injected at a dosage of 10 mg/kg body wetight. Rabbit IgG (10 mg/kg; Zymed Laboratories Inc.) was administered in a similar fashion as a control. We used a minimum of 6 mice per genotype for morphometric analyses.
RNA preparation.
For quantitative real-time PCR and microarray analyses, hearts were removed from euthanized mice at P6.5 and immersed immediately in RNALater (QIAGEN Inc.). Both mitral valve leaflets were dissected in RNALater and stored at –80°C in Trizol (Invitrogen Corp.) until genotypes were determined. Eight valves for each genotype were pooled and homogenized using syringe lysis. RNA extraction was performed according to the standard Trizol protocol, using 10 μg of glycogen carrier.
Quantitative real-time PCR.
First-strand cDNA was synthesized using the SuperScript First-Strand Synthesis System for RT-PCR kit (Invitrogen Corp.). When possible, Assays on Demand (Applied Biosystems) ready-made primer and Taqman (Applied Biosystems) probes were used. Both the Assays on Demand and custom primers were designed to span introns to prevent detection of genomic DNA contaminants. Singleplex experiments, with samples run in triplicate, were carried out on the MJ Research Opticon system (MJ Research). Relative quantification using the standard curve method was used to compare gene expression between genotypes. Samples without cDNA and samples containing RNA but missing reverse transcriptase were run as controls for each gene tested. The housekeeping gene β-2 microglobulin was included to control for loading error. P values were obtained using ANOVA analyses with 2 factors (genotype and run variation) in an additive model.
Expression profiling using DNA microarrays.
Four samples of RNA derived from murine mitral valves were submitted for microarray analyses (1 Fbn1+/+, 2 Fbn1C1039G/+, and 1 Fbn1C1039G/C1039G). Samples were processed using the 2-round RNA amplification protocol described by Affymetrix (Affymetrix GeneChip Eukaryotic Small Sample Target Labeling Assay Version II; http://www.affymetrix.com/support/technical/technotes/smallv2_technote.pdf). Briefly, 100 ng of starting total RNA was used to synthesize first-strand cDNA using oligonucleotide probes with 24 oligo-dT plus T7 promoter as primer (Proligo LLC) and the SuperScript Choice System (Invitrogen Corp.). Following the double-stranded cDNA synthesis, the product was purified by phenol-chloroform extraction, and unlabeled ribonucleotides were used in a first round of in vitro transcription cRNA amplification (MegaScript, Ambion). The subsequent cycle of cDNA synthesis was started with random primers, and the oligo-dT with T7 promoter was again used as a primer for the second-strand cDNA synthesis step. The double-stranded cDNA product was then purified again by phenol-chloroform extraction. Biotinylated antisense cRNA was subsequently generated through in vitro transcription using the BioArray RNA HighYield Transcript Labeling kit (ENZO Life Sciences Inc). Then, 15 μg of the biotinylated labeled cRNA was fragmented at 94°C for 35 minutes (100 mM Trix-acetate, pH 8.2, 500 mM KOAc, 150 mM MgOAC), and 10 μg of total fragmented cRNA was hybridized to the Affymetrix murine genome GeneChip array MOE430A for 16 hours at 45°C with constant rotation. Affymetrix Fluidics Station 400 was then used to wash and stain the chips, removing the unhybridized target and incubating with a streptavidin-phycoerythrin conjugate to stain the biotinylated cRNA. The staining was then amplified using goat IgG as a blocking reagent and biotinylated anti-streptavidin antibody (goat), followed by a second staining step with a streptavidin-phycoerythrin conjugate. Fluorescence was detected using the Affymetrix GS3000 GeneArray Scanner, and image analysis of each GeneChip was done through the GeneChip Operating System software from Affymetrix (GCOS1.1.1), using the standard default settings. For comparison between different chips, we used global scaling, which scaled all probe sets to a user-defined target intensity (TGT) of 150.
To ascertain the quality of the total RNA from the samples, we used the Agilent Bioanalyzer Lab on a Chip technology and confirmed the rRNA ratios and clean run patterns of the samples. Likewise, this technology is used to confirm the quality of the RNA in the form of cRNA and fragmented cRNA. To assess the quality of the hybridization, GeneChip image, and comparison between chips, we confirmed the following parameters: scaling factor values within comparable range (between 0.7 and 1.4); low background values (between 51 and 65); high percentage of present calls (between 47 and 52); consistent 3′:5′ ratios of GAPDH as representation of housekeeping genes; and presence or absence of Bio B and C as internal spike controls.
These descriptions include all information currently considered under the Minimum Information About a Microarray Experiment (MIAME) supportive guidelines, with which the Johns Hopkins Medical Institution Microarray Core Facility abides in all its procedures.
Microarray data analysis.
The mRNA samples were analyzed using the Affymetrix MOE430A gene chip. Under the expectation that only a small fraction of genes would be differentially expressed between samples of different genotypes, we adjusted the brightness of the sample chips to a comparable level by normalizing the cell intensity file (.CEL) probe signal values at the Affymetrix probe pair (perfect match [PM]; mismatch [MM]) data level with the method of “invariant set normalization,” (37) which chooses a subset of PM probes with small within-subset rank difference between 2 chips through an iterative procedure to serve as the basis for fitting a normalization curve. The normalized .CEL data were then used to estimate the PM/MM-model-based expression index (with standard errors) for the probe sets, leading to further computation of the fold changes and their 90% confidence intervals (38). The lower bound of a 90% confidence interval, a conservative estimate of the fold change, was used to initially identify differentially expressed genes such that there was 90% confidence that the true fold change in expression was greater than or equal to 1.5. A difference of at least 50 in the magnitude of the signal intensity was set as a supplemental criterion to minimize the effects of “noise” (or nondifferentially expressed) genes. The computing was performed with dChip 1.2 and 1.3 (test version; http://www.dchip.org). Microarray data is posted online (http://www.ncbi.nlm.nih.gov/geo, accession number GSE1852).
In situ hybridization.
A 305-bp sequence from the central coding region of bovine βIGH3 was prepared as previously described (39). Seventy-day gestation bovine embryos were fixed in 10% buffered neutral formalin in a volume 20 times the weight of the tissue for 24–72 hours at room temperature. Sections of fixed embryos were cut at a thickness of 5 μm, and in situ hybridization was performed as previously described (39). Slides were counterstained with hematoxylin-eosin, and silver grains were visualized with the use of dark-field microscopy.
Acknowledgments
We thank members of H.C. Dietz’s laboratory for their support and advice. This work was supported by the Howard Hughes Medical Institute (to H.C. Dietz); NIH grants AR41135 and AR049698 (to H.C. Dietz), HL53325 (to R.P. Mecham), and HL067056 (to D.P. Judge); the National Marfan Foundation (to H.C. Dietz and A. Cheng); the William S. Smilow Center for Marfan Syndrome Research (to H.C. Dietz); the Dana and Albert “Cubby” Broccoli Center for Aortic Diseases (to D.P. Judge); and the Stanley J. Sarnoff Endowment for Cardiovascular Science, Inc. (to C.M. Ng).
Footnotes
Nonstandard abbreviations used: AV, atrioventricular; BMP, bone morphogenetic protein; EDN1, endothelin-1; FLNA, filamin α; βig-h3, TGF-β–inducible gene H3; LAP, latency-associated peptide; LTBP, latent TGF-β–binding protein; MFS, Marfan syndrome; MM, mismatch; MVP, mitral valve prolapse; PM, perfect match; P-Smad2, phosphorylated form of Smad2; TIMP1, tissue inhibitor of metalloproteinase 1.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Pyeritz RE. The Marfan syndrome. Annu. Rev. Med. 2000;51:481–510. doi: 10.1146/annurev.med.51.1.481. [DOI] [PubMed] [Google Scholar]
- 2.Dietz HC, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 3.Tamura K, et al. Abnormalities in elastic fibers and other connective-tissue components of floppy mitral valve. Am. Heart J. 1995;129:1149–1158. doi: 10.1016/0002-8703(95)90397-6. [DOI] [PubMed] [Google Scholar]
- 4.Durbin AD, Gotlieb AI. Advances towards understanding heart valve response to injury. Cardiovasc. Pathol. 2002;11:69–77. doi: 10.1016/s1054-8807(01)00109-0. [DOI] [PubMed] [Google Scholar]
- 5.Weber H, et al. Endothelin-1 and angiotensin-II stimulate delayed mitogenesis in cultured rat aortic smooth muscle cells: evidence for common signaling mechanisms. Mol. Endocrinol. 1994;8:148–158. doi: 10.1210/mend.8.2.8170471. [DOI] [PubMed] [Google Scholar]
- 6.Rizvi MAD, Katwa L, Spadone DP, Myers PR. The effects of endothelin-1 on collagen type I and type III synthesis in cultured porcine coronary artery vascular smooth muscle cells. J. Mol. Cell. Cardiol. 1996;28:243–252. doi: 10.1006/jmcc.1996.0023. [DOI] [PubMed] [Google Scholar]
- 7.Mow T, Pedersen HD. Increased endothelin-receptor density in myxomatous canine mitral valve leaflets. J. Cardiovasc. Pharmacol. 1999;34:254–260. doi: 10.1097/00005344-199908000-00011. [DOI] [PubMed] [Google Scholar]
- 8.Olsen LH, et al. Increased NADPH-diaphorase activity in canine myxomatous mitral valve leaflets. J. Comp. Path. 2003;129:120–130. doi: 10.1016/s0021-9975(03)00019-7. [DOI] [PubMed] [Google Scholar]
- 9.Boulanger C, Luscher TF. Release of endothelin from the porcine aorta. Inhibition by endothelium-derived nitric oxide. J. Clin. Invest. 1990;85:587–590. doi: 10.1172/JCI114477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dallas SL, Miyazono K, Skerry TM, Mundy GR, Bonewald LF. Dual role for the latent transforming growth factor-β binding protein in storage of latent TGF-β in the extracellular matrix and as a structural matrix protein. J. Cell Biol. 1995;131:539–549. doi: 10.1083/jcb.131.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isogai Z, et al. Latent transforming growth factor-β binding protein 1 interacts with fibrillin and is a microfibril-associated protein. J. Biol. Chem. 2003;278:2750–2757. doi: 10.1074/jbc.M209256200. [DOI] [PubMed] [Google Scholar]
- 12.Neptune ER, et al. Dysregulation of TGF-β activation contributes to pathogenesis in Marfan syndrome. Nat. Genet. 2003;33:407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 13.Judge DP, et al. Evidence for a critical contribution of haploinsufficiency in the complex pathogenesis of Marfan syndrome. J. Clin. Invest. 2004;114:172–181. doi:10.1172/JCI200420641. doi: 10.1172/JCI20641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flanders KC, et al. Transforming growth factor-β1: histochemical localization with antibodies to different epitopes. J. Cell Biol. 1989;108:653–660. doi: 10.1083/jcb.108.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomita H, et al. Early induction of transforming growth factor-β via angiotensin II type 1 receptors contributes to cardiac fibrosis induced by long-term blockade of nitric oxide synthesis in rats. Hypertension. 1998;32:273–279. doi: 10.1161/01.hyp.32.2.273. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto T, Takagawa S, Katayama I, Nishioka K. Anti-sclerotic effect of transforming growth factor-β antibody in a mouse model of bleomycin-induced scleroderma. Clin. Immunol. 1999;92:6–13. doi: 10.1006/clim.1999.4720. [DOI] [PubMed] [Google Scholar]
- 17.Munier FL, et al. Kerato-epithelin mutations in four 5q31-linked corneal dystrophies. Nat. Genet. 1997;15:247–251. doi: 10.1038/ng0397-247. [DOI] [PubMed] [Google Scholar]
- 18.Skonier J, et al. Beta ig-h3: a transforming growth factor-beta-reponsive gene encoding a secreted protein that inhibits cell attachment in vitro and suppresses the growth of CHO cells in nude mice. DNA Cell Biol. 1994;13:571–584. doi: 10.1089/dna.1994.13.571. [DOI] [PubMed] [Google Scholar]
- 19.Bae JS, et al. βig-h3 supports keratinocyte adhesion, migration, and proliferation through α3β1 integrin. Biochem. Biophys. Res. Commun. 2002;294:940–948. doi: 10.1016/S0006-291X(02)00576-4. [DOI] [PubMed] [Google Scholar]
- 20.Schorderet DF, et al. Genomic characterization and embryonic expression of the mouse Bigh3 (Tgfbi) gene. Biochem. Biophys. Res. Commun. 2000;274:267–274. doi: 10.1006/bbrc.2000.3116. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez W, Chen Z, Damon DH. Transforming growth factor-β regulation of endothelin expression in rat vascular cell and organ cultures. J. Cardiovasc. Pharmacol. 2001;37:219–226. doi: 10.1097/00005344-200102000-00009. [DOI] [PubMed] [Google Scholar]
- 22.Araki M, et al. Endothelin-1 as a protective factor against beta-adrenergic agonist induced apoptosis in cardiac myocytes. J. Am. Coll. Cardiol. 2000;36:1411–1418. doi: 10.1016/s0735-1097(00)00822-6. [DOI] [PubMed] [Google Scholar]
- 23.Yang YY, Tsai HF, Lu SC, Huang YF, Chang YC. Regulation of tissue inhibitors of metalloproteinase-1 gene expression by cytokines in human gingival fibroblasts. J. Endod. 2002;28:803–805. doi: 10.1097/00004770-200212000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki A, Masuda Y, Ohta Y, Ikeda K, Watanabe K. Filamin associates with smads and regulates transforming growth factor-β signaling. J. Biol. Chem. 2001;276:17871–17877. doi: 10.1074/jbc.M008422200. [DOI] [PubMed] [Google Scholar]
- 25.Robertson SP, et al. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat. Genet. 2003;33:487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- 26.Camenisch TD, et al. Temporal and distinct TGF-β ligand requirements during mouse and avian endocardial cushion morphogenesis. Dev. Biol. 2002;248:170–181. doi: 10.1006/dbio.2002.0731. [DOI] [PubMed] [Google Scholar]
- 27.Galvin KM, et al. A role for smad6 in development and homeostasis of the cardiovascular system. Nat. Genet. 2000;24:171–174. doi: 10.1038/72835. [DOI] [PubMed] [Google Scholar]
- 28.Sugi Y, Yamamura H, Okagawa H, Markwald RR. Bone morphogenetic protein-2 can mediate myocardial regulation of atriventricular cushion mesenchymal cell formation in mice. Dev. Biol. 2004;269:505–518. doi: 10.1016/j.ydbio.2004.01.045. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima Y, Yamagishi T, Hokari S, Nakamura H. Mechanisms involved in valvuloseptal endocardial cushion formation in early cardiogenesis: roles of transforming growth factor (TGF)-β and bone morphogenetic protein (BMP) Anat. Rec. 2000;258:119–127. doi: 10.1002/(SICI)1097-0185(20000201)258:2<119::AID-AR1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Jiao K, et al. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim RY, Robertson EJ, Solloway MJ. Bmp6 and Bmp7 are required for cushion formation and septation in the developing mouse heart. Dev. Biol. 2001;235:449–466. doi: 10.1006/dbio.2001.0284. [DOI] [PubMed] [Google Scholar]
- 32.Jackson LF, et al. Defective valvulogenesis in HB-EGF and TACE-null mice is associated with aberrant BMP signaling. EMBO J. 2003;2:2704–2716. doi: 10.1093/emboj/cdg264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delot EC. Control of endocardial cushion and cardiac valve maturation by BMP signaling pathways. Mol. Genet. Metab. 2003;80:27–35. doi: 10.1016/j.ymgme.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Bascom CC, et al. Complex regulation of transforming growth factor β1, β2, and β3 mRNA expression in mouse fibroblasts and keratinocytes by transforming growth factors β1 and β2. Mol. Cell. Biol. 1989;9:5508–5515. doi: 10.1128/mcb.9.12.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charbonneau NL, Ono RN, Corson GM, Keene DR, Sakai LY. Fine tuning of growth factor signals depends on fibrillin microfibril networks. Birth Defects Res., Part C, Embryo Today. 2004;72:37–50. doi: 10.1002/bdrc.20000. [DOI] [PubMed] [Google Scholar]
- 36.Arteaga-Solis E, et al. Regulation of limb patterning by extracellular microfibrils. J. Cell Biol. 2001;154:275–281. doi: 10.1083/jcb.200105046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection, Proc. Natl. Acad. Sci. U. S. A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:research0032.1–0032. doi: 10.1186/gb-2001-2-8-research0032. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prosser IW, et al. Regional heterogeneity of elastin and collagen gene expression in intralobar arteries in response to hypoxic pulmonary hypertension as demonstrated by in situ hybridization. Am. J. Pathol. 1989;135:1073–1088. [PMC free article] [PubMed] [Google Scholar]