Abstract
Background
We designed this study to investigate the influence of different ratios of n-6/n-3 polyunsaturated fatty acid in the diet of reflux esophagitis (RE) rats’ and the effect on the PI3K/Akt pathway.
Material/Methods
RE rats were randomly divided into a sham group and modeling groups of different concentrations of n-6/n-3 polyunsaturated fatty acid (PUFA): 12:1 group, 10:1 group, 5:1 group, and 1:1 group. RT-PCR and Western-blot were used to detect the expression of PI3K, Akt, p-Akt, NF-κBp50, and NF-κBp65 proteins in esophageal tissue.
Results
In the n-6/n-3 PUFAs groups the expression of PI3K, Akt, p-Akt, nf-kbp50, and NF-κBp65 mRNA decreased with the decrease in n-6/n-3 ratios in the diet. The lowest expression of each indicator occurred in the 1:1 n-6/n-3 group compared with other n-6/n-3 groups, the difference was statistically significant (p<0.05).
Conclusions
The inhibition of n-3 PUFAs in the development of esophageal inflammation in rats with RE was attributed to the function of PI3K/Akt-NF-κB signaling pathway.
MeSH Keywords: Esophagitis, Peptic; Fatty Acids, Omega-3; Rats, Inbred ACI
Background
Domestic as well as overseas scholars perceive that cellular inflammatory factors play important roles in reflux esophagitis (RE); nevertheless, the activation of inflammatory factors relies on the conduction of multiple signaling pathways inside and outside the cells. Some studies have confirmed that the phosphatidylinositol-3-kinase/serine/threonine kinase (PI3K/Akt) pathway is involved in most inflammatory reactions in vivo. The n-3 polyunsaturated fatty acid (PUFA) can modulate the gene expression of inflammatory factors through exerting effects on activation of related transcription factors in inflammatory pathways [1–5]. In recent decades, studies have illustrated that n-3 PUFAs as well as its metabolites (resolvins and protectin D1) are able to inhibit the generation of inflammatory factors and lessen cytokine response by inhibiting NF-κB, subsequently activating its immune regulation function [6,7].
Our study aimed to detect the expression of PI3K/Akt-NF-κB signaling pathway critical target protein in esophageal tissue of rats with reflux esophagitis by feeding reflux esophagitis rats with different ratio of n-6/n-3PUFAs in their diet, exploring its possible intervention mechanism.
Material and Methods
Experimental animals and forage
Experimental animal: sterile male Sprague Dawley (SD) rats weighing 230±20 g were purchased from SLRC Laboratory Animal Center in Shanghai, and kept at 25°C with 12 hour dark/light cycle. There were five rats in each cage.
Food: n-6PUFAs was supplied by “Golden Dragon Fish” brand sunflower seed oil; n-3 PUFAs was extracted from deep sea fish oil. The PUFAs were formulated into the pellet food in accordance with standard experimental protocols. Except for the PUFAs, other ingredients of the pellets were the same. The rats were fed at 9 am, as for the proxima luce. Remaining pellets were discarded, and subsequently, new food was added at a rate of 100 g/kg per rat every day.
Preparation of animal models
Lodophor was used to disinfect rats’ middle abdomen. About 1–2 cm laparotomy was performed at the xiphoid abdominal midline: the fur and abdominal muscles were with layered-cut. The peritoneum in the pylorus duodenal junction was isolated, avoiding blood vessels; loop ligature of nylon cable ties were used to create a predetermined inner diameter after the nylon tie passed through the pylorus duodenal junction. the redundant tie was snipped. The junction between forestomach and glandular stomach was ligated with 5-0 sutures. The stomach and duodenum were checked to ensure there was no hemorrhaging. The abdominal cavity was perfused using 0.5% metronidazole (1 mL) along with gentamicin (20,000 IU) and then closed using 5-0 sutures. Then 75% alcohol was used to disinfect the skin and the wound area. In the sham operation, the abdomen was opened, the stomach and duodenum were dissociated; and the abdominal cavity was closed after 1 minute. All surgery was done using asepsis conditions. The care of the animals conformed to the regulations of friendly treating experimental animals issued by the Ministry of Science and Technology in 2006.
Animal grouping
Seventy-five SD rats were randomly divided into sham operation and modeling intervention groups. The 15 rats that composed the sham operation group, were raised by common forage diet after receiving a sham operation, while the rest of the 60 rats in modeling intervention group were raised on different ratios of n-6/n-3PUFAs in their forage diet after they underwent modeling. According to different proportions of n-3PUFAs in the forage diet, we divided the 60 rats into four groups: 12: 1n-6/n-3 (12:1), 10:1 n-6/n-3 (10:1), 5: 1 n-6/n-3 (5:1) and 1: 1 n-6/n-3 (1:1) group. There were 15 rats in each group, subsequently they were fed with the forage diet with set ratios, and the esophageal tissue was removed after two weeks for testing. In the common forage diet, the ratio of n-6/n-3PUFAs reached 12: 1; therefore, we selected this group fed by common forage diet as the controls in our study.
The quantitative detection of relevant indicators’ protein in esophageal tissue
The total protein of 80 μg was extracted and SDS-PAGE gel electrophoresis used to transfer protein to PVDF film, that was then sealed using dried skimmed milk. The first antibody (1:100) was added for overnight incubation at 4°C; then the second antibody incubated for 2 hours at room temperature and subsequently developed using DAB. Quantity One Imaging Analysis Software was used to analyze the strip absorbance value.
The detection of correlated indicators’ mRNA in esophageal tissue
Total RNA was extracted using the Trizol method according to TaKaRa reverse transcription and amplification kit instructions. Related primer sequences are shown in Table 1.
Table 1.
Related primer sequences.
| Primer | Sequence (5′-3′) | Product length |
|---|---|---|
| PI3K | Upstream 5′ TGGTTCTTGCGAAGTGAGATAG3′ | 117 bp |
| Downstream 5′ CTGCTGCGTGAAGTCCTGTA 3′ | ||
| Akt | Upstream 5′ TAGGCATCCCTTCCTTACAGC 3′ | 114 bp |
| Downstream 5′ CGCTCACGAGACAGGTGGA 3′ | ||
| NF-κBp50 | Upstream 5′ GGCAGAAGTCAACGCTCAG 3′ | 142 bp |
| Downstream 5′ TGTCGTCCCATCGTAGGT 3′ | ||
| NF-κBp65 | Upstream 5′ AGCGAGACCTGGAGCAAG 3′ | 105 bp |
| Downstream 5′ GGACCGCATTCAAGTCATAG 3′ | ||
| β-actin | Upstream 5′ TTCCAGCCTTCCTTCCTG 3′ | 102 bp |
| Downstream 5′ GGCATAGAGGTCTTTACGG 3′ |
Statistical analysis
SPSS 19.0 software was used for the analysis, mean ± standard deviation (χ̄±s) was used to describe the data which were represented by case (n); single factor analysis of variance was used to compare the data between multiple groups; LSD method was used to compare differences between two groups; and rank sum test was used to compare the data. The criterion of the test was a=0.05, p<0.05 indicating the differentiation had statistical significance.
Results
The expression of PI3K, Akt, p-Akt, NF-κBp50 and NF-κBp65 protein in RE rats’ esophageal tissue with different ratio of n-6/n-3PUFAs in the diet
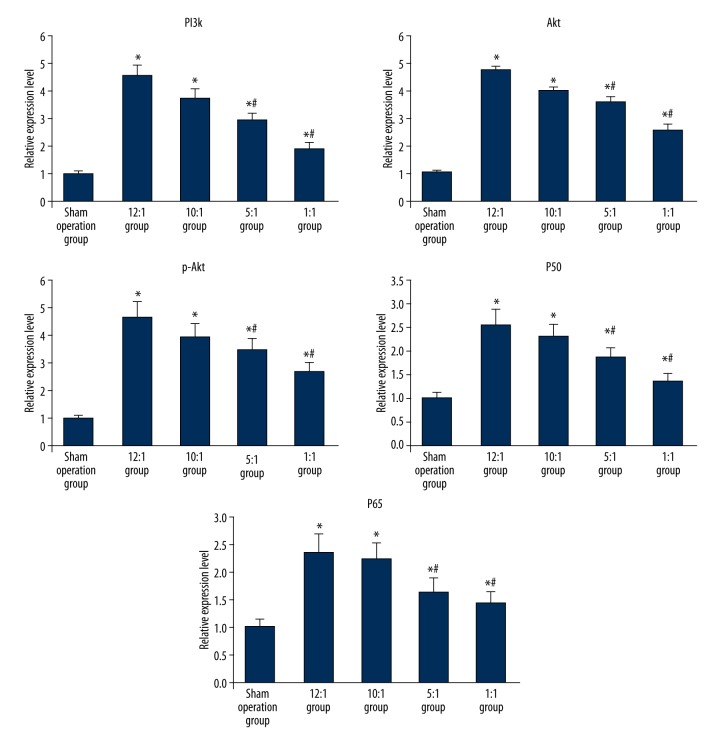
Western blot was utilized to detect the expression of PI3K, Akt, p-Akt, NF-κBp50, and NF-κBp65 protein in inflammation of esophageal mucosa of rats fed different ratio of n-6/n-3 PUFAs in the forage diet. Compared with sham operation group, the expression of PI3K, Akt, p-Akt, NF-κBp50, and NF-κBp65 protein in the modeling groups all were significantly increased with the differences being statistically significant (p<0.05); In the modeling groups with different concentration ratios of n-6/n-3PUFAs in the forage diets, the expression of PI3K, Akt, p-Akt, NF-κBp50, and NF-κBp65 protein kept pace with the proportion of n-6/n-3, in other words, if the ratio of n-6/n-3 decreased, the expression of PI3K, Akt, p-Akt, NF-κBp50, and NF-κBp65 protein decreased. The lowest expression of each indicator occurred when the proportion of n-6/n-3 reached 1: 1 in the forage diet; the difference was statistical significant compared with other ratios of n-6/n-3 in the forage diet of the modeling group (p<0.05) (Figures 1, 2).
Figure 1.
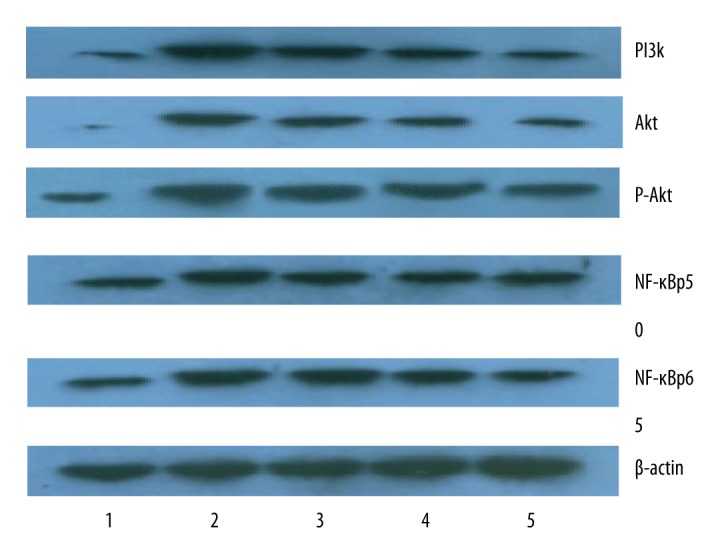
The expression of PI3K, Akt, p-Akt, NF-κBp50, and NF-κBp65 in esophageal tissue were tested by Western blot. 1 – The sham operation group; 2 – The 12:1 group; 3 – The 10:1 group; 4 – The 5:1 group; 5 – The 1:1 group.
Figure 2.
The comparison of different protein (PI3K, Akt, p-Akt, NF-κBp50, and NF-κBp65) expression in esophageal tissues of each group. * Represented p<0.05 vs. sham operation group; # standed for p<0.05 vs. blank control.
The expression of PI3K, Akt, NF-κBp50, and NF-κBp65 mRNA in reflux esophagitis rats’ esophageal tissue with different ratio of n-6/n-3PUFAs in the forage diet
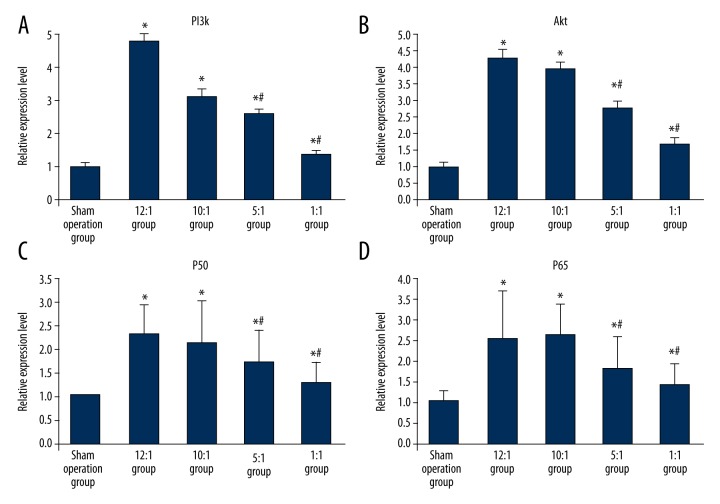
Compared with the sham operation group, the PI3K, Akt, NF-κBp50, and NF-κBp65 RNA in each modeling group all significantly increased, the difference was statistically significance (p<0.05); in the modeling group with different ratios of n-6/n-3PUFAs in the forage diet, if the ratio of n-6/n-3 decreased, subsequently the amount of PI3K, Akt, NF-κBp50, and NF-κBp65 RNA decreased. When the ratio of n-6/n-3 in the forage diet was 1:1, the expression of each indicator was the lowest. The difference was statistically significant compared with the other ratios in the forage diet (p<0.05) (Figure 3A–3D).
Figure 3.
(A) The comparison of the amount of PI3KmRNA in the esophageal tissue of RE rats with different ratios of n-6/n-3PUFAs in the forage diet. (B) The comparison of the amount of AktmRNA in the esophageal tissue of RE rats with different ratios of n-6/n-3PUFAs in the forage diet. (C) The comparison of the amount of NF-κBp50mRNA in the esophageal tissue of RE rats with different ratios of n-6/n-3PUFAs in the forage diet. (D) The comparison of the amount of NF-κBp65mRNA in the esophageal tissue of RE rats with different ratios of n-6/n-3PUFAs in the forage diet. * Represented p<0.05 vs. sham operation group; # standed for p<0.05 vs. blank control.
Discussion
There have been many studies investigating PI3K/Akt signaling pathway in tumorigenesis, tumor development, and metastasis; however, in most neoplastic diseases, the long-term and reduplicated stimulation is the potential element that may accelerate tumorigenesis, tumor development, as well as metastasis, which demonstrates that PI3K/Akt signaling pathway is associated with inflammation. Consequently, there have been more recent reports about the role of PI3K/Akt signaling pathway in inflammatory immune response. Some studies [8–11] have suggested that the inhibition of PI3K/AKT reduced the release of multiple inflammatory factors. Samuel et, al. [12] discovered that the expression of p-Akt and NF-κB in injured pancreatic tissue significantly increased with pancreatic duct ligation to induce animal models to experience acute pancreatitis, while expression of p-Akt and NF-κB significantly decreased after the ligation was loosened. Studies [13,14] on progression of esophageal lesions also reported that Akt was associated with esophageal lesions [15,16], meanwhile, the expression of p-Akt in pancreatic cancer and Barrett’s esophagus with serious dysplasia significantly increased compared with that in esophagus without dysplasia [17–22].
Conclusions
Our study illustrated that the expression of the principle target molecules PI3K, Akt, p-Akt, and NF-κB genes and proteins involved in PI3K/Akt-NF-κB signaling pathway significantly increased, implying that the signaling pathway would likely play a lead role in the occurrence and development of reflux esophagitis. Our study also demonstrated through the detection of the critical target protein of PI3K/Akt-NF-κB signaling pathway in esophageal tissue, n-3 PUFAs could inhibit the expression of PI3K, p-Akt, and NF-κB, moreover, with the increasing ratio of n-3PUFAs in the forage diet, its inhibitory effect had the tendency to increase. The result indicated that the inhibition of n-3 PUFAs in the development of esophageal inflammation in rats with reflux esophagitis was attributed to the function of PI3K/Akt-NF-κB signaling pathway.
Footnotes
Source of support: Departmental sources
References
- 1.Abliz A, Deng W, Sun R, et al. Wortmannin, PI3K/Akt signaling pathway inhibitor, attenuates thyroid injury associated with severe acute pancreatitis in rats. Int J Clin Exp Pathol. 2015;8(11):13821–33. [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Y, Tu C, Zhao Y, et al. Placental growth factor enhances angiogenesis in human intestinal microvascular endothelial cells via PI3K/Akt pathway: Potential implications of inflammation bowel disease. Biochem Biophys Res Commun. 2016;470(4):967–74. doi: 10.1016/j.bbrc.2016.01.073. [DOI] [PubMed] [Google Scholar]
- 3.Liu HB, Meng QH, Huang C, et al. Nephroprotective effects of polydatin against ischemia/reperfusion injury: A role for the PI3K/Akt signal pathway. Oxid Med Cell Longev. 2015;2015:362158. doi: 10.1155/2015/362158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malemud CJ. The PI3K/Akt/PTEN/mTOR pathway: A fruitful target for inducing cell death in rheumatoid arthritis? Future Med Chem. 2015;7(9):1137–47. doi: 10.4155/fmc.15.55. [DOI] [PubMed] [Google Scholar]
- 5.Zhou LT, Wang KJ, Li L, et al. Pinocembrin inhibits lipopolysaccharide-induced inflammatory mediators production in BV2 microglial cells through suppression of PI3K/Akt/NF-kappaB pathway. Eur J Pharmacol. 2015;761:211–16. doi: 10.1016/j.ejphar.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Jones ML, Mark PJ, Keelan JA, et al. Maternal dietary omega-3 fatty acid intake increases resolvin and protectin levels in the rat placenta. J Lipid Res. 2013;54(8):2247–54. doi: 10.1194/jlr.M039842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Wang X, Li N, Li J. The study of n-3PUFAs protecting the intestinal barrier in rat HS/R model. Lipids Health Dis. 2014;13:146. doi: 10.1186/1476-511X-13-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stiles BL. PI-3-K and AKT: Onto the mitochondria. Adv Drug Deliv Rev. 2009;61(14):1276–82. doi: 10.1016/j.addr.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 9.Falasca M. PI3K/Akt signalling pathway specific inhibitors: A novel strategy to sensitize cancer cells to anti-cancer drugs. Curr Pharm Des. 2010;16(12):1410–16. doi: 10.2174/138161210791033950. [DOI] [PubMed] [Google Scholar]
- 10.Huang JB, Ding Y, Huang DS, et al. Inhibition of the PI3K/AKT pathway reduces tumor necrosis factor-alpha production in the cellular response to wear particles in vitro. Artif Organs. 2013;37(3):298–307. doi: 10.1111/j.1525-1594.2012.01568.x. [DOI] [PubMed] [Google Scholar]
- 11.Saleem M, Afaq F, Adhami VM, Mukhtar H. Lupeol modulates NF-kappaB and PI3K/Akt pathways and inhibits skin cancer in CD-1 mice. Oncogene. 2004;23(30):5203–14. doi: 10.1038/sj.onc.1207641. [DOI] [PubMed] [Google Scholar]
- 12.Samuel I, Yorek MA, Zaheer A, Fisher RA. Bile-pancreatic juice exclusion promotes Akt/NF-kappaB activation and chemokine production in ligation-induced acute pancreatitis. J Gastrointest Surg. 2006;10(7):950–59. doi: 10.1016/j.gassur.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Beales IL, Ogunwobi O, Cameron E, et al. Activation of Akt is increased in the dysplasia-carcinoma sequence in Barrett’s oesophagus and contributes to increased proliferation and inhibition of apoptosis: A histopathological and functional study. BMC Cancer. 2007;7:97. doi: 10.1186/1471-2407-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sagatys E, Garrett CR, Boulware D, et al. Activation of the serine/threonine protein kinase Akt during the progression of Barrett neoplasia. Hum Pathol. 2007;38(10):1526–31. doi: 10.1016/j.humpath.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Du Z, Luo J, et al. Inhibition of heat shock protein 90 suppresses squamous carcinogenic progression in a mouse model of esophageal cancer. J Cancer Res Clin Oncol. 2015;141(8):1405–16. doi: 10.1007/s00432-014-1896-8. [DOI] [PubMed] [Google Scholar]
- 16.Fassan M, Realdon S, Pizzi M, et al. Programmed cell death 4 nuclear loss and miR-21 or activated Akt overexpression in esophageal squamous cell carcinogenesis. Dis Esophagus. 2012;25(3):263–68. doi: 10.1111/j.1442-2050.2011.01236.x. [DOI] [PubMed] [Google Scholar]
- 17.Xiaoping L, Xiaowei Z, Leizhen Z, Weijian G. Expression and significance of CD44 and p-AKT in pancreatic head cancer. World J Surg Oncol. 2015;13:334. doi: 10.1186/s12957-015-0746-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohtsubo K, Yamada T, Zhao L, et al. Expression of Akt kinase-interacting protein 1, a scaffold protein of the PI3K/PDK1/Akt pathway, in pancreatic cancer. Pancreas. 2014;43(7):1093–100. doi: 10.1097/MPA.0000000000000168. [DOI] [PubMed] [Google Scholar]
- 19.Jung KH, Yan HH, Fang Z, et al. HS-104, a PI3K inhibitor, enhances the anticancer efficacy of gemcitabine in pancreatic cancer. Int J Oncol. 2014;45(1):311–21. doi: 10.3892/ijo.2014.2435. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Cheng Sun SH, Sun SJ, et al. Phosph-Akt1 expression is associated with a favourable prognosis in pancreatic cancer. Ann Acad Med Singapore. 2010;39(7):548–47. [PubMed] [Google Scholar]
- 21.Chadha KS, Khoury T, Yu J, et al. Activated Akt and Erk expression and survival after surgery in pancreatic carcinoma. Ann Surg Oncol. 2006;13(7):933–39. doi: 10.1245/ASO.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Ozaki Y, Tatebe S, Ikeguchi M. [Molecular mechanism in pathogenesis of pancreatic neoplasms: p-Akt, PTEN]. Nihon Rinsho. 2006;64(Suppl 1):41–43. [in Japanese] [PubMed] [Google Scholar]