Abstract
Background
We analyzed placental DNA methylation levels at repeated sequences (LINE1 elements) and all CCGG sites (the LUMA assay) to study the effect of modifiable clinical or laboratory procedures involved in in vitro fertilization. We included four potential modifiable factors: oxygen tension during embryo culture, fresh embryo transfer vs frozen embryo transfer, intracytoplasmic sperm injection (ICSI) vs conventional insemination or day 3 embryo transfer vs day 5 embryo transfer.
Results
Global methylation levels differed between placentas from natural conceptions compared to placentas conceived by IVF. Placentas from embryos cultured at 20% oxygen showed significant differences in LINE1 methylation compared to in vivo conceptions, while those from embryos cultured at 5% oxygen, did not have significant differences. In addition, placentas from fresh embryo transfer had significantly different LINE1 methylation compared to placentas from in vivo conceptions, while embryos resulting from frozen embryos were not significantly different from controls. On sex-stratified analysis, only males had significant methylation differences at LINE1 elements stratified for the modifiable factors. As expected, LINE1 methylation was significantly different between males and females in the control population. However, we did not observe sex-specific differences in the IVF group. We validated this sex-specific observation in an additional cohort and in opposite sex IVF twins.
Conclusion
We show that two clinically modifiable factors (embryo culture in 5 vs 20% oxygen tension and fresh vs frozen embryo transfer) are associated with global placental methylation differences. Interestingly, males appear more vulnerable to such treatment-related global changes in DNA methylation than do females.
Electronic supplementary material
The online version of this article (doi:10.1186/s13148-017-0318-6) contains supplementary material, which is available to authorized users.
Keywords: Methylation, Modifiable factors, IVF, Oxygen tension, Fresh transfer, Frozen transfer, LINE1
Background
Assisted reproductive technologies (ART) have been associated with multiple epigenetic changes including alteration in DNA methylation in both the placenta and the offspring [1–7]. Several explanations have been proposed as contributing to the etiology of these changes, including infertility itself, superovulation, in vitro fertilization (IVF), gamete/embryo manipulation, and embryo culture [8, 9]. We have recently demonstrated, using an analysis of placentas from donor oocyte recipients, that site-specific DNA methylation differences observed between ART and in vivo conceptions are associated, at least in part, with the ART procedure itself, and not the underlying infertility [10]. Animal studies have confirmed that multiple techniques utilized during IVF may play a role in these methylation changes [11–19].
Collectively, these observations suggest strongly that some of the clinical or laboratory procedures used in ART are responsible for altering DNA methylation levels at multiple sites in the genome. Given this likelihood, we chose to compare modifications of several ART procedures, individually, for an association with DNA methylation differences. The clinical and laboratory procedures compared were embryo culture in 5 vs 20% O2, fresh embryo transfer vs frozen embryo transfer, intracytoplasmic sperm injection (ICSI) vs conventional insemination and day 3 embryo transfer vs day 5 embryo transfer. We used surrogate measures of genome-wide DNA methylation as biomarkers of whether exposure to individual clinical or laboratory protocols used in ART might alter epigenetic marks in the placenta. The rationale behind using measures of methylation level that are averaged over hundreds-of-thousands-to-millions of CpG sites rather than methylation levels at specific sites is that a surrogate genome-wide average is more likely to be representative of the degree to which a particular factor is capable of disrupting epigenetic marks. In addition, multi-site methylation differences that are cumulative and observed between groups stratified on the basis of particular exposures are more likely to be reproducible than differences at individual sites because the exposures being investigated are not hypothesized to have effects that are targeted at specific CpGs. Moreover, the collection of CpGs usually interrogated by site-specific, array-based profiling methods are highly-selected and enriched for gene promoters and gene bodies. Interrogating methylation levels at repeated sequences that make up a large fraction of the human genome or all sites recognized by a particular restriction endonuclease is more likely to be representative of the genome, as a whole, than even large numbers of CpG sites selected to be within or adjacent to coding sequences. Finally, searching for procedure-specific, site-specific methylation differences using array-based epigenome-wide profiling methods requires significant statistical penalties because of the requirement of correction for multiple testing.
The aim of this study was to analyze the placental DNA methylation levels at repeated sequences (LINE1 elements) and all CCGG sites (the LUMA assay) to study the effect of modifiable clinical or laboratory procedures involved in in vitro fertilization. We included four potential modifiable factors: oxygen tension during embryo culture, fresh embryo transfer vs frozen embryo transfer, intracytoplasmic sperm injection (ICSI) vs conventional insemination or day 3 embryo transfer vs day 5 embryo transfer.
Methods
Samples and clinical protocols
The present study was approved by the University of Pennsylvania Institutional Review Board (approval number 804530). Placentas were acquired from live-born deliveries resulting from IVF pregnancies and naturally conceived pregnancies (controls). For IVF pregnancies, all patients had undergone in vitro fertilization at Penn Fertility Care (between 2006–2012 for original cohort and 2013 onwards for validation cohort) using standard protocols. Superovulation was performed using recombinant or purified-urinary follicle stimulating hormone and/or human menopausal gonadotropin. Gonadotropin dose was chosen based on patient characteristics and was adjusted during stimulation as clinically indicated based on patient response. Oocyte maturation was induced with human chorionic gonadotropin or leuprolide acetate followed by transvaginal egg retrieval 35–36 h later. Fertilization, by intracytoplasmic sperm injection (ICSI) was performed for either male factor or unexplained infertility as clinically indicated. Conventional insemination or ICSI and embryo culture were performed utilizing appropriate media (VitroLife; Gothenburg, Sweden) in microdroplets under oil in either 20% oxygen tension (5% CO2 in air) or 5% oxygen tension (5% O2, 5% CO2, and 90% N2), and transferred to the uterus on either day 3 (cleavage) or day 5 (blastocyst) of embryo development. Luteal support was provided by intramuscular progesterone (50 mg). Frozen embryos were cryopreserved at either the pronuclear or blastocyst stage using a slow-cooling protocol and subsequently thawed and transferred in a hormonally programmed cycle utilizing increasing doses of oral micronized estradiol (2–6 mg) followed by intramuscular progesterone (25–50 mg) to induce a receptive endometrium and to determine the timing of transfer.
DNA preparation and bisulfite conversion
Placentas were collected at delivery and processed for DNA analysis within 5 h as previously described [1, 20]. Briefly, placental tissue (1.5–2.5 cm3) was excised from the fetal surface of the placenta, directly behind the cord insertion site. The sample was rinsed extensively with sterile saline solution to minimize maternal blood contamination. The tissue was transferred to a 15-ml Falcon tube for DNA extraction and initially stored at 4 °C; nucleic acid extractions were performed within 2–4 days of collection. Placenta genomic DNA was extracted using standard phenol-chloroform extraction methods. The isolated DNA was dissolved in TrisCl (10 mM, pH 8.0) and stored at −80 °C until further use. Unmethylated cytosine in genomic DNA (0.5–1 μg) was converted to uracil by treatment with sodium bisulfite using the EZ DNA Methylation KitTM (Zymo Research Corp., USA), following the manufacturer’s guidelines. The bisulfite-converted DNA was dissolved in 20-μl TrisCl (10 mM, pH 8.0) buffer and stored at −20 °C until further use.
Luminometric methylation assay (LUMA)
Luminometric methylation assay (LUMA) was used to estimate global methylation levels of placental samples by sampling the fraction of the 2.3 million CCGG sites that are methylated. The protocol has been described previously [21]. We used a modification of this assay [22]. In short, placental genomic DNA (300 ng) was subjected to parallel digestions by methylation insensitive restriction endonuclease (MspI), and its methylation sensitive isoschizomer (HpaII) to produce 5′-CG overhangs. EcoRI was used in both the reactions for normalization, thus, resulting in 5′AATT overhangs. The digested samples were pyrosequenced (PyroMark Q96 ID, Qiagen) using the nucleotide dispensation order: GTGTCACATGTGTG. The peak heights corresponding to HpaII or MspI and EcoRI were then used to calculate the genomic DNA methylation fraction using the formula 1-[HpaII(G)/EcoRI(T)]/[MspI(G)/EcoRI(T)].
LINE1 methylation
Genomic DNA was treated with sodium bisulfite using the EZ DNA methylation kit (Zymo Research). Bisulfite converted DNA was amplified using primers and pyrosequenced as described previously [23]. LINE1 methylation levels were estimated by taking the average of the first three CpGs.
Statistical analysis
Statistical analysis was performed using GraphPad Prism (ver. 6.0 for Mac OS). Between-group differences in continuous data were analyzed using a two-tailed unpaired t test for two groups and one-way ANOVA for more than two groups. Tukey’s post hoc test was applied for multiple comparisons. Categorical data was analyzed using Fisher’s exact test for 2 × 2 contingency tables and a chi-square test for higher order contingency tables. Differences between males and females were analyzed using one-tailed unpaired t test due to prior expectation [24] for controls and singleton IVF; and one-tailed paired t test for opposite sex twins.
Results
Sample characteristics
Placental samples from 259 singleton pregnancies (182 ART- and 77 in vivo-conceived-controls) were analyzed by LUMA. High quality LINE1 pyrosequencing assay data were also obtained for a subset of the 259 singleton pregnancies (126 ART and 65 control placental samples). Samples that did not pass pyrosequencing quality control (absent peaks or broad peaks) were not included for LINE1 analysis. LINE1 pyrosequencing was also done in the validation cohort (57 controls and 55 ART) and opposite sex twins (39 pairs). The demographic profile and relevant clinical characteristics of all samples are shown in Table 1.
Table 1.
Demographic profile and clinical characteristics of the study subjects
| Original cohort | Validation cohort | Twin cohort | |||||
|---|---|---|---|---|---|---|---|
| ART (n = 182) | Controls (n = 77) | P value | ART (n = 55) | Controls (n = 57) | P value | ART (n = 39 pairs) | |
| Maternal age (years, mean ± SD) | 34.7 ± 3.6 | 32.2 ± 4.8 | < 0.0001 a | 35.8 ± 3.9 | 29.5 ± 6.1 | < 0.0001 a | 37.2 ± 5.2 |
| Paternal age (years, mean ± SD) | 36.9 ± 5.7 | 33.3 ± 5.2 | < 0.0001 a | 38.3 ± 6.5 | 34.8 ± 4.6 | 0.0610 | 39.0 ± 6.9 |
| Mode of egg fertilization (ICSI/non ICSI) | 54/127 | NA | – | 30/23 | NA | – | 14/24 |
| Oxygen tension (20/5%) | 123/59 | NA | – | 16/38 | NA | – | 19/19 |
| Type of embryo transfer (fresh/frozen) | 128/54 | NA | – | 33/22 | NA | – | 26/13 |
| Embryo transfer day (day 3/day 5) | 110/72 | NA | – | 17/37 | NA | – | 18/20 |
| Gestational age (weeks, mean ± SD) | 39.2 ± 1.3 | 39.1 ± 1.2 | 0.4306a | 38.2 ± 3.6 | 38.2 ± 6.4 | 0.9990a | 36.5 ± 2.1 |
| Birth weight (grams, mean ± SD) | 3441.5 ± 498.6 | 3318.8 ± 559.3 | 0.0837a | 3196 ± 918.4 | 3411 ± 541.7 | 0.1324a | 2653.1 ± 576.9 |
| Males (%) | 83(45.6) | 39 (50.6) | 0.4972b | 26 | 29 | 0.7103b | 39(50.0) |
| Females (%) | 99(54.4) | 38 (49.4) | 29 | 28 | 39(50.0) | ||
ART assisted reproductive technology; SD standard deviation;
ICSI intracytoplasmic sperm injection; NA not applicable
aUnpaired two-tailed t test
bFisher’s exact test
Values in italics denote significance
The control and ART groups differed significantly in both maternal and paternal age (p < 0.0001), with ART parents being 3–4 years older. Given the significant difference in parental age, we stratified each population to test for methylation differences associated with parental age (Additional file 1), based on a prior definition of “advanced parental age” (>35 vs ≤35 years; [25]). We tested four potentially modifiable clinical and laboratory factors used in ART for an effect on methylation level (Table 1 and Additional file 2). These modifiable factors were as follows: (1) mode of egg fertilization (ICSI or conventional insemination); (2) oxygen tension during embryo culture (20 or 5%); (3) type of embryo transfer (fresh or frozen); (4) embryo transfer day (day 3 or day 5).
Surrogate measures of global methylation in ART vs controls
The LUMA assay measures methylation level averaged over 2.3 million CCGG sites (approximately 8% of the 28 million CpG sites in the human genome), and the LINE1 assay measures average methylation levels at three CpG sites in 516,000 copies of LINE1 elements (approximately 5.5% of the CpG sites in the genome). CCGG site methylation levels were significantly different (p = 0.0002) between controls and ART (Fig. 1a and Table 2) as were LINE1 element methylation levels (p = 0.0034) (Fig. 1b and Table 3). Given these global differences between the control and ART groups, we stratified each group according to parental age and stratified the ART group by four clinical/laboratory procedures to determine whether differences were associated with one or more of these factors.
Fig. 1.
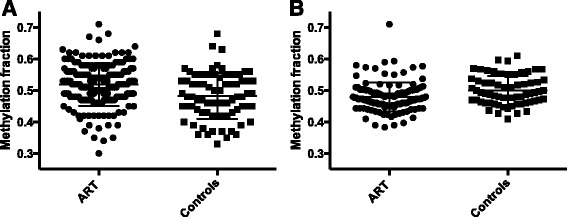
Average global methylation fraction of placental samples in the original cohort: methylation fraction of CCGG sites using LUMA assay (a); methylation fraction of LINE-1 elements using pyrosequencing assay (b)
Table 2.
Luminometric methylation assay (LUMA)
| Criteria for comparison | Groups (n) | Global methylation fraction (mean ± SD) | P value | |
|---|---|---|---|---|
| ANOVA | Tukey’s post hoc test (vs controls) | |||
| Controls (77) | 0.4827 ± 0.0723 | – | – | |
| Total ART | ART (182) | 0.5182 ± 0.0670 | – | 0.0002 a |
| Mode of egg fertilization | ICSI (54) | 0.5224 ± 0.0533 | 0.0009 | < 0.01 |
| Non ICSI (127) | 0.5158 ± 0.0720 | < 0.01 | ||
| Oxygen tension | 20%(123) | 0.5176 ± 0.0689 | 0.0009 | < 0.01 |
| 5%(59) | 0.5195 ± 0.0634 | < 0.01 | ||
| Type of embryo transfer | Fresh (128) | 0.5109 ± 0.0684 | < 0.0001 | < 0.05 |
| Frozen (54) | 0.5357 ± 0.0608 | < 0.0001 | ||
| Embryo transfer day | Day 3 (110) | 0.5212 ± 0.0689 | 0.0007 | < 0.001 |
| Day 5 (72) | 0.5138 ± 0.0643 | < 0.05 | ||
ANOVA analysis of variance; ART assisted reproductive technology; SD standard deviation; ICSI intracytoplasmic sperm injection
aUnpaired two-tailed t test
Values in italics denote significance
Table 3.
LINE1 methylation pyrosequencing assay
| Criteria for comparison | Groups (n) | Global methylation fraction (mean ± SD) | P value | |
|---|---|---|---|---|
| ANOVA | Tukey’s post hoc test (vs controls) | |||
| Controls (65) | 0.4997 ± 0.0463 | – | – | |
| Total ART | ART (126) | 0.4789 ± 0.0459 | – | 0.0034 a |
| Mode of egg fertilization | ICSI (39) | 0.4732 ± 0.0322 | 0.0090 | < 0.05 |
| Non ICSI (87) | 0.4815 ± 0.0508 | < 0.05 | ||
| Oxygen tension | 20%(73) | 0.4781 ± 0.0526 | 0.0135 | < 0.05 |
| 5%(53) | 0.4799 ± 0.0352 | ns | ||
| Type of embryo transfer | Fresh (90) | 0.4784 ± 0.0487 | 0.0137 | < 0.05 |
| Frozen (36) | 0.4800 ± 0.0385 | ns | ||
| Embryo transfer day | Day 3 (67) | 0.4783 ± 0.0529 | 0.0137 | < 0.05 |
| Day 5 (59) | 0.4796 ± 0.0463 | < 0.05 | ||
ANOVA analysis of variance; ART assisted reproductive technology; SD standard deviation; ICSI intracytoplasmic sperm injection; ns non significant
aUnpaired two-tailed t test
Values in italics denote significance
Note on assay reproducibility and directional difference between LUMA and LINE1 methylation levels
Though the absolute differences observed in methylation between ART and control patients is small (Tables 2 and 3), the differences are highly statistically significant. In addition, multiple studies, particularly from the oncology literature, have shown a lack of correlation between methylation levels measured by LUMA and LINE1 assays [26, 27]. This lack of correlation between the two assays is likely due to the fact that they interrogate different compartments of the epigenome (see Discussion).
In terms of the reproducibility of the techniques, many of the samples have been run multiple times. We have calculated the standard deviation and also used Bland–Altman plots to test for reproducibility. According to Bland–Altman [28], if 95% of the samples are within the limits of agreement then the assay is reproducible. In our data, 96.5% of samples are within the limits for the LUMA assay, and 100% of samples are in range for the LINE1 assay.
Parental age
We divided the samples on the basis of maternal and paternal age (> or ≤35 years of age; Additional file 1). The paternal age data were missing for some ART samples where anonymous sperm donors were used (n = 7). In the control group, paternal age data were missing for 14 samples. For both maternal and paternal age, global methylation differences between ART and controls, as measured by both LUMA and LINE1, were highly significant in the younger parental age group (≤35 years), suggesting that the difference in methylation between ART and control groups (Additional file 1) is not a result of the older average age of ART patients. We did not observe a significant difference in global methylation between ART and controls in the older parental age group though the number of controls in this comparison was small (LUMA: n = 22 for maternal age and n = 26 for paternal age; LINE1: n = 16 for maternal age and n = 21 for paternal age) (Additional file 1).
Mode of egg fertilization
Nearly 30% of the ART samples resulted from eggs that were fertilized using intra cytoplasmic sperm injection (ICSI) (Table 1). We compared methylation of CCGG sites in the placentas between ICSI and conventional insemination subgroups but did not observe any significant differences (data not shown). On the other hand, each subgroup was significantly different from controls in methylation levels (p < 0.01). Overall, ANOVA indicated that the three groups showed a significant difference in methylation levels (p = 0.0009) (Table 2).
Similarly, LINE1 methylation levels differed significantly (p = 0.009) between the three groups (ICSI, conventional insemination and controls). Furthermore, both the ART subgroups showed significant difference from the controls on post hoc analysis (Table 3).
Oxygen tension
The embryos in the ART group were cultured either at atmospheric oxygen tension (20%) or physiological oxygen tension (5%). Both oxygen tension ART groups differ significantly in CCGG methylation levels from controls (p < 0.01). ANOVA indicated a significant difference between the groups (p = 0.0009) (Table 2). However, samples from individuals cultured in 20% oxygen did not differ from those cultured in 5% oxygen.
The LINE1 methylation levels between the three groups (5% oxygen tension, 20% oxygen tension, and controls) also differed significantly (p = 0.0135). However, in this instance, the control group showed a significant difference with the 20% oxygen group (p < 0.05) but not the physiologic oxygen group (Table 3).
Type of embryo transfer
ART samples were stratified into fresh and frozen subgroups.
Overall, these subgroups and controls have a significant difference in CCGG methylation levels (p < 0.0001). Post hoc analysis showed significant methylation differences in both the ART groups compared to controls (p < 0.05) (Table 2).
Similarly, we observed significant differences (p = 0.0137) in LINE1 methylation levels between the three groups (fresh, frozen, and controls) (Table 3). Furthermore, the fresh group differed significantly from controls (p < 0.05) but the frozen group did not.
Embryo transfer day
We analyzed global methylation levels based on the stage at which the embryo was transferred back to the uterus; cleavage stage (day 3) or blastocyst stage (day 5).
Again, these subgroups and controls were significantly different in CCGG methylation levels (Table 2; p = 0.0007), but these ART subgroups did not differ significantly from each other (data not shown). However, each of these subgroups had significantly different methylation levels compared to controls (p < 0.05) (Table 2).
We found a significant difference in LINE1 methylation between controls, day 3 and day 5 embryo transfers (p = 0.0137). On applying Tukey’s post hoc test, both the embryo transfer subgroups showed significant methylation difference from the controls (p < 0.05) (Table 3).
Sex of newborns
Although there was no significant difference in the sex ratio of ART vs control offspring, it is appropriate to ask whether the methylation differences observed between the ART and control groups are restricted to one sex because sex differences in both global and site-specific methylation levels have been observed [24, 29–34]. Stratifying the samples according to sex demonstrates that significant methylation differences are found in both the female (p = 0.0226) and the male (p = 0.0030) subgroups for CCGG sites (Additional file 1). However, LINE1 elements showed significant difference in methylation levels only for the male (p = 0.0015) subgroup. Furthermore, female ART placentas did not show significant differences for any of the modifiable factors (data not shown). On the other hand, both male ART groups were significantly different (p < 0.05) from controls based on the mode of egg fertilization and embryo transfer day (Table 4). In addition, the 20% oxygen tension group (p < 0.01) and placentas from fresh embryo transfer (p < 0.05) showed significantly different LINE1 methylation levels compared to the controls. Such differences were not evident for physiologic oxygen tension or frozen embryo transferred groups (Table 4).
Table 4.
LINE1 methylation pyrosequencing assay for males
| Criteria for comparison | Groups (n) | Global methylation fraction (mean ± SD) | P value | |
|---|---|---|---|---|
| ANOVA | Tukey’s post hoc test (vs controls) | |||
| Controls (33) | 0.5091 ± 0.0463 | – | – | |
| Mode of egg fertilization | ICSI (19) | 0.4739 ± 0.0339 | 0.0057 | < 0.05 |
| Non ICSI (37) | 0.4809 ± 0.0434 | < 0.05 | ||
| Oxygen tension | 20%(31) | 0.4738 ± 0.0404 | 0.0044 | < 0.01 |
| 5%(25) | 0.4843 ± 0.0399 | ns | ||
| Type of embryo transfer | Fresh (45) | 0.4751 ± 0.0386 | 0.0033 | < 0.01 |
| Frozen (11) | 0.4924 ± 0.0457 | ns | ||
| Embryo transfer day | Day 3 (28) | 0.4779 ± 0.0435 | 0.0067 | < 0.05 |
| Day 5 (28) | 0.4791 ± 0.0375 | < 0.05 | ||
ANOVA analysis of variance; SD standard deviation; ICSI intracytoplasmic sperm injection; ns non significant
Values in italics denote significance
Sex differences
We observed methylation differences in LINE1 elements between male and female controls (p = 0.0494). However, ART placentas did not show (p = 0.4727) any sex-related methylation differences (Table 5). We obtained similar results in the validation cohort; sex-related differences were observed in controls (p = 0.0078) but not in ART placentas (p = 0.2149). The lack of a sex-difference among the ART population was confirmed by examining placentas from opposite sex twins (Table 5). The sex-specific methylation differences observed in controls do not appear among ART opposite sex twins (Table 5).
Table 5.
Sex differences for LINE1 methylation pyrosequencing assay
| Cohort | Group | Males | Females | P valuesa | ||
|---|---|---|---|---|---|---|
| Number of placentas | Global methylation fraction (mean ± SD) | Number of placentas | Global methylation fraction (mean ± SD) | |||
| Original cohort | Controls | 33 | 0.5091 ± 0.0463 | 32 | 0.4902 ± 0.0448 | 0.0494 |
| ART | 56 | 0.4785 ± 0.0402 | 70 | 0.4791 ± 0.0503 | 0.4727 | |
| Validation cohort | Controls | 29 | 0.4266 ± 0.0401 | 28 | 0.3936 ± 0.0583 | 0.0078 |
| ART | 26 | 0.4227 ± 0.0382 | 29 | 0.4386 ± 0.09538 | 0.2149 | |
| Opposite sex twins | ART | 39 | 0.4076 ± 0.0294 | 39 | 0.4098 ± 0.0320 | 0.3199 |
ART assisted reproductive technology; SD standard deviation
aUnpaired one-tailed t test
Values in italics denote significance [59]
Discussion
We have observed that placentas from ART conceptions differ from control placentas in DNA methylation levels, whether examined CpG site-specifically [1, 35] or at hundreds-of-thousands-to millions of sites, simultaneously (Fig. 1; Tables 2 and 3). Notably, both hypomethylation and hypermethylation of ART samples are observed in both site-specific assays [1, 35] and at multiple-interrogated sites in different sequence contexts (Fig. 1; Tables 2 and 3). Given that the range of methylation values observed in ART and control populations overlaps to a great degree (Fig. 1), one may legitimately ask what is the source of the statistically significant differences observed. Our hypothesis is that the differences are due to a relatively small fraction of individuals in the ART group who have highly disrupted, so-called “outlier”, DNA methylation levels at a significant fraction of interrogated CpG sites [35]. We have observed that such individuals are more prevalent in “low-birth-weight” ART children [35] and multiple individuals who have hypermethylation at CCGG sites, compared with the most extreme controls, can be seen in Fig. 1a. An excess of such outliers is less obvious for hypomethylation of LINE1 elements (Fig. 1b) but there are, in fact, many more ART individuals with LINE1 methylation values less than 0.45 than control individuals.
Assisted reproductive technologies have been linked to epigenetic disorders and adverse neonatal outcomes [36, 37]. These anomalies may be the effect of parental factors (infertility or advanced age) [38], clinical and laboratory procedures used in ART or both parental factors and ART. We have observed that the site-specific DNA methylation differences found in autologous in vitro fertilized groups are also observed in children conceived with the aid of donor oocytes [10]. This observation suggests a role for ART procedures in causing these discrepancies but does not eliminate a role for parental factors. In addition to infertility, the most distinguishing characteristic of parents of ART children is older age. The present study clearly demonstrates that the global methylation differences between ART and control population are present even among the offspring of younger parents (≤35 years). This suggests that one or more of the interventions involved in ART could be responsible for these epigenetic differences, potentially leading to adverse outcomes in ART-conceived children.
The clinically modifiable ART interventions, such as superovulation, in vitro fertilization (conventional or ICSI), embryo culture (in 5 or 20% O2), and embryo transfer (cleavage or blastocyst stage), take place during the preimplantation embryonic stage during a time of extensive epigenetic reprogramming. We hypothesized that some or all of these interventions might influence the establishment and maintenance of epigenetic marks, leading to abnormal DNA methylation levels.
While both CCGG site methylation and LINE1 methylation differed between ART and control groups, only LINE1 methylation discriminated between different ART interventions. The reasons for this difference in discriminatory power between surrogate global measures of DNA methylation are unclear. CCGG sites are enriched in hypomethylated CpG islands [39, 40], which are found near the promoters of approximately half of all genes, while LINE1 elements are dispersed in repeated sequences that are generally hypermethylated [40]. ART is associated with an increase in methylation in the usually hypomethylated CCGG site (Table 2) and a decrease in methylation in the usually hypermethylated LINE1 sites (Table 3). In this regard, LINE1 elements are normally protected from the wave of genome-wide demethylation that occurs during preimplantation development [41]. The maintenance of LINE1 elements in the methylated state in preimplantation embryos has been suggested to suppress retrotransposition [23]. The fact that ART placentas, overall, and each ART intervention, individually, leads to lower levels of LINE1 methylation (Table 3) suggest the possibility that ART interventions may result in hypomethylation of some LINE1 elements and a corresponding level of new LINE1 retrotranspositions in the genome.
CCGG site methylation did not reveal any differences among the different interventions, as both ART groups within an intervention differ significantly from the controls, without significant differences between the interventions being compared. However, LINE1 methylation did show differences between the ART groups in the oxygen tension of embryo culture and embryo transfer type.
It has been argued earlier that embryo culture at physiologic oxygen tension (5%) has a better success rate [42]. Studies in mice have shown that 20% O2 delays oocyte maturation, thereby impairing the development of oocytes [43] and adversely affecting the developmental potential of blastocysts [44]. Low concentrations of reactive oxygen species have been observed in mouse embryos cultured in 5% O2 compared to those from 20% O2 [45]. In human IVF studies, physiologic oxygen concentration has been found to improve the blastulation rate [46], blastocyst yield and embryo quality [47], and increase live births [42]. Cochrane Database Review [48] also supports the view that physiologic oxygen tension has a better success rate. Our data (Table 3) also suggest a detrimental effect of 20% oxygen over 5% oxygen tension on the epigenome, in comparison with controls, as assayed by global LINE1 methylation.
Superovulation is an integral part of IVF procedure wherein women undergoing IVF are administered exogenous gonadotropins to stimulate the production of multiple follicles. This results in serum estradiol levels 10 times greater in a fresh IVF cycles than observed during a natural cycle [49]. This hormonally imbalanced uterine environment has been shown to affect embryo implantation [50] and fetal growth [51] in mice. Frozen embryo transfers (FET) have been found to overcome these adverse effects of supraphysiologic hormonal levels and improve endometrial receptivity [52]. A recent meta-analysis of 13 cohort studies concluded that FET reduces the risks of preterm birth and low birth weight [53]. At the expression level, frozen embryos showed more consistent gene expression over morphologically matched fresh embryos [54]. Our data suggests that global methylation levels of frozen transfers are consistent with the control group but these are significantly variable for the fresh transfers.
The other interventions studied were the effects of ICSI vs conventional IVF and day 3 vs day 5 of embryo transfer. Our data are inconclusive on whether either modification of either of these practices is less disruptive to the epigenome. Others have found that ICSI is associated with more epigenetic alterations compared to conventional IVF [55], but there is no clear indication as to whether day 3 or day 5 embryo transfer is better. A recent Cochrane Database Review found moderately high clinical pregnancy and live birth rates in blastocyst fresh transfers, but the data was inconclusive in regards to cumulative live birth and pregnancy rates [56].
Our sex-stratified analysis suggests that sex of the newborns could have a major role in influencing the observed methylation differences between ART and control placentas. In our study, CCGG methylation showed significant differences for both male and female subgroups, but the LINE1 methylation data showed differences in male subgroups only. On further analysis based on the modifiable factors, the males showed similar results as observed in the combined analysis (males and females). On the other hand, we did not see any differences in the female group. This led us to investigate the methylation difference between the sexes. We found significant methylation difference between males and females in controls, but this difference is not present in the ART population. The absence of sex-influenced methylation differences in ART was validated in an additional cohort and in opposite sex twins conceived by ART.
There are reports showing that LINE1 methylation is influenced by sex [24, 57]. LINE1 hypomethylation in females was attributed to dietary differences and decreased circulatory folate levels due to menstruation in these studies. However, it is unclear why this sex difference is not evident in the ART children.
Males are more sensitive to maternal obesity induced inflammation [58], suggesting a greater impact of in utero environment on male fetuses. Such a sex bias in sensitivity to in utero environmental exposures could provide a possible explanation for the sex-specific effect of ART on expunging male/female epigenetic differences that exist in in vivo-conceived offspring. Further studies are needed to evaluate these sex specific differences in depth.
There are limitations to this study. Clearly, other factors could be affecting DNA methylation, including culture media, response to superovulation, maternal BMI as well as the many other unknown factors that might affect the epigenome. In addition, long-term clinical significance of these changes in global methylation must still be investigated. However, these findings strengthen previous data, from our group and others, that techniques utilized during ART lead to changes in DNA methylation.
Conclusions
We conclude that two clinically modifiable factors (5 vs 20% oxygen tension of embryo culture and fresh vs frozen embryo transfer) are associated with global placental methylation differences. In both interventions, the subgroup associated with better clinical outcomes (5% O2 and frozen embryo transfer; [42–48, 52–54]) is also associated with global DNA methylation levels that are closer to those of children conceived in vivo. This suggests not only that DNA methylation may be responsible for (at least some) the adverse clinical outcomes associated with IVF, but that modification of our current practices may decrease the incidence of these adverse outcomes. Further investigations correlating changes in DNA methylation and adverse perinatal outcomes are necessary to establish the protocols that can minimize epigenetic changes following ART and reduce complications associated with IVF.
Acknowledgements
We are grateful to May Truongcao for technical assistance.
Funding
This work was supported by the National Institute of Health (Grant number P50-HD-068157 to C.C. and C.S.).
Availability of data and materials
Not applicable.
Authors’ contributions
JG participated in the sample preparation, methylation assays, and data analysis. JG, CC, CS, and MM participated in data interpretation and manuscript preparation. MM and CC participated in patient selection, collection and evaluation of clinical data for subgroup analysis. CC, CS, and MM designed the study and supervised all aspects of the work. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The present study was approved by the University of Pennsylvania Institutional Review Board (approval number 804530). Written informed consent was obtained in advance from the mother of each newborn.
Additional files
Effect of parental age and sex of newborns. (DOCX 87 kb)
Distribution of global methylation fractions based on modifiable factors: mode of egg fertilization by LUMA (A) and LINE1 assay (B); oxygen tension by LUMA (C) and LINE1 assay (D); type of embryo transfer by LUMA (E) and LINE1 assay (F) and; embryo transfer day by LUMA (G) and LINE1 assay (H). (PPTX 385 kb)
References
- 1.Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, et al. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet. 2009;18:3769–78. doi: 10.1093/hmg/ddp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelissen EC, Dumoulin JC, Daunay A, Evers JL, Tost J, van Montfoort AP. Placentas from pregnancies conceived by IVF/ICSI have a reduced DNA methylation level at the H19 and MEST differentially methylated regions. Hum Reprod. 2013;28:1117–26. doi: 10.1093/humrep/des459. [DOI] [PubMed] [Google Scholar]
- 3.Santos F, Hyslop L, Stojkovic P, Leary C, Murdoch A, Reik W, et al. Evaluation of epigenetic marks in human embryos derived from IVF and ICSI. Hum Reprod. 2010;25:2387–95. doi: 10.1093/humrep/deq151. [DOI] [PubMed] [Google Scholar]
- 4.Gomes MV, Huber J, Ferriani RA, AmaralNeto AM, Ramos ES. Abnormal methylation at the KvDMR1 imprinting control region in clinically normal children conceived by assisted reproductive technologies. Mol Hum Reprod. 2009;15:471–7. doi: 10.1093/molehr/gap038. [DOI] [PubMed] [Google Scholar]
- 5.Katagiri Y, Aoki C, Tamaki-Ishihara Y, Fukuda Y, Kitamura M, Matsue Y, et al. Effects of assisted reproduction technology on placental imprinted gene expression. Obstet Gynecol Int. 2010;2010:437528. [DOI] [PMC free article] [PubMed]
- 6.Zheng HY, Tang Y, Niu J, Li P, Ye DS, Chen X, et al. Aberrant DNA methylation of imprinted loci in human spontaneous abortions after assisted reproduction techniques and natural conception. Hum Reprod. 2013;28:265–73. doi: 10.1093/humrep/des358. [DOI] [PubMed] [Google Scholar]
- 7.Lou H, Le F, Zheng Y, Li L, Wang L, Wang N, et al. Assisted reproductive technologies impair the expression and methylation of insulin-induced gene 1 and sterol regulatory element-binding factor 1 in the fetus and placenta. Fertil Steril. 2014;101:974–80. doi: 10.1016/j.fertnstert.2013.12.034. [DOI] [PubMed] [Google Scholar]
- 8.Pinborg A, Wennerholm UB, Romundstad LB, Loft A, Aittomaki K, Soderstrom-Anttila V, et al. Why do singletons conceived after assisted reproduction technology have adverse perinatal outcome? Systematic review and meta-analysis. Hum Reprod Update. 2013;19:87–104. doi: 10.1093/humupd/dms044. [DOI] [PubMed] [Google Scholar]
- 9.Ceelen M, vanWeissenbruch MM, Vermeiden JP, van Leeuwen FE, Delemarre-van de Waal HA. Growth and development of children born after in vitro fertilization. Fertil Steril. 2008;90:1662–73. doi: 10.1016/j.fertnstert.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Song S, Ghosh J, Mainigi M, Turan N, Weinerman R, Truongcao M, et al. DNA methylation differences between in vitro- and in vivo-conceived children are associated with ART procedures rather than infertility. Clin Epigenetics. 2015;7:41. doi: 10.1186/s13148-015-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le F, Wang LY, Wang N, Li L, le Li J, Zheng YM, et al. In vitro fertilization alters growth and expression of Igf2/H19 and their epigenetic mechanisms in the liver and skeletal muscle of newborn and elder mice. Biol Reprod. 2013;88:75. doi: 10.1095/biolreprod.112.106070. [DOI] [PubMed] [Google Scholar]
- 12.Li T, Vu TH, Ulaner GA, Littman E, Ling JQ, Chen HL, et al. IVF results in de novo DNA methylation and histone methylation at an Igf2-H19 imprinting epigenetic switch. Mol Hum Reprod. 2005;11:631–40. doi: 10.1093/molehr/gah230. [DOI] [PubMed] [Google Scholar]
- 13.Wright K, Brown L, Brown G, Casson P, Brown S. Microarray assessment of methylation in individual mouse blastocyst stage embryos shows that in vitro culture may have widespread genomic effects. Hum Reprod. 2011;26:2576–85. doi: 10.1093/humrep/der201. [DOI] [PubMed] [Google Scholar]
- 14.Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, Schultz RM, et al. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131:3727–35. doi: 10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- 15.Young LE, Fernandes K, McEvoy TG, Butterwith SC, Gutierrez CG, Carolan C, et al. Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet. 2001;27:153–4. doi: 10.1038/84769. [DOI] [PubMed] [Google Scholar]
- 16.Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. BiolReprod. 2000;62:1526–35. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- 17.Rivera RM, Stein P, Weaver JR, Mager J, Schultz RM, Bartolomei MS. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum Mol Genet. 2008;17:1–14. doi: 10.1093/hmg/ddm280. [DOI] [PubMed] [Google Scholar]
- 18.de Waal E, Mak W, Calhoun S, Stein P, Ord T, Krapp C, et al. In vitro culture increases the frequency of stochastic epigenetic errors at imprinted genes in placental tissues from mouse concepti produced through assisted reproductive technologies. BiolReprod. 2014;90:22. doi: 10.1095/biolreprod.113.114785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fortier AL, Lopes FL, Darricarrere N, Martel J, Trasler JM. Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet. 2008;17:1653–65. doi: 10.1093/hmg/ddn055. [DOI] [PubMed] [Google Scholar]
- 20.Turan N, Ghalwash MF, Katari S, Coutifaris C, Obradovic Z, Sapienza C. DNA methylation differences at growth related genes correlate with birth weight: a molecular signature linked to developmental origins of adult disease? BMC Med Genomics. 2012;5:10. doi: 10.1186/1755-8794-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karimi M, Johansson S, Stach D, Corcoran M, Grander D, Schalling M, et al. LUMA (LUminometricMethylan Assay)- A high throughput method to the analysis of genomic DNA methylation. Exp Cell Res. 2006;312: 1989:95. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Pilsner JR, Lazarus AL, Nam DH, Letcher RJ, Sonne C, Dietz R, et al. Mercure- associated DNA hypomethylation in polar bear brains via the Luminometric Methylation Assay: a sensitive method to study epigenetics in wildlife. Mol Ecol. 2010;19:307–14. doi: 10.1111/j.1365-294X.2009.04452.x. [DOI] [PubMed] [Google Scholar]
- 23.Bae JM, Shin SH, Kwon HJ, Park SY, Kook MC, Kim YW, et al. ALU and LINE-1 hypomethylations in multistep gastric carcinogenesis and their prognostic implications. Int J Cancer. 2012;131:1323–31. doi: 10.1002/ijc.27369. [DOI] [PubMed] [Google Scholar]
- 24.El-Maarri O, Walier M, Behne F, Uum JV, Singer H, Diaz-Lacava A, et al. Methylation at global LINE-1 repeats in human blood are affected by gender but not by age or natural hormone cycles. PLoS One. 2011;6(1):e16252. doi: 10.1371/journal.pone.0016252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bray I, Gunnell D, Davey SG. Advanced paternal age: how old is too old? J Epidemiol Community Health. 2006;60(10):851–3. doi: 10.1136/jech.2005.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lisanti S, Omar WAW, Tomaszewski B, De Prins S, Jacobs G, Koppen G, et al. Comparison of methods for quantification of global DNA methylation in human cells and tissues. PLoS One. 2013;8(11):e79044. doi: 10.1371/journal.pone.0079044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delgado-Cruzata L, Vin-Raviv N, Tehranifar P, Flom J, Reynolds D, Gonzalez K, et al. Correlations in global DNA methylation measures in peripheral blood mononuclear cells and granulocytes. Epigenetics. 2014;9(11):1504–10. doi: 10.4161/15592294.2014.983364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bland JM, Altman DG. Measuring agreement in method comparison studies. Stat Methods Med Res. 1999;8:135–60. doi: 10.1191/096228099673819272. [DOI] [PubMed] [Google Scholar]
- 29.vanDongen J, Nivard MG, Willemsen G, Hottenga JJ, Helmer Q, Dolan CV, et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat Commun. 2016; doi: 10.1038/ncomms11115. [DOI] [PMC free article] [PubMed]
- 30.Reizel Y, Spiro A, Sabag O, Skversky Y, Hecht M, Keshet I, et al. Gender-specific postnatal demethylation and establishment of epigenetic memory. Genes Dev. 2015;29:923–33. doi: 10.1101/gad.259309.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallou-Kabani C, Gabory A, Tost J, Karimi M, Mayeur S, Lesage J, et al. Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS One. 2010;5:e14398. doi: 10.1371/journal.pone.0014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gabory A, Ferry L, Fajardy I, Jouneau L, Gothie JD, Vige A, et al. Maternal diets trigger sex-specific divergent trajectories of gene expression and epigenetic systems in mouse placenta. PLoS One. 2012;7:e47986. doi: 10.1371/journal.pone.0047986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Maarri O, Becker T, Junen J, Manzoor SS, Diaz-Lacava A, Schwaab R, et al. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet. 2007;122:505–14. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- 34.Singmann P, Shem-Tov D, Wahl S, Grallert H, Fiorito G, Shin SY, et al. Characterization of whole-genome autosomal differences of DNA methylation between men and women. Epigenetics Chromatin. 2015;8:43. doi: 10.1186/s13072-015-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh J, Mainigi M, Coutifaris C, Sapienza C. Outlier DNA methylation levels as an indicator of environmental exposure and risk of undesirable birth outcome. Hum Mol Genet. 2016;25(1):123–9. doi: 10.1093/hmg/ddv458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lazaraviciute G, Kauser M, Bhattacharya S, Haggarty P, Bhattacharya S. A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously. Hum Reprod Update. 2014;20(6):840–52. doi: 10.1093/humupd/dmu033. [DOI] [PubMed] [Google Scholar]
- 37.Qin J, Liu X, Sheng X, Wang H, Gao S. Assisted reproductive technology and the risk of pregnancy-related complications and adverse pregnancy outcomes in singleton pregnancies: a meta-analysis of cohort studies. Fertil Steril. 2016;105:73–85.e1-6. doi: 10.1016/j.fertnstert.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Simpson JL. Birth defects and assisted reproductive technologies. Semin Fetal Neonatal Med. 2014;19(3):177–82. doi: 10.1016/j.siny.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat Rev Genet. 2004;5(6):446–55. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- 40.Knothe C, Shiratori H, Resch E, Ultsch A, Geisslinger G, Doehring A. Disagreement between two common biomarkers of global DNA methylation. Clin Epigenetics. 2016;8:60. doi: 10.1186/s13148-016-0227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Messerschmidt DM, Knowles BB, Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014;28(8):812–28. doi: 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meintjes M, Chantilis SJ, Douglas JD, Rodriguez AJ, Guerami AR, Bookout DM, et al. A controlled randomized trial evaluating the effect of lowered incubator oxygen tension on live births in a predominantly blastocyst transfer program. Hum Reprod. 2009;24(2):300–7. doi: 10.1093/humrep/den368. [DOI] [PubMed] [Google Scholar]
- 43.Adam AA, Takahashi Y, Katagiri S, Nagano M. Effects of oxygen tension in the gas atmosphere during in vitro maturation, in vitro fertilization and in vitro culture on the effeiciency of in vitro production of mouse embryos. Jpn J Vet Res. 2004;52(2):77–84. [PubMed] [Google Scholar]
- 44.Karagenc L, Sertkaya Z, Ciray N, Ulug U, Bahceci M. Impact of oxygen concentration on embryonic development of mouse zygotes. Reprod Biomed Online. 2004;9(4):409–17. doi: 10.1016/S1472-6483(10)61276-X. [DOI] [PubMed] [Google Scholar]
- 45.Kwon HC, Yang HW, Hwang KJ, Yoo JH, Kim MS, Lee CH, et al. Effects of low oxygen condition on the generation of reactive oxygen species and the development in mouse embryos cultured in vitro. J Obstet Gynaecol Res. 1999;25(5):359–66. doi: 10.1111/j.1447-0756.1999.tb01177.x. [DOI] [PubMed] [Google Scholar]
- 46.Kovacic B, Vlaisavljevic V. Influence of atmospheric versus reduced oxygen concentration on development of human blastocysts in vitro: a prospective study on sibling oocytes. Reprod Biomed Online. 2008;17(2):229–36. doi: 10.1016/S1472-6483(10)60199-X. [DOI] [PubMed] [Google Scholar]
- 47.Ciray HN, Aksoy T, Yaramanci K, Karavaka I, Bahceci M. In vitro culture under physiologic oxygen concentration improves blastocyst yield and quality: a prospective randomized survey on sibling oocytes. FertilSteril. 2009;91(Suppl 4):1459–61. doi: 10.1016/j.fertnstert.2008.07.1707. [DOI] [PubMed] [Google Scholar]
- 48.Bontekoe S, Mantikou E, van Wely M, Seshadri S, Repping S, Mastenbroek S. Low oxygen concentrations for embryo culture in assisted reproductive technologies. Cochrane Database Syst Rev. 2012;7:CD008959. doi: 10.1002/14651858.CD008950.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joo BS, Park SH, An BM, Kim KS, Moon SE, Moon HS. Serum estradiol levels during controlled ovarian hyperstimulation influence the pregnancy outcome of in vitro fertilization in a concentration-dependent manner. Fertil Steril. 2010;93(2):442–6. doi: 10.1016/j.fertnstert.2009.02.066. [DOI] [PubMed] [Google Scholar]
- 50.Fossum GT, Davidson A, Paulson RJ. Ovarian hyperstimulation inhibits embryo implantation in the mouse. J In Vitro Fert Embryo Transf. 1989;6:7–10. doi: 10.1007/BF01134574. [DOI] [PubMed] [Google Scholar]
- 51.Bonakdar E, Edriss MA, Bakhtari A, Jafarpour F, Asgari V, Hosseini SM, et al. A physiological, rather than superovulated, post-implantation environment can attenuate the compromising effect of assisted reproductive techniques on gene expression in developing mice embryos. Mol Reprod Dev. 2015;82(3):191–206. doi: 10.1002/mrd.22461. [DOI] [PubMed] [Google Scholar]
- 52.Roque M, Valle M, Guimaraes F, Sampaio M, Geber S. Freeze-all policy: fresh vs. frozen-thawed embryo transfer. Fertil Steril. 2015;103(5):1190–3. doi: 10.1016/j.fertnstert.2015.01.045. [DOI] [PubMed] [Google Scholar]
- 53.Zhao J, Xu B, Zhang Q, Li YP. Which one has a better obstretic and perinatal outcome in singleton pregnancy, IVF/ICSI or FET?: a systemic review and meta-analysis. Reprod Biol Endocrinol. 2016;14(1):51. doi: 10.1186/s12958-016-0188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaw L, Sneddon SF, Brison DR, Kimber SJ. Comparison of gene expression in fresh and frozen-thawed human preimplantation embryos. Reproduction. 2012;144(5):569–82. doi: 10.1530/REP-12-0047. [DOI] [PubMed] [Google Scholar]
- 55.Ventura-Junca P, Irarrazaval I, Rolle AJ, Gutierrez JI, Moreno RD, Santos MJ. In vitro fertilization (IVF) in mammals: epigenetic and developmental alterations. Scientific and bioethical implications for IVF in humans. Biol Res. 2015;48:68. doi: 10.1186/s40659-015-0059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glujovsky D, Farquhar C, QuinteiroRetamar AM, Alvarez Sedo CR, Blake D. Cleavage stage versus blastocyst stage embryo transfer in assisted reproductive technology. Cochrane Database Syst Rev. 2016;6:CD002118. doi: 10.1002/14651858.CD002118.pub5. [DOI] [PubMed] [Google Scholar]
- 57.Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16(1):108–14. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 58.Graf AE, Lallier SW, Waidyaratne G, Thompson MD, Tipple TE, Hester ME, et al. Maternal high fat diet exposure is associated with increased hepcidin levels, decreased myelination, and neurobehavioral changes made in male offspring. Brain Behav Immune. 2016;58:369–78. doi: 10.1016/j.bbi.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giavarina D. Understanding bland altman analysis. Biochemia medica. 2015;25:141–51. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


