Abstract
Background
Feeding higher-energy prepartum is a common practice in the dairy industry. However, recent data underscore how it could reduce performance, deepen negative energy balance, and augment inflammation and oxidative stress in fresh cows. We tested the effectiveness of rumen-protected methionine in preventing the negative effect of feeding a higher-energy prepartum. Multiparous Holstein cows were fed a control lower-energy diet (CON, 1.24 Mcal/kg DM; high-straw) during the whole dry period (~50 d), or were switched to a higher-energy (OVE, 1.54 Mcal/kg DM), or OVE plus Smartamine M (OVE + SM; Adisseo NA) during the last 21 d before calving. Afterwards cows received the same lactation diet (1.75 Mcal/kg DM). Smartamine M was top-dressed on the OVE diet (0.07% of DM) from -21 through 30 d in milk (DIM). Liver samples were obtained via percutaneous biopsy at -10, 7 and 21 DIM. Expression of genes associated with energy and lipid metabolism, hepatokines, methionine cycle, antioxidant capacity and inflammation was measured.
Results
Postpartal dry matter intake, milk yield, and energy-corrected milk were higher in CON and OVE + SM compared with OVE. Furthermore, milk protein and fat percentages were greater in OVE + SM compared with CON and OVE. Expression of the gluconeogenic gene PCK1 and the lipid-metabolism transcription regulator PPARA was again greater with CON and OVE + SM compared with OVE. Expression of the lipoprotein synthesis enzyme MTTP was lower in OVE + SM than CON or OVE. Similarly, the hepatokine FGF21, which correlates with severity of negative energy balance, was increased postpartum only in OVE compared to the other two groups. These results indicate greater liver metabolism and functions to support a greater production in OVE + SM. At 7 DIM, the enzyme GSR involved in the synthesis of glutathione tended to be upregulated in OVE than CON-fed cows, suggesting a greater antioxidant demand in overfed cows. Feeding OVE + SM resulted in lower similar expression of GSR compared with CON. Expression of the methionine cycle enzymes SAHH and MTR, both of which help synthesize methionine endogenously, was greater prepartum in OVE + SM compared with both CON and OVE, and at 7 DIM for CON and OVE + SM compared with OVE, suggesting greater Met availability. It is noteworthy that DNMT3A, which utilizes S-adenosylmethionine generated in the methionine cycle, was greater in OVE and OVE + SM indicating higher-energy diets might enhance DNA methylation, thus, Met utilization.
Conclusions
Data indicate that supplemental Smartamine M was able to compensate for the negative effect of prepartal energy-overfeeding by alleviating the demand for intracellular antioxidants, thus, contributing to the increase in production. Moreover Smartamine M improved hepatic lipid and glucose metabolism, leading to greater liver function and better overall health.
Electronic supplementary material
The online version of this article (doi:10.1186/s40104-017-0147-7) contains supplementary material, which is available to authorized users.
Keywords: Energy, Methionine, Nutrigenomics, Transition period
Background
The transition period, defined as last 3 weeks prepartum through 3 weeks postpartum, is one of the most important stages of lactation in dairy cattle. Years of strong genetic selection and improvement have allowed modern dairy cows to reach high production performance, both in quantity and quality. However, this has made the transition between late pregnancy to early lactation a significant period of metabolic and immune challenges [1–3]. Because failure to adequately meet these challenges can compromise production, induce metabolic diseases, and increase rates of culling in early lactation [4], the management of the transition cow remains a focal point for dairy producers.
Following the “steaming up” concept of RB Boutflour [5], transition cows during the dry period were first traditionally offered a high fiber/low energy density ration, to then increase the energy density of the ration with a lower fiber content in the last month of gestation (i.e. “close-up” period). This early century practice is still embedded in the modern dairy industry. However, multiple studies have consistently reported negative effects of prepartum energy overfeeding on cow health and productivity. Among these, prepartum hyperglycemia and hyperinsulinemia together with marked postpartum adipose tissue mobilization (i.e., greater blood NEFA concentration) [6–11] have strong negative impact on postpartal health indices [12–15].
Our general hypothesis was that supplementation with rumen-protected methionine (Smartamine M, Adisseo NA) could ameliorate the transition to lactation and the health status of the cows, while controlling and reducing the negative effects of prepartal excess energy. In fact, methionine (Met) itself was able to increase both quantity and quality of production [16, 17], controlling the inflammatory and the oxidative stress status that characterize the transition period [18–20]. These outcomes are partly due to Met’s ability to enhance liver function, reducing triacylglycerol accumulation and improving the metabolic capacity of the liver to orchestrate the metabolic transition into lactation [16–20]. Furthermore, Met itself, and several of its metabolites, display an immunonutritional role both in humans [21–24] and in dairy cows [16]. Therefore, in the present study we used serum and plasma biomarkers coupled with targeted hepatic transcriptome analysis from transition cows fed prepartum either a control low energy, a higher-energy, or a higher-energy diet supplemented with rumen-protected Met. Production and immune responses have been published elsewhere [25].
Methods
Experimental design and dietary treatments
All procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Illinois. Complete details of the experimental design and animal management have been reported previously [25]. Briefly, 65 multiparous Holstein were enrolled and completed the trail remaining healthy throughout the length of the study. All cows were fed ad libitum the same control lower-energy diet (CON; NEL = 1.24 Mcal/kg DM; no Met supplementation) during the far-off dry period (i.e., -50 to -21 d relative to parturition). Consequently, during the close-up period (i.e. -21 d to calving), cows were randomly allocated to either a higher-energy diet (OVE; NEL = 1.54 Mcal/kg DM), OVE plus Smartamine M (OVE + SM; Adisseo, NA) or remained on CON. The same basal lactation diet (NEL = 1.75 Mcal/kg DM) was fed to all cows postpartum until d 30 relative to parturition. Smartamine M was top-dressed during the entire experiment over the OVE or lactation diet from -21 through 30 d relative to parturition at a rate of 0.07% of offered DM. For the current study, only a subset of cows were considered for blood biomarker (n = 10 per group) and hepatic gene expression (n = 8 per group) analyses.
Blood sampling and biomarker analysis
Blood was sampled at -26, -21, -10, 7, 14 and 21 d relative to parturition by coccygeal venipuncture using evacuated tubes (BD Vacutainer; BD and Co., Franklin Lakes, NJ) containing either clot activator or lithium heparin for serum and plasma, respectively. Blood was used for determination of (i) metabolic biomarkers: cholesterol, creatinine, growth hormone (GH), insulin-like growth factor 1 (IGF-1), leptin, urea; (ii) liver health biomarkers: albumin, bilirubin, ceruloplasmin, gamma-glutamyl-transpeptidase (GGT), glutamic oxaloacetic transaminase (GOT), haptoglobin, interleukin 6, serum amyloid A (SAA); (iii) and oxidative status biomarkers: β-carotene, glutathione, nitric oxides (NOx, NO2, NO3), paraoxonase, antioxidant capacity (oxygen radical absorbance capacity, ORAC), total reactive oxygen metabolites (ROM), tocopherol.
Concentration of albumin, cholesterol, bilirubin, creatinine, urea, GOT, and GGT were assessed using kits purchased from Instrumentation Laboratory (Lexington, MA) using a clinical auto-analyzer (ILAB 600, Instrumentation Laboratory). Concentrations of ROM were analyzed with the d-ROMs-test, purchased from Diacron (Grosseto, Italy). Concentrations of haptoglobin, ceruloplasmin, paraoxonase and NOx were analyzed using the methods previously described [26–28], adapting the procedures to a clinical auto-analyzer (ILAB 600, Instrumentation Laboratory). SAA and ORAC determinations were performed using the Synergy 2 Multi-Detection Microplate Reader (BioTek Instruments, Inc., Winooski, VT). SAA concentration was assessed with a commercial ELISA immunoassay kit (Tridelta Development Ltd., Maynooth, Co. Kildare, Ireland), while ORAC was determined measuring the fluorescent signal from a probe (fluorescein) that decreases in the presence of radical damage [29]. Quantification of GH, IGF-1, and leptin concentration was as previously described [14]. Bovine IL-6 (Cat. No. ESS0029; Thermo Scientific, Rockford, IL) plasma concentration was determined using commercial ELISA kits, while plasma vitamin A, vitamin E, and β-carotene were extracted with hexane and analyzed by reverse- phase HPLC using an Allsphere ODS-2 column (3 μm, 150 × 4.6 mm; Grace Davison Discovery Sciences, Deerfield, IL), a UV detector set at 325 nm (for vitamin A), 290 nm (for vitamin E), or 460 nm (for β-carotene), and 80:20 methanol:tetrahydrofurane as the mobile phase.
Hepatic gene expression analysis
Liver tissue was harvested via percutaneous biopsy under local anesthesia at -10, 7 and 21 d relative to parturition. Tissue samples were immediately snap frozen in liquid nitrogen and then stored at -80 °C. Complete information about RNA extraction and qPCR procedures can be found in Additional file 1. Briefly, RNA samples were extracted from the frozen tissue and used for cDNA synthesis using established protocols in our laboratory [30]. The qPCR performed was SYBR Green-based, using a 6-point standard curve. Genes selected for transcript profiling are associated with (i) energy metabolism: insulin like growth factor-1 (IGF1), pyruvate carboxylase (PC), phosphoenolpyruvate carboxykinase 1 (PCK1), pyruvate dehydrogenase kinase 4 (PDK4); (ii) fatty acid metabolism: acyl-CoA oxidase 1 (ACOX1), apolipoprotein B (APOB), γ-butyrobetaine hydroxylase 1 (BBOX1), carnitine palmitoyltransferase 1A (CPT1A), 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2), microsomal triglyceride transfer protein (MTTP), peroxisome proliferator activated receptor α (PPARA), solute carrier family 22 member 5 (SLC22A5), trimethyllysine hydroxylase, ε (TMLHE); (iii) hepatokines: angiopoietin like 4 (ANGPTL4), fibroblast growth factor 21 (FGF21); (iv) the methionine cycle: betaine--homocysteine S-methyltransferase (BHMT), betaine--homocysteine S-methyltransferase 2 (BHMT2), DNA (cytosine-5-)-methyltransferase 1 (DNMT1), DNA (cytosine-5-)-methyltransferase 3 α (DNMT3A), methionine adenosyltransferase 1A (MAT1A), 5-methyltetrahydrofolate-homocysteine methyltransferase (MTR), phosphatidylethanolamine N-methyltransferase (PEMT), S-adenosylhomocysteine hydrolase (SAHH); (v) the antioxidant system: cystathionine-beta-synthase (CBS), cysteine sulfinic acid decarboxylase (CSAD), cystathionine gamma-lyase (CTH), glutamate-cysteine ligase catalytic subunit (GCLC), glutathione peroxidase 1 (GPX1), glutathione reductase (GSR), glutathione synthetase (GSS), superoxide dismutase 1, soluble (SOD1), superoxide dismutase 2, mitochondrial (SOD2); (vi) and the inflammatory response: ceruloplasmin (CP), haptoglobin (HP), nuclear factor κB subunit 1 (NFKB1), retinoid X receptor α (RXRA), serum amyloid A2 (SAA2), suppressor of cytokine signaling 2 (SOCS2), signal transducer and activator of transcription 3 (STAT3), signal transducer and activator of transcription 5B (STAT5B). Primer sequences and qPCR performances are reported in Additional file 1.
Statistical analysis
After normalization with the geometric mean of the internal control genes, qPCR data were log2 transformed prior to statistical analysis to obtain a normal distribution. Statistical analysis was performed with SAS (v9.3). Both datasets (blood and qPCR) were subjected to ANOVA and analyzed using repeated measures ANOVA with PROC MIXED. The statistical model included diet (D; CON, OVE, and OVE + SM), time (T; d -26, -21, -10, 7, 14, and 21 for blood biomarkers, d -10, 7, and 21 for qPCR analysis) and their interaction (D*T) as fixed effect. Cow, nested within treatment, was the random effect. For blood data, data pre-treatment at d-26 relative to parturition, when available, were used as a covariate. The Kenward-Roger statement was used for computing the denominator degrees of freedom, while spatial power was used as the covariance structure. Data were considered significant at a P ≤ 0.05 using the PDIFF statement in SAS. For ease of interpretation, expression data reported in Table 1 and Fig. 1 are the log2 back-transformed LSM that resulted from the statistical analysis. Standard errors were also adequately back-transformed.
Table 1.
Effect of feeding a control lower-energy diet (CON, 1.24 Mcal/kg DM; high-straw) during the whole dry period (~50 d), a higher-energy (1.54 Mcal/kg DM) diet without (OVE) rumen-protected methionine during the last 21 d before calving, or OVE plus rumen-protected methionine (Smartamine M; OVE + SM; Adisseo NA) from -21 d before calving through the first 30 d postpartum on hepatic gene expression (relative mRNA abundance, log2 back-transformed LSM) in Holstein cows
| Diet1 | P-value3 | ||||||
|---|---|---|---|---|---|---|---|
| CON | OVE | OVE + SM | SE2 | D | T | D*T | |
| Energy metabolism | |||||||
| IGF1 | 1.91 | 1.97 | 2.41 | 0.22 | 0.19 | <.0001 | 0.17 |
| PC | 0.24 | 0.23 | 0.20 | 0.02 | 0.24 | <.0001 | 0.09 |
| PCK1 | 0.33a | 0.25b | 0.31a | 0.02 | 0.03 | 0.10 | 0.59 |
| PDK4 | 0.34 | 0.31 | 0.78 | 0.37 | 0.35 | 0.03 | 0.98 |
| Fatty acid oxidation, Lipoprotein and Cholesterol synthesis | |||||||
| ACOX1 | 1.34 | 1.21 | 1.27 | 0.07 | 0.41 | 0.75 | 0.61 |
| APOB | 1.93 | 1.67 | 1.82 | 0.11 | 0.22 | 0.32 | 0.42 |
| BBOX1 | 0.39 | 0.36 | 0.38 | 0.02 | 0.61 | 0.04 | 0.96 |
| CPT1A | 0.13 | 0.13 | 0.14 | 0.01 | 0.89 | <.0001 | 0.49 |
| HMGCS2 | 1.13 | 1.00 | 0.93 | 0.10 | 0.32 | 0.22 | 0.56 |
| MTTP | 1.48a | 1.50a | 1.28b | 0.08 | 0.05 | 0.02 | 0.84 |
| PPARA | 0.43 | 0.40 | 0.46 | 0.02 | 0.16 | <.0001 | 0.03 |
| SLC22A5 | 2.89 | 2.92 | 3.08 | 0.29 | 0.88 | <.0001 | 0.14 |
| TMLHE | 0.42 | 0.40 | 0.41 | 0.02 | 0.68 | 0.003 | 0.39 |
| Hepatokines | |||||||
| ANGPTL4 | 0.01 | 0.01 | 0.02 | 0.002 | 0.34 | <.0001 | 0.03 |
| FGF21 | 0.27 | 0.13 | 0.22 | 0.07 | 0.15 | 0.005 | 0.0006 |
| Methionine cycle and methylation | |||||||
| BHMT | 1.13 | 0.96 | 1.02 | 0.11 | 0.54 | 0.002 | 0.70 |
| BHMT2 | 0.38 | 0.45 | 0.37 | 0.04 | 0.20 | 0.21 | 0.57 |
| DNMT1 | 0.02 | 0.02 | 0.02 | 0.001 | 0.53 | 0.03 | 0.64 |
| DNMT3A | 0.99a | 1.26b | 1.20b | 0.09 | 0.04 | 0.05 | 0.91 |
| MAT1A | 1.46 | 1.55 | 1.52 | 0.08 | 0.71 | 0.09 | 0.86 |
| MTR | 0.05a | 0.03b | 0.04ab | 0.002 | 0.01 | 0.31 | 0.65 |
| PEMT | 0.33 | 0.29 | 0.30 | 0.02 | 0.31 | 0.07 | 0.95 |
| SAHH | 1.39 | 1.25 | 1.39 | 0.06 | 0.17 | 0.0003 | 0.0009 |
| Antioxidant system | |||||||
| CBS | 1.41 | 1.66 | 1.51 | 0.11 | 0.27 | 0.08 | 0.93 |
| CSAD | 0.23 | 0.26 | 0.22 | 0.04 | 0.64 | 0.0004 | 0.36 |
| CTH | 0.45 | 0.46 | 0.45 | 0.02 | 0.93 | 0.24 | 0.86 |
| GCLC | 0.06 | 0.06 | 0.05 | 0.004 | 0.80 | 0.01 | 0.35 |
| GPX1 | 1.39 | 1.37 | 1.39 | 0.10 | 0.99 | 0.04 | 0.77 |
| GSR | 0.28 | 0.30 | 0.26 | 0.02 | 0.11 | <.0001 | 0.11 |
| GSS | 0.45 | 0.49 | 0.46 | 0.02 | 0.34 | 0.78 | 0.95 |
| SOD1 | 3.56 | 3.64 | 3.40 | 0.12 | 0.27 | 0.07 | 0.52 |
| SOD2 | 4.06 | 4.08 | 4.10 | 0.21 | 0.99 | 0.92 | 0.53 |
| Inflammatory response | |||||||
| CP | 2.48 | 2.13 | 2.37 | 0.22 | 0.49 | 0.003 | 0.85 |
| HP | 0.18 | 0.13 | 0.14 | 0.09 | 0.89 | 0.01 | 0.48 |
| NFKB1 | 2.29 | 2.14 | 2.48 | 0.14 | 0.25 | 0.04 | 0.83 |
| RXRA | 0.57 | 0.56 | 0.58 | 0.03 | 0.86 | 0.20 | 0.66 |
| SAA2 | 0.01 | 0.01 | 0.02 | 0.002 | 0.68 | 0.02 | 0.95 |
| SOCS2 | 2.09 | 1.84 | 2.26 | 0.24 | 0.44 | 0.16 | 0.60 |
| STAT3 | 1.38 | 1.42 | 1.39 | 0.11 | 0.96 | 0.05 | 0.43 |
| STAT5B | 2.30 | 2.26 | 2.45 | 0.07 | 0.15 | 0.002 | 0.07 |
1Prepartum dietary treatment: CON = control energy, OVE = moderate energy, OVE + SM = OVE supplemented with rumen-protected methionine (Smartamine M, Adisseo Inc.)
2SE = greatest standard error of the mean
3D = diet, T = time, D*T = diet by time interaction
a, bSignificant difference among dietary groups (P ≤ 0.05). Differences reported for genes with a significant (P ≤ 0.05) Diet effect
Fig. 1.
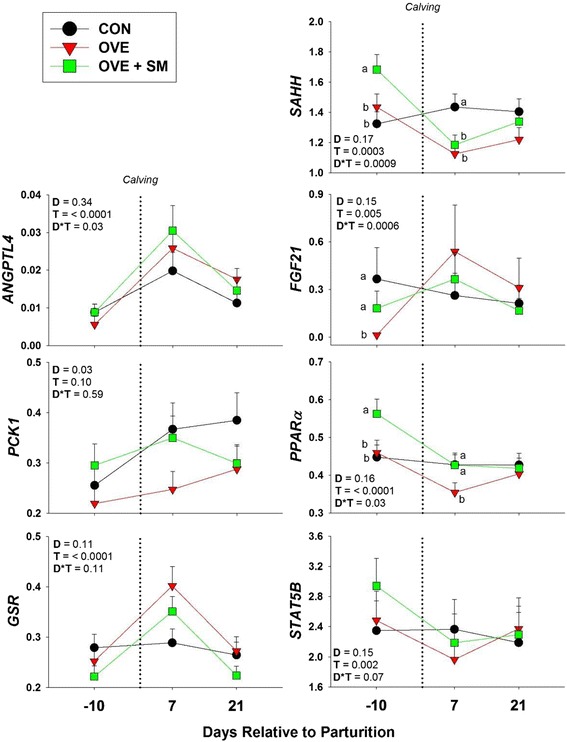
Effect of feeding a control lower-energy diet (CON, 1.24 Mcal/kg DM; high-straw) during the whole dry period (~50 d), a higher-energy (1.54 Mcal/kg DM) diet without (OVE) rumen-protected methionine during the last 21 d before calving, or OVE plus rumen-protected methionine (Smartamine M; OVE + SM; Adisseo NA) from -21 d before calving through the first 30 d postpartum on hepatic gene expression (log2 back-transformed LSM) in Holstein cows
Results
Blood biomarkers
Metabolism
Time affected all metabolic biomarkers (cholesterol, creatinine, GH, IGF1, leptin, urea; T, P < 0.001). However, no effect of diet or its interaction with time was detected (D, D*T, P > 0.05) (Fig. 2).
Fig. 2.
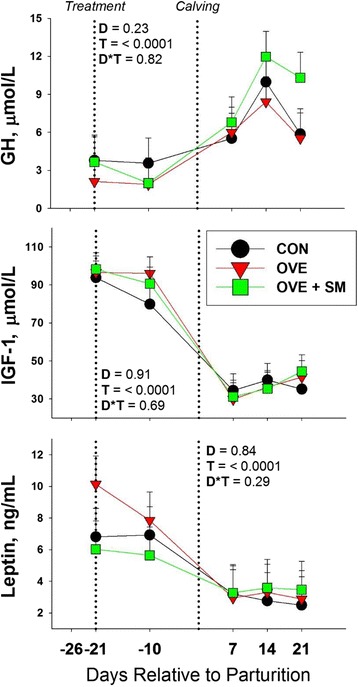
Effect of feeding a control lower-energy diet (CON, 1.24 Mcal/kg DM; high-straw) during the whole dry period (~50 d), a higher-energy (1.54 Mcal/kg DM) diet without (OVE) rumen-protected methionine during the last 21 d before calving, or OVE plus rumen-protected methionine (Smartamine M; OVE + SM; Adisseo NA) from -21 d before calving through the first 30 d postpartum on endocrine profiles in Holstein cows
Health status
No effects of diet or its interaction with time were significant for haptoglobin or IL-6 concentration (D, D*T, P > 0.05). Diet affected albumin concentration (D*T, P < 0.05), with greater (P < 0.05) postpartum concentrations in OVE + SM compared with both other groups. The overall concentration of total bilirubin, ceruloplasmin, and serum amyloid A tended to be affected by diet (D, P < 0.10), with greater levels (P < 0.05) in OVE cows compared with CON (total bilirubin), OVE + SM (ceruloplasmin), or both other groups (SAA). Diet also affected GGT and GOT concentration, as OVE + SM had greater (P < 0.05) overall GGT concentration (D, P < 0.10), especially postpartum (d 14 and 21), compared with OVE, and lower (P < 0.05) GOT concentration postpartum (d 7 and 14) (D*T, P < 0.05) compared with CON cows. Time affected the concentration of all previous health biomarkers (T, P < 0.01).
Antioxidant and oxidative status
No effect of diet was detected for ORAC, total ROM and tocopherol (D, P > 0.05). Total NOx also were not affected by diet, despite the fact that concentrations of both NO2 and NO3 had significant interactions or diet effects (NO2, D*T, P < 0.10; NO3, D, P < 0.05, D*T, P < 0.10). Diet had a strong effect on GSH concentration (D, P < 0.001), with greatest concentration (P < 0.05) in OVE + SM cows compared with both other groups. When interacting with time, diet tended to affect blood concentration of β-carotene and retinol (D*T, P < 0.10). For the first, the response was due to a greater (P < 0.05) concentration in OVE + SM cows compared with CON at -21 and -10 d, and to a lower (P < 0.05) concentration in OVE compared with CON at 14 d relative to parturition. In the case of retinol, the interaction was due the increasing (P < 0.05) concentration postpartum from 7 to 21 d in OVE + SM cows, while in CON and OVE cows the concentration remained constant (P > 0.05). This led to a greater (P < 0.05) retinol concentration in OVE + SM at 21 d postpartum compare with OVE. Diet also affected paraoxonase concentration, with overall greater level (P < 0.05) in CON compared with OVE and OVE + SM (D, P < 0.05). This difference was due to greater (P < 0.05) concentration in CON cows at -21, -10 and 7 d relative to parturition (D*T, P < 0.05).
Gene expression
Energy metabolism
Cows fed the CON or OVE + SM diets had greater PCK1 expression compared with OVE cows (D, P < 0.05). Diet also affected the expression of the fatty acid metabolism related genes MTTP (D, P < 0.05) and PPARA (D*T, P < 0.05). Expression of MTTP was in fact greater (P < 0.05) in CON and OVE cows, compared with OVE + SM, while PPARA expression was greater (P < 0.05) prepartum (-10 d) for OVE + SM compared with CON and OVE, and lower (P < 0.05) early postpartum (7 d) for OVE compared with the other two groups.
Hepatokines and inflammation
Diet alone did not affect genes related to hepatokiens and the inflammatory response (D, P > 0.05). However, the hepatokines ANGPTL4 and FGF21 had a significant interaction with time (D*T, P < 0.05). For FGF21 this significance was due to a greater (P < 0.05) prepartal expression in CON and OVE + SM compared with OVE cows, while for ANPTL4 no differences among dietary groups were detected across the analyzed time points (P > 0.05).
Methionine cycle and antioxidant system
No genes concerning the antioxidant system were significantly affected by diet, or its interaction with time (D, D*T, P > 0.05). However, MTR and DNMT3A, genes of the methionine cycle, had an overall effect of diet (D, P < 0.05). Expression of MTR was greater (P < 0.05) in CON compared with OVE, with OVE + SM having an intermediate level of expression, while DNMT3A expression was greater (P < 0.05) in OVE and OVE + SA compared with CON cows. Furthermore, SAHH expression was greater (D*T, P < 0.05) prepartum in OVE + SM cows compared with the other dietary groups; whereas, expression was greater (P < 0.05) early postpartum (7 d) in CON cows compared with OVE and OVE + SM.
Discussion
Overfeeding dairy cows in the weeks prior parturition (e.g. close up period) has been previously linked with a more pronounced negative energy balance postpartum, due to bigger drops in voluntary dry matter intake (DMI) along with sustained lipid mobilization and possible accumulation of triacylglycerol (TAG) in the liver [25]. The present study confirmed the overfeeding-induced depression of DMI postpartum and hepatic TAG accumulation [25]. Furthermore, despite previous studies reporting that overfed cows were always able to maintain similar levels of milk production as the control-fed counterparts [31], these changes led to worse milk performance including lower milk and energy corrected milk yield [25].
As hypothesized, supplementation of rumen-protected Met to a moderate energy diet was able to overcome the detrimental effects of energy overfeeding. In fact, OVE + SM cows compared with OVE had greater postpartal DMI and better milk production, matching the performance of the control-fed group [25]. Despite the fact that the improved DMI, likely a consequence of the improved health status, could easily explain the improved production performance, other cellular and physiologic also likely were contributing factors.
The hepatic transcriptome revealed how Met supplementation restored PCK1 expression (an important gluconeogenic gene) to the level of control-fed cows. At least postpartum this could be explained by the higher insulin concentration in OVE + SM [25], as hepatic PCK1 mRNA expression is directly related to insulin level [32]. The increased insulin concentration also could explain why circulating glucose was lower in OVE + SM cows [25] compared with CON, i.e. overfeeding alone does not affect peripheral insulin resistance [9], and the increased insulin concentration was not followed by changes in GH or IGF1, hence, the improved milk production with OVE + SM also might have resulted from an increase in glucose availability directly channeled to peripheral tissues and the mammary gland. In the latter case it would have contributed to greater lactose production. Peripheral tissues, i.e. adipose and muscle, rely mainly on GLUT4 (an insulin-dependent transporter) for glucose uptake, while the mammary gland uses mainly GLUT1 (usually described as insulin-independent) as the preferred glucose transporter [33]. However, a recent study revealed that insulin increases GLUT1 expression in bovine mammary explants, thus, providing evidence of a functional link between circulating insulin and mammary glucose uptake [34].
Supplementing Met also increased both fat and protein percentage during the first week of lactation [25]. Because biomarkers of muscle catabolism were not affected by diet (e.g. urea and creatinine) and DMI was similar in CON and OVE + SM, we speculate that Met itself, combined with higher circulating insulin, might have been the primary cause of the improved protein percentage. In fact, previous research demonstrated that an increase in amino acid supply (e.g. abomasal casein infusion) could markedly improve milk protein yield, especially when the circulating level of insulin was artificially raised through a clamp [35, 36]. The lower inflammation status and greater liver function around calving in the OVE + SM cows (lower concentrations of albumin and greater bilirubin, ceruloplasmin, GGT, GOT, and SAA) would have guaranteed higher availability of plasma amino acids [37] to the mammary gland for protein synthesis. The increase in fat content, which agrees with several previous studies [16, 38–41], might have been related to cellular pathways involving Met and its methylated compounds (e.g. choline [42]), which some data indicate are important for supporting milk fat synthesis in cows [43].
As previously mentioned, overfeeding energy prepartum led to hepatic TAG accumulation [25], a condition that, if excessive, could become a potential burden for proper liver function [2]. OVE cows, in fact, had signs of impaired liver function and inflammatory condition postpartum including lower concentrations of albumin and greater bilirubin, ceruloplasmin, GGT, GOT, and SAA (Table 2, Fig. 3). As hypothesized, supplemental Met was able to correct these effects of the OVE diet. Thus, as a primary outcome, OVE + SM cows had less liver TAG accumulation [25] despite similar NEFA concentration between OVE and OVE + SM [25]. This was at least in part due to greater PPARA expression with Met supplementation.
Table 2.
Effect of feeding a control lower-energy diet (CON, 1.24 Mcal/kg DM; high-straw) during the whole dry period (~50 d), a higher-energy (1.54 Mcal/kg DM) diet without (OVE) rumen-protected methionine during the last 21 d before calving, or OVE plus rumen-protected methionine (Smartamine M; OVE + SM; Adisseo NA) from -21 d before calving through the first 30 d postpartum on biomarker concentrations of metabolism, liver health, and oxidative status in Holstein cows
| Diet1 | P-values3 | ||||||
|---|---|---|---|---|---|---|---|
| Items | CON | OVE | OVE + SM | SE2 | D | T | D*T |
| Metabolism | |||||||
| Cholesterol, mmol/L | 3.24 | 3.16 | 3.26 | 0.11 | 0.76 | <.0001 | 0.71 |
| Creatinine, μmol/L | 97.60 | 98.88 | 97.68 | 1.53 | 0.77 | <.0001 | 0.17 |
| GH, ng/mL | 5.75 | 4.79 | 6.95 | 1.08 | 0.23 | <.0001 | 0.82 |
| IGF-1, ng/mL | 56.65 | 60.03 | 59.98 | 6.64 | 0.91 | <.0001 | 0.69 |
| Leptin, ng/mL | 4.44 | 5.42 | 4.40 | 1.62 | 0.84 | <.0001 | 0.29 |
| Urea, mmol/L | 5.20 | 5.05 | 5.05 | 0.18 | 0.77 | <.0001 | 0.30 |
| Liver health | |||||||
| Albumin, g/L | 35.41 | 35.54 | 36.32 | 0.41 | 0.24 | 0.0002 | 0.05 |
| Bilirubin , μmol/L | 2.29a | 3.38b | 2.57ab | 0.41 | 0.10 | <.0001 | 0.57 |
| Ceruloplasmin, μmol/L | 2.77ab | 2.91b | 2.61a | 0.09 | 0.006 | <.0001 | 0.51 |
| GGT, U/L | 22.96a | 25.21ab | 26.85b | 1.17 | 0.07 | <.0001 | 0.008 |
| GOT, U/L | 84.76 | 90.30 | 81.71 | 5.61 | 0.48 | <.0001 | 0.04 |
| Haptoglobin, g/L | 0.42 | 0.46 | 0.41 | 0.06 | 0.76 | 0.003 | 0.86 |
| IL-6, pg/mL | 530.63 | 586.37 | 412.76 | 98.56 | 0.37 | 0.001 | 0.67 |
| SAA, μg/mL | 35.55a | 54.00b | 34.77a | 7.79 | 0.10 | 0.0005 | 0.58 |
| Oxidative status | |||||||
| β-carotene, mg/100 mL | 0.20 | 0.19 | 0.23 | 0.02 | 0.14 | 0.04 | 0.06 |
| Liver GSH, mmol/L | 953a | 1281b | 1693c | 120 | 0.0002 | 0.05 | 0.14 |
| NO2, μmol/L | 6.03 | 6.66 | 6.80 | 0.45 | 0.44 | 0.01 | 0.09 |
| NO3, μmol/L | 18.65a | 16.90b | 16.77b | 0.40 | 0.002 | <.0001 | 0.08 |
| NOx, μmol/L | 24.61 | 23.54 | 23.67 | 0.56 | 0.31 | <.0001 | 0.18 |
| ORAC, TE mol/L | 12,731 | 12,359 | 12,739 | 198 | 0.25 | <.0001 | 0.66 |
| Paraoxonase, U/mL | 77.96a | 68.41b | 66.74b | 2.68 | 0.01 | <.0001 | 0.02 |
| Retinol, μg/100 mL | 46.39 | 41.79 | 43.42 | 3.10 | 0.44 | 0.0009 | 0.08 |
| ROM, mg of H2O2/100 mL | 14.01 | 12.99 | 13.44 | 0.49 | 0.31 | <.0001 | 0.79 |
| Tocopherol, μg/mL | 3.67 | 3.68 | 3.16 | 0.44 | 0.46 | <.0001 | 0.31 |
1Prepartum dietary treatment: CON = control energy, OVE = moderate energy, OVE + SM = OVE supplemented with rumen-protected methionine (Smartamine M, Adisseo Inc.)
2SE = greatest standard error of the mean
3D = diet, T = time, D*T = diet by time interaction
a, b, cSignificant difference among dietary groups (P ≤ 0.05). Differen reported for biomarkers with a tendency (P ≤ 0.10) or a significan (P ≤ 0.05) Diet effect
Fig. 3.
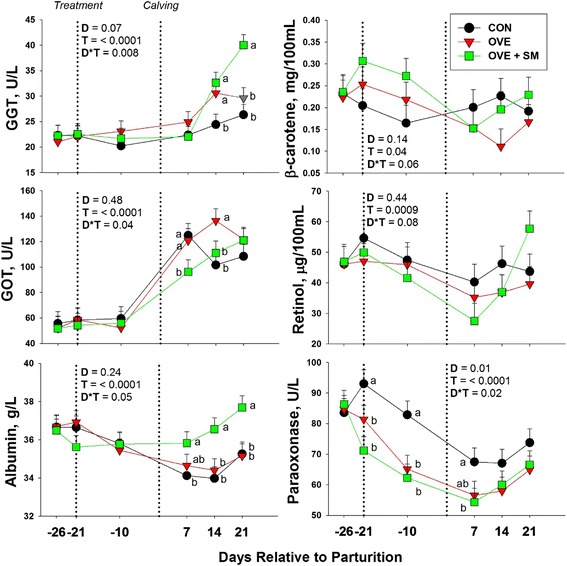
Effect of feeding a control lower-energy diet (CON, 1.24 Mcal/kg DM; high-straw) during the whole dry period (~50 d), a higher-energy (1.54 Mcal/kg DM) diet without (OVE) rumen-protected methionine during the last 21 d before calving, or OVE plus rumen-protected methionine (Smartamine M; OVE + SM; Adisseo NA) from -21 d before calving through the first 30 d postpartum on blood biomarkers of liver function and antioxidant status in Holstein cows
Among the most important metabolic functions coordinated by PPARα are LCFA uptake, intracellular activation, oxidation, and ketogenesis [44]. Thus its greater expression in OVE + SM cows could have improved NEFA handling, i.e. through greater oxidation. Furthermore, PCK1 is also involved in glyceroneogenesis, as it can catalyze the production of glycerol-3-phospate for use during fatty acid esterification [45]. Thus the increase of its expression could have further improved NEFA handling by the liver. The lower expression of MTTP in the OVE + SM cows was lower indicated a potentially lower capacity of these cows to synthesize and export VLDL. However, the data from Bernabucci et al. [46] indicated that apolipoprotein mRNA transcription rather than MTTP might be the limiting step in the repackaging of TAG into lipoproteins, hence, explaining the increase in concentration of plasma VLDL in OVE + SM cows [25]. As a subsequent outcome, the improved fatty acid metabolism in liver with Met supplementation reduces the risk of liver dysfunction, an idea supported by the biomarkers of liver function (e.g. greater albumin and VLDL, and lower bilirubin) in OVE + SM cows [47].
Metabolic dysfunction and inflammatory events are often linked through oxidative stress, a common outcome to both scenarios [48–50]. The present study partly confirmed the possible molecular mechanisms through which prepartum overfeeding could cause an increased concentration of oxidants proposed by Loor et al. [51]. OVE did not cause changes in total ROM and NOx, however, these cows had an impairment of the antioxidant system. Despite similar blood antioxidant capacity, paraoxonase concentration was in fact lower in OVE cows, a condition that not only indicates liver dysfunction, but one that has been proven to lead to an increase in the inflammatory status (confirmed by higher ceruloplasmin and SAA), which notoriously causes an increase in oxidative stress, and a reduction of antioxidative protection during the early postpartum period [27, 52]. As for paraoxonase, postpartum (d 14) concentration of β-carotene, a precursor of vitamin A, which exerts antioxidant effects [53], also was reduced in OVE compared with CON.
Supplementation of rumen-protected Met has been proven to benefit the oxidative status of periparturient cows [19, 20], in large part because it is a precursor for the biosynthesis of glutathione and taurine, two of the most important cellular antioxidants [54, 55]. In the present study, Met supplementation to cows fed a higher energy diet prepartum was able to improve their compromised antioxidant status. In fact, despite the lack of changes in ROM or paraoxonase compared with OVE, OVE + SM cows had greater glutathione concentrations, even compared with CON, together with higher retinol concentrations up to the level of control-fed cows. Concerning retinol, its concentration is also regulated by the hepatic synthesis of its carrier, retinol binding protein [56]. Thus, a greater plasma retinol concentration, besides suggesting a better antioxidant status, could also be a response to the better liver functionality detected in OVE + SM cows. Furthermore, GSR expression was decreased in OVE + SM cows to a similar level than OVE. GSR encodes the protein glutathione reductase, a central enzyme of cellular antioxidant defense, which reduces oxidized glutathione disulfide to the sulfhydryl form [57]. This further suggests a lesser oxidative status in cows fed methionine, which despite having a greater glutathione concentration seemed to have less of a need to restore the pool of its active form.
Other health benefits of methionine supplementation could also be noticed in the lower somatic cell count in milk. For instance, OVE + SM cows compared with both CON and OVE had lower milk SCC [25], a result that further highlights the immunometabolic effects of methionine and its metabolites [16, 21–23].
At a molecular level, the greater expression SAHH prepartum in OVE + SM cows underscores that the increased Met supply to the liver through supplementation was directed through the methionine cycle, leading to the higher glutathione concentrations. However, overfeeding energy prepartum (e.g. OVE and OVE + SM) seemed to reduce the overall expression of MTR, as if regenerating Met was not a hepatic priority. This becomes relevant in early lactation, because after calving the decrease in expression of both MTR and SAHH in all groups indicated that cows might redirect the circulating Met to the mammary gland for milk production. To further complicate this scenario, the greater DNMT3A expression in both OVE and OVE + SM cows indicated a role of overfeeding in its regulation. Its greater expression could indicate a higher need of methyl groups from methionine by the liver, hence, in light of the lower hepatic regeneration (e.g. lower MTR) but greater utilization (e.g. higher DNMT3A) Met supplementation (e.g. OVE + SM) favored the mammary demand. The fact that milk production was restored to the level of CON cows in the OVE + SM cows supports this scenario.
The mechanisms by which prepartal overfeeding causes a greater DNMT3A expression, increasing DNA methylation and leading to greater consumption of methyl groups from Met, are not clear. Insulin sensitivity was previously associated with increased global methylation [58], but overfeeding cows prepartum never led to its impairment in our previous experiments [9, 10]. On the other hand, levels of hepatic methylation were associated with fatty liver disease in humans [59, 60]. Because OVE cows had a greater hepatic TAG content [25], DNMT3A expression regulation could be explained by the alterations in lipid metabolism.
Conclusions
Current results confirm the detrimental outcome (e.g., reduced DMI, compromised liver function, and higher inflammatory status) of a higher-energy diet during the close up period in dairy cows, thus, supporting the need for energy restriction in the close-up period. However, if the practice persists, dairy producers should improve the diet methionine supply. In fact, supplemental rumen-protected methionine was effective in reducing the aforementioned effects, by (i) stimulating DMI and milk production, (ii) improving hepatic fatty acid metabolism and reducing TAG accumulation, (iii) improving general biomarkers of liver function, and (iv) limiting the postpartal negative effect of inflammation on the cow antioxidant system. Further investigation is needed to assess the effect of methionine supplementation to a prepartal energy restricted diet during the close-up.
Acknowledgements
The authors thank Travis Michels and Mike Katterhenry of the University of Illinois Dairy Research Unit (Urbana) staff for help with animal management.
Funding
Financial support for the research was provided in part by Adisseo (Commentry, France) and Hatch funds under project ILLU-538–914, National Institute of Food and Agriculture, Washington, DC, USA.
Availability of data and materials
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
MVR performed the qPCR statistical analysis and wrote the main draft of the manuscript, with inputs from DL, ET and JJL. JLL and DL designed the study. JSO performed the animal study and qPCR. ET and JSO performed the blood biomarker analysis. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
All procedures for this study (protocol no. 09214) were approved by the Institutional Animal Care and Use Committee of the University of Illinois.
Abbreviations
- ACOX1
Acyl-CoA oxidase 1
- ANGPTL4
Angiopoietin like 4
- APOB
Apolipoprotein B
- BBOX1
γ-butyrobetaine hydroxylase 1
- BHMT
BHMT2, Betaine--homocysteine S-methyltransferase 1, and 2
- CBS
Cystathionine-beta-synthase
- CP
Ceruloplasmin
- CPT1A
Carnitine palmitoyltransferase 1A
- CSAD
Cysteine sulfinic acid decarboxylase
- CTH
Cystathionine gamma-lyase
- DMI
Dry matter intake
- DNMT1
DNMT3A, DNA (cytosine-5-)-methyltransferase 1, and 3 α
- FGF21
Fibroblast growth factor 21
- GCLC
Glutamate-cysteine ligase catalytic subunit
- GGT
Gamma-glutamyl-transpeptidase
- GH
Growth hormone
- GLUT1
GLUT4, Glucose transporter 1, and 4
- GOT
Glutamic oxaloacetic transaminase
- GPX1
Glutathione peroxidase 1
- GSR
Glutathione reductase
- GSS
Glutathione synthetase
- HMGCS2
3-hydroxy-3-methylglutaryl-CoA synthase 2
- HP
Haptoglobin
- IGF1
Insulin like growth factor-1
- IL6
Interleukin 6
- MAT1A
Methionine adenosyltransferase 1A
- Met
Methionine
- MTR
5-methyltetrahydrofolate-homocysteine methyltransferase
- MTTP
Microsomal triglyceride transfer protein
- NEFA
Non-esterified fatty acids
- NEL
Net energy for lactation
- NFKB1
Nuclear factor κB subunit 1
- NOx
NO2, NO3, Nitric oxides
- ORAC
Oxygen radical absorbance capacity
- PC
Pyruvate carboxylase
- PCK1
Phosphoenolpyruvate carboxykinase 1
- PDK4
Pyruvate dehydrogenase kinase 4
- PEMT
Phosphatidylethanolamine N-methyltransferase
- PPARA
Peroxisome proliferator activated receptor α
- ROM
Reactive oxygen metabolites
- RXRA
Retinoid X receptor α
- SAA
SAA2, Serum amyloid A, and A2
- SAHH
S-adenosylhomocysteine hydrolase
- SLC22A5
Solute carrier family 22 member 5
- SOCS2
Suppressor of cytokine signaling 2
- SOD1
SOD2, Superoxide dismutase 1 (soluble), and 2 (mitochondrial)
- STAT3
STAT5B, Signal transducer and activator of transcription 3, and 5B
- TAG
Triacylglycerol
- TMLHE
Trimethyllysine hydroxylase, ε
- VLDL
Very low density lipoprotein
Additional file
Complete gene expression methodology, including primer sequences and qPCR performance. Figures S1-S5 include the two-way interaction graphs not shown in the manuscript. (DOCX 8109 kb)
Contributor Information
Mario Vailati-Riboni, Email: vailati2@illinois.edu.
Johan S. Osorio, Email: johan.osorio@sdstate.edu
Erminio Trevisi, Email: erminio.trevisi@unicatt.it.
Daniel Luchini, Email: daniel.luchini@adisseo.com.
Juan J. Loor, Email: jloor@illinois.edu
References
- 1.Bell AW. Regulation of organic nutrient metabolism during transition from late pregnancy to early lactation. J Anim Sci. 1995;73(9):2804–19. doi: 10.2527/1995.7392804x. [DOI] [PubMed] [Google Scholar]
- 2.Drackley JK. ADSA Foundation Scholar Award. Biology of dairy cows during the transition period: the final frontier? J Dairy Sci. 1999;82(11):2259–73. doi: 10.3168/jds.S0022-0302(99)75474-3. [DOI] [PubMed] [Google Scholar]
- 3.van Knegsel ATM, Hammon HM, Bernabucci U, Bertoni G, Bruckmaier RM, Goselink RMA, et al. Metabolic adaptation during early lactation: key to cow health, longevity and a sustainable dairy production chain. CAB Rev. 2014;9(002):1–15. doi: 10.1079/PAVSNNR20149002. [DOI] [Google Scholar]
- 4.Curtis CR, Erb HN, Sniffen CJ, Smith RD, Kronfeld DS. Path analysis of dry period nutrition, postpartum metabolic and reproductive disorders, and mastitis in Holstein cows. J Dairy Sci. 1985;68(9):2347–60. doi: 10.3168/jds.S0022-0302(85)81109-7. [DOI] [PubMed] [Google Scholar]
- 5.Boutflour RB. Limiting factors in the feeding and management of milk cows. In: Report of proceedings of 8th world’s dairy congress. London; 1928. p. 15-20.
- 6.Holtenius K, Agenas S, Delavaud C, Chilliard Y. Effects of feeding intensity during the dry period. 2. Metabolic and hormonal responses. J Dairy Sci. 2003;86(3):883–91. doi: 10.3168/jds.S0022-0302(03)73671-6. [DOI] [PubMed] [Google Scholar]
- 7.Janovick NA, Boisclair YR, Drackley JK. Prepartum dietary energy intake affects metabolism and health during the periparturient period in primiparous and multiparous Holstein cows. J Dairy Sci. 2011;94(3):1385–400. doi: 10.3168/jds.2010-3303. [DOI] [PubMed] [Google Scholar]
- 8.Ji P, Drackley JK, Khan MJ, Loor JJ. Overfeeding energy upregulates peroxisome proliferator-activated receptor (PPAR)gamma-controlled adipogenic and lipolytic gene networks but does not affect proinflammatory markers in visceral and subcutaneous adipose depots of Holstein cows. J Dairy Sci. 2014;97(6):3431–40. doi: 10.3168/jds.2013-7295. [DOI] [PubMed] [Google Scholar]
- 9.Ji P, Osorio JS, Drackley JK, Loor JJ. Overfeeding a moderate energy diet prepartum does not impair bovine subcutaneous adipose tissue insulin signal transduction and induces marked changes in peripartal gene network expression. J Dairy Sci. 2012;95(8):4333–51. doi: 10.3168/jds.2011-5079. [DOI] [PubMed] [Google Scholar]
- 10.Khan MJ, Jacometo CB, Graugnard DE, Correa MN, Schmitt E, Cardoso F, et al. Overfeeding Dairy Cattle During Late-Pregnancy Alters Hepatic PPARalpha-Regulated Pathways Including Hepatokines: Impact on Metabolism and Peripheral Insulin Sensitivity. Gene Regul Syst Bio. 2014;8:97–111. doi: 10.4137/GRSB.S14116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rukkwamsuk T, Wensing T, Geelen MJ. Effect of overfeeding during the dry period on the rate of esterification in adipose tissue of dairy cows during the periparturient period. J Dairy Sci. 1999;82(6):1164–9. doi: 10.3168/jds.S0022-0302(99)75339-7. [DOI] [PubMed] [Google Scholar]
- 12.Soliman M, Kimura K, Ahmed M, Yamaji D, Matsushita Y, Okamatsu-Ogura Y, et al. Inverse regulation of leptin mRNA expression by short- and long-chain fatty acids in cultured bovine adipocytes. Domest Anim Endocrinol. 2007;33(4):400–9. doi: 10.1016/j.domaniend.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Dann HM, Litherland NB, Underwood JP, Bionaz M, D’Angelo A, McFadden JW, et al. Diets during far-off and close-up dry periods affect periparturient metabolism and lactation in multiparous cows. J Dairy Sci. 2006;89(9):3563–77. doi: 10.3168/jds.S0022-0302(06)72396-7. [DOI] [PubMed] [Google Scholar]
- 14.Graugnard DE, Moyes KM, Trevisi E, Khan MJ, Keisler D, Drackley JK, et al. Liver lipid content and inflammometabolic indices in peripartal dairy cows are altered in response to prepartal energy intake and postpartal intramammary inflammatory challenge. J Dairy Sci. 2013;96(2):918–35. doi: 10.3168/jds.2012-5676. [DOI] [PubMed] [Google Scholar]
- 15.Shahzad K, Bionaz M, Trevisi E, Bertoni G, Rodriguez-Zas SL, Loor JJ. Integrative analyses of hepatic differentially expressed genes and blood biomarkers during the peripartal period between dairy cows overfed or restricted-fed energy prepartum. PLoS One. 2014;9(6):e99757. doi: 10.1371/journal.pone.0099757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osorio JS, Ji P, Drackley JK, Luchini D, Loor JJ. Supplemental Smartamine M or MetaSmart during the transition period benefits postpartal cow performance and blood neutrophil function. J Dairy Sci. 2013;96(10):6248–63. doi: 10.3168/jds.2012-5790. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Z, Vailati-Riboni M, Trevisi E, Drackley JK, Luchini DN, Loor JJ. Better postpartal performance in dairy cows supplemented with rumen-protected methionine compared with choline during the peripartal period. J Dairy Sci. 2016;99(11):8716–32. doi: 10.3168/jds.2015-10525. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Z, Bulgari O, Vailati-Riboni M, Trevisi E, Ballou MA, Cardoso FC, et al. Rumen-protected methionine compared with rumen-protected choline improves immunometabolic status in dairy cows during the peripartal period. J Dairy Sci. 2016;99(11):8956–69. doi: 10.3168/jds.2016-10986. [DOI] [PubMed] [Google Scholar]
- 19.Osorio JS, Ji P, Drackley JK, Luchini D, Loor JJ. Smartamine M and MetaSmart supplementation during the peripartal period alter hepatic expression of gene networks in 1-carbon metabolism, inflammation, oxidative stress, and the growth hormone-insulin-like growth factor 1 axis pathways. J Dairy Sci. 2014;97(12):7451–64. doi: 10.3168/jds.2014-8680. [DOI] [PubMed] [Google Scholar]
- 20.Osorio JS, Trevisi E, Ji P, Drackley JK, Luchini D, Bertoni G, et al. Biomarkers of inflammation, metabolism, and oxidative stress in blood, liver, and milk reveal a better immunometabolic status in peripartal cows supplemented with Smartamine M or MetaSmart. J Dairy Sci. 2014;97(12):7437–50. doi: 10.3168/jds.2013-7679. [DOI] [PubMed] [Google Scholar]
- 21.Grimble RF. The effects of sulfur amino acid intake on immune function in humans. J Nutr. 2006;136(6 Suppl):1660S–5. doi: 10.1093/jn/136.6.1660S. [DOI] [PubMed] [Google Scholar]
- 22.Grimble RF, Grimble GK. Immunonutrition: role of sulfur amino acids, related amino acids, and polyamines. Nutrition. 1998;14(7-8):605–10. doi: 10.1016/S0899-9007(98)80041-5. [DOI] [PubMed] [Google Scholar]
- 23.Li P, Yin YL, Li D, Kim SW, Wu G. Amino acids and immune function. Br J Nutr. 2007;98(2):237–52. doi: 10.1017/S000711450769936X. [DOI] [PubMed] [Google Scholar]
- 24.Redmond HP, Stapleton PP, Neary P, Bouchier-Hayes D. Immunonutrition: the role of taurine. Nutrition. 1998;14(7-8):599–604. doi: 10.1016/S0899-9007(98)00097-5. [DOI] [PubMed] [Google Scholar]
- 25.Li C, Batistel F, Osorio JS, Drackley JK, Luchini D, Loor JJ. Peripartal rumen-protected methionine supplementation to higher energy diets elicits positive effects on blood neutrophil gene networks, performance and liver lipid content in dairy cows. J Anim Sci Biotechnol. 2016;7:18. doi: 10.1186/s40104-016-0077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacometo CB, Osorio JS, Socha M, Correa MN, Piccioli-Cappelli F, Trevisi E, et al. Maternal consumption of organic trace minerals alters calf systemic and neutrophil mRNA and microRNA indicators of inflammation and oxidative stress. J Dairy Sci. 2015;98(11):7717–29. doi: 10.3168/jds.2015-9359. [DOI] [PubMed] [Google Scholar]
- 27.Bionaz M, Trevisi E, Calamari L, Librandi F, Ferrari A, Bertoni G. Plasma paraoxonase, health, inflammatory conditions, and liver function in transition dairy cows. J Dairy Sci. 2007;90(4):1740–50. doi: 10.3168/jds.2006-445. [DOI] [PubMed] [Google Scholar]
- 28.Trevisi E, Amadori M, Cogrossi S, Razzuoli E, Bertoni G. Metabolic stress and inflammatory response in high-yielding, periparturient dairy cows. Res Vet Sci. 2012;93(2):695–704. doi: 10.1016/j.rvsc.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Lucini L, Kane D, Pellizzoni M, Ferrari A, Trevisi E, Ruzickova G, et al. Phenolic profile and in vitro antioxidant power of different milk thistle [Silybum marianum (L.) Gaertn.] cultivars. Ind Crop Prod. 2016;83:11–6. doi: 10.1016/j.indcrop.2015.12.023. [DOI] [Google Scholar]
- 30.Vailati Riboni M, Kanwal M, Bulgari O, Meier S, Priest NV, Burke CR, et al. Body condition score and plane of nutrition prepartum affect adipose tissue transcriptome regulators of metabolism and inflammation in grazing dairy cows during the transition period. J Dairy Sci. 2016;99(1):758–70. doi: 10.3168/jds.2015-10046. [DOI] [PubMed] [Google Scholar]
- 31.Graugnard DE, Bionaz M, Trevisi E, Moyes KM, Salak-Johnson JL, Wallace RL, et al. Blood immunometabolic indices and polymorphonuclear neutrophil function in peripartum dairy cows are altered by level of dietary energy prepartum. J Dairy Sci. 2012;95(4):1749–58. doi: 10.3168/jds.2011-4579. [DOI] [PubMed] [Google Scholar]
- 32.Aschenbach JR, Kristensen NB, Donkin SS, Hammon HM, Penner GB. Gluconeogenesis in dairy cows: the secret of making sweet milk from sour dough. IUBMB Life. 2010;62(12):869–77. doi: 10.1002/iub.400. [DOI] [PubMed] [Google Scholar]
- 33.Zhao FQ, Keating AF. Expression and regulation of glucose transporters in the bovine mammary gland. J Dairy Sci. 2007;90(Suppl 1):E76–86. doi: 10.3168/jds.2006-470. [DOI] [PubMed] [Google Scholar]
- 34.Shao Y, Wall EH, McFadden TB, Misra Y, Qian X, Blauwiekel R, et al. Lactogenic hormones stimulate expression of lipogenic genes but not glucose transporters in bovine mammary gland. Domest Anim Endocrinol. 2013;44(2):57–69. doi: 10.1016/j.domaniend.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Mackle TR, Dwyer DA, Ingvartsen KL, Chouinard PY, Lynch JM, Barbano DM, et al. Effects of insulin and amino acids on milk protein concentration and yield from dairy cows. J Dairy Sci. 1999;82(7):1512–24. doi: 10.3168/jds.S0022-0302(99)75378-6. [DOI] [PubMed] [Google Scholar]
- 36.Griinari JM, McGuire MA, Dwyer DA, Bauman DE, Barbano DM, House WA. The role of insulin in the regulation of milk protein synthesis in dairy cows. J Dairy Sci. 1997;80(10):2361–71. doi: 10.3168/jds.S0022-0302(97)76187-3. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Z, Loor JJ, Piccioli-Cappelli F, Librandi F, Lobley GE, Trevisi E. Circulating amino acids during the peripartal period in cows with different liver functionality index. J Dairy Sci. 2016;99(3):2257-67. [DOI] [PubMed]
- 38.Wang C, Liu HY, Wang YM, Yang ZQ, Liu JX, Wu YM, et al. Effects of dietary supplementation of methionine and lysine on milk production and nitrogen utilization in dairy cows. J Dairy Sci. 2010;93(8):3661–70. doi: 10.3168/jds.2009-2750. [DOI] [PubMed] [Google Scholar]
- 39.Broderick GA, Muck RE. Effect of alfalfa silage storage structure and rumen-protected methionine on production in lactating dairy cows. J Dairy Sci. 2009;92(3):1281–9. doi: 10.3168/jds.2008-1303. [DOI] [PubMed] [Google Scholar]
- 40.Broderick GA, Stevenson MJ, Patton RA. Effect of dietary protein concentration and degradability on response to rumen-protected methionine in lactating dairy cows. J Dairy Sci. 2009;92(6):2719–28. doi: 10.3168/jds.2008-1277. [DOI] [PubMed] [Google Scholar]
- 41.Lundquist RG, Otterby DE, Linn JG. Influence of three concentrations of DL-methionine or methionine hydroxy analog on milk yield and milk composition. J Dairy Sci. 1985;68(12):3350–4. doi: 10.3168/jds.S0022-0302(85)81245-5. [DOI] [PubMed] [Google Scholar]
- 42.Pinotti L, Baldi A, Dell’Orto V. Comparative mammalian choline metabolism with emphasis on the high-yielding dairy cow. Nutr Res Rev. 2002;15(2):315–32. doi: 10.1079/NRR200247. [DOI] [PubMed] [Google Scholar]
- 43.Guretzky NA, Carlson DB, Garrett JE, Drackley JK. Lipid metabolite profiles and milk production for Holstein and Jersey cows fed rumen-protected choline during the periparturient period. J Dairy Sci. 2006;89(1):188–200. doi: 10.3168/jds.S0022-0302(06)72083-5. [DOI] [PubMed] [Google Scholar]
- 44.Mandard S, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. Cell Mol Life Sci. 2004;61(4):393–416. doi: 10.1007/s00018-003-3216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beale EG, Harvey BJ, Forest C. PCK1 and PCK2 as candidate diabetes and obesity genes. Cell Biochem Biophys. 2007;48(2-3):89–95. doi: 10.1007/s12013-007-0025-6. [DOI] [PubMed] [Google Scholar]
- 46.Bernabucci U, Ronchi B, Basirico L, Pirazzi D, Rueca F, Lacetera N, et al. Abundance of mRNA of apolipoprotein b100, apolipoprotein e, and microsomal triglyceride transfer protein in liver from periparturient dairy cows. J Dairy Sci. 2004;87(9):2881–8. doi: 10.3168/jds.S0022-0302(04)73418-9. [DOI] [PubMed] [Google Scholar]
- 47.Bertoni G, Trevisi E, Calamari L, Bionaz M. The inflammation could have a role in the liver lipidosis occurence in dairy cows. In: Joshi N, Herdt TH (Eds.) Production Diseases in Farm Animals. 12th International Conference. Wageningen: Wageningen Academic Publ; 2006. p. 157–58.
- 48.Sordillo LM, Mavangira V. The nexus between nutrient metabolism, oxidative stress and inflammation in transition cows. Anim Prod Sci. 2014;54(9):1204–14. [Google Scholar]
- 49.Sordillo LM, Raphael W. Significance of metabolic stress, lipid mobilization, and inflammation on transition cow disorders. Vet Clin North Am Food Anim Pract. 2013;29(2):267–78. doi: 10.1016/j.cvfa.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 50.Bradford BJ, Yuan K, Farney JK, Mamedova LK, Carpenter AJ. Invited review: Inflammation during the transition to lactation: New adventures with an old flame. J Dairy Sci. 2015;98(10):6631-50. [DOI] [PubMed]
- 51.Loor JJ, Dann HM, Guretzky NA, Everts RE, Oliveira R, Green CA, et al. Plane of nutrition prepartum alters hepatic gene expression and function in dairy cows as assessed by longitudinal transcript and metabolic profiling. Physiol Genomics. 2006;27(1):29–41. doi: 10.1152/physiolgenomics.00036.2006. [DOI] [PubMed] [Google Scholar]
- 52.Turk R, Juretic D, Geres D, Turk N, Rekic B, Simeon-Rudolf V, et al. Serum paraoxonase activity in dairy cows during pregnancy. Res Vet Sci. 2005;79(1):15–8. doi: 10.1016/j.rvsc.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 53.Kartha VN, Krishnamurthy S. Antioxidant function of vitamin A. Int J Vitam Nutr Res. 1977;47(4):394–401. [PubMed] [Google Scholar]
- 54.Pompella A, Visvikis A, Paolicchi A, De Tata V, Casini AF. The changing faces of glutathione, a cellular protagonist. Biochem Pharmacol. 2003;66(8):1499–503. doi: 10.1016/S0006-2952(03)00504-5. [DOI] [PubMed] [Google Scholar]
- 55.Shimada K, Jong CJ, Takahashi K, Schaffer SW. Role of ROS Production and Turnover in the Antioxidant Activity of Taurine. Adv Exp Med Biol. 2015;803:581–96. doi: 10.1007/978-3-319-15126-7_47. [DOI] [PubMed] [Google Scholar]
- 56.Goodman DS. Plasma retinol-binding protein. Ann N Y Acad Sci. 1980;348:378–90. doi: 10.1111/j.1749-6632.1980.tb21314.x. [DOI] [PubMed] [Google Scholar]
- 57.Meister A. On the cycles of glutathione metabolism and transport. Curr Top Cell Regul. 1981;18:21–58. doi: 10.1016/B978-0-12-152818-8.50009-8. [DOI] [PubMed] [Google Scholar]
- 58.Zhao JY, Goldberg J, Bremner JD, Vaccarino V. Global DNA Methylation Is Associated With Insulin Resistance A Monozygotic Twin Study. Diabetes. 2012;61(2):542–6. doi: 10.2337/db11-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gallego-Duran R, Romero-Gomez M. Epigenetic mechanisms in non-alcoholic fatty liver disease: An emerging field. World J Hepatol. 2015;7(24):2497–502. doi: 10.4254/wjh.v7.i24.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pirola CJ, Gianotti TF, Burgueno AL, Rey-Funes M, Loidl CF, Mallardi P, et al. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62(9):1356–63. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.


