Abstract
Seven new marine 11-acetoxy-9,11-secosterols, pinnisterols D–J (1–7), with a 1,4-quinone moiety, were discovered from the gorgonian coral Pinnigorgia sp. In this study, the structures of secosterols 1–7 were revealed by spectroscopic analysis. Bioactivity study showed that secosterol 1 treatment inhibited cell viability in a hepatic stellate cell line, HSC-T6, with an IC50 value of 3.93 μM; and secosterols 2, 5, and 7 reduced elastase enzyme release, and 3, 5, and 7 decreased the production of superoxide anions from human neutrophils.
Keywords: 9,11-secosterol; gorgonian; Pinnigorgia; cytotoxicity; hepatic stellate cell line (HSC-T6); anti-inflammatory; superoxide anion; elastase
1. Introduction
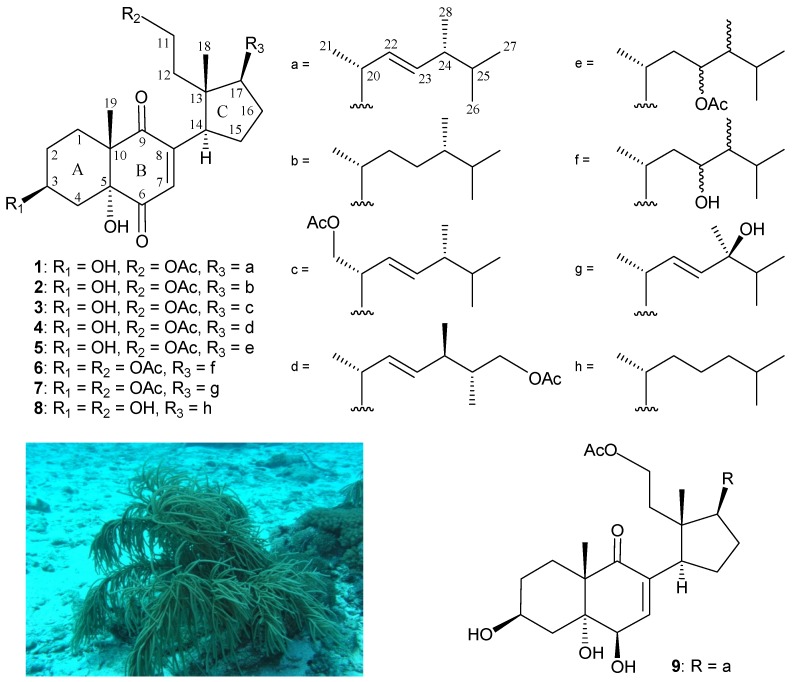
Since the isolation in 1972 of the first marine 9,11-secosteroid, 9-oxo-9,11-secogorgost-5-ene-3β, 11-diol 11-acetate, from the gorgonian coral Pseudopterogorgia americana [1], a series of compounds of this group has been prepared from various marine invertebrates, particularly sponges and octocorals, with complex structures and interesting bioactivities [2]. Our continued investigations of gorgonian coral Pinnigorgia sp. (phylum Cnidaria, class Anthozoa, subclass Octocorallia, order Alcyonacea, family Gorgoniidae) have yielded various interesting secondary metabolites [3,4,5,6,7,8], including 9,11-secosterols [3,4,5], some of which have been shown to possess bioactivity, such as anti-inflammatory and cytotoxic properties [3,4,5,6,7,8]. With the aim of discovering bioactive marine metabolites for new drug development in the future, we carried out an investigation of the chemical composition of the gorgonian coral Pinnigorgia sp. In this study, we performed compound preparation and structure determination, and investigated the cytotoxicity and anti-inflammatory activities of seven new 9,11-secosterols, pinnisterols D–J (1–7), following further study of Pinnigorgia sp. (Figure 1).
Figure 1.
Chemical structures of pinnisterols D–J (1–7), sterol 8, and pinnisterol A (9), and an image of gorgonian coral Pinnigorgia sp.
2. Results and Discussion
A new metabolite was isolated as a colorless oil, and was named pinnisterol D (1). The high-resolution electrospray ionization mass spectrum (HRESIMS) showed a signal at m/z 525.31853 (calcd. for C30H46O6 + Na, 525.31921), and therefore the molecular formula of 1 was determined to be C30H46O6 (8° of unsaturation). 13C and distortionless enhancement polarization transfer (DEPT) experimental data indicated that 1 had 30 carbons, including seven methyls, seven sp3 methylenes (including one oxymethylene), six sp3 methines (including one oxymethine), two sp3 quaternary carbons, one sp3 tertiary alcohol, three sp2 methines, one sp2 tertiary carbon, two ketonic carbonyls, and one ester carbonyl (Table 1). In addition, IR spectroscopy demonstrated that the compound contained hydroxy (νmax 3446 cm−1), ester (νmax 1741 cm−1), and α,β-unsaturated ketonic carbonyl (νmax 1685 cm−1) groups. The latter structural feature of 1 was further proven by the presence of signals at δC 197.5 (C-6), 134.1 (CH-7), 152.0 (C-8), and 201.6 (C-9) in the 13C NMR spectrum. Signals of carbons at δC 133.8 (CH-22) and 133.5 (CH-23) suggested the existence of a disubstituted olefin, and this was confirmed by two olefin proton signals at δH 5.26 (1H, dd, J = 14.8, 6.8 Hz, H-23) and 5.23 (1H, dd, J = 14.8, 6.8 Hz, H-22) (Table 1). The presence of Me-21, Me-28, Me-26, and Me-27 groups resulted in four doublets located at δH 1.05 (3H, J = 6.8 Hz), 0.91 (3H, J = 6.8 Hz), 0.84 (3H, J = 7.2 Hz), and 0.82 (3H, J = 7.2 Hz), respectively, and the existence of H3-18 and H3-19 resulted in two sharp singlets at δH 0.73 and δH 1.21, respectively. In the 1H NMR spectrum, one acetyl methyl signal (δH 2.02, 3H, s) was observed. Based on the aforementioned findings, metabolite 1 was determined to be a tricyclic compound.
Table 1.
Results of 1H (400 MHz, CDCl3) and 13C (100 MHz, CDCl3) NMR experiments and 1H–1H COSY and HMBC correlations for secosterol 1.
| Position | δH (J in Hz) | δC, Multiple | 1H–1H COSY | HMBC |
|---|---|---|---|---|
| 1 | 2.21 m; 1.78 m | 25.9, CH2 | H2-2 | C-2, -10 |
| 2 | 2.00 m; 1.49 m | 29.9, CH2 | H2-1, H-3 | n. o. a |
| 3 | 4.03 m | 66.7, CH | H2-2, H2-4 | n. o. |
| 4 | 2.17 m; 1.79 m | 35.7, CH2 | H-3 | C-2, -3, -5, -6, -10 |
| 5 | 80.6, C | |||
| 6 | 197.5, C | |||
| 7 | 6.45 s | 134.1, CH | C-5, -8, -9, -14 | |
| 8 | 152.0, C | |||
| 9 | 201.6, C | |||
| 10 | 52.0, C | |||
| 11 | 4.18 m | 61.0, CH2 | H2-12 | C-12, -13, acetate carbonyl |
| 12 | 1.68 m; 1.26 ddd (14.8, 9.2, 5.6) | 37.1, CH2 | H2-11 | C-11, -13, -14, -17, -18 |
| 13 | 47.4, C | |||
| 14 | 3.37 dd (10.8, 8.0) | 43.9, CH | H2-15 | C-7, -8, -9, -13, -15, -18 |
| 15 | 1.75 m; 1.66 m | 27.1, CH2 | H-14, H2-16 | C-16 |
| 16 | 1.78 m; 1.57 m | 25.6, CH2 | H2-15, H-17 | C-15 |
| 17 | 1.81 m | 51.0, CH | H2-16, H-20 | C-15, -16, -18, -21 |
| 18 | 0.73 s | 18.0, CH3 | C-12, -13, -14, -17 | |
| 19 | 1.21 s | 20.5, CH3 | C-1, -5, -9, -10 | |
| 20 | 2.24 m | 38.3, CH | H-17, H3-21, H-22 | C-16, -17, -22, -23 |
| 21 | 1.05 d (6.8) | 21.8, CH3 | H-20 | C-17, -20, -22 |
| 22 | 5.23 dd (14.8, 6.8) | 133.8, CH | H-20, H-23 | C-20, -24 |
| 23 | 5.26 dd (14.8, 6.8) | 133.5, CH | H-22, H-24 | C-20, -24 |
| 24 | 1.86 m | 43.0, CH | H-23, H-25, H3-28 | C-22, -23, -25, -26, -27, -28 |
| 25 | 1.46 m | 33.1, CH | H-24, H3-26, H3-27 | C-23, -24, -26, -27, -28 |
| 26 | 0.84 d (7.2) | 20.0, CH3 | H-25 | C-24, -25, -27 |
| 27 | 0.82 d (7.2) | 19.7, CH3 | H-25 | C-24, -25, -26 |
| 28 | 0.91 d (6.8) | 17.5, CH3 | H-24 | C-23, -24, -25 |
| 11-OAc | 171.1, C | |||
| 2.02 s | 21.1, CH3 | Acetate carbonyl |
a n. o. = not observed.
From the 1H NMR spectrum and 1H–1H correlation spectroscopy (COSY) of 1 (Table 1), the following correlations were revealed: H2-1/H2-2/H-3/H2-4, H2-11/H2-12, H-14/H2-15/H2-16/H-17/H-20/H-22/H-23/H-24/H-25/H3-26, H-20/H3-21, H-24/H3-28, and H-25/H3-27. Together with data of key heteronuclear multiple bond coherence (HMBC) correlations between H2-4, H-7, H3-19/C-5; H2-4/C-6; H-7, H-14/C-8; H-7, H-14, H3-19/C-9; H2-1, H2-4, H3-19/C-10; and H2-11, H2-12, H-14, and H3-18/C-13, all the information allowed determination of the main carbon skeleton of 1 (Table 1).
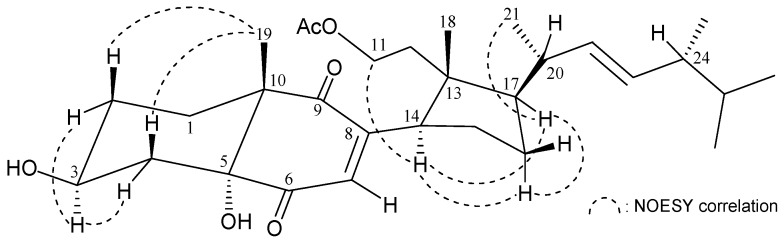
The correlations identified using nuclear Overhauser effect spectroscopy (NOESY) in addition to comparison of NMR data with those of known 9,11-secosterol 8, isolated from Korean marine sponge Ircinia sp. [9], and pinnisterol A (9) [4], enabled clarification of the configuration of 1 (Figure 1). The stereochemistries of stereogenic centers C-3, C-5, C-10, C-13, C-14, and C-17 in 1 were the same as those of 8. In addition, the main NOE correlations for 1 were interactions between H-3/H-2α (δH 2.00), H-2β (δH 1.49)/H3-19, H-3/H-4α (δH 2.17), H-4β (δH 1.79)/H3-19; thus, H-3 and the 5-hydroxy group in 1 should be positioned on the α-face (Figure 2).
Figure 2.
Selected NOESY correlations observed for 1.
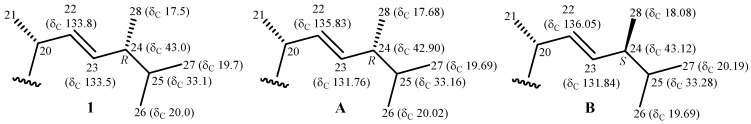
There was a greater coupling constant between H-22 and H-23 (J = 14.8 Hz), which supported a trans relationship between H-22 and H-23. This implied that the configuration of C-24 should be R according to the 13C NMR chemical shift of C-28 (δC 17.5). A previous study showed that, for a known sterol, (22E,24R)-24-methylcholesta-5,22-dien-3β-ol, with an identical chain, and the 24S epimer, (22E,24S)-24-methylcholesta-5,22-dien-3β-ol, the 13C NMR value of C-28 resonates at δC 17.68 ppm in the 24R epimer, with a relative 0.4 ppm downfield chemical shift (Figure 3) [10].
Figure 3.
Schematic diagrams of 13C NMR chemical shift data of the side-chain of pinnisterol D (1), (22E,24R)-24-methylcholesta-5,22-dien-3β-ol (A), and (22E,24S)-24-methylcholesta-5,22-dien-3β-ol (B) [10].
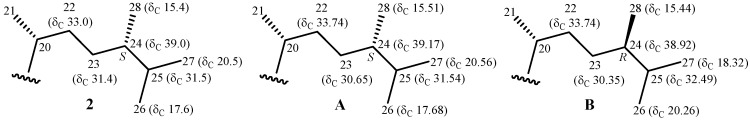
Pinnisterol E (2) was present as a colorless oil. From HRESIMS analysis, the signal at m/z 527.33410 (calcd. for C30H48O6 + Na, 527.33486) suggested the molecular formula of 2 to be C30H48O6 (7° of unsaturation), and the IR spectrum demonstrated the existence of hydroxy (νmax 3381 cm−1), ester (νmax 1740 cm−1), and α,β-unsaturated ketonic carbonyl (νmax 1686 cm−1) groups. The whole series of spectroscopic data demonstrated that secosterols 2 and 1 had an identical core structure, the difference being limited to the absence in 2 of the carbon-carbon double bond between C-22/23. The complete assignments of 1H and 13C NMR of 2 (Table 2 and Table 3) were compared with the values of 1, and secosterol 2 was assigned as having structure 2, with the same configurations of the core rings A–C. In addition, both compounds had identical stereogenic centers at C-3, C-5, C-10, C-13, C-14, and C-17, and their 1H and 13C NMR chemical shifts and proton coupling constants were in concurrence also. Based on the 13C NMR chemical shifts of C-25 (δC 31.5), C-26 (δC 17.6), and C-27 (δC 20.5), the configuration of the stereogenic center at C-24 was assigned as S. Previous study also showed that the 13C NMR values of C-25, C-26, and C-27 resonates at δC 31.54, 17.68, and 20.56 ppm in a 24S epimer of a known sterol, (24S)-24-methylcholest-5-en-3β-ol, with an identical side chain, and the 13C NMR values of C-25, C-26, and C-27 in a 24R epimer, (24R)-24-methylcholest-5-en-3β-ol, were observed at δC 32.49, 20.26, and 18.32 ppm, respectively (Figure 4) [10].
Table 2.
1H NMR data for secosterols 2–4.
| δH | 2 a | 3 b | 4 b |
|---|---|---|---|
| 1 | 2.21 m; 1.76 m | 2.21 m; 1.74 m | 2.20 m; 1.72 m |
| 2 | 2.00 m; 1.47 m | 2.00 m; 1.47 m | 2.00 m; 1.49 m |
| 3 | 4.04 m | 4.04 m | 4.04 m |
| 4 | 2.15 ddd (14.4, 5.2, 2.0) c; 1.79 m | 2.14 m; 1.77 m | 2.13 m; 1.79 m |
| 7 | 6.47 s | 6.46 s | 6.46 s |
| 11 | 4.16 m | 4.17 t (6.3) | 4.18 m |
| 12 | 1.73 m; 1.23 m | 1.59 m; 1.30 m | 1.69 m; 1.25 m |
| 14 | 3.39 dd (10.8, 8.4) | 3.37 t (9.8) | 3.38 dd (11.2, 8.4) |
| 15 | 1.76 m; 1.68 m | 1.77 m; 1.69 m | 1.76 m; 1.65 m |
| 16 | 1.89 m; 1.53 m | 1.75 m; 1.49 m | 1.76 m; 1.55 m |
| 17 | 1.76 m | 1.97 m | 1.77 m |
| 18 | 0.74 s | 0.73 s | 0.73 s |
| 19 | 1.22 s | 1.21 s | 1.22 s |
| 20 | 1.42 m | 1.46 m | 2.23 m |
| 21 | 0.99 d (6.8) | 4.01 dd (10.5, 7.0); 3.96 dd (10.5, 7.0) | 1.06 d (7.0) |
| 22 | 1.44 m; 0.95 m | 5.36 dd (15.4, 8.4) | 5.24 dd (15.4, 8.4) |
| 23 | 1.40 m; 0.96 m | 5.22 dd (15.4, 8.4) | 5.30 dd (15.4, 8.4) |
| 24 | 1.20 m | 1.90 m | 2.19 m |
| 25 | 1.61 m | 1.45 m | 1.71 m |
| 26 | 0.79 d (6.8) | 0.84 d (7.0) | 0.85 d (7.0) |
| 27 | 0.86 d (6.8) | 0.82 d (7.0) | 3.98 dd (10.5, 6.3); 3.85 dd (10.5, 6.3) |
| 28 | 0.78 d (6.8) | 0.93 d (7.0) | 0.98 d (7.0) |
| 11-OAc | 2.02 s | 2.02 s | 2.03 s |
| 21-OAc | 2.04 s | ||
| 27-OAc | 2.07 s |
a Spectra recorded at 400 MHz in CDCl3; b Spectra recorded at 700 MHz in CDCl3; c J values (in Hz) in parentheses.
Table 3.
13C NMR data for secosterols 2–7.
| δC | 2 a | 3 b | 4 b | 5 a | 6 a | 7 a |
|---|---|---|---|---|---|---|
| 1 | 25.9, CH2 | 25.8, CH2 | 25.9, CH2 | 25.9, CH2 | 25.7, CH2 | 25.7, CH2 |
| 2 | 29.9, CH2 | 29.9, CH2 | 29.9, CH2 | 29.9, CH2 | 25.9, CH2 | 25.9, CH2 |
| 3 | 66.7, CH | 66.7, CH | 66.7, CH | 66.7, CH | 69.7, CH | 69.8, CH |
| 4 | 35.8, CH2 | 35.7, CH2 | 35.7, CH2 | 35.6, CH2 | 32.0, CH2 | 31.9, CH2 |
| 5 | 80.6, C | 80.6, C | 80.6, C | 80.5, C | 80.1, C | 80.0, C |
| 6 | 197.4, C | 197.4, C | 197.5, C | 197.5, C | 197.3, C | 197.5, C |
| 7 | 134.1, CH | 134.1, CH | 134.1, CH | 134.2, CH | 134.2, CH | 134.2, CH |
| 8 | 152.1, C | 151.6, C | 151.8, C | 151.7, C | 151.8, C | 151.6, C |
| 9 | 201.5, C | 201.6, C | 201.6, C | 201.6, C | 201.2, C | 201.3, C |
| 10 | 52.0, C | 52.0, C | 52.0, C | 52.0, C | 51.9, C | 51.9, C |
| 11 | 61.0, CH2 | 60.8, CH2 | 61.0, CH2 | 60.9, CH2 | 61.0, CH2 | 61.0, CH2 |
| 12 | 37.2, CH2 | 36.7, CH2 | 37.0, CH2 | 37.0, CH2 | 37.2, CH2 | 37.2, CH2 |
| 13 | 47.5, C | 47.5, C | 47.3, C | 47.4, C | 47.4, C | 47.4, C |
| 14 | 43.7, CH | 43.5, CH | 43.8, CH | 43.7, CH | 43.7, CH | 43.8, CH |
| 15 | 27.2, CH2 | 27.1, CH2 | 26.9, CH2 | 27.0, CH2 | 27.1, CH2 | 27.1, CH2 |
| 16 | 26.2, CH2 | 25.8, CH2 | 25.7, CH2 | 26.2, CH2 | 26.4, CH2 | 25.8, CH2 |
| 17 | 50.7, CH | 46.3, CH | 50.7, CH | 51.4, CH | 51.6, CH | 50.9, CH |
| 18 | 17.6, CH3 | 18.5, CH3 | 18.0, CH3 | 17.6, CH3 | 17.8, CH3 | 18.0, CH3 |
| 19 | 20.5, CH3 | 20.4, CH3 | 20.5, CH3 | 20.4, CH3 | 20.3, CH3 | 20.2, CH3 |
| 20 | 34.9, CH | 42.2, CH | 38.4, CH | 33.2, CH | 33.9, CH | 38.2, CH |
| 21 | 19.2, CH3 | 67.2, CH2 | 21.7, CH3 | 20.4, CH3 | 21.1, CH3 | 21.6, CH3 |
| 22 | 33.0, CH2 | 127.5, CH | 135.4, CH | 35.4, CH2 | 39.4, CH2 | 132.9, CH |
| 23 | 21.4, CH2 | 138.1, CH | 131.1, CH | 76.7, CH | 74.6, CH | 134.5, CH |
| 24 | 39.0, CH | 43.4, CH | 38.2, CH | 42.7, CH | 45.5, CH | 75.0, C |
| 25 | 31.5, CH | 32.9, CH | 37.4, CH | 28.5, CH | 27.7, CH | 38.1, CH |
| 26 | 17.6, CH3 | 20.0, CH3 | 13.1, CH3 | 21.6, CH3 | 21.7, CH3 | 17.5, CH3 |
| 27 | 20.5, CH3 | 19.7, CH3 | 67.9, CH2 | 18.6, CH3 | 17.6, CH3 | 17.1, CH3 |
| 28 | 15.4, CH3 | 17.5, CH3 | 18.3, CH3 | 11.0, CH3 | 10.5, CH3 | 25.2, CH3 |
| 3-OAc | 170.8, C | 170.9, C | ||||
| 21.3, CH3 | 21.1, CH3 | |||||
| 11-OAc | 171.1, C | 171.1, C | 171.1, C | 171.1, C | 171.0, C | 170.9, C |
| 21.1, CH3 | 21.1, CH3 | 21.1, CH3 | 21.1, CH3 | 21.1, CH3 | 21.1, CH3 | |
| 21-OAc | 171.1, C | |||||
| 21.0, CH3 | ||||||
| 23-OAc | 170.7, C | |||||
| 21.4, CH3 | ||||||
| 27-OAc | 171.3, C | |||||
| 21.1, CH3 |
a Spectra recorded at 100 MHz in CDCl3; b Spectra recorded at 175 MHz in CDCl3.
Figure 4.
Schematic diagrams of 13C NMR chemical shift data of the side-chain of pinnisterol E (2), (24S)-24-methylcholest-5-en-3β-ol (A), and (24R)-24-methylcholest-5-en-3β-ol (B) [9].
Pinnisterol F (3) was present as a colorless oil. From HRESIMS analysis, the signal at m/z 583.32406 (calcd. for C32H48O8 + Na, 583.32469) suggested the molecular formula of 3 to be C32H48O8 (9° of unsaturation). The NMR signals of 3 (Table 2 and Table 3) were similar to those of 1, except that the signals related to the C-21 methyl in 1 were substituted by signals for an acetoxymethylene group in 3. From the HMBC spectrum of 3, it was revealed that an ester carbonyl carbon at δC 171.1 correlated with a methyl signal at δH 2.04 and a pair of oxygenated methylene protons at δH 4.01 (1H, dd, J = 10.5, 7.0 Hz) and 3.96 (1H, dd, J = 10.5, 7.0 Hz), which revealed that an acetoxy group was at the position C-21 in the side chain of 3. Thus, pinnisterol F (3) was found to be the 21-acetoxy derivative of 1.
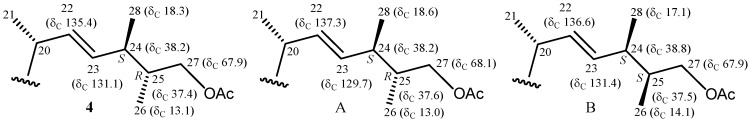
Pinnisterol G (4) had a molecular formula identical to that of 3, C32H48O8, with a HRESIMS signal located at m/z 583.32432 (calcd. for C32H48O8 + Na, 583.32469) with nine degrees of unsaturation, indicating that secosterols 3 and 4 were isomers. Comparison of the NMR data of 4 with those of 3 (Table 2 and Table 3) showed that both compounds possessed the same sterol nucleus and a similar side chain, but differed in terms of the location of one acetoxy group. From an HMBC experiment, it was revealed that one ester carbonyl carbon at δC 171.3 correlated with one methyl signal at δH 2.07 and a pair of oxymethylene protons signals at δH 3.98 (dd, J = 10.5, 6.3 Hz) and 3.85 (dd, J = 10.5, 6.3 Hz), which indicated that an acetoxy group was located at C-27 in the side chain. The configurations at C-24 and C-25 were therefore designated as S- and R-forms, respectively, on the basis of the 13C NMR chemical shifts of C-24 (δC 38.2), C-25 (δC 37.4), C-26 (δC 13.1), C-27 (δC 67.9), and C-28 (δC 18.3). It was reported that the 13C NMR values of C-24, C-25, C-26, C-27, and C-28 resonate at δC 38.2, 37.6, 13.0, 68.1, and 18.6 ppm in a 24S and a 25R epimer of a known sterol, echrebsteroid C, with the same side chain, and the 13C NMR values of C-24, C-25, C-26, C-27, and C-28 in a 24S and 25S epimer, echrebsteroid B, appeared at δC 38.8, 37.5, 14.1, 67.9, and 17.1 ppm (Figure 5) [11].
Figure 5.
Schematic diagrams of 13C NMR chemical shift data of the side-chain of pinnisterol G (4), echrebsteroid C (A), and echrebsteroid B (B) [11].
Pinnisterol H (5) was isolated as a colorless oil. Based on the HRESIMS signal located at m/z 585.33988 (calcd. for C32H50O8 + Na, 585.34034), it was concluded that the molecular formula of 5 was C32H50O8 (8° of unsaturation). The IR spectrum of 5 indicated the presence of hydroxy (νmax 3448 cm−1), ester (νmax 1736 cm−1) and α,β-unsaturated ketonic carbonyl (νmax 1686 cm−1) groups. The whole series of spectroscopic data showed that secosterol 5 and secosterol 1 shared the same core structure, with the exception of the addition of an acetoxy group to substitute the alkene at C-23 in 5. The complete assignments of the 13C and 1H NMR of pinnisterol H (5) (Table 3 and Table 4) were compared with the values of 1, and the HMBC correlations fully supported the positions of the functional groups of 5, indicating that it had a structure of the same configuration as secosterols 1–4 in the core rings A–C. The proton coupling constants and NMR chemical shift data also further supported this finding, though the configurations of C-23 and C-24 were not determined at this stage.
Table 4.
1H NMR (400 MHz, CDCl3) data for secosterols 5–7.
| δH | 5 | 6 | 7 |
|---|---|---|---|
| 1 | 2.22 m; 1.76 m | 2.27 m; 1.78 m | 2.28 m; 1.76 m |
| 2 | 1.97 m; 1.48 m | 1.98 m; 1.60 m | 1.96 m; 1.61 m |
| 3 | 4.02 m | 5.05 m | 5.05 m |
| 4 | 2.12 m; 1.78 m | 2.24 m; 1.81 m | 2.24 m; 1.79 m |
| 7 | 6.45 s | 6.46 s | 6.45 s |
| 11 | 4.14 m | 4.18 m | 4.17 m |
| 12 | 1.63 m; 1.23 m | 1.70 m; 1.27 m | 1.67 m; 1.26 m |
| 14 | 3.36 dd (11.2, 8.4) a | 3.39 dd (10.8, 8.0) | 3.39 dd (10.4, 8.0) |
| 15 | 1.76 m; 1.65 m | 1.80 m; 1.69 m | 1.76 m; 1.69 m |
| 16 | 1.91 m; 1.52 m | 1.93 m; 1.53 m | 1.99 m; 1.53 m |
| 17 | 1.69 m | 1.81 m | 1.81 m |
| 18 | 0.73 s | 0.74 s | 0.73 s |
| 19 | 1.20 s | 1.22 s | 1.21 s |
| 20 | 1.54 m | 1.63 m | 2.28 m |
| 21 | 1.02 d (6.8) | 1.13 d (6.8) | 1.08 d (6.8) |
| 22 | 1.67 m; 1.24 m | 1.72 m; 1.07 m | 5.53 dd (15.6, 8.4) |
| 23 | 5.01 m | 3.59 m | 5.48 (15.6) |
| 24 | 1.47 m | 1.32 m | |
| 25 | 1.57 m | 1.84 m | 1.67 m |
| 26 | 0.93 d (6.8) | 0.94 d (7.2) | 0.89 d (6.8) |
| 27 | 0.83 d (6.8) | 0.84 d (7.2) | 0.88 d (6.8) |
| 28 | 0.81 d (6.8) | 0.79 d (7.2) | 1.21 s |
| 3-OAc | 2.04 s | 2.04 s | |
| 11-OAc | 2.02 s | 2.02 s | 2.02 s |
| 23-OAc | 2.03 s |
a J values (in Hz) in parentheses.
Pinnisterol I (6) was obtained as a colorless oil. The HRESIMS signal at m/z 585.33988 (calcd. for C32H50O8 + Na, 585.34034) suggested the molecular formula of 6 to be C32H50O8 (8° of unsaturation). The NMR signals of 6 (Table 3 and Table 4) were very similar to those of 5, with the exception that 5 had signals corresponding to 3-hydroxy and 23-acetoxy groups, which were substituted by signals for acetoxy and hydroxy groups, respectively, in 6. From a NOESY experiment, the correlations of data of 5 and 6 demonstrated that the configurations of the stereogenic centers in the core rings A–C were identical to those of 1. The configurations of stereogenic centers C-23 and C-24 of 6 were also not determined at this stage.
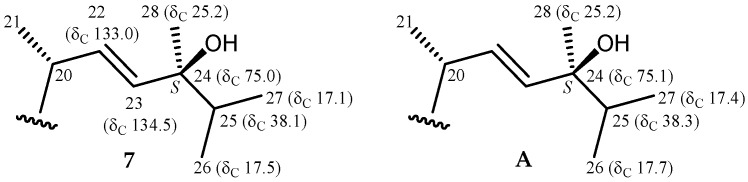
Pinnisterol J (7) was obtained as a colorless oil and had the molecular formula C32H48O8, as determined by the HRESIMS signal at m/z 583.32433 (calcd. for C32H48O8 + Na, 583.32469) (9° of unsaturation). According to the NMR spectroscopic data (Table 3 and Table 4), compound 7 showed the same nuclear structure as that of compound 6. In the 13C NMR data of 7, one additional disubstituted olefin was identified from signals of carbons at δC 132.9 (CH-22) and 134.5 (CH-23). The presence of a 24-hydroxy group was evidenced by HMBC correlations between H-22, H-23, H-25, H3-26, H3-27, and H3-28/C-24 (δC 75.0), a methyl-containing oxygenated tertiary carbon. There was a greater coupling constant between H-22 and H-23 (J = 15.6 Hz), suggesting that a trans relationship existed between H-22 and H-23. The configuration of the C-24 stereogenic center was assigned as S on the basis of the 13C NMR chemical shifts of C-24 (δC 75.0), C-25 (δC 38.1), C-26 (δC 17.5), C-27 (δC 17.1), and C-28 (δC 25.2). It was reported that the 13C NMR values of C-24, C-25, C-26, C-27, and C-28 resonates at δC 75.1, 38.3, 17.7, 17.4, and 25.2 ppm in a 24S epimer of a known synthetic product, 24(S)-hydroxyvitamin D2, with the same side chain (Figure 6) [12].
Figure 6.
Schematic diagrams of 13C NMR chemical shift data of the side-chain of pinnisterol J (7) and 24(S)-hydroxyvitamin D2 (A) [12].
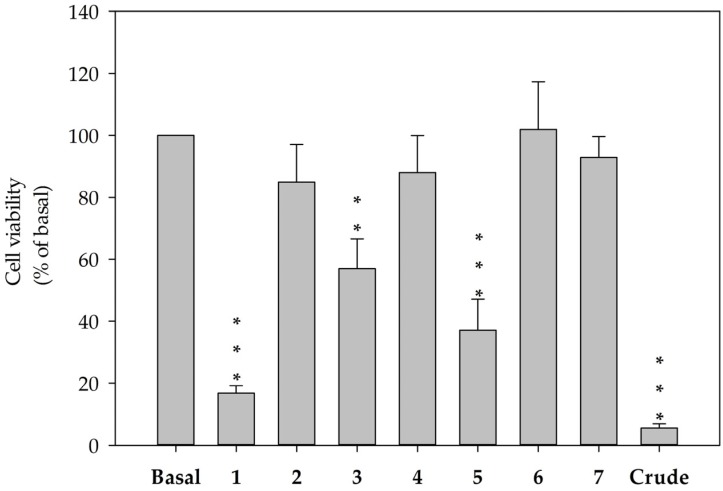
The hepatic stellate cell, a major cell type involved in liver fibrosis, is also responsible for liver damage by increasing proliferation and protein secretion associated with the formation of scar tissue. In cytotoxicity testing, secosterols 1–7 were examined in terms of their cytotoxic effects on HSC-T6, an immortalized rat hepatic stellate cell lines. At a concentration of 10 μM, secosterols 1, 3, and 5 significantly decreased the viability of HSC-T6 cells to 16.8 (IC50 = 3.93 μM), 56.9 and 37.1%, respectively (Figure 7). These results implied that the functional groups in the side chain of secosterols 1–7 play important roles in determining the activity of the compounds.
Figure 7.
Decreased cell viability in HSC-T6 cells treated with secosterols 1–7 for 24 h at a concentration of 10 μM. Cells were treated with secosterols, DMSO (control) or coral crude extract at 6 μg/mL. Cytotoxicity was monitored spectrophotometrically at OD 450 nm. Quantitative data are expressed as the mean ± S.E.M. (n = 3–4). ** p < 0.01, *** p < 0.001 compared to basal.
In anti-inflammatory testing, secosterols 2, 5, and 7 displayed inhibitory effects on the release of elastase (IC50 = 2.33, 2.59 and 3.89 μM, respectively), and secosterols 3, 5, and 7 showed inhibitory effects on human neutrophils in terms of the generation of superoxide anions (IC50 = 5.52, 3.26, and 3.71 μM, respectively) (Table 5).
Table 5.
Inhibitory effects of secosterols 2–7 on elastase release and superoxide anion generation by human neutrophils in response to fMet-Leu-Phe/Cytochalastin B.
| Compound | Elastase Release | Superoxide Anions |
|---|---|---|
| IC50 (μM) | IC50 (μM) | |
| 2 | 2.33 ± 0.27 | >10 |
| 3 | >10 | 5.52 ± 1.06 |
| 4 | >10 | >10 |
| 5 | 2.59 ± 0.29 | 3.26 ± 0.33 |
| 6 | >10 | >10 |
| 7 | 3.89 ± 1.16 | 3.71 ± 0.51 |
3. Experimental Section
3.1. General Experimental Procedures
Optical rotations were measured with a digital polarimeter (P-1010; Japan Spectroscopic Corporation, Tokyo, Japan); and infrared spectra were recorded on a spectrometer (FT/IR-4100; Japan Spectroscopic Corporation); peaks are reported in cm−1. NMR spectra were obtained on a 400 MHz NMR spectrometer (Mercury Plus; Varian, Palo Alto, CA, USA) and a 700 MHz NMR spectrometer (AVIIIHD700X; Bruker, Bremen, Germany), using the residual CHCl3 signal (δH 7.26 ppm) and CDCl3 (δC 77.1 ppm) as internal standards for 1H NMR and 13C NMR, respectively. Coupling constant values (J) are given in Hz. ESIMS and HRESIMS were performed using mass spectrometry (Tesla solariX FTMS system, Bruker). TLC was carried out on Kieselgel 60 F254 precoated plates (0.25 mm, Merck, Darmstadt, Germany), and spots were visualized by the standard method. Column chromatography was performed on silica gel at a size of 230–400 mesh (Merck). HPLC experiments were performed using the following systems: normal-phase HPLC (NP-HPLC) injection port, 7725 (Rheodyne, Rohnert Park, CA, USA); pump, L-7110 (Hitachi, Tokyo, Japan); and semi-preparative normal-phase column (Supelco Ascentis Si Cat #:581515-U, 25 cm × 21.2 mm, 5 μm, Sigma-Aldrich, St. Louis, MO, USA). Reverse-phase HPLC (RP-HPLC) injection port (7725; Rheodyne); pump, L-2130 (Hitachi); photodiode array detector (L-2455; Hitachi); and reverse-phase column (25 cm × 21.2 mm, Luna 5 μm C18(2) 100 Å, AXIA Packed; Phenomenex, Torrance, CA, USA).
3.2. Animal Material
Specimens of gorgonian coral Pinnigorgia sp. were collected in August 2012 by hand while scuba diving off the coast of Green Island located near the southeast of Taiwan. The samples were then stored in a freezer until extraction. A voucher specimen was deposited in the National Museum of Marine Biology & Aquarium, Taiwan (specimen No.: NMMBA-TW-GC-2012-130). Identification of the species of this organism was done by comparison as described in the previous publication [13].
3.3. Extraction and Separation
Extraction of compounds was performed at room temperature unless otherwise specified. Pinnigorgia sp. (wet weight 1.98 kg; dry weight 0.86 kg) was sliced, and the sliced bodies were then extracted with ethyl acetate (EtOAc). The EtOAc extract (84.9 g) was partitioned with n-hexane and methanol (MeOH). The MeOH layer (12.6 g) was separated on a Sephadex LH-20 column and elution was performed using a solvent mixture dichloromethane (DCM):MeOH (1:1); the separation yielded 7 subfractions A–G. Fraction F was separated by silica gel column chromatography and then eluted with n-hexane/acetone (stepwise, 50/50 %v/v to 100% acetone) to afford eight subfractions F1–F8. Fraction F2 was purified by silica gel column chromatography and then eluted with n-hexane/acetone (stepwise, 90/10 %v/v to 100% acetone) to yield 13 subfractions F2A–F2M. Fraction F2F was purified by NP-HPLC using a mixture of n-hexane/EtOAc (5:2) to afford 14 subfractions F2F1–F2F14. Fractions F2F5 and F2F6 were repurified by RP-HPLC using a mixture of MeOH/H2O (90/10 %v/v at 5.0 mL/min flow rate) to yield 6 (1.8 mg) and 7 (4.8 mg), respectively. Fraction F2H was purified by NP-HPLC using a mixture of n-hexane/EtOAc (50/50 %v/v) to yield 14 subfractions F2H1–F2H14. Fractions F2H9 and F2H10 were repurified by RP-HPLC using a mixture of MeOH/H2O (90/10 %v/v at 5.0 mL/min flow rate) to yield 1 (3.8 mg) and 2 (1.5 mg), respectively. Fraction F2I was repurified by NP-HPLC using a mixture of n-hexane/EtOAc (50/50 %v/v at 3.0 mL/min flow rate) to afford 15 subfractions F2I1–F2I15, including compound 5 (F2I9, 6.4 mg). Fraction F2I10 was purified by RP-HPLC using a mixture of MeOH/H2O (85/15 %v/v at 5.0 mL/min, flow rate) to afford 3 (1.0 mg) and 4 (1.0 mg), respectively.
Pinnisterol D (1): colorless oil; [α] +44 (c 1.3, CHCl3); IR (neat) νmax 3446, 1741, 1685 cm−1; 1H (400 MHz, CDCl3) and 13C (100 MHz, CDCl3) NMR data (see Table 1); ESIMS m/z 525 [M + Na]+; HRESIMS m/z 525.31853 (calcd. for C30H46O6 + Na, 525.31921).
Pinnisterol E (2): colorless oil; [α] −35 (c 0.5, CHCl3); IR (neat) νmax 3381, 1740, 1686 cm−1; 1H (400 MHz, CDCl3) and 13C (100 MHz, CDCl3) NMR data (see Table 2 and Table 3); ESIMS m/z 527 [M + Na]+; HRESIMS m/z 527.33410 (calcd. for C30H48O6 + Na, 527.33486).
Pinnisterol F (3): colorless oil; [α] −152 (c 0.3, CHCl3); IR (neat) νmax 3446, 1736, 1686 cm−1; 1H (700 MHz, CDCl3) and 13C (175 MHz, CDCl3) NMR data (see Table 2 and Table 3); ESIMS m/z 583 [M + Na]+; HRESIMS m/z 583.32406 (calcd. for C32H48O8 + Na, 583.32469).
Pinnisterol G (4): colorless oil; [α] −176 (c 0.3, CHCl3); IR (neat) νmax 3447, 1739, 1686 cm−1; 1H (700 MHz, CDCl3) and 13C (175 MHz, CDCl3) NMR data (see Table 2 and Table 3); ESIMS m/z 583 [M + Na]+; HRESIMS m/z 583.32432 (calcd. for C32H48O8 + Na, 583.32469).
Pinnisterol H (5): colorless oil; [α] −20 (c 0.3, CHCl3); IR (neat) νmax 3448, 1736, 1686 cm−1; 1H (400 MHz, CDCl3) and 13C (100 MHz, CDCl3) NMR data (see Table 3 and Table 4); ESIMS m/z 585 [M + Na]+; HRESIMS m/z 585.33988 (calcd. for C32H50O8 + Na, 585.34034).
Pinnisterol I (6): colorless oil; [α] −129 (c 0.5, CHCl3); IR (neat) νmax 3446, 1736, 1685 cm−1; 1H (400 MHz, CDCl3) and 13C (100 MHz, CDCl3) NMR data (see Table 3 and Table 4); ESIMS m/z 585 [M + Na]+; HRESIMS m/z 585.33988 (calcd. for C32H50O8 + Na, 585.34034).
Pinnisterol J (7): colorless oil; [α] −5 (c 0.2, CHCl3); IR (neat) νmax 3455, 1736, 1687 cm−1; 1H (400 MHz, CDCl3) and 13C (100 MHz, CDCl3) NMR data (see Table 3 and Table 4); ESIMS m/z 583 [M + Na]+; HRESIMS m/z 583.32433 (calcd. for C32H48O8 + Na, 583.32469).
3.4. Anti-Hepatofibric Assay
The anti-hepatofibric effects of secosterols 1–7, at a concentration of 10 μM, were analyzed using a colorimetric assay (WST-1-based method). The steps of the assay were performed according to a previously-published method [14].
3.5. Generation of Superoxide Anions and Release of Elastase by Human Neutrophils
Human neutrophils were prepared by Ficoll centrifugation followed by dextran sedimentation. Measurements of superoxide anion generation and elastase release from neutrophils were performed based on published procedures [15,16]. Briefly, MeO-Suc-Ala-Ala-Pro-Valp-nitroanilide was used as the elastase substrate for the elastase release assay, and the method using superoxide dismutase-inhibitable reduction of ferricytochrome c was employed to measure superoxide anion production.
4. Conclusions
Our ongoing investigations of coral metabolites showed that gorgonian corals belonging to the genus Pinnigorgia are rich in 9,11-secosterols. In cytotoxicity tests, secosterol 1 showed significant cytotoxicity against HSC-T6 cells. In anti-inflammatory activity tests on human neutrophils, secosterols 2, 5, and 7 displayed inhibitory effects on elastase release, and 3, 5, and 7 showed inhibitory effects on the generation of superoxide anions. Our findings suggested that these new 9,11-secosterols could be developed as promising bioactive agents, and further biomedical study is necessary in order to identify their potential applications in the treatment of disease.
Acknowledgments
This research was supported by grants from several institutions, including the National Museum of Marine Biology & Aquarium; the National Dong Hwa University; the National Sun Yat-sen University; and the National Research Program for Biopharmaceuticals, Ministry of Science and Technology (Grant No. MOST 105-2325-B-291-001, 105-2811-B-291-003, 104-2325-B-291-001, 103-2325-B-291-001 and 104-2320-B-291-001-MY3) awarded to P.-J.S.
Supplementary Materials
HRESIMS, 1H and 13C spectra of new compounds 1–7 and 2D NMR data (HSQC, 1H−1H COSY, HMBC and NOESY spectra) of new compound 1 are available online at www.mdpi.com/1660-3397/15/1/11/s1.
Author Contributions
P.-J.S. designed the whole experiment and contributed to manuscript preparation. Y.-C.C. and T.-L.H. researched data. L.-M.K. analyzed the data and performed data acquisition.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Enwall E.L., van der Helm D., Hsu I.N., Pattabhiraman T., Schmitz F.J., Spraggins R.L., Weinheimer A.J. Crystal structure and absolute configuration of two cyclopropane containing marine steroids. J. Chem. Soc. Chem. Commun. 1972:215–216. doi: 10.1039/c39720000215. [DOI] [Google Scholar]
- 2.Sica D., Musumeci D. Secosteroids of marine origin. Steroids. 2004;69:743–756. doi: 10.1016/j.steroids.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Chang Y.-C., Kuo L.-M., Su J.-H., Hwang T.-L., Kuo Y.-H., Lin C.-S., Wu Y.-C., Sheu J.-H., Sung P.-J. Pinnigorgiols A–C, 9,11-secosterols with a rare ring arrangement from a gorgonian coral Pinnigorgia sp. Tetrahedron. 2016;72:999–1004. doi: 10.1016/j.tet.2015.12.072. [DOI] [Google Scholar]
- 4.Chang Y.-C., Kuo L.-M., Hwang T.-L., Yeh J., Wen Z.-H., Fang L.-S., Wu Y.-C., Lin C.-S., Sheu J.-H., Sung P.-J. Pinnisterols A–C, new 9,11-secosterols from a gorgonian Pinnigorgia sp. Mar. Drugs. 2016;14:12. doi: 10.3390/md14010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su Y.-D., Cheng C.-H., Wen Z.-H., Wu Y.-C., Sung P.-J. New anti-inflammatory sterols from a gorgonian Pinnigorgia sp. Bioorg. Med. Chem. Lett. 2016;26:3060–3063. doi: 10.1016/j.bmcl.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 6.Chang Y.-C., Chen N.-F., Hwang T.-L., Tseng C.-C., Wu T.-Y., Peng B.-R., Wen Z.-H., Fang L.-S., Wu Y.-C., Sheu J.-H., et al. New marine sterols from an algal-bearing gorgonian coral Pinnigorgia sp. Steroids. 2016;115:123–129. doi: 10.1016/j.steroids.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 7.Chang H.-H., Chang Y.-C., Chen W.-F., Hwang T.-L., Fang L.-S., Wen Z.-H., Chen Y.-H., Wu Y.-C., Sung P.-J. Pubinernoid A and apo-9′-fucoxanthinone, secondary metabolites from a gorgonian coral Pinnigorgia sp. Nat. Prod. Commun. 2016;11:707–708. [PubMed] [Google Scholar]
- 8.Chang Y.-C., Hwang T.-L., Sheu J.-H., Wu Y.-C., Sung P.-J. New anti-inflammatory 9,11-secosterols with a rare tricyclo[5,2,1,1]decane ring from a Formosan gorgonian Pinnigorgia sp. Mar. Drugs. 2016;14:218. doi: 10.3390/md14120218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang I., Choi H., Won D.H., Nam S.-J., Kang H. An antibacterial 9,11-secosterol from a marine sponge Ircinia sp. Bull. Korean Chem. Soc. 2014;35:3360–3362. doi: 10.5012/bkcs.2014.35.11.3360. [DOI] [Google Scholar]
- 10.Wright J.L.C., McInnes A.G., Shimizu S., Smith D.G., Walter J.A., Idler D., Khalil W. Identification of C-24 alkyl epimers of marine sterols by 13C nuclear magnetic resonances spectroscopy. Can. J. Chem. 1978;56:1898–1903. [Google Scholar]
- 11.Cao F., Shao C.-L., Chen M., Zhang M.-Q., Xu K.-X., Meng H., Wang C.-Y. Antiviral C-25 epimers of 26-acetoxy steroids from the South China Sea gorgonian Echinogorgia rebekka. J. Nat. Prod. 2014;77:1488–1493. doi: 10.1021/np500252q. [DOI] [PubMed] [Google Scholar]
- 12.Coutts L.D., Geiss W.B., Gregg B.T., Helle M.A., King C.-H.R., Itov Z., Mateo M.E., Meckler H., Zettler M.W., Knutson J.C. A stereospecific synthesis of 24(S)-hydroxyvitamin D2, a prodrug for 1α,24(S)-dihydroxyvitamin D2. Org. Process Res. Dev. 2002;6:246–255. doi: 10.1021/op010229e. [DOI] [Google Scholar]
- 13.Fabricius K., Alderslade P. Soft Corals and Sea Fans—A Comprehensive Guide to the Tropical Shallow-Water Genera of the Central-West Pacific, the Indian Ocean and the Red Sea. 1st ed. Australian Institute of Marine Science; Townsville, Australia: 2001. pp. 218–219. [Google Scholar]
- 14.Kuo L.-M., Kuo C.-Y., Lin C.-Y., Hung M.-F., Shen J.-J., Hwang T.-L. Intracellular glutathione depletion by oridonin leads to apoptosis in hepatic stellate cells. Molecules. 2014;19:3327–3344. doi: 10.3390/molecules19033327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S.-C., Chung P.-J., Ho C.-M., Kuo C.-Y., Hung M.-F., Huang Y.-T., Chang W.-Y., Chang Y.-W., Chan K.-H., Hwang T.-L. Propofol inhibits superoxide production, elastase release, and chemotaxis in formyl peptide-activated human neutrophils by blocking formyl peptide receptor 1. J. Immunol. 2013;190:6511–6519. doi: 10.4049/jimmunol.1202215. [DOI] [PubMed] [Google Scholar]
- 16.Yu H.-P., Hsieh P.-W., Chang Y.-J., Chung P.-J., Kuo L.-M., Hwang T.-L. 2-(2-Fluorobenzamido) benzoate ethyl ester (EFB-1) inhibits superoxide production by human neutrophils and attenuates hemorrhagic shock-induced organ dysfunction in rats. Free Radic. Biol. Med. 2011;50:1737–1748. doi: 10.1016/j.freeradbiomed.2011.03.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.