Abstract
A novel β-1,3-galactosidase, designated as MaBGA (β-galactosidase from Marinomonas sp. BSi20414), was successfully purified to homogeneity from Marinomonas sp. BSi20414 isolated from Arctic sea ice by ammonium sulfate precipitation and anion exchange chromatography, resulting in an 8.12-fold increase in specific activity and 9.9% recovery in total activity. MaBGA displayed its maximum activity at pH 6.0 and 60 °C, and maintained at least 90% of its initial activity over the pH range of 5.0–8.0 after incubating for 1 h. It also exhibited considerable thermal stability, which retained 76% of its initial activity after incubating at 50 °C for 6 h. In contrast to other β-galactosidases, MaBGA displayed strict substrate specificity, not only for the glycosyl group, but also for the linkage type. To better understand the structure–function relationship, the encoding gene of MaBGA was obtained and subject to bioinformatics analysis. Multiple alignments and phylogenetic analysis revealed that MaBGA belonged to the glycoside hydrolase family 42 and had closer genetic relationships with thermophilic β-galactosidases of extremophiles. With the aid of homology modeling and molecular docking, we proposed a reasonable explanation for the linkage selectivity of MaBGA from a structural perspective. On account of the robust stability and 1,3-linkage selectivity, MaBGA would be a promising candidate in the biosynthesis of galacto-oligosaccharide with β1–3 linkage.
Keywords: β-galactosidase, Marinomonas, thermostable, purification, gene cloning, linkage selectivity
1. Introduction
The enzyme β-galactosidases (EC 3.2.1.23, BGA), which are widely distributed in various organisms, including animals, plants, bacteria, archaea, yeasts and fungi, are capable of catalyzing the hydrolysis of molecules containing the β-glycosidic bond, to release their terminal non-reducing galactose molecules. In some cases, β-galactosidases can catalyze the reverse reaction of the hydrolysis, transglycosylation, when receptors of galactosyl are monosaccharides, disaccharides or oligosaccharides, instead of water molecules [1]. Due to the catalytic characteristic, β-galactosidases are important for the dairy industry to produce milk with low/no lactose for people who suffer from lactose intolerance [2]. Moreover, β-galactosidases are also widely utilized for enzymatic synthesis of galacto-oligosaccharides, which can be employed to stimulate the growth of beneficial bacteria selectively in the gut, as prebiotics [3].
Based on the similarity of the amino acid sequences, β-galactosidases are mainly divided into four glycoside hydrolase (GH) families [4]—GH1, GH2, GH35 and GH42—according to the carbohydrate-active enzymes database (CAZy) [5]. All of these four families belong to the GH-A superfamily, of which members have two glutamic acid residues as catalytic active sites located in an (α/β)8 TIM (the triosephosphate isomerase) barrel domain [6]. Generally, GH1 and GH2 β-galactosidases are mainly found in mesophiles and display high lactase activity [7]. GH35 β-galactosidases are usually found in pathogens such as Streptococcus pneumoniae [8,9,10], with specific activity toward β-1,3-linkages. β-galactosidases belonging to the GH42 family are mostly stemmed from extremophiles, including thermophiles [11,12,13,14,15], halophiles [16,17] and alkaliphiles [18].
Owing to the attractive properties such as heat resistance and salt tolerance, GH42 β-galactosidases have received extensive attention in recent years. It is expected to obtain new enzymes with excellent properties from microorganisms living in extreme environments [19]. The Arctic is one of the most extreme regions to be inhabited by plenty of microorganisms, which have been proven to be the natural treasure house for screening novel enzymes [20,21]. In our previous study, a strain designated as BSi20414 with high β-galactosidase activity was isolated from Arctic sea ice and identified as Marinomonas [22]. The optimal catalytic temperature of the crude enzyme was determined as 60 °C, indicating that it might be a thermophilic enzyme. Generally, robust thermal-stability is indispensable for the practical application of enzymes. Thus, to obtain a promising thermal-stable β-galactosidase and provide a comprehensive evaluation of its potential in practical application, the enzyme that possessed β-galactosidase activity from Marinomonas sp. BSi20414 was purified to homogeneity and characterized extensively in the present work. In addition to biochemical characterization, the encoding gene of MaBGA was cloned by degenerate PCR and chromosome walking, and was further subject to bioinformatics analysis to investigate its structure–function relationships.
2. Results
2.1. Purification of Wild-Type MaBGA
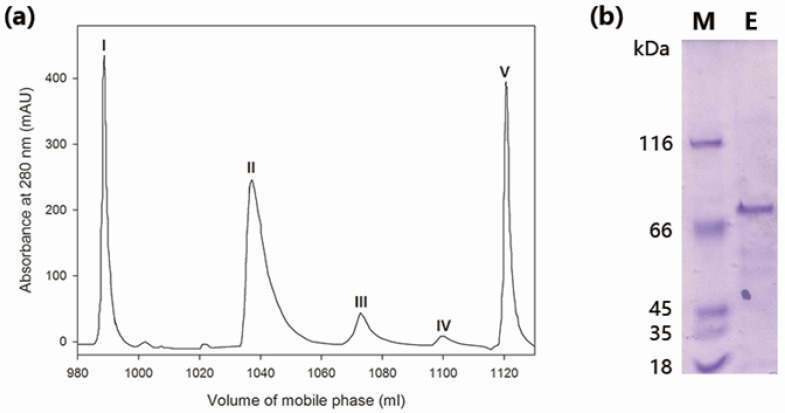
The crude enzyme was concentrated by 60% of ammonium sulfate and then separated into five components, peak I–V (Figure 1a), by anion exchange chromatography. Among these five peaks, only peak IV exhibited β-galactosidase activity toward o-nitrophenyl-β-galactoside (ONPG). The purity of peak IV was examined by SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) analysis, which showed a single band corresponding to about 70 kDa (Figure 1b), indicating that MaBGA had been successfully purified. As shown in Table 1, the two-step purification procedure yielded an 8.12-fold increase in specific activity and a recovery of 9.9% in total activity.
Figure 1.
Purification of wild-type MaBGA (β-galactosidase from Marinomonas sp. BSi20414). (a) Ion exchange chromatography. Peak I was unbound proteins; Peak II and III were proteins eluted by 0.1–0.2 M of NaCl; Peak IV was protein eluted by 0.20–0.24 M of NaCl; Peak V was protein eluted by 0.24–0.6 M of NaCl; (b) SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) analysis of purified MaBGA. Lane M: protein molecular weight marker; Lane E: purified MaBGA.
Table 1.
Purification of MaBGA.
| Purification Steps | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Recovery (%) | Purification Fold |
|---|---|---|---|---|---|
| Cell lysate | 162.54 | 1818.18 | 11.19 | 100 | 1 |
| Ammonium sulfate precipitation | 29.9 | 927.27 | 31.01 | 51 | 2.77 |
| HiTrap DEAE FF | 1.01 | 91.80 | 90.89 | 9.9 | 8.12 |
2.2. Enzymatic Characterization of MaBGA
2.2.1. Effect of pH on the Activity and Stability of MaBGA
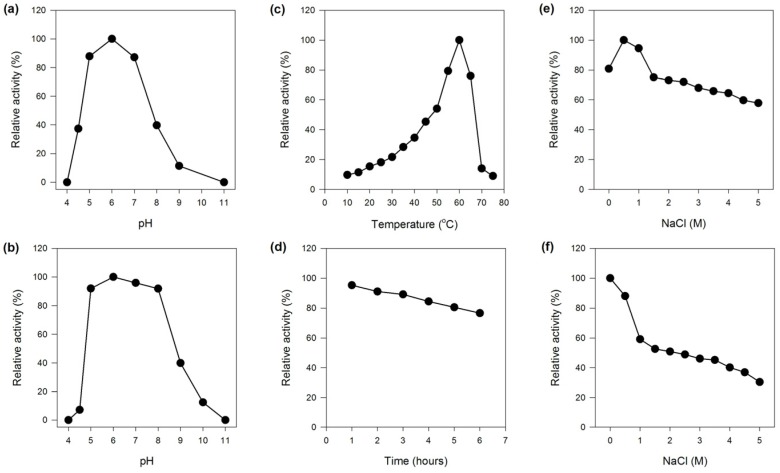
The optimum pH of MaBGA was determined as 6.0, and it exhibited more than 80% of its maximum activity over the pH range of 5.0–7.0, outside of which the activity decreased sharply (Figure 2a). The stability of MaBGA showed a similar pattern with that of the activity response to pH, which was stable around the neural condition, and could maintain at least 90% of its initial activity over the pH ranging from 5.0 to 8.0, after incubating in Britton–Robinson buffer with different pH values for 1 h (Figure 2b).
Figure 2.
Effects of pH, temperature and NaCl on the activity and stability of MaBGA. (a) Effect of pH on the activity of MaBGA; (b) Effect of pH on the stability of MaBGA; (c) Effect of temperature on the activity of MaBGA; (d) Effect of time on the stability of MaBGA; (e) Effect of NaCl on the activity of MaBGA; (f) Effect of NaCl on the stability of MaBGA.
2.2.2. Effect of Temperature on the Activity and Stability of MaBGA
MaBGA exhibited the highest activity at 60 °C, and less than 50% of the maximum activity was measured at temperatures below 45 °C (Figure 2c). Generally, an enzyme with a relatively high optimal reaction temperature often possessed superior thermal stability. With no exception, MaBGA was stable at 50 °C, which could maintain 76% of its initial activity after incubating for 6 h (Figure 2d). In addition, the half-life of MaBGA at 50 °C was determined as 16 h.
2.2.3. Effect of NaCl on the Activity and Stability of MaBGA
MaBGA showed the highest activity with 0.5 M NaCl contained in the reaction buffer. Although the activity decreased along with the increase in the concentration of NaCl, MaBGA still displayed 55% of its maximum activity with 5 M NaCl added (Figure 2e). MaBGA was unstable while incubated in buffers containing NaCl above 0.5 M, and it could only maintain 30% of its initial activity after incubating in buffer with 5 M NaCl added for 1 h (Figure 2f).
2.2.4. Effects of Metal Ions and Chemicals on the Activity of MaBGA
As shown in Table 2, K+, Na+ and Mn2+ displayed no significant effects on the activity of MaBGA, as well as EDTA. Interestingly, Fe2+ is capable of improving the activity of MaBGA by 111%, whereas other bivalent cations—Mg2+, Co2+, Ni2+ and Zn2+—slightly inhibited the activity of the enzyme. Moreover, reducing agents, such as l-cysteine, l-glutathion and dithiotreitol showed no notable effect on the activity of MaBGA, indicating that no disulfide bond was indispensable to the enzyme.
Table 2.
Effects of metal ions and chemicals on the activity of MaBGAL.
| Metal Ions | Relative Activity (%) | Chemicals | Relative Activity (%) |
|---|---|---|---|
| K+ | 96 | EDTA | 98 |
| Na+ | 95 | l-Cysteine | 110 |
| Fe2+ | 211 | l-Glutathion | 103 |
| Mn2+ | 98 | Dithiotreitol | 106 |
| Mg2+ | 89 | ||
| Co2+ | 88 | ||
| Ni2+ | 76 | ||
| Zn2+ | 74 |
2.2.5. Substrate Specificity and Steady-State Kinetic Analysis
MaBGA possessed a narrow substrate spectrum, which showed no activity toward p-nitrophenyl-β-d-glucopyranoside, p-nitrophenyl-β-d-xylopyranoside, p-nitrophenyl-β-d-lactopyranoside, p-nitrophenyl-β-d-glucuronide, p-nitrophenyl-a-d-galactopyranoside, p-nitrophenyl-β-l-arabinopyranoside and p-nitrophenyl-β-d-cellobioside. Moreover, MaBGA showed not only group selectivity, but also showed linkage selectivity in the substrate recognition process, of which the activity toward p-nitrophenyl-β-d-galactopyranoside was 4.22-fold greater than that of ONPG (Table 3).
Table 3.
Substrate specificity of MaBGA.
| Substrate | Relative Activity (%) |
|---|---|
| o-nitrophenyl-β-d-galactopyranoside | 100 |
| p-nitrophenyl-β-d-galactopyranoside | 422 |
| p-nitrophenyl-β-d-glucopyranoside | <1 |
| p-nitrophenyl-β-d-xylopyranoside | <1 |
| p-nitrophenyl-β-d-lactopyranoside | <1 |
| p-nitrophenyl-β-d-glucuronide | <1 |
| p-nitrophenyl-a-d-galactopyranoside | <1 |
| p-nitrophenyl-β-l-arabinopyranoside | <1 |
| p-nitrophenyl-β-d-cellobioside | <1 |
The steady-state kinetic constants of MaBGA were determined by using a nonlinear fitting plot. The apparent Michaelis–Menten constant Km and the maximum reaction velocity Vmax were calculated as 14.19 mM and 1.049 μM·min−1, respectively.
2.2.6. Linkage Selectivity Analysis
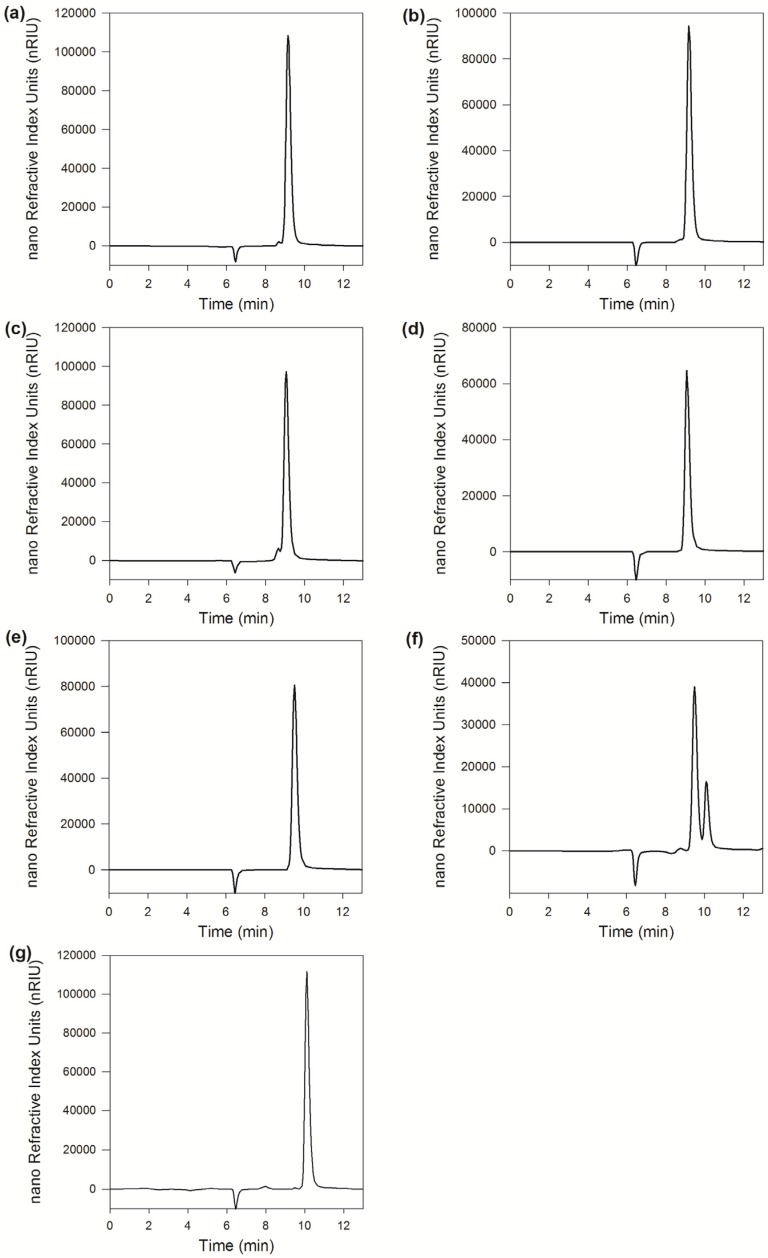
As shown in Figure 3, the chromatograms of Galβ1–4GlcNAc showed no change before and after the reaction catalyzed by MaBGA, as well as Galβ1–6GlcNAc, suggesting that MaBGA was unable to hydrolyze both Galβ1–4GlcNAc and Galβ1–6GlcNAc. With regard to Galβ1–3GlcNAc, the product chromatogram generated a new peak corresponding to the standard of β-galactose, with an identical retention time of 10.1 min, indicating that MaBGA was capable of degrading Galβ1–3GlcNAc selectively.
Figure 3.
Linkage selectivity analysis. (a), (c), (e) and (g) were chromatograms of Galβ1–4GlcNAc, Galβ1–6GlcNAc, Galβ1–3GlcNAc and galactose, respectively; (b), (d) and (f) were chromatograms of Galβ1–4GlcNAc, Galβ1–6GlcNAc and Galβ1–3GlcNAc hydrolyzed by MaBGA, respectively.
2.3. Gene Cloning and Sequence Analysis
2.3.1. Gene Cloning
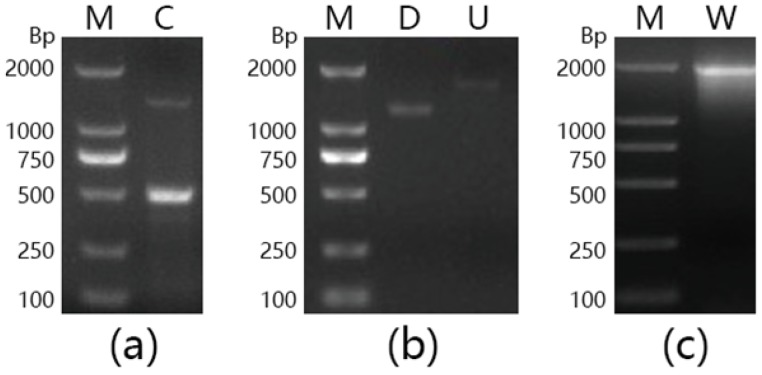
A 500-bp fragment was amplified from the genomic DNA of Marinomonas sp. BSi20414, by using the degenerate primers F1 and R2 (Figure 4a). The nucleotide sequence of the fragment showed an identity of 84% with a putative β-galactosidase gene of Marinomonas sp. MWYL1, suggesting that the partial β-galactosidase gene sequence of Marinomonas sp. BSi20414 had been successfully obtained. After chromosome walking, 1800-bp and a 1600-bp DNA fragments (Figure 4b), corresponding to the upstream and downstream sequence of the target gene, were amplified and sequenced. The intermediate and flanking sequences were utilized for assembly and subsequent amplification of the intact mabga gene, with a size of 2000 bp (Figure 4c).
Figure 4.
Agarose gel electrophoresis analysis of DNA fragments amplified by PCR. (a) Degenerated PCR; (b) Chromosome walking PCR; (c) Full length amplification. Lane M: DNA marker; Lane C: Fragment amplified by F1/R2; Lane D: Downstream fragment; Lane U: Upstream fragment; Lane W: Full-length fragment of gene mabga.
2.3.2. Sequence Analysis
Gene mabga encodes a peptide consisting of 656 amino acids with a calculated molecular weight of 74.28 kDa in accordance with the results of SDS-PAGE. Significant Pfam-A matches revealed that MaBGA belonged to the glycoside hydrolase family 42. The protein sequence of MaBGA showed 54.6%, 54.0%, 52.5%, 52.0%, 40.6%, and 38.2% identities with the well-characterized galactosidases from Thermus sp. IB-21 (Q8GEA9) [13], Thermus thermophilus A4 (O69315) [23], Thermus sp. T2 (O54315) [14], Thermus brockianus ITI360 (Q9X6C6) [24], Halorubrum lacusprofundi ATCC 49239 (B9LW38) [16], and Haloferax lucentense DSM 14919 (P94804) [17], respectively. In addition, MaBGA exhibited the highest identity of 90% with a putative BGA from Marinomonas sp. MWYL1.
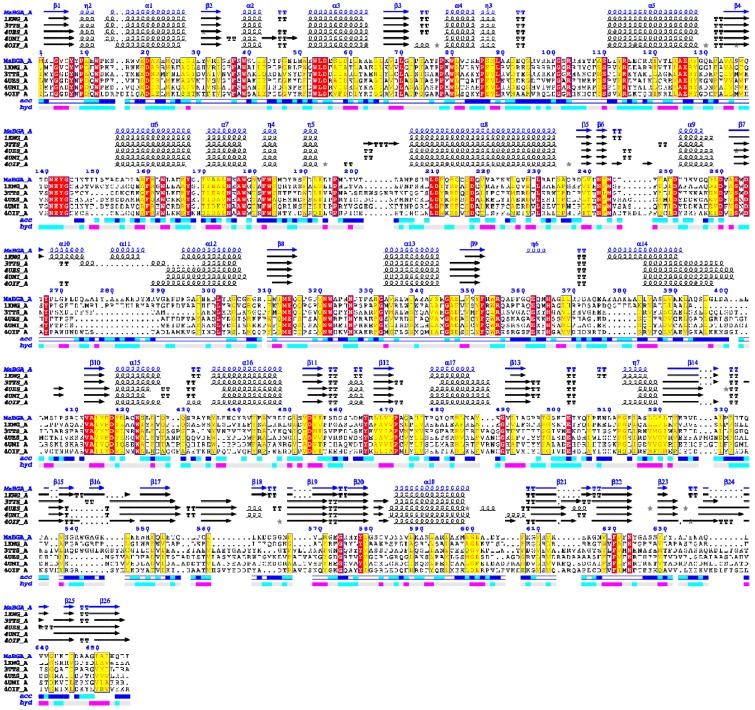
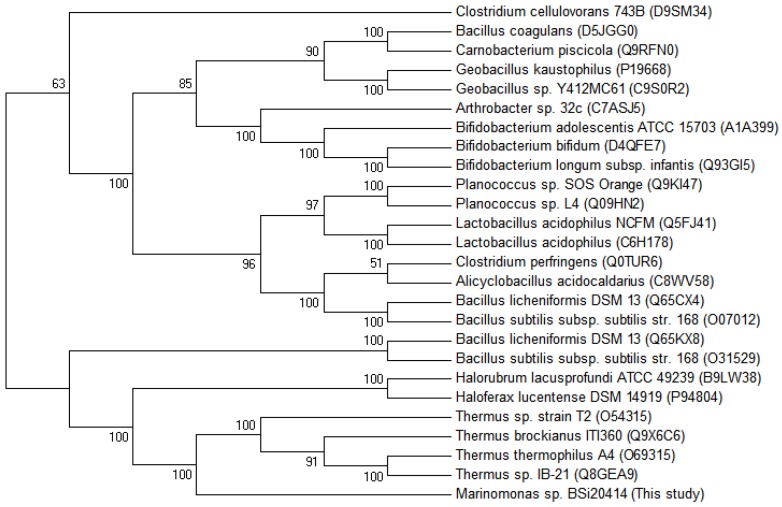
Multiple alignments of protein sequences of structure-solved GH42 β-galactosidase showed that MaBGA shared the conserved catalytic residues, Glu142 and Glu314, as well as other GH42 BGAs (Figure 5). Phylogenetic analysis of characterized BGAs showed that these BGAs diverged into two clusters, and MaBGA affiliated to the branch consisting of thermophilic BGAs (Figure 6) that exhibited considerable stability against heat in previous studies, suggesting that these BGAs, including MaBGA, might be originated from the same ancestral sequence.
Figure 5.
Multiple alignment of structure-solved β-galactosidases of the GH42 family (glycoside hydrolase 42 family). Identical residues and conserved substitutions are shaded red and yellow, respectively. Secondary structures of β-galactosidases are presented on the top: helices with squiggles, β-strands with arrows, turns with TT letters.
Figure 6.
Unrooted phylogenetic tree of β-galactosidase belonging to the GH42 family. The phylogenetic tree was built using the neighbor joining method [31] in MEGA 6 [32], with a bootstrap test of 1000 replicates. The GenBank accession numbers were provided in the bracket followed by the species names.
2.4. Structural Analysis of MaBGA
2.4.1. Homology Modelling
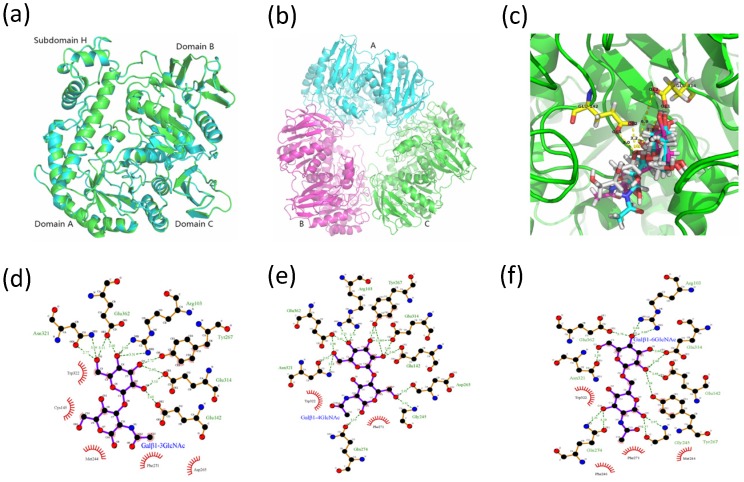
The tertiary structures of MaBGA were constructed by various software or online servers, including SWISS-MODEL [25], Robetta [26], MODELLER [27] and I-TASSER [28], then evaluated by ProSA [29] and PROCHECK [30]. Both of the Z-score and Ramachandran plot statistics indicated that the three-dimensional structures of MaBGA had been modeled reasonably (Table 4), especially for the model constructed by MODELLER, which scored highest and was selected for the docking study. The superposition of the MaBGA monomer structure onto the structure of β-galactosidase from Thermus thermophilus A4 [23] demonstrated the relatively high similarity between them, with a root mean square deviation value of 0.17 (Figure 7a). As with other GH42 BGAs [11,18,23], the quaternary structure of MaBGA was predicted as a homo-trimer, which resembles a flowerpot, with a cone-shaped tunnel in the center of the flowerpot surrounded by three subunits (Figure 7b).
Table 4.
Evaluation of models generated by different modeling approaches.
| Model | Z-Score 1 | Ramachandran Plot 2 | |||
|---|---|---|---|---|---|
| Most Favored (%) | Additional Allowed (%) | Generously Allowed (%) | Disallowed (%) | ||
| Template (4oif) | −12.19 | 88.3 | 10.7 | 0.6 | 0.3 |
| Robetta | −10.19 | 87.9 | 10.2 | 1.6 | 0.4 |
| Template (1kwk) | −12.24 | 90.6 | 8.2 | 0.7 | 0.4 |
| SWISS-MODEL | −9.93 | 89.7 | 8.1 | 1.5 | 0.7 |
| MODELLER | −10.08 | 91.9 | 6.8 | 0.4 | 0.9 |
| Template (1kwg) | −12.27 | 91.2 | 7.9 | 0.6 | 0.4 |
| I-TASSER | −10.14 | 79.1 | 15.8 | 3.2 | 1.9 |
1 Calculated by ProSA-web; 2 Calculated by PROCHECK.
Figure 7.
Three-dimensional structures of MaBGA. (a) Superposition of the MaBGA monomer structure (green) on the structures of β-galactosidase from Thermus thermophilus A4 (cyan; PDB entry 1kwk); (b) Ribbon representation of the trimer structure of MaBGA; (c) Ball and stick representation of the docking models of MaBGA with Galβ1–3GlcNAc (white), Galβ1–4GlcNAc (cyan) and Galβ1–6GlcNAc (magenta), respectively; (d) Schematic diagram of MaBGA/Galβ1–3GlcNAc interactions; (e) Schematic diagram of MaBGA/Galβ1–4GlcNAc interactions; (f) Schematic diagram of MaBGA/Galβ1–6GlcNAc interactions.
2.4.2. Molecular Docking Analysis
The model of MaBGA generated by MODELLER [27] was subject to GROMACS [33] software packages for energy minimization, to remove steric clashes. The refined model was employed for molecular docking with Galβ1–3GlcNAc, Galβ1–4GlcNAc and Galβ1–6GlcNAc by Autodock 4.2 [34], respectively. Cluster analysis was performed on different conformations with a root mean square deviation (RMSD) tolerance of 2.0 Å. Conformation with the lowest estimated binding free energy was utilized for analysis. As shown in Figure 7c, the galactosyl group of these three substrates adopts similar conformations, including the oxygen atom which links the acetylglucosamine group. However, the acetylglucosamine group of the substrates adopts a varied conformation corresponding to their lowest free energy. The two-dimensional projection of the interaction of the enzyme/substrate complex showed that no hydrogen bond was generated between the enzyme and the glucosyl group of Galβ1–3GlcNAc (Figure 7d), in contrast to those of Galβ1–4GlcNAc (Figure 7e) and Galβ1–6GlcNAc (Figure 7f), which formed three and four pairs with the enzyme, respectively.
3. Discussion
In the present study, a thermostable β-1,3-galactosidase MaBGA was successfully purified to homogeneity from Marinomonas sp. BSi20414 isolated from Arctic sea ice by ammonium sulfate precipitation and anion exchange chromatography, resulting in an 8.12-fold increase in specific activity and 9.9% recovery in total activity. The purification results showed that the two-step purification method is efficient for separating MaBGA from the wild-type strain of Marinomonas sp. BSi20414, which also provides a reference for extracting other proteins from strains belonging to the genus of Marinomonas.
Interestingly, as an enzyme stemmed from a strain living in permanently low-temperature marine environments, MaBGA displayed extraordinary stability against heat, with the half-life determined as 16 h at 50 °C. Phylogenetic analysis of characterized GH42 BGAs also revealed that MaBGA had closer genetic relationships with thermophilic BGAs derived from extremophiles, including thermophiles [11,12,13,14,15] and halophiles [16,17]. On account of the enzymatic and phylogenetic analyses, MaBGA was considered as a thermophilic enzyme, although the thermal stability of MaBGA is weaker than those of its thermophilic counterparts. Additionally, MaBGA only shared high identity (>70%) with BGAs of the genus Marinomonas, and no sequence with identity more than 55% was found in their related marine species. On the basis of the above evidence, a putative explanation was proposed to illustrate the mismatch of enzyme stability and circumstance. It is supposed that the ancestor of the genus Marinomonas acquired the gene encoding thermophilic β-galactosidase from other thermophiles by occasional horizontal transfer, then experienced adaptive evolution under low-temperature marine environments for a long period, which led to a decrease in thermal stability without selection pressures.
Another point worth mentioning is that MaBGA has a strict substrate specificity, unlike other GH42 BGAs. Furthermore, it displayed not only group selectivity, but also linkage selectivity in the substrate recognition process. As indicated above, MaBGA was able to hydrolyze Galβ1–3GlcNAc, but was unable to hydrolyze Galβ1–4GlcNAc and Galβ1–6GlcNAc. To better understand the linkage selectivity of MaBGA, it is essential to put MaBGA against its structural contexts. Thus, the three-dimensional structure of MaBGA was constructed and subject to docking analysis after energy minimization by molecular dynamics. As shown in Figure 7c, for all these three substrates, although the galactosyl group adopts similar lowest energy conformations, the distance is a bit long for the reaction between the oxygen atom linking the acetylglucosamine group and the carboxyl group of catalytic residues (Glu142/Glu314). Therefore, the substrate molecule needs to fine-tune its geometry to shorten the distance mentioned above by overcoming the energy barrier. However, the planar representation of the interaction of the enzyme/substrate complex indicated that the strong interaction between the glucosyl group of Galβ1–4GlcNAc (Figure 7e)/Galβ1–6GlcNAc (Figure 7f) and the enzyme might lead to the failure of the substrates to adjust their conformation for an optimal fit. Therefore, we proposed that the favored binding conformation with lowest free binding energy of the substrate is not close enough to the catalytic residues to let the reaction occur, thus the substrate might be fine-tuning its conformation to achieve an optimal geometry for the reaction. However, due to the different binding energy between the glucosyl group and enzyme, Galβ1–4GlcNAc and Galβ1–6GlcNAc cannot readily overcome the energy barrier, other than Galβ1–3GlcNAc. In general, further experiments, such as enzyme/substrate complex co-crystallization and site-directed mutagenesis, are still needed to test the hypothesis.
A previous study had proven that galacto-oligosaccharides with β1–3 linkage have a stronger bifidogenic effect than those with β1–4 and β1–6 linkages [35], indicating that the former would be more popular as prebiotics than the latter two. Generally, the production of galacto-oligosaccharides is implemented by the transglycosylation activity of β-galactosidase [36], therefore, the linkage of galacto-oligosaccharides will depend on the linkage recognition ability of β-galactosidase. Since β-galactosidases that existed in the nature which are capable of recognizing β1–3 galactoside linkage are very few, the β-1,3-galactosidase MaBGA studied in the present work not only could provide a promising candidate for the biosynthesis of galacto-oligosaccharides with β1–3 linkage, but also would offer a good model for research on the substrate recognition mechanism of β-galactosidase.
4. Materials and Methods
4.1. Strains, Plasmids, and Culture Conditions
Strain BSi20414, used as the source of β-galactosidase, was isolated from a core sample of sea ice collected from Canada Basin, Arctic Ocean, and was characterized as Marinomonas in our previous study [22]. The strain was cultivated in medium (pH 7.0) containing MgCl2 (0.5%, w/v), MgSO4·7H2O (0.4%, w/v), KCl (0.1%, w/v), CaCl2 (0.06%, w/v), lactose (1.5%, w/v) and Tryptone (0.5%, w/v), on a shaking incubator at 180 rev·min−1 at 30 °C for 96 h. Escherichia coli DH5α used for gene cloning was cultivated at 37 °C in Luria–Bertani medium. Plasmid pMD18-T (Takara) was used to conduct TA cloning for sequencing. All chemicals used in this study were of analytical grade.
4.2. Purification of Wild-Type MaBGA
All purification steps were conducted at 4 °C. Cells were harvested by centrifugation at 10,000× g for 10 min. The pellet was washed three times with normal saline and was suspended by pre-cooling PBS buffer (pH 7.0, 50 mM). The suspension was lysed by sonication (burst of 2 s followed by intervals of 5 s for 30 min). The cell debris was removed by centrifugation at 10,000× g for 15 min and the supernatant was precipitated with ammonium sulfate (60%, w/v). The precipitate was collected by centrifugation at 10,000× g for 10 min, then dissolved and dialyzed using PBS buffer (pH 7.0, 50 mM) overnight. Subsequently, the protein solution was filtered by cellulose acetate film with pore size of 0.22 μm, and the filtrate was loaded onto an anion exchange column HiTrap DEAE FF, which was pre-equilibrated with PBS buffer (pH 8.0, 50 mM). The column was first washed with PBS buffer (pH 8.0, 50 mM) for tenfold resin volumes, then was eluted by PBS buffer (pH 8.0, 50 mM) containing NaCl with a linear gradient from 0.1 M to 0.6 M. Every eluting peak was collected and measured by standard activity assay. The protein concentration was assayed by the method of Bradford using BSA (bovine serum albumin) as a standard [37].
4.3. SDS-PAGE Analysis
The purified MaBGA was analyzed by denaturing discontinuous SDS-PAGE on a 5% stacking gel and a 10% separating gel as described by Laemmli [38]. Gels were stained with Coomassie Brilliant Blue R-250. The molecular weight of MaBGA was determined by comparing its electrophoretic mobility with Protein Molecular Weight Marker (MBI).
4.4. β-galactosidase Activity Assay
The β-galactosidase activity was assayed by measuring the absorbance of ONP (o-nitrophenyl) at 420 nm in 50 mM PBS buffer (pH 7.0) with 10 mM ONPG as substrate. The ONP concentration was calculated from the standard curve obtained under the same experimental condition. One unit of enzyme activity was defined as the amount of the enzyme that catalyzed the formation of 1 μmol of ONP per minute.
4.5. Effect of pH on the Activity and Stability of MaBGA
The optimum pH for MaBGA was determined by measuring the activity in Britton–Robinson buffer with different pH ranging from 3.0 to 12.0. The pH stability was assayed by measuring the residual activity after incubating MaBGA in different pH buffers at 37 °C for 1 h.
4.6. Effect of Temperature on the Activity and Stability of MaBGA
To study the effect of temperature on the activity of MaBGA, the enzyme activity was assayed at different temperatures from 10 to 70 °C with 5 °C intervals at pH 7.0. The thermal stability was determined by assaying the residual activity after incubating the enzyme at 50 °C for 6 h with 1 h intervals.
4.7. Effect of NaCl on the Activity and Stability of MaBGA
In order to determine the effects of NaCl on the activity of MaBGA, the enzyme activity was assayed with 0.5, 1, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, 5 M NaCl added individually. The NaCl tolerance of MaBGA was determined by measuring the residual activity after incubating the enzyme in buffers containing diverse concentration of NaCl from 0.5 M to 5M at 37 °C for 1 h.
4.8. Effect of Metal Ions and Chemicals on the Activity of MaBGA
To investigate the effects of metal ions and chemicals on the MaBGA activity, 1 mM of KCl, NaCl, FeCl2, MnCl2, MgCl2, CoCl2, NiCl2, ZnCl2, EDTA and 10 mM of l-cysteine, l-glutathion and dithiotreitol were added to the reaction system individually, and the activity of MaBGA was then measured under the standard assay as described above. No chemical was added in the control.
4.9. Substrate Specificity
The substrates’ specificity of MaBGA was measured by the standard assay, except that ONPG was replaced by p-nitrophenyl-β-d-galactopyranoside, p-nitrophenyl-β-d-glucopyranoside, p-nitrophenyl-β-d-xylopyranoside, p-nitrophenyl-β-d-lactopyranoside, p-nitrophenyl-β-d-glucuronide, p-nitrophenyl-a-d-galactopyranoside, p-nitrophenyl-β-l-arabinopyranoside, p-nitrophenyl-β-d-cellobioside, respectively.
4.10. Steady-State Kinetic Analysis
For steady-state kinetic analysis, the activity of MaBGA was measured by using various concentrations of ONPG from 0.1 mM to 19 mM. The kinetic constants of the enzyme were determined by using a nonlinear fitting of the Michaelis–Menten equation: v = Vmax·[S]/(Km + [S]), where [S] and Km are the concentration and Michaelis constants of ONPG, respectively.
4.11. Linkage Selectivity Analysis
To determine the activity of MaBGA toward different linkage types, Galβ1–3GlcNAc, Galβ1–4GlcNAc and Galβ1–6GlcNAc were used as substrates, respectively. The reaction products were filtered by nitrocellulose membrane with pore size of 0.22 μm, in advance of being subject to detection by HPLC equipped with an Aminex HPX-87P column and differential detector. The column temperature and flow rate were set as 85 °C and 0.5 mL·min−1.
4.12. Gene Cloning and Sequence Analysis
The partial sequence of gene mabga was amplified by using degenerate primer pairs F1/R1, F1/R2, F1/R3, F2/R1, F2/R2, F2/R3, F3/R1, F3/R2, F3/R3, A208/B1, A208/C1, A208/C2, A76/B1, A76/C1, A76/C2, A195/B1, A195/C1 and A195/C2 (Table 1), respectively, which were designed according to the conservative regions of the protein sequence of β-galactosidases. PCR was performed as follows: 95 °C for 4 min; followed by 30 cycles of 95 °C for 1 min, 50 °C for 1 min, and 72 °C for 2 min; with a final extension at 72 °C for 10 min. The amplified fragment was inserted into the pMD18-T vector and then transformed into E. coli DH5α for sequencing.
The 5′ and 3′ flanking regions of the known partial sequence were obtained by using DNA Walking SpeedUp™ Kit of Seegene, which adopted the thermal asymmetric interlaced PCR (TAIL-PCR) strategy [39]. The nested specific primers for upstream and downstream regions were designed based on the obtained partial sequence (Table 5). TAIL-PCR was performed as described by the kit. The amplified fragments were purified and ligated into the pMD18-T vector for sequencing. The upstream, downstream and obtained partial sequences were assembled to obtain a full-length mabga gene.
Table 5.
Primers used for gene cloning.
| Primers | Sequence (5′ to 3′) | Purpose |
|---|---|---|
| F1 | GCNTGGGGNAAYGTNTTYT | Degenerated PCR |
| F2 | TNTGGACNTGGGARGCNTT | Degenerated PCR |
| F3 | GGARCARCARCCNGGNCCNGT | Degenerated PCR |
| R1 | CCARCANGCRTCRTARTCRAA | Degenerated PCR |
| R2 | RAANGCYTCCCANGTCCA | Degenerated PCR |
| R3 | GGRTTRTGNGGNGCCARTT | Degenerated PCR |
| A208 | TGGATHATGGAGGAGCCC | Degenerated PCR |
| A76 | CGGGACCTGGTGCAYAAYTAY | Degenerated PCR |
| A195 | CAYAAYTAYATGGGCTTCTTC | Degenerated PCR |
| B1 | CAGACCCAGAACGAGTAYKGN | Degenerated PCR |
| C1 | GCACCACAAGTACCACCARGA | Degenerated PCR |
| C2 | GTYCTRDWNCTGCACCGGCCG | Degenerated PCR |
| U1 | CCGTAAAGAATCCCATGAGT | DNA Walking (1st-round upstream) |
| D1 | GGACATTTTGCGTGCG | DNA Walking (1st-round downstream) |
| U2 | AACGCTGAAAGTCCAACCCGAT | DNA Walking (2nd-round upstream) |
| D2 | GGACACTTATCCGCTGGGTTT | DNA Walking (2nd-round downstream) |
| U3 | GATTGGCTTCGGTCACGGT | DNA Walking (3rd-round upstream) |
| D3 | CCCGATTTTGGTGCTTTTCA | DNA Walking (3rd-round downstream) |
| DW-ACP 1 | ACP-AGGTC | DNA Walking (1st-round) |
| DW-ACP 2 | ACP-TGGTC | DNA Walking (1st-round) |
| DW-ACP 3 | ACP-GGGTC | DNA Walking (1st-round) |
| DW-ACP 4 | ACP-CGGTC | DNA Walking (1st-round) |
| DW-ACP N | ACPN-GGTC | DNA Walking (2nd-round) |
| Uni-primer | TCACAGAAGTATGCCAAGCGA | DNA Walking (3rd-round) |
| MaBGA-F | CGGAATTCAAGTTAGGTGTATGTTACTACCCAG | Full-length amplification |
| MaBGA-R | GTTCGCGCTCGAGGATTTCTTGCCAAATGGC | Full-length amplification |
Homologous search in GenBank was performed using the BLAST server (http://www.ncbi.nlm.nih.gov/BLAST). Alignment of multiple protein sequences was conducted using the Clustal X 2.0 program [40] and rendered by ESPript [41]. A phylogenetic tree of multiple β-galactosidase was constructed using the neighbor-joining method [31] in MEGA6 [32], with a bootstrap test of 1000 replicates.
4.13. Homology Modelling and Molecular Docking Analysis
The three-dimensional model of MaBGA was constructed by using MODELLER [27], Robetta [26], I-TASSER [28] and SWISS-MODEL [25], respectively. Precise evaluation of the model quality was performed using ProSA-web [29] and PROCHECK [30]. To remove steric clashes, the constructed model was subject to an energy minimization process in vacuum by using the steepest descent method for about 5000 iterations in GROMACS 4.5 [33].
The refined model was used for docking with Galβ1–3GlcNAc, Galβ1–4GlcNAc and Galβ1–6GlcNAc, respectively, using Autodock 4.2 [34] with default parameters. The representation of the protein structure was achieved using the program PyMOL (The PyMOL Molecular Graphics System, Version 1.7 Schrödinger, LLC., New York, NY, USA).
5. Conclusions
In this work, a thermostable β-1,3-galactosidase MaBGA derived from Marinomonas sp. BSi20414, was first purified to homogeneity and characterized extensively. MaBGA displayed robust stability against heat and strict substrate specificity toward both the glycosyl group and the linkage type. Although further experiments are required to decipher its substrate recognition mechanism, our study provided an attractive alternative for biosynthesis of galacto-oligosaccharide with β1–3 linkage and laid the groundwork for the protein engineering to modify the linkage preference of β-galactosidase.
Acknowledgments
This study was supported by Chinese Polar Environment Comprehensive Investigation and Assessment Program (CHINARE-01-06, CHINARE-04-02, CHINARE-02-01), Open Fund of Key Laboratory of Biotechnology and Bioresources Utilization (Dalian Minzu University, KF2015009), State Ethnic Affairs Commission & Ministry of Education of China, and National Natural Science Foundation of China (31200599).
Author Contributions
Y.Y. and B.C. conceived and designed the experiments; Q.Z. and L.Z. performed the experiments; H.D., Q.Z. and L.Z. analyzed the data; Y.Y. and B.C. contributed reagents/materials/analysis tools; H.D. and B.C. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Juers D.H., Matthews B.W., Huber R.E. LacZ β-galactosidase: Structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 2012;21:1792–1807. doi: 10.1002/pro.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliveira C., Guimaraes P.M., Domingues L. Recombinant microbial systems for improved β-galactosidase production and biotechnological applications. Biotechnol. Adv. 2011;29:600–609. doi: 10.1016/j.biotechadv.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Park A.R., Oh D.K. Galacto-oligosaccharide production using microbial β-galactosidase: Current state and perspectives. Appl. Microbiol. Biotechnol. 2010;85:1279–1286. doi: 10.1007/s00253-009-2356-2. [DOI] [PubMed] [Google Scholar]
- 4.Henrissat B., Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997;7:637–644. doi: 10.1016/S0959-440X(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 5.Lombard V., Ramulu H.G., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (cazy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies G., Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 7.Adam A.C., Rubio-Texeira M., Polaina J. Lactose: The milk sugar from a biotechnological perspective. BFSN. 2005;44:553–557. doi: 10.1080/10408690490931411. [DOI] [PubMed] [Google Scholar]
- 8.Singh A.K., Pluvinage B., Higgins M.A., Dalia A.B., Woodiga S.A., Flynn M., Lloyd A.R., Weiser J.N., Stubbs K.A., Boraston A.B., et al. Unravelling the multiple functions of the architecturally intricate streptococcus pneumoniae β-galactosidase, BgaA. PLoS Pathog. 2014;10:e1004364. doi: 10.1371/journal.ppat.1004364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu D., Zhang F., Zhang H., Hao L., Gong X., Geng M., Cao M., Zheng F., Zhu J., Pan X., et al. The β-galactosidase (BgaC) of the zoonotic pathogen streptococcus suis is a surface protein without the involvement of bacterial virulence. Sci. Rep. 2014;4:4140. doi: 10.1038/srep04140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng W., Wang L., Jiang Y.L., Bai X.H., Chu J., Li Q., Yu G., Liang Q.L., Zhou C.Z., Chen Y. Structural insights into the substrate specificity of streptococcus pneumoniae β(1,3)-galactosidase BgaC. J. Biol. Chem. 2012;287:22910–22918. doi: 10.1074/jbc.M112.367128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solomon H.V., Tabachnikov O., Lansky S., Salama R., Feinberg H., Shoham Y., Shoham G. Structure-function relationships in Gan42B, an intracellular GH42 β-galactosidase from geobacillus stearothermophilus. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015;71:2433–2448. doi: 10.1107/S1399004715018672. [DOI] [PubMed] [Google Scholar]
- 12.Solomon H.V., Tabachnikov O., Feinberg H., Govada L., Chayen N.E., Shoham Y., Shoham G. Crystallization and preliminary crystallographic analysis of ganb, a GH42 intracellular β-galactosidase from geobacillus stearothermophilus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013;69:1114–1119. doi: 10.1107/S1744309113023609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang S.K., Cho K.K., Ahn J.K., Bok J.D., Kang S.H., Woo J.H., Lee H.G., You S.K., Choi Y.J. Three forms of thermostable lactose-hydrolase from Thermus sp. IB-21: Cloning, expression, and enzyme characterization. J. Biotechnol. 2005;116:337–346. doi: 10.1016/j.jbiotec.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 14.Vian A., Carrascosa A.V., Garcia J.L., Cortes E. Structure of the β-galactosidase gene from thermus sp. Strain T2: Expression in escherichia coli and purification in a single step of an active fusion protein. Appl. Environ. Microbiol. 1998;64:2187–2191. doi: 10.1128/aem.64.6.2187-2191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohtsu N., Motoshima H., Goto K., Tsukasaki F., Matsuzawa H. Thermostable β-galactosidase from an extreme thermophile, Thermus sp. A4: Enzyme purification and characterization, and gene cloning and sequencing. Biosci. Biotechnol. Biochem. 1998;62:1539–1545. doi: 10.1271/bbb.62.1539. [DOI] [PubMed] [Google Scholar]
- 16.Karan R., Capes M.D., DasSarma P., DasSarma S. Cloning, overexpression, purification, and characterization of a polyextremophilic β-galactosidase from the antarctic haloarchaeon halorubrum lacusprofundi. BMC Biotechnol. 2013;13:3. doi: 10.1186/1472-6750-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes M.L., Scopes R.K., Moritz R.L., Simpson R.J., Englert C., Pfeifer F., Dyall-Smith M.L. Purification and analysis of an extremely halophilic β-galactosidase from haloferax alicantei. Biochim. Biophys. Acta. 1997;1337:276–286. doi: 10.1016/S0167-4838(96)00174-4. [DOI] [PubMed] [Google Scholar]
- 18.Maksimainen M., Paavilainen S., Hakulinen N., Rouvinen J. Structural analysis, enzymatic characterization, and catalytic mechanisms of β-galactosidase from bacillus circulans sp. Alkalophilus. FEBS J. 2012;279:1788–1798. doi: 10.1111/j.1742-4658.2012.08555.x. [DOI] [PubMed] [Google Scholar]
- 19.Ferrer M., Golyshina O., Beloqui A., Golyshin P.N. Mining enzymes from extreme environments. Curr. Opin. Microbiol. 2007;10:207–214. doi: 10.1016/j.mib.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Lee H.S., Kwon K.K., Kang S.G., Cha S.-S., Kim S.-J., Lee J.-H. Approaches for novel enzyme discovery from marine environments. Curr. Opin. Biotechnol. 2010;21:353–357. doi: 10.1016/j.copbio.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Ferrer M., Beloqui A., Timmis K.N., Golyshin P.N. Metagenomics for mining new genetic resources of microbial communities. J. Mol. Microbiol. Biotechnol. 2008;16:109–123. doi: 10.1159/000142898. [DOI] [PubMed] [Google Scholar]
- 22.Zeng Q., Wang Y., Sun K., Yu Y., Chen B. Preliminary studies on the screening, identification and optimum fermentative conditions of a strain Marinomonas sp. Bsi20414 isolated from arctic sea ice producing β-galactosidase. Chin. J. Polar Res. 2011:108–114. [Google Scholar]
- 23.Hidaka M., Fushinobu S., Ohtsu N., Motoshima H., Matsuzawa H., Shoun H., Wakagi T. Trimeric crystal structure of the glycoside hydrolase family 42 β-galactosidase from thermus thermophilus a4 and the structure of its complex with galactose. J. Mol. Biol. 2002;322:79–91. doi: 10.1016/S0022-2836(02)00746-5. [DOI] [PubMed] [Google Scholar]
- 24.Fridjonsson O., Watzlawick H., Gehweiler A., Rohrhirsch T., Mattes R. Cloning of the gene encoding a novel thermostable α-galactosidase from thermus brockianus ITI360. Appl. Environ. Microbiol. 1999;65:3955–3963. doi: 10.1128/aem.65.9.3955-3963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., et al. Swiss-model: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webb B., Sali A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinform. 2014 doi: 10.1002/0471250953.bi0506s15. [DOI] [PubMed] [Google Scholar]
- 28.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-tasser suite: Protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiederstein M., Sippl M.J. Prosa-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. Procheck: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 31.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M.R., Smith J.C., Kasson P.M., van der Spoel D. Gromacs 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. Autodock4 and autodocktools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Depeint F., Tzortzis G., Vulevic J., I’Anson K., Gibson G.R. Prebiotic evaluation of a novel galactooligosaccharide mixture produced by the enzymatic activity of bifidobacterium bifidum ncimb 41171, in healthy humans: A randomized, double-blind, crossover, placebo-controlled intervention study. Am. J. Clin. Nutr. 2008;87:785–791. doi: 10.1093/ajcn/87.3.785. [DOI] [PubMed] [Google Scholar]
- 36.Legler G. Glycoside hydrolases: Mechanistic information from studies with reversible and irreversible inhibitors. Adv. Carbohydr. Chem. Biochem. 1990;48:319–384. doi: 10.1016/s0065-2318(08)60034-7. [DOI] [PubMed] [Google Scholar]
- 37.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 38.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage t4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y.G., Whittier R.F. Thermal asymmetric interlaced pcr: Automatable amplification and sequencing of insert end fragments from p1 and yac clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-J. [DOI] [PubMed] [Google Scholar]
- 40.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 41.Gouet P., Robert X., Courcelle E. Espript/endscript: Extracting and rendering sequence and 3d information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]