Abstract
Cell-size homeostasis entails a fundamental balance between growth and division. The budding yeast Saccharomyces cerevisiae establishes this balance by enforcing growth to a critical cell size prior to cell cycle commitment (Start) in late G1 phase. Nutrients modulate the critical size threshold, such that cells are large in rich medium and small in poor medium. Here, we show that two potent negative regulators of Start, Sfp1 and Sch9, are activators of the ribosomal protein (RP) and ribosome biogenesis (Ribi) regulons, the transcriptional programs that dictate ribosome synthesis rate in accord with environmental and intracellular conditions. Sfp1 and Sch9 are required for carbon-source modulation of cell size and are regulated at the level of nuclear localization and abundance, respectively. Sfp1 nuclear concentration responds rapidly to nutrient and stress conditions and is regulated by the Ras/PKA and TOR signaling pathways. In turn, Sfp1 influences the nuclear localization of Fhl1 and Ifh1, which bind to RP gene promoters. Starvation or the absence of Sfp1 causes Fhl1 and Ifh1 to localize to nucleolar regions, concomitant with reduced RP gene transcription. These findings suggest that nutrient signals set the critical cell-size threshold via Sfp1 and Sch9-mediated control of ribosome biosynthetic rates.
Keywords: Ribosome, nutrients, nucleolus, size, Start
Cell-size homeostasis requires that proliferating cells coordinate growth and the cell cycle, such that each division is matched by a doubling of mass. Cell growth and the cell cycle are, however, separate processes that can be uncoupled, either by endogenous developmental mechanisms, for instance, during oogenesis and the rapid cleavage cycles following fertilization, or within the laboratory. In eukaryotic cells, doubling time is usually limited, not by the time required to duplicate and divide the genome, but rather by the time required to double cell mass. Consistently, cell cycle progression is typically dependent on cell growth. In contrast, mass accumulation continues unabated during most cell cycle arrests (Saucedo and Edgar 2002).
In the unicellular budding yeast, growth and the cell cycle are coupled at Start, the short interval during late G1 phase after which cells are committed to division (Hartwell et al. 1974). Passage through Start has several requirements as follows: (1) growth to a critical cell size (Johnston et al. 1977), (2) nutrient sufficiency (Hartwell et al. 1974), (3) attainment of a critical translation rate (Hartwell and Unger 1977; Moore 1988), and for haploids, (4) absence of mating pheromone (Hartwell et al. 1974). The first three conditions are likely to be interrelated. The critical size requirement and minimum translation rate explain why slowing growth rate increases the length of G1 phase, whereas the time required to transit the rest of the cell cycle is relatively constant (Hartwell and Unger 1977). The critical-size threshold maintains uniform cell size over many generations, and under minimal nutrient conditions forces cells to accumulate the energy stores required to complete the division cycle. At constant nutrient levels, the critical-size requirement couples growth and division only in daughter cells, as mother cells by definition have already attained critical size.
Start depends on activation of the SBF and MBF transcription factor complexes that bind the promoters of G1/S-regulated genes (Nasmyth 1996). SBF and MBF are composed of related DNA-binding proteins Swi4 and Mbp1, respectively, which interact with a common regulatory subunit Swi6 to drive expression of a massive suite of ∼200 genes. Among these, the key transcripts are the G1 cyclins CLN1 and CLN2 and the B-type cyclins CLB5 and CLB6. Cln1 and Cln2 activate Cdc28, the cyclin-dependent kinase (CDK) that controls all cell cycle transitions in budding yeast. Cln1/2–Cdc28 phosphorylation events trigger bud emergence and inactivate Sic1 and Cdh1, two key inhibitors of B-type cyclin–Cdc28 activity. Once unleashed, Clb5/6–Cdc28 complexes initiate DNA replication (Nasmyth 1996). Start can therefore be viewed as the short interval required to accumulate sufficient Cln1/2–Cdc28 activity to phosphorylate Cdh1 and Sic1, and perhaps other substrates.
The connection between cell size and SBF/MBF activation remains enigmatic. It has recently been discovered that SBF, and likely MBF, activation occurs upon dissociation of the Start-repressor Whi5 (Jorgensen et al. 2002; Costanzo et al. 2004; de Bruin et al. 2004). Loss of the interaction between SBF/MBF and Whi5, which correlates with movement of Whi5 to the cytoplasm, is driven by phosphorylation of Whi5 by Cln3–Cdc28 (Costanzo et al. 2004; de Bruin et al. 2004). Cln3 appears to act in parallel to Bck2, a poorly understood activator of SBF/MBF, as cells lacking CLN3 and BCK2 are inviable due to permanent G1 arrest (Wijnen and Futcher 1999). Remarkably, deletion of WHI5 bypasses this arrest (Costanzo et al. 2004; de Bruin et al. 2004). Because Start activation is highly sensitive to Cln3 dosage, it has been presumed that a critical concentration of Cln3–Cdc28 activity triggers Start. Although Cln3 protein and Cln3–Cdc28 kinase activity do not increase during G1 phase (Tyers et al. 1993; McInerny et al. 1997), a size-dependent increase in Cln3 nuclear abundance may help activate Start (Futcher 1996; H. Wang et al. 2004).
Nutrients modulate the critical cell-size threshold in proportion to the proliferation rate (Johnston et al. 1979; Lorincz and Carter 1979; Tyson et al. 1979). The effects of nutrients on critical cell size are conveyed rapidly, as cells are not committed to Start at a given size threshold until just before the threshold is reached (Lorincz and Carter 1979). Thus, shifting cells from poor to rich medium temporarily increases the fraction of unbudded G1-phase cells as these cells grow to the new threshold (Johnston et al. 1979). Nutrient effects are mediated in part by the Ras/PKA pathway; decreased Ras/PKA signaling, as in a cdc25-1 strain, decreases critical cell size, whereas hyperactive Ras/PKA signaling, as in a RAS2Val19 strain, increases critical cell size (Baroni et al. 1989). Strains that exhibit constitutive PKA activity do not adjust cell size in response to carbon source quality (Tokiwa et al. 1994).
Ribosome biogenesis is a chief occupation of growing cells, accounting for >50% of total transcription in yeast and mammalian cells (Warner 1999; Moss and Stefanovsky 2002). In yeast, the rate of ribosome synthesis is dictated by the rate of transcription of the RNA and protein subunits of the ribosome (Warner 1999). rRNA and RP gene transcription is exquisitely sensitive to the growth potential of the cell and is rapidly repressed in response to a wide variety of internal and external stresses (Ju and Warner 1994; Warner 1999; Gasch et al. 2000). The 137 RP genes, referred to here as the RP regulon, are tightly coregulated. The vast majority of genes in the RP regulon have promoter-binding sites for Rap1, whereas a few have sites for Abf1 (Warner 1999). As Rap1 silences telomeric repeats but activates glycolytic and RP genes (Warner 1999), its activity depends on context, presumably generated by combinations of associated co-factors. The RP-specific transcription factors that cooperate with Rap1 have not yet been identified.
A large number (>200) of other genes, which we term the Ribi regulon, show nearly identical transcriptional responses as RP genes to environmental or genetic perturbations (Gasch et al. 2000; Hughes et al. 2000; Wade et al. 2001; Jorgensen et al. 2002; Miyoshi et al. 2003). The promoters of these coexpressed genes are strongly enriched for the presence of two motifs, termed RRPE and PAC, and therefore appear to constitute a distinct regulon (Gasch et al. 2000; Hughes et al. 2000; Wade et al. 2001; Jorgensen et al. 2002). Most of these genes encode proteins involved in ribosome biogenesis, a process involving more than 100 accessory factors that assemble and modify rRNA and RPs in the nucleolus (Hughes et al. 2000; Wade et al. 2001; Fatica and Tollervey 2002; Jorgensen et al. 2002). A number of additional functional categories are present in this regulon, including subunits of RNA Polymerase I and III, enzymes involved in ribo-nucleotide metabolism, tRNA synthetases, and translation factors (Gasch et al. 2000; Wade et al. 2001; Jorgensen et al. 2002). The Ribi regulon thus consists of non-RP genes that boost translational capacity. Two central nutrient-signaling conduits, the Ras/PKA and TOR signaling pathways, can activate rRNA, RP, and Ribi transcription (Klein and Struhl 1994; Neuman-Silberberg et al. 1995; Cardenas et al. 1999; Hardwick et al. 1999; Powers and Walter 1999; Y. Wang et al. 2004). Of these, the control of rRNA synthesis is best understood, as it is known to depend largely on phosphorylation of the critical initiation factor TIF-1A/Rrn3 (Grummt 2003). Both Rap1-binding sites and RRPE elements can render gene transcription sensitive to Ras/PKA signaling, although the presumed kinase targets have not been identified (Klein and Struhl 1994; Neuman-Silberberg et al. 1995; Y. Wang et al. 2004).
Systematic determination of cell-size distributions for all yeast deletion strains has recently identified many new potential Start regulators (Jorgensen et al. 2002; Zhang et al. 2002). Many of the genes that encode potential Start repressors are implicated in ribosome biogenesis, suggesting not only a link between these two seemingly disparate systems, but also that cell-size mutants may identify new regulators of ribosome synthesis (Jorgensen et al. 2002). Beginning with the most potent size regulators, Sfp1 and Sch9, we have elaborated a dynamic transcriptional network that dictates the ribosome synthesis rate and the critical cell-size threshold.
Results
Sfp1 and Sch9 strongly influence cell size
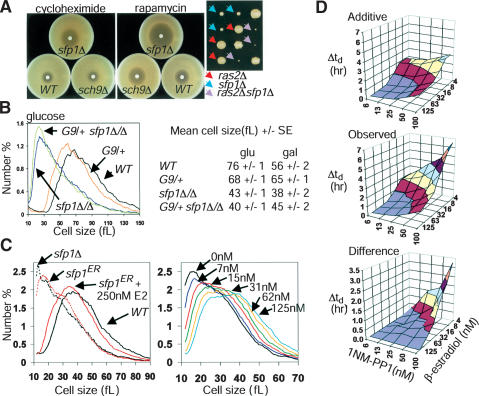
Of all nonessential genes, deletions in either SFP1 or SCH9 caused the greatest decrease in cell size (Whi phenotype). These deletion strains also proliferate slowly and accumulate cells in G1 phase, suggesting SFP1 and SCH9 are important for mass accumulation (Fig. 1A) (Jorgensen et al. 2002). Sfp1 is a putative transcription factor with a split zinc-finger domain at its C terminus that is essential for function (Fingerman et al. 2003). Sch9 is an AGC family kinase that possesses a C2 lipid-binding domain and is the closest yeast homolog of the metazoan pro-survival kinase Akt/PKB (Fabrizio et al. 2001; Jorgensen et al. 2002). Sch9 kinase activity is required for its function (data not shown). To conditionally overexpress Sch9, we constructed a strain bearing a galactose-inducible allele integrated at the endogenous locus (GAL1–SCH9). Like SFP1, overexpression of SCH9 caused a large cell phenotype, but unlike SFP1, SCH9 overexpression was not toxic (Fig. 1A). To enable conditional control of Sch9 activity at endogenous levels without confounding nutrient source shifts, we engineered an analog-sensitive (as) allele of SCH9 at the endogenous locus (sch9as). Substititution of a bulky amino acid with either alanine or glycine in the ATP-binding pocket of kinases often permits highly specific inhibition by analogs of the cell-permeable kinase inhibitor PP1, such as C3-1′-naphthyl-methyl PP1 (1NM-PP1) (Bishop et al. 2000). Unexpectedly, the integrated sch9as allele separated the proliferation and size functions of SCH9, conferring a nearly wild-type proliferation rate, but a strong Whi phenotype (Fig. 1B). Sch9as was almost fully inhibited in vivo by 100 nM 1NM-PP1, as determined by effects on doubling time and cell size (Fig. 1B). At this concentration, 1NM-PP1 had no effect on wild-type cells (data not shown) (Bishop et al. 2000).
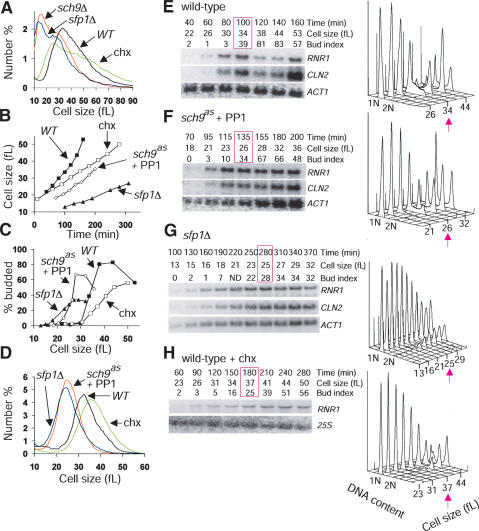
Figure 1.
Sch9 and Sfp1 regulate cell size and RP and Ribi regulon transcription. (A) Galactose-regulated alleles of SFP1 and SCH9. (B) Inactivation of an analog-sensitive (as) allele of SCH9 by 1NM-PP1. A total of 100 nM 1NM-PP1 or DMSO solvent was added at 0 min to log phase, glucose cultures of sch9as and sch9Δ. Cell-size distributions were measured after 6 h and compared with reference distributions. Without 1NM-PP1, the sch9as allele is hypomorphic, with nearly wild-type doubling time (td), but a strong Whi phenotype (td: 96 min, median cell size: 30 fL, vs. 87 min, 42 fL for wild type and 153 min, 28 fL for sch9Δ). (C) Sch9 and Sfp1 activate the RP and Ribi regulons. Expression profiles for GAL1–SFP1 and sfp1Δ/Δ were repetitions of experiments described previously (Jorgensen et al. 2002). Expression profiles were determined for sch9as and wild-type cultures harvested at indicated times after addition of 100 nM 1NM-PP1; expression ratios are relative to untreated cells from the same culture. The presence of RAP1, PAC, RRPE, or PAC + RRPE (P+R) promoter elements is indicated (Pilpel et al. 2001; Jorgensen et al. 2002). (D) Induction of representative genes in the Ribi (NOP1, NSR1) and RP (RPPO, RPL11B) regulons upon restoration of GAL1–SCH9 or GAL1–SFP1 expression. An ACT1 loading control and GAL7, SFP1, and SCH9 induction controls are shown. (E) Comparison of gene sets regulated by Sfp1 and Sch9. Expression profiles of sfp1Δ/Δ and inactivated sch9as (1NM-PP1, 90 min) were plotted against one another. The number of genes in each functional group is indicated.
Sfp1 and Sch9 activate the RP and Ribi regulons
We have previously shown that Sfp1 activates the RP and Ribi regulons (Jorgensen et al. 2002; Fingerman et al. 2003). Further expression profiles of GAL1–SFP1 and sfp1Δ/Δ strains elaborated the Ribi regulon to 236 genes (Fig. 1C; Supplementary Table 1). We next asked whether Sch9 also controls these genes. Addition of 100 nM 1NM-PP1 resulted in a rapid repression (<30min) of the RP regulon (∼2.5-fold) and the Ribi regulon (∼1.5-fold) in an sch9as strain, followed by a slight but reproducible recovery of both regulons (Fig. 1C). These transcriptional effects preceded effects of 1NM-PP1 on proliferation rate and cell size (Fig. 1B). Northern analysis confirmed that representative members of the RP (RPP0, RPL11B) and Ribi (NOP1, NSR1) regulons were induced upon expression of GAL1–SCH9 (Fig. 1D). Sfp1 and Sch9 are thus both required for maximal RP and Ribi expression. However, inhibition of sch9as also induced 38 genes not altered in sfp1Δ/Δ cells (Fig. 1E), almost all of which were strongly induced as cultures deplete glucose and enter stationary phase (DeRisi et al. 1997). Many stationary phase genes are also repressed by Ras/PKA, consistent with the genetic placement of Sch9 parallel to Ras/PKA (Toda et al. 1988).
Sfp1 and Sch9 are negative regulators of Start
A number of observations suggested that Sfp1 and Sch9 might regulate the critical cell size threshold at Start. First, sfp1Δ and sch9Δ cells are much smaller than other deletion mutants with the same proliferation rate (Jorgensen et al. 2002). Second, the hypomorphic alleles sfp1MYC13 (Jorgensen et al. 2002) and sch9as caused marked decreases in cell size with little increase in doubling time, similar to mutations in canonical Start regulators (Fig. 1A,B). Third, increasing the doubling time of wild-type cells to that of sfp1Δ or sch9Δ cells by treatment with sublethal doses of cycloheximide reduced the size of new-born daughter cells, but not average cell size (Fig. 2A). We directly tested the role of Sfp1 and Sch9 at Start by synchronous release of G1-phase daughter cells obtained by centrifugal elutriation. The slow rate at which daughter cells lacking Sfp1 or Sch9 activity accumulated volume underscored the role of these gene products in cell growth (Fig. 2B). All aspects of Start, including the activation of SBF- and MBF-dependent transcription, bud emergence, and DNA replication initiation were accelerated with respect to cell size in strains that lacked Sfp1 or Sch9 activity (Fig. 2C–G). For example, whereas wild-type cells expressed the MBF-regulated gene RNR1 at a size of ∼30 fL, cells that lack Sfp1 or Sch9 activity do so at sizes of ∼16 fL and ∼21 fL, respectively. These results indicate that Sfp1 and Sch9 lie upstream of the G1/S transcriptional machinery. In contrast, treatment of wild-type cells with a sublethal dose of cycloheximide that slowed growth to the same extent as loss of Sch9 actually delayed Start, as measured by all of the above parameters (Fig. 2B–D,H). We note that the absence of Sfp1, and to a lesser extent Sch9, caused considerable heterogeneity in traversal through Start, as shown by the extended window of G1/S transcription and the slow accumulation of cells with buds and G2 DNA content. This effect may arise from a reduced translation rate, as cycloheximide-treated wild-type G1 phase cultures behaved in a similar manner. The heterogeneity in Start progression is not, however, caused by heterogenous cell growth rate (Fig. 2D).
Figure 2.
Sfp1 and Sch9 are negative regulators of Start. (A) Size distributions of sfp1Δ and sch9Δ strains. Doubling times were as follows: sfp1Δ 220 ± 13 min, sch9Δ 153 ± 3 min, wild type (WT) 89 ± 2 min, wild type (WT) + 200 nM cycloheximide 149 ± 4 min. (B–H) Determination of critical cell size. Small G1-phase daughter cells (>97% unbudded) were isolated from late log-phase cultures (3–4 × 107 cells/mL, raffinose medium with no drugs) of wild-type, sfp1Δ and sch9as strains by centrifugal elutriation and reinoculated in glucose medium. A total of 100 nM 1NM-PP1 was added to the sch9as culture upon reinoculation; 200 nM cycloheximide was added to a wild-type culture upon reinoculation. (B) Daughter cell size at various times after reinoculation for each culture. (C) Bud index as a function of cell size for each culture. (D) Cell-size distributions at the >25% budded time point for each culture. Passage through Start was monitored by cell size, bud index, and expression of SBF (CLN2)- and MBF (RNR1)-specific transcripts for wild-type cells (E), sch9as cells in 100 nM 1NM-PP1 (F), sfp1Δ cells (G), and wild-type cells in 200 nM cycloheximide (H). The time point at which cultures were >25% budded is highlighted in pink. All data was reproduced in duplicate experiments. 1NM-PP1 (100 nM) had no effect on the critical cell size of a wild-type strain (data not shown).
Strains lacking SFP1 or SCH9 are impervious to carbon source effects on cell size
If Sfp1 and Sch9 mediate the effects of nutrients on the critical cell size threshold, strains lacking these gene products should be largely impervious to nutrient modulation of cell size. The size distributions of sfp1Δ/Δ and sch9Δ/Δ diploid cells were, in fact, barely reduced on poor carbon sources, unlike wild-type and whi5Δ/Δ cells (Fig. 3A,B). sfp1Δ/Δ cells appeared to be at near minimal cell size, as overexpression of the hyperactive CLN3-1 allele had little impact on the mean size of these cells (Fig. 3B). Importantly, sfp1Δ and sch9Δ strains proliferated much more slowly than wild type on all carbon sources examined (data not shown), unlike strains that lack the large ribosomal subunit RPL11B gene, which is only rate-limiting for growth when cells are in glucose medium (Zhao et al. 2003).
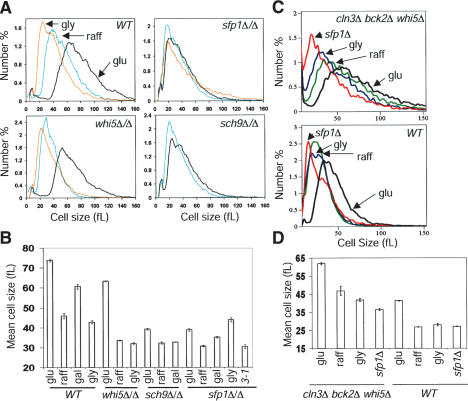
Figure 3.
Cells lacking SFP1 or SCH9 fail to adjust cell size in response to carbon source. (A) Representative size distributions of log-phase cultures of the indicated strains in synthetic glucose (black), raffinose (blue), or glycerol (orange) medium. (B) Mean cell sizes of the indicated cultures (n = 6, range: 10–180 fL). Error bars extend one S.E. in each direction. The sfp1Δ/Δ GAL1–CLN3-1 (3-1) strain was propagated in synthetic galactose medium. (C) Nutrient and SFP1-dependent control of size in cells that lack known upstream regulators of SBF/MBF. Wild-type and cln3Δ bck2Δ whi5Δ strains were in rich glucose, raffinose, or glycerol medium. The sfp1Δ and sfp1Δ cln3Δ bck2Δ whi5Δ strains were in rich glucose medium. (D) Mean cell sizes of the indicated cultures (n ≥ 2, range: 10–180 fL). Error bars extend one S.E. in each direction.
In contrast to sfp1Δ and sch9Δ cells, the size of cln3Δ bck2Δ whi5Δ cells was fully responsive to carbon source (Fig. 3C,D). Introduction of sfp1Δ into the cln3Δ bck2Δ whi5Δ background rendered cells very small, but not as small as the sfp1Δ mutation alone (Fig. 3C,D). Because sfp1Δ is not fully epistatic for cell size to the triple cln3Δ bck2Δ whi5Δ mutant or to any of the single mutants (data not shown), Whi5, Cln3, and Bck2 must still play a role in sfp1Δ cells. These results argue that nutrient signaling, via Sfp1 and Sch9, influences the activation of SBF and MBF by a mechanism independent of known upstream regulators.
Sch9 abundance and phosphorylation are altered by nutrient signals
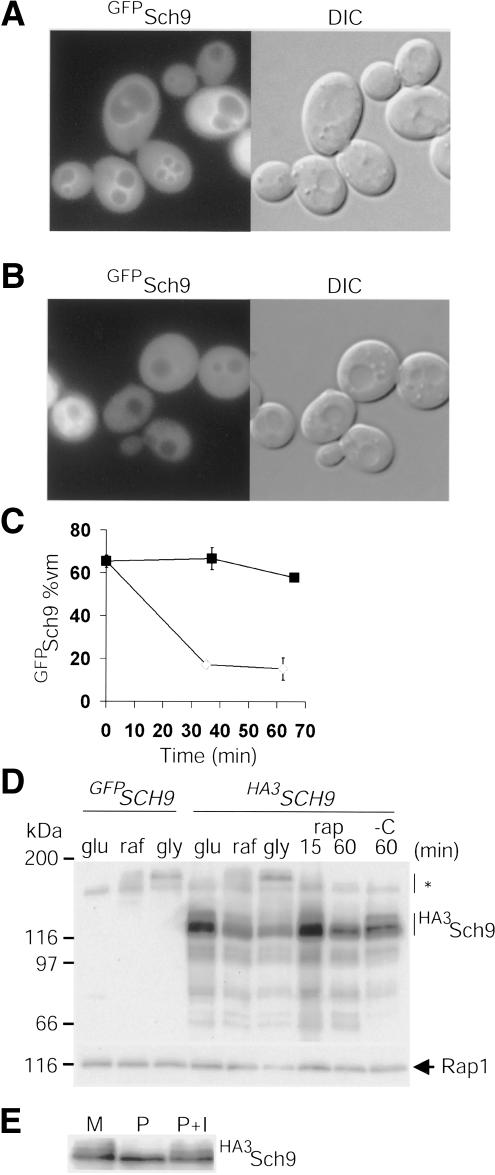
As the RP and Ribi regulons respond dynamically to the environment, we examined Sfp1 and Sch9 abundance and localization under different nutrient and stress conditions using functional fusions to epitope tags and GFP variants. In log phase, GFPSch9 localized throughout the cell, but was unexpectedly enriched at the vacuolar membrane (Fig. 4A). We note that a C-terminal Sch9GFP fusion protein was not functional (data not shown). GFPSch9 localization was dynamic, as the vacuolar membrane signal largely disappeared following carbon starvation (Fig. 4B,C). GFPSch9 localization was not altered by different carbon sources or by rapamycin (data not shown). The abundance of HA3Sch9 was, however, dependent on TOR activity as all forms, and in particular HA3Sch9 phospho-isoforms, decreased upon rapamycin treatment (Fig. 4D,E). Under steady-state proliferation on different carbon sources, HA3Sch9 abundance correlated with cell growth rate, RP/Ribi transcription, and cell size (Fig. 4D) (Gasch et al. 2000).
Figure 4.
Sch9 abundance and localization is modulated by nutrients. (A) Enrichment of GFPSch9 at the vacuolar membrane (vm). (B) Depletion of GFPSch9 from the vm after carbon starvation (37 min). (C) Kinetics of GFPSch9 depletion from the vm. (Closed squares) +Glucose; (open diamonds) -glucose. Error bars extend one S.E. in each direction. (D) HA3Sch9 abundance and phosphorylation status are altered by nutrient conditions. Cells that expressed HA3SCH9 from the SCH9 promoter were propagated in rich glucose, raffinose, or glycerol medium. Cells propagated in glucose medium were treated with rapamycin (rap, 200 ng/mL) or carbon starved (-C). HA3Sch9 was visualized by immunoblot with anti-HA antibody. GFPSch9 served as a no-tag control. Asterisk indicates nonspecific cross-reactive species. (E) Sch9 is a phosphoprotein. HA3Sch9 expressed at endogenous levels was immunoprecipitated with anti-HA antibody and either mock treated (M), treated with alkaline phosphatase (P), or treated with alkaline phosphatase in the presence of phosphatase inhibitors (P+I) and detected by anti-HA immunoblotting.
Sfp1 nuclear concentration is regulated by multiple nutrient and stress signals
As expected for a transcription factor, Sfp1YFP was localized predominantly to the nucleus in growing cells (Fig. 5A). Strikingly, Sfp1YFP rapidly (∼5 min) relocalized to the cytoplasm upon exposure to a broad range of stresses. Cytoplasmic relocalization was also observed with a GFPSfp1 fusion protein (data not shown). Quantitation of the Sfp1YFP nuclear:cytoplasmic (N:C) ratio revealed that under some conditions (carbon starvation, oxidative stress), Sfp1YFP became uniformly distributed (i.e., an N:C ratio of unity), whereas in others (rapamycin, tunicamycin, nitrogen starvation), a greater fraction of Sfp1YFP was retained in the nucleus (Fig. 5A,B). Neither the abundance nor the electrophoretic mobility of Sfp1MYC13 was altered upon carbon starvation or rapamycin treatment (Fig. 5C). Sfp1YFP relocalization correlated temporally with RP/Ribi repression as carbon starvation, nitrogen starvation, rapamycin, and H202 treatment all provoked rapid Sfp1YFP export and RP/Ribi gene repression (Fig. 5B,D). Because most RP and Ribi transcripts are highly unstable, their abundance precisely mirrors transcription rate (Warner 1999; Grigull et al. 2004). An exception to this rapid response was secretory pathway stress induced by tunicamycin, which is known to gradually inhibit RP gene expression by a PKC-dependent pathway (Li et al. 2000). Correspondingly, tunicamycin caused a gradual decline in both nuclear Sfp1YFP and RP/Ribi transcription (Fig. 5B) (Miyoshi et al. 2003). In log phase proliferation on different carbon sources, the extent of Sfp1YFP nuclear localization was also proportional to cell-growth rate, RP/Ribi expression, and cell size (Fig. 5E) (Gasch et al. 2000).
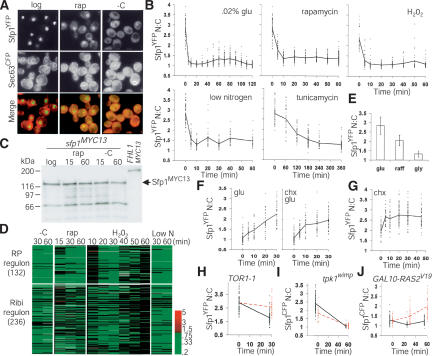
Figure 5.
Sfp1 nuclear concentration responds rapidly to environmental stimuli and is regulated by the Ras/PKA and TOR signaling pathways. (A) Sfp1 localization is regulated by nutrient signals. An SFP1YFP SEC63CFP strain was visualized; cell and nuclear membranes were demarcated by Sec63CFP. Carbon starvation (-C) was for 15 min, and rapamycin treatment (rap, 200 ng/mL) for 20 min. (B) Sfp1 exits the nucleus in response to various stress conditions. An SFP1YFP SEC63CFP strain was depleted for carbon or nitrogen or treated with H202 (0.30 mM), rapamycin (200 ng/mL), or tunicamycin (2 μg/mL). The Sfp1YFP nuclear:cytoplasmic ratio (N:C) of each cell at each time point was plotted (dashes, 30–100 cells), as was the average ratio (thick line). (C) Sfp1 abundance and electrophoretic mobility are not altered by carbon starvation (-C) or rapamycin (rap, 200 ng/mL) treatment. Cells were harvested after the indicated time (min). Sfp1MYC13 in cell lysates was visualized by immunoblot with anti-MYC antibody. Fhl1MYC13 served as a specificity control. (D) Sfp1 relocalization correlates withre-pression of the RP and Ribi regulons. Genome-wide expression profiles comparing mRNA abundance before and after carbon starvation (-C) were obtained de novo. Expression profiles for rapamycin addition (rap), oxidative stress (H202), and nitrogen starvation (-N) were derived from published data (Hardwick et al. 1999; Gasch et al. 2000). Scale indicates fold change. (E) Nuclear localization of Sfp1 in different carbon sources. The average Sfp1YFP N:C ratio was determined under steady-state proliferation in glucose, raffinose, and glycerol medium. Error bars extend one S.D. in each direction. (F) Sfp1 re-enters the nucleus rapidly in response to glucose. Stationary phase SFP1YFP SEC63CFP cells were re-fed with glucose in the presence or absence of cycloheximide (chx, 10 μM) and Sfp1YFP N:C ratios determined. (G) Sfp1 may effect a feedback response to ribosome shortage. Sfp1YFP N:C ratio was measured in SFP1YFP SEC63CFP cells proliferating in raffinose medium before and after addition of chx (10 μM). (H) The rapamycin-resistant allele TOR1-1 blocks Sfp1 relocalization in response to rapamycin but not nitrogen starvation. A TOR1-1 SFP1YFP strain was treated with rapamycin (red dotted line, 200 ng/mL) or starved for nitrogen (black, offset for visualization) for 30 min. (I) Compromised Ras activity lowers the nuclear concentration of Sfp1. The N:C ratio of Sfp1CFP was quantitated before and after 60 min of carbon starvation in a tpk1wimp strain (red dotted line, SFP1CFP tpk1wimp bcy1Δ tpk2Δ tpk3Δ) and in a control strain (black line, SFP1CFP, offset for visualization). At t = 0 min, the difference between the wild-type and wimp strain was significant (Student's t-test, p = 4.4 × 10-11). Due to higher cell autofluorescence, Sfp1CFP N:C ratios are less than Sfp1YFP N:C ratios. (J) Hyperactive Ras signaling drives Sfp1 into the nucleus. A GAL10–RAS2V19 SFP1CFP strain (red dotted line) and a control SFP1CFP strain (black, offset for visualization), proliferating in synthetic raffinose medium, were induced with galactose at t = 0, and visualized after 30 and 60 min. Both strains were deleted for GAL1 and are incapable of metabolizing galactose.
Importantly, Sfp1 localization responded quickly to improved nutrient conditions. Addition of glucose to stationary phase cells rapidly (∼5 min) increased the nuclear concentration of Sfp1YFP (Fig. 5F). This nuclear import of Sfp1YFP occurred even in the presence of a lethal concentration of cycloheximide, and so did not require protein synthesis (Fig. 5F). In fact, cycloheximide treatment of cells in suboptimal raffinose medium rapidly increased the nuclear concentration of Sfp1YFP, but had no effect on Ste12YFP, an unrelated transcription factor (Fig. 5G; data not shown). Sfp1 may thus effect the compensatory feedback loop that induces RP transcription within 15 min of cycloheximide treatment (Cardenas et al. 1999).
Sfp1 localization also depended on TOR and Ras/PKA activity. The rapamycin-resistant TOR1-1 allele blocked Sfp1YFP nuclear depletion in response to rapamycin (Fig. 5H). However, the partial nuclear retention of Sfp1YFP after rapamycin treatment of wild-type cells argues that repression of the TOR pathway does not suffice to explain the complete relocalization upon carbon starvation (Fig. 5B). A tpk1wimp strain, which has weak constitutive PKA activity (Cameron et al. 1988), had reduced nuclear Sfp1CFP (Fig. 5I), whereas overexpression of a hyperactive RAS2Val19 allele stimulated nuclear accumulation of Sfp1CFP (Fig. 5J) and RP/Ribi induction with the same kinetics (Y. Wang et al. 2004). However, because a tpk1wimp strain still relocalized Sfp1CFP upon carbon starvation, diminished Ras/PKA signaling was also not sufficient to account for the totality of Sfp1CFP export (Fig. 5I). Likewise, a high level of Ras/PKA activity caused by either of the hyperactive GAL10–RAS2Val19 or GPA2Ala273 alleles could not drive Sfp1 into the nucleus of carbon-starved cells (data not shown). Deletion of Snf1, an AMP-activated kinase that signals many stress responses, also had no effect on Sfp1 relocalization upon carbon starvation (data not shown). Multiple signals must therefore regulate nuclear localization of Sfp1. We note that Sfp1 cannot completely account for the pronounced effect of TOR and Ras/PKA signaling on RP transcription, as loss of Sfp1 activity causes an approximately threefold repression of RP genes as opposed to the >10-fold decreases caused by loss of TOR or PKA activity (Fig. 1C) (Neuman-Silberberg et al. 1995; Powers and Walter 1999).
Sfp1 and Sch9 act in parallel
Consistent with the above Sfp1 localization results, sfp1Δ strains are sensitive to cycloheximide (Fingerman et al. 2003) and to decreases in Ras/PKA and TOR signaling (Fig. 6A), arguing that these pathways regulate additional activators of RP and Ribi expression. The incomplete overlap in expression profiles (Fig. 1E), lack of genetic epistasis (Fig. 6B), and the inviability of the sfp1Δ sch9Δ double mutant indicate that Sfp1 and Sch9 act in parallel pathways (Jorgensen et al. 2002). Consistently, inhibition of sch9as did not affect the localization of Sfp1CFP (data not shown). However, because both sfp1Δ and sch9Δ strains have substantial growth defects, the inviability of the double mutant might simply reflect nonspecific additive growth defects. To precisely modulate Sfp1 function, we constructed an sfp1ER allele in which the genomic SFP1 sequence is fused to the ligand-binding domain of the estrogen receptor (ER). In the absence of the natural ligand β-estradiol, the ER domain is sequestered in an inactive complex with Hsp90 (Mattioni et al. 1994). The activity of Sfp1ER in vivo exhibits a smooth dose dependence on β-estradiol concentration (Fig. 6C). To quantitatively assess genetic synergy, an sfp1ER sch9as double-mutant strain was challenged with increasing concentrations of β-estradiol and 1NM-PP1, in effect applying overlapping gradients of Sfp1 and Sch9 activity. Severe proliferation defects were observed at combined concentrations of β-estradiol and 1NM-PP1 that individually caused little defect, consistent with a parallel role for each in RP/Ribi transcription (Fig. 6D). As Sfp1 and Sch9 both appear to be downstream of TOR signaling (Figs. 4D, 5A,H), their combined action may account for the TOR requirement in RP/Ribi gene expression.
Figure 6.
Sfp1 and Sch9 function in parallel pathways. (A) sfp1Δ cells are sensitive to cycloheximide and to decreases in TOR or PKA pathway activity. (Left) Filter disks containing 3 nmole cycloheximide or 15 μg rapamycin were incubated on lawns of the indicated strains for 2 d. (Right) Synthetic proliferation defects between ras2Δ and sfp1Δ. Spore clones were scored after 4 d. (B) Sch9 does not control cell size strictly via Sfp1. Size distributions of an sfp1Δ/Δ strain in the presence or absence of a functionally null heterozygous GAL1–SCH9 (G9/+) allele are shown. Mean cell sizes of the indicated strains in either rich glucose or galactose medium are indicated to the right (n = 4). (C) An sfp1ER allele is modulated by β-estradiol (E2). Size distributions of log phase sfp1ER cultures with or without 250 nM E2 (left) or in the presence of various E2 concentrations (right). (D) Progressively compromised SCH9 and SFP1 activity reveals synergistic proliferation defects. An sch9as sfp1ER strain was inoculated into varying concentrations of 1NM-PP1 and E2 in synthetic glucose medium. Doubling times (td) were determined for each culture and the increase relative to cultures proliferating in 6.25 nM 1NM-PP1 and 125 nM E2 (concentrations at which Sch9as and Sfp1ER were fully active) was calculated. (Top) The increase in doubling time (Δtd) resulting from individually increasing 1NM-PP1 or decreasing E2 was used to calculate the predicted additive effects. Actual increases in doubling time (middle) and the difference (bottom) are plotted.
Sfp1 appears to act at RP promoters via Fhl1 and Ifh1
Genome-wide chromatin immunoprecipitation (ChIP) analysis of many potential transcriptional regulators in yeast, including Sfp1, has been recently reported (Lee et al. 2002). Of the 211 intergenic regions most enriched in Sfp1 complexes at p < 0.02, 42 corresponded to RP genes, a proportion not due to chance (p < 10-14). But, the less than twofold promoter enrichment in the reported ChIP data for Sfp1 (Lee et al. 2002), and our own ChIP analysis (data not shown), suggests that the Sfp1–RP promoter interaction is at the limit of detection. Sfp1–Ribi promoter interactions have not yet been detected by ChIP (Lee et al. 2002; Fingerman et al. 2003; P. Jorgensen and M. Tyers, unpubl.). To determine how Sfp1 and Sch9 might regulate RP promoters, we examined two novel transcription factors that also bind to these promoters, the forkhead/FHA domain protein Fhl1 and the zinc-finger protein Rgm1 (Lee et al. 2002). Fhl1 and a genetically interacting factor, Ifh1, are required for an unknown aspect of ribosome synthesis, suggesting that they may be activators of RP transcription (Hermann-Le Denmat et al. 1994; Cherel and Thuriaux 1995).
We detected several strong genetic interactions within this network of putative RP gene regulators. In light of the severe proliferation defects of sfp1Δ and fhl1Δ strains, we were surprised to find that a sfp1Δ fhl1Δ double-mutant strain proliferated as well as a fhl1Δ strain; in contrast, sch9Δ caused additive defects with fhl1Δ (Fig. 7A). However, deletion of FHL1 did not cause an obvious cell size phenotype (Fig. 7B). The sfp1Δ size defect was epistatic to fhl1Δ as the sfp1Δ fhl1Δ double mutant was as small as the sfp1Δ single mutant (Fig. 7B). The control of Sfp1 over cell size may thus be distinct from its control over the RP regulon. Conversely, the colony size of an sch9Δ, but not an sfp1Δ strain was severely compromised by an allele of IFH1 (ifh1ER) and by rgm1Δ, neither of which alone caused noticeable defects (Fig. 7C; data not shown).
Figure 7.
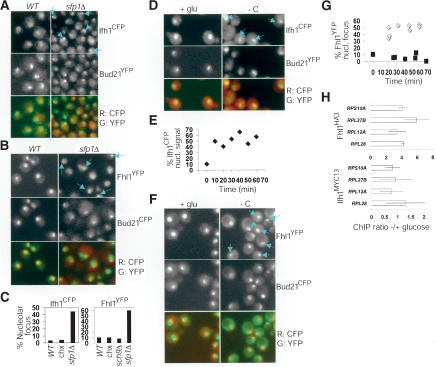
Sfp1 influences Fhl1 and Ifh1 interactions with RP promoters. (A) FHL1 deletion is epistatic to SFP1 deletion for colony size. Spore clones were imaged after 7 d (top) and 5 d (bottom). (B) FHL1 deletion is not epistatic to SFP1 deletion for cell size. Strains recently derived from a tetratype tetrad dissection (as in A) were sized in log phase in rich glucose medium. These representative distributions were highly reproducible. (C) Synthetic proliferation defects between sch9Δ and rgm1Δ and an allele of IFH1. Spore clones were imaged after 3 and 2 d, respectively. (D) Fhl1 and Ifh1 bind specifically to RP promoters. Real-time PCR was used to quantitate the efficiency with which the promoter regions of the indicated genes were captured in Fhl1HA3 (F7 anti-HA antibody) or Ifh1MYC13 (9E10 anti-MYC antibody) complexes. Binding to Ribi promoters (URA7, RPA190, NSR1) and control promoters (PGK1, ACT1) was not observed. (E) Ifh1MYC13 and Fhl1HA3 bind poorly to RP promoters in sfp1Δ cells. The sfp1Δ/wild-type ratio of ChIP efficiency for Ifh1MYC13 and Fhl1HA3 was calculated for two individual experiments. Error bars extend one S.E. in each direction.
These genetic interactions suggested that Sfp1 might influence RP transcription via Fh1l and Ifh1. To investigate Fhl1 and Ifh1 regulation, we created various C-terminal epitope tag or GFP variant fusions by integration of fusion cassettes at the genomic FHL1 and IFH1 loci, all of which provided full function in vivo. We first recapitulated the association of Fhl1HA3 with RP promoters by ChIP analysis; as controls, Fhl1 did not locate to another highly active promoter regulated by Rap1 (PGK1) or to Ribi promoters (Fig. 7D). We then determined that Ifh1MYC13 also bound specifically to RP promoters (Fig. 7D). The association of Ifh1MYC13 and Fhl1HA3 with RP promoters was reduced in sfp1Δ cells by approximately fourfold and approximately twofold, respectively (Fig. 7E). Neither the sfp1Δ mutation nor carbon starvation detectably altered Ifh1MYC13 or Fhl1HA3 abundance or electrophoretic mobility (data not shown).
Sfp1 dramatically influenced the localization of Ifh1 and Fhl1. In wild-type cells in glucose medium, Ifh1CFP and Fhl1YFP were predominantly nuclear, but excluded from the nucleolus (Fig. 8A,B). In sfp1Δ strains, however, a high percentage of cells showed striking enrichment of Ifh1CFP and Fhl1YFP in a subnuclear focus that corresponded to the nucleolus, as shown by colocalization with Bud21YFP or Bud21CFP (Fig. 8A,B). These foci were not observed in strains lacking Sch9 activity or after treatment with a sublethal concentration of cycloheximide (Fig. 8C; data not shown), indicating that reduced growth or translation rate was not sufficient to induce nucleolar relocalization. We then examined whether Ifh1CFP and Fhl1YFP changed localization in response to a nutrient stress that causes Sfp1 to exit the nucleus. Upon carbon starvation, Ifh1CFP rapidly infiltrated the nucleolus of most cells (Fig. 8D,E). Likewise, in most cells, almost all Fhl1YFP relocalized near the nucleolus after carbon-source depletion (Fig. 8F,G). As controls, two unrelated transcription factors, Ste12YFP and Rst1YFP, did not form foci in sfp1Δ cells or upon carbon starvation (data not shown). Fhl1YFP and Ifh1CFP nucleolar localization thus inversely correlated with the nuclear concentration of Sfp1. Surprisingly, in spite of the transcriptional repression of RP genes and Ifh1 and Fhl1 relocalization to the nucleolus, Ifh1 and Fhl1 were readily detected at RP promoters under conditions of carbon source limitation (Fig. 8H). The activity of Fhl1/Ifh1 may therefore be dictated by the nuclear environment of promoter regions.
Figure 8.
Ifh1 and Fhl1 localize to the nucleolus in cells lacking SFP1 and upon carbon starvation. (A) An sfp1Δ IFH1CFP BUD21YFP strain and a control IFH1CFP BUD21YFP strain were visualized in glucose medium. Blue arrows indicate cells with nucleolar foci of Ifh1CFP. (B) An sfp1Δ FHL1YFP BUD21CFP strain and a control FHL1YFP BUD21CFP strain were visualized in glucose medium. Blue arrows indicate cells with nucleolar foci of Fhl1YFP. Wild-type and sfp1Δ cells are not presented to scale to improve the visualization of Fhl1YFP in the small nuclei and nucleoli of sfp1Δ cells. (C) Quantitation of Ifh1CFP and Fhl1YFP nucleolar foci. CFP and YFP fluorescence was visualized in a single plane in the indicated strains proliferating in glucose medium. If a clear nucleolar (Bud21Y/CFP) crescent or dot was evident in this plane, it was determined whether or not an adjacent or overlapping focus of Ifh1CFP or Fhl1YFP was present. (chx) Cells in 400 nM cycloheximide. For each condition, >165 in focus nucleoli were scored. (D) Ifh1CFP often relocalizes to the nucleolus upon carbon starvation. Carbon starvation was for 25 min (-C). Blue arrows indicate cells with nucleolar foci of Ifh1CFP. (E) Ifh1CFP nucleolar relocalization is rapid and sustained. At the indicated times after carbon starvation, Ifh1CFP and Bud21YFP fluorescence was visualized in five planes and the percentage of cells with nucleolar Ifh1CFP fluorescence determined. In total, 290 cells were analyzed. (F) Fhl1 relocalizes to a perinucleolar focus upon carbon starvation. Carbon starvation was for 55 min (-C). Blue arrows indicate cells with nucleolar foci of Fhl1YFP. (G) Fhl1YFP perinucleolar relocalization upon carbon starvation is rapid and sustained. Cells were resuspended in synthetic medium with (black squares) or without (gray diamonds) glucose. Images were processed as in C. At least 240 in-focus nucleoli were examined for each time course. (H) Carbon starvation does not dissociate Ifh1 or Fhl1 from RP promoters. ChIP efficiency was determined in glucose medium and after 30 min of carbon starvation. Average ratio of -/+ glucose ChIP efficiency was determined for two individual experiments. Error bars extend one S.E. in each direction.
Discussion
An unexpected but fundamental connection has emerged between two ostensibly disparate systems, ribosome biogenesis and Start. Phenotypic identification of potential Start repressors isolated not only 15 ribosome biogenesis factors but, more significantly, upstream activators of the mRNA transcriptional program that sets the rate of ribosome synthesis, namely Sfp1 and Sch9 (Jorgensen et al. 2002). We have elaborated these initial connections to uncover a dynamic transcription factor network that responds to nutrient signals in part through altered localization of critical regulators. Other uncharacterized potential Start repressors may well participate in this emerging network that controls RP and Ribi expression (Jorgensen et al. 2002).
A dynamic transcriptional network at RP promoters
Sfp1 is a primary element in RP/Ribi regulation. Given that the Ribi regulon responds more rapidly than the RP regulon in response to Sfp1 (Jorgensen et al. 2002), it is puzzling that Sfp1 binds weakly to RP promoters, but apparently not to Ribi promoters (Lee et al. 2002; Fingerman et al. 2003). Nevertheless, Sfp1 activates Ribi transcription via the RRPE element (Fingerman et al. 2003). Consistent with a primary regulatory role, Sfp1 nuclear concentration responds within minutes to environmental conditions and appears to dictate the nuclear localization of Fhl1 and Ifh1, two novel regulators of RP promoters (Fig. 9A). Additional activators of the RP and Ribi regulons include the Sch9, TOR, and Ras/PKA pathways, all of which respond to nutrient signals. Sfp1 is downstream of TOR and Ras/PKA kinases, but the observed genetic redundancies indicate that these signaling pathways must modify multiple elements of the RP promoter network. Sfp1, Fhl1, and Rgm1 are likely to bind directly to RP promoter elements, such as the T-rich sequence or more recently described cis elements (Warner 1999; Pilpel et al. 2001; Beer and Tavazoie 2004), thereby placing the RP regulon under complex combinatorial control (Fig. 9A). As Rap1-binding sites mediate nearly all transcriptional activation at RP promoters (Warner 1999), it seems likely that most other components will subserve Rap1, perhaps by switching Rap1 between activation and repression modes. In support of this model, overexpression of the N terminus of IFH1 disrupts telomeric and mating-type silencing, both of which require Rap1 (Singer et al. 1998).
Figure 9.
(A) Initial model for the transcription system at RP promoters. Sfp1 localization is controlled by various signals and in turn influences the localization of Ifh1 and Fhl1. As suggested by synthetic lethal interactions, Sch9 modulates RP transcription by a pathway parallel to Sfp1/Fhl1/Ifh1/Rgm1, perhaps by phosphorylating Rap1. Sfp1, Fhl1, and Rgm1 likely bind to the promoter directly via still uncharacterized sequence elements (gray box) (Warner 1999; Pilpel et al. 2001; Beer and Tavazoie 2004). Ifh1 and Fhl1 may activate RP transcription by switching Rap1 (or Abf1) between intrinsic transcriptional activation (A) and repressive (R) functions. (B) Summary of transcription factor localization correlated with active and inactive (carbon starvation) RP transcription. Two possible nonmutually exclusive models explain the nucleolar relocalization of Ifh1 and Fhl1 upon carbon starvation. (Left) In the first model, unbound Ifh1 and Fhl1 are sequestered in or near the nucleolus (nucl), where they may have additional functions, such as repressing rRNA transcription. In the second model, Ifh1 and Fhl1 and repressed RP promoters are drawn into the nucleolar region upon carbon starvation. (C) A model of Start entry. Ribosome synthesis rates and ploidy establish the critical cell size threshold, which represses the SBF and MBF transcription factor complexes by an unknown mechanism. Cell size or a parameter that correlates with size, such as translation rate, signals to Cln3 and/or Bck2 to activate SBF and MBF. The model is complicated by the effects of ribosome synthesis on translation and by increased Cln3 abundance in rich nutrient conditions (dashed line).
The dynamic nature of the RP/Ribi control network is manifest at several levels. First, the ability of the two regulons to respond quickly to environmental conditions rests on adept transcriptional responses coupled with mRNA instability (Warner 1999; Grigull et al. 2004). Second, Sfp1 relocalization is remarkably responsive to the environment. As it is never completely excluded from the nucleus, the RP and Ribi promoters may be quite sensitive to the nuclear concentration of Sfp1, as might befit weak interactions of Sfp1 with RP promoters. Third, Fhl1 and Ifh1 are unexpectedly subject to nucleolar relocalization. Ifh1 and/or Fhl1 may potentially have dedicated nucleolar roles, such as repression of RNA Polymerase I and III, and thereby constitute an unsuspected line of communication between RP and rRNA transcription (Fig. 9B). In an alternate, but not mutually exclusive model, the nucleolar focus of Ifh1 and Fhl1 in sfp1Δ and carbon-starved cells may signify a cluster of repressed RP genes, as Ifh1 and Fhl1 continue to bind to RP promoters in these cells (Fig. 9B). Intriguingly, physical interactions have been found between Fhl1 and the rDNA-binding factor Hmo1 and between Ifh1 and the nucleolar protein Utp22 (data not shown), suggesting that Fhl1/Ifh1 may tether inactive RP genes to nucleolar structures. Spatial control at the level of gene compartmentalization is evident in nucleolar tRNA gene clusters in rapidly growing cells and in Rap1-dependent clustering of silenced loci at the nuclear periphery (Gasser 2001; Thompson et al. 2003). Furthermore, the relocalization of transcription factors appears to be a common feature in ribosome biogenesis as the limiting regulatory factor for rRNA transcription, Rrn3, as well as RNA PolI subunits, are released from the nucleolus upon rapamycin treatment (Tsang et al. 2003; Mayer et al. 2004).
Sch9 relays nutrient signals
An unexpected level of spatio-dynamic control has emerged with the localization of Sch9 to the vacuole. Despite its discovery as a high-copy suppressor of the Ras/PKA pathway in yeast (Toda et al. 1988), and its recently discovered roles in cell longevity and size (Fabrizio et al. 2001; Jorgensen et al. 2002), the presumed components that lie upstream and downstream of Sch9 have proven elusive. Budding yeast expresses two bona fide orthologs of the PDK1 kinase that activates Akt/PKB in metazoans, called Pkh1 and Pkh2, although these kinases are activated by sphingolipids, not phosphati-dylinositol trisphosphate (Casamayor et al. 1999). Pkh1 and Pkh2 activate Ypk1 and Ykr2, the yeast orthologs of the metazoan kinase SGK (Casamayor et al. 1999). As its sequence contains the predicted activating phosphorylation sites in the activation loop and hydrophobic motif, Sch9 may also be a Pkh1/2 substrate. As the vacuole is an important reservoir of amino acids, phosphate, and other metabolites, Sch9 may communicate the status of these internal nutrient pools to RP/Ribi transcription and Start. Elaboration of the Sch9 pathway in yeast may provide insights into the regulation of cell size by Akt/PKB in metazoans.
Ribosome biogenesis and nutrient modulation of critical cell size
On the basis of the above observations, we propose a refined model for nutrient modulation of the critical cell-size threshold at Start. Sfp1, Sch9, and Ras/PKA function in a nonlinear network that dictates both critical cell size and expression of the Ribi and RP regulons. Critical cell size at Start is decreased when any of these components is crippled, whereas either constitutive activation (Ras/PKA) or inactivation (Sfp1, Sch9) renders cell size impervious to carbon source control (Baroni et al. 1989; Tokiwa et al. 1994). Each component of this trio is needed for proper RP and Ribi gene expression (Klein and Struhl 1994; Neuman-Silberberg et al. 1995; Y. Wang et al. 2004). Like Ras/PKA signaling, we have found that Sfp1 and Sch9 are sensitive to nutrient conditions, at the level of localization and abundance, respectively. In addition, strains deleted for numerous genes implicated in the actual events of ribosome biogenesis, as well as ribosome structural genes, are similarly, if less dramatically, uncoupled for growth and division (Jorgensen et al. 2002). Finally, the cell size at which SBF/MBF activation, budding, and DNA replication initiation occur is diminished in cells proliferating in poor nutrients (Johnston et al. 1979; Lorincz and Carter 1979; Tyson et al. 1979; Stuart and Wittenberg 1995; Flick et al. 1998). All of these observations can be unified by a model in which nutrient control of the critical cell-size threshold at Start is communicated by rates of ribosome production (Fig. 9C).
In this model, nutrient status influences the Start machinery via proximal events in ribosome biogenesis, rather than by downstream changes in protein synthetic rate. This configuration would effectively anticipate future changes in translation rate and adjust the cell-size threshold accordingly. In support of this idea, upon shift of cells from ethanol to glucose medium, rates of rRNA transcription reach maximal levels within a few minutes, whereas protein synthetic rate does not achieve a maximum until nearly an hour after the shift (Kief and Warner 1981). Concordantly, upon nutrient shifts, cells adjust their critical cell-size threshold very rapidly (Lorincz and Carter 1979).
This model begs the question of how ribosome biogenesis might impinge on the Start machinery. As ribosome biogenesis factors implicated in size control lie along the entire pre-60S assembly pathway, the cell may monitor flux through this branch (Fatica and Tollervey 2002). Notably, inhibition of ribosome biogenesis by secretory defects also appears to proceed via the 60S branch (Zhao et al. 2003). Dissection of the ribosome biogenesis signal is complicated by redundancy among the many components, the existence of feedback loops, and its essential role in the cell. As G1/S transcription is greatly accelerated in cells lacking SFP1 or SCH9, it is possible that SBF/MBF are a target of the presumed signal. Other less-direct models in which Ras/PKA, Sch9, and Sfp1 impinge on a second common target might also be considered. For instance, decreasing the overall rate of RP and Ribi transcription in nutrient-limited cells might free up RNA Polymerase II and other core transcriptional regulators for recruitment by SBF/MBF, and presumably all other promoters (Thomas 2000).
Two mechanisms to explain nutrient modulation of critical cell size have been proposed previously, both of which invoke changes in the abundance of G1 cyclins (Baroni et al. 1994; Tokiwa et al. 1994; Polymenis and Schmidt 1997; Flick et al. 1998). However, such models do not account for all of the data presented here and elsewhere. Carbon source control of Start occurs in part via transcriptional regulation of CLN1, because cln1Δ strains fail to appropriately increase critical cell size when shifted from poor to glucose medium; moreover, glucose and cAMP repress CLN1 even relative to other G1/S transcripts like CLN2 (Tokiwa et al. 1994; Flick et al. 1998). However, in these studies, critical cell size at Start was inferred indirectly by bud emergence, rather than by measurement of SBF/MBF-dependent transcription. Because SBF/MBF activation is highly dependent on CLN3, but independent of CLN1 and CLN2 (Tyers et al. 1993; Dirick et al. 1995; Stuart and Wittenberg 1995), Cln1 likely controls the length of the Start interval rather than the timing of Start entry.
A second model postulates that Cln3 couples the critical cell size threshold to nutrients by virtue of Cln3 translational control and protein instability (Polymenis and Schmidt 1997; Hall et al. 1998). It is clear that carbon source controls critical cell size at the level of SBF/MBF activation, an effect that in principle might be transmitted through Cln3 (Stuart and Wittenberg 1995). As shown here, however, carbon source and SFP1 strongly impact cell size even in cells that lack the known upstream regulators of SBF/MBF, thereby obviating the Cln3-based model. Furthermore, the observation that in poor nutrients cells have very low levels of Cln3 and translational capacity yet pass Start at a small size, is also at odds with this model (Tokiwa 1995; Hall et al. 1998). That is, nutrient upshifts delay Start, despite increases in Cln3 abundance (Johnston et al. 1977; Lorincz and Carter 1979; Tokiwa et al. 1994). From this perspective, nutrient modulation of the critical cell-size threshold is quite remarkable, as not only must the yeast cell growing in poor nutrients pass Start with less translational capacity, but it must do so with much less Cln3. That is, in poor nutrients, less Cln–Cdc28 activity seems to be required to pass Start. Consistently, Start-defective cdc28-4 mutants at a semipermissive temperature cycle threefold faster in pyruvate medium than in glucose medium (Shuster 1982).
To synposize, a sharp distinction must be drawn between the critical cell-size threshold, which is altered by nutrients and ploidy, and the mechanism by which cells gage their size, perhaps through translation rate. A sizing role for translation rate is suggested by the increased critical size caused by sublethal doses of cycloheximide. Furthermore, even cells beyond the critical cell-size threshold require a minimum rate of protein synthesis before budding can occur (Moore 1988). Given these observations, the critical cell-size threshold and the critical translation-rate requirement may be one and the same. As argued above, the mechanisms that set this threshold may be entirely distinct from the processes that measure size.
Ribosome biogenesis and cell cycle control
Several recent findings suggest that primordial links between ribosome biogenesis and the cell cycle may couple growth and division in metazoan systems (Saucedo and Edgar 2002; Ruggero and Pandolfi 2003). As in yeast, disruption of ribosome biogenesis, but not the translational machinery, in flies causes a small cell-size phenotype (Montagne et al. 1999; Thomas 2000). In mice, blocking the synthesis of new ribosomes allows hepatocytes to grow, but not enter the cell cycle (Volarevic et al. 2000). Similarly, in human cells, overexpression of a dominant-negative version of a conserved ribosome biogenesis factor called Bop1 causes G1-phase arrest in a p53-dependent manner (Pestov et al. 2001). Many additional connections between p53 and the nucleolus have been unearthed (Ruggero and Pandolfi 2003). For example, the critical activator of p53, the Arf tumor suppressor, is a nucleolar protein and an inhibitor of ribosomal RNA processing (Sugimoto et al. 2003). It has recently been proposed that all stresses that stabilize p53 do so by disrupting the nucleolus (Rubbi and Milner 2003).
As in yeast, mechanisms exist in mammalian cells to ensure that ribosomal content can be doubled with each cell cycle. Delineation of the signals that emanate from ribosome biogenesis to the cell cycle machinery in yeast should thus illuminate analogous processes in metazoans as well as the evolutionary history of size control processes. Like Sfp1, Myc is a direct modulator of RP transcription and cell size (Eisenman 2001), whereas Sch9 and Akt are highly related kinases that regulate ribosome synthesis and cell size (Saucedo and Edgar 2002). Understanding these connections has been lent new urgency by the discovery of numerous links between ribosome biogenesis and cancer (Ruggero and Pandolfi 2003), including the recent demonstration that RP genes are haploinsufficient tumor suppressors in zebrafish (Amsterdam et al. 2004).
Materials and methods
Yeast strains, plasmids, and medium
Extensive descriptions of all methods are provided as Supplemental Material. Yeast culture and size analysis was as described previously (Jorgensen et al. 2002). All experiments were performed with log phase cells at O.D.600 < 0.5 and cell concentration <3 × 107 cells/mL to minimize repression of the Ribi and RP regulons by inadvertent nutrient depletion (Ju and Warner 1994; DeRisi et al. 1997). Strains used in this study are listed in Supplementary Table 2 and were typically generated by genomic integration of standard C-terminal tagging cassettes, including GFP variant cassettes obtained from the Yeast Resource Center (University of Washington, Seattle). Centrifugal elutriation, Northern analysis, genome-wide expression profiles, chromatin immunoprecipitation, and as allele construction were carried out as described (Tyers et al. 1993; Bishop et al. 2000; Jorgensen et al. 2002; Costanzo et al. 2004).
Quantitative microscopy
Live-cell microscopy was performed with an Eclipse E600FN microscope (Nikon) and an Orca II CCD camera (Hamamatsu). Cultures in synthetic medium were rapidly concentrated and immediately visualized to mitigate starvation effects. For quantitation of Sfp1YFP or Sfp1CFP nuclear:cytoplasmic ratio, five serial sections were taken of each cell field. Sec63CFP or Sec63YFP was used to delineate the nuclear envelope. Metamorph (Universal Imaging) was used to capture and quantitate the fluorescence signal from an identically sized region from the nucleus and the cytoplasm. To induce carbon starvation, cells were washed once and resuspended in synthetic medium with 0.02% glucose. For nitrogen starvation, cells were resuspended in synthetic glucose medium with 1/50th the standard concentration of amino acids and ammonium sulfate. Ifh1CFP and Fhl1YFP signals were visualized in the same manner, except that nucleolar regions were demarcated with Bud21YFP or Bud21CFP. Because allowing cells to remain concentrated for even short periods of time caused redistribution of Sfp1, Fhl1, and Ifh1, all results were confirmed with directly mounted dilute cultures.
Acknowledgments
We thank David Bellows, Jeanne Hirsch, Didier Picard, Charles Di Como, Peggy Baudouin-Cornu for reagents, and David Shore, Jon Warner, Erin O'Shea, Mike Hall, and Bruce Futcher for communicating unpublished results. This work was supported by NIH grant P01 Ca41086 to J.R.B., a Canadian Institutes of Health Research (CIHR) grant to M.T., a CIHR Doctoral Award to P.J., a Natural Sciences and Engineering Research Council Studentship to J.R.S., and a Canada Research Chair to M.T.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1228804.
References
- Amsterdam A., Sadler, K.C., Lai, K., Farrington, S., Bronson, R.T., Lees, J.A., and Hopkins, N. 2004. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2: 690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroni M.D., Martegani, E., Monti, P., and Alberghina, L. 1989. Cell size modulation by CDC25 and RAS2 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroni M.D., Monti, P., and Alberghina, L. 1994. Repression of growth-regulated G1 cyclin expression by cyclic AMP in budding yeast. Nature 371: 339-342. [DOI] [PubMed] [Google Scholar]
- Beer M.A. and Tavazoie, S. 2004. Predicting gene expression from sequence. Cell 117: 185-198. [DOI] [PubMed] [Google Scholar]
- Bishop A.C., Ubersax, J.A., Petsch, D.T., Matheos, D.P., Gray, N.S., Blethrow, J. Shimizu, E., Tsien, J.Z., Schultz, P.G., Rose, M.D., et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407: 395-401. [DOI] [PubMed] [Google Scholar]
- Cameron S., Levin, L., Zoller, M., and Wigler, M. 1988. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell 53: 555-566. [DOI] [PubMed] [Google Scholar]
- Cardenas M.E., Cutler, N.S., Lorenz, M.C., Di Como, C.J., and Heitman, J. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes & Dev. 13: 3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A., Torrance, P.D., Kobayashi, T., Thorner, J., and Alessi, D.R. 1999. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9: 186-197. [DOI] [PubMed] [Google Scholar]
- Cherel I. and Thuriaux, P. 1995. The IFH1 gene product interacts with a fork head protein in Saccharomyces cerevisiae. Yeast 11: 261-270. [DOI] [PubMed] [Google Scholar]
- Costanzo M., Nishikawa, J.L., Tang, X., Millman, J.S., Shub, O., Breitkreuz, K., Dewar, D., Rupes, I., Andrews B., and Tyers, M. 2004. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117: 899-913. [DOI] [PubMed] [Google Scholar]
- de Bruin R.A.M., McDonald, W.H., Kalashnikova, T.I., Yates III, J., and Wittenberg, C. 2004. Cln3 activates G1-specific transcription via phosphorylation of the SBF-bound repressor Whi5. Cell 117: 887-898. [DOI] [PubMed] [Google Scholar]
- DeRisi J.L., Iyer, V.R., and Brown, P.O. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680-686. [DOI] [PubMed] [Google Scholar]
- Dirick L., Bohm, T., and Nasmyth, K. 1995. Roles and regulation of Cln–Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 14: 4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman R.N. 2001. Deconstructing myc. Genes & Dev. 15: 2023-2030. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Pozza, F., Pletcher, S.D., Gendron, C.M., and Longo, V.D. 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292: 288-290. [DOI] [PubMed] [Google Scholar]
- Fatica A. and Tollervey, D. 2002. Making ribosomes. Curr. Opin. Cell. Biol. 14: 313-318. [DOI] [PubMed] [Google Scholar]
- Fingerman I., Nagaraj, V., Norris, D., and Vershon, A.K. 2003. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot. Cell 2: 1061-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick K., Chapman-Shimshoni, D., Stuart, D., Guaderrama, M., and Wittenberg, C. 1998. Regulation of cell size by glucose is exerted via repression of the CLN1 promoter. Mol. Cell. Biol. 18: 2492-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B. 1996. Cyclins and the wiring of the yeast cell cycle. Yeast 12: 1635-1646. [DOI] [PubMed] [Google Scholar]
- Gasch A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D., and Brown, P.O. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S.M. 2001. Positions of potential: Nuclear organization and gene expression. Cell 104: 639-642. [DOI] [PubMed] [Google Scholar]
- Grigull J., Mnaimneh, S., Pootoolal, J., Robinson, M.D., and Hughes, T.R. 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals post-transcriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 24: 5534-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. 2003. Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes & Dev. 17: 1691-1702. [DOI] [PubMed] [Google Scholar]
- Hall D.D., Markwardt, D.D., Parviz, F., and Heideman, W. 1998. Regulation of the Cln3–Cdc28 kinase by cAMP in Saccharomyces cerevisiae. EMBO J. 17: 4370-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick J.S., Kuruvilla, F.G., Tong, J.K., Shamji, A.F., and Schreiber, S.L. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. 96: 14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H. and Unger, M.W. 1977. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J. Cell Biol. 75: 422-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., Culotti, J., Pringle, J.R., and Reid, B.J. 1974. Genetic control of the cell division cycle in yeast. Science 183: 46-51. [DOI] [PubMed] [Google Scholar]
- Hermann-Le Denmat S., Werner, M., Sentenac, A., and Thuriaux, P. 1994. Suppression of yeast RNA polymerase III mutations by FHL1, a gene coding for a fork head protein involved in rRNA processing. Mol. Cell. Biol. 14: 2905-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.D., Estep, P.W., Tavazoie, S., and Church, G.M. 2000. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296: 1205-1214. [DOI] [PubMed] [Google Scholar]
- Johnston G.C., Pringle, J.R., and Hartwell, L.H. 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105: 79-98. [DOI] [PubMed] [Google Scholar]
- Johnston G.C., Ehrhardt, C.W., Lorincz, A., and Carter, B.L. 1979. Regulation of cell size in the yeast Saccharomyces cerevisiae. J. Bacteriol. 137: 1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Nishikawa, J.L., Breitkreutz, B.J., and Tyers, M. 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297: 395-400. [DOI] [PubMed] [Google Scholar]
- Ju Q. and Warner, J.R. 1994. Ribosome synthesis during the growth cycle of Saccharomyces cerevisiae. Yeast 10: 151-157. [DOI] [PubMed] [Google Scholar]
- Kief D.R. and Warner, J.R. 1981. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol. Cell. Biol. 1: 1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C. and Struhl, K. 1994. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating Rap1 transcriptional activity. Mol. Cell. Biol. 14: 1920-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Rinaldi, N.J., Robert, F., Odom, D.T., Bar-Jospeh, Z., Gerber, G.K., Hannett, N.M., Harbison, C.T., Thompson, C.N., Simon, I., et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799-804. [DOI] [PubMed] [Google Scholar]
- Li Y., Moir, R.D., Sethy-Coraci, I.K., Warner, J.R., and Willis, I.M. 2000. Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol. Cell. Biol. 20: 3843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz A. and Carter, B.L.A. 1979. Control of cell size at bud initiation in Saccharomyces cerevisiae. J. Gen. Microbiol. 113: 287-295. [Google Scholar]
- Mattioni T., Louvion, J.F., and Picard, D. 1994. Regulation of protein activities by fusion to steroid binding domains. Methods Cell Biol. 43 Pt A: 335-352. [DOI] [PubMed] [Google Scholar]
- Mayer C., Zhao, J., Yuan, X., and Grummt, I. 2004. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes & Dev. 18: 423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerny C.J., Partridge, J.F., Mikesell, G.E., Creemer, D.P., and Breeden, L.L. 1997. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes & Dev. 11: 1277-1288. [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Shirai, C., and Mizuta, K. 2003. Transcription of genes encoding trans-acting factors required for rRNA maturation/ribosomal subunit assembly is coordinately regulated with ribosomal protein genes and involves Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 31: 1969-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne J., Stewart, M.J., Stocker, H., Hafen, E., Kozma, S.C., and Thomas, G. 1999. Drosophila S6 kinase: A regulator of cell size. Science 285: 2126-2129. [DOI] [PubMed] [Google Scholar]
- Moore S.A. 1988. Kinetic evidence for a critical rate of protein synthesis in the Saccharomyces cerevisiae yeast cell cycle. J. Biol. Chem. 263: 9674-9681. [PubMed] [Google Scholar]
- Moss T. and Stefanovsky, V.Y. 2002. At the center of eukaryotic life. Cell 109: 545-548. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. 1996. At the heart of the budding yeast cell cycle. Trends Genet. 12: 405-412. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg F.S., Bhattacharya, S., and Broach, J.R. 1995. Nutrient availability and the RAS/cyclic AMP pathway both induce expression of ribosomal protein genes in Saccharomyces cerevisiae but by different mechanisms. Mol. Cell. Biol. 15: 3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov D.G., Strezoska, Z., and Lau, L.F. 2001. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: Effects of nucleolar protein Bop1 on G1/S transition. Mol. Cell. Biol. 21: 4246-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel Y., Sudarsanam, P., and Church, G.M. 2001. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat. Genet. 29: 153-159. [DOI] [PubMed] [Google Scholar]
- Polymenis M. and Schmidt, E.V. 1997. Coupling of cell division to cell growth by translational control of the G1 cyclin Cln3 in yeast. Genes & Dev. 11: 2522-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T. and Walter, P. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10: 987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi C.P. and Milner, J. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22: 6068-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D. and Pandolfi, P.P. 2003. Does the ribosome translate cancer? Nat. Rev. Cancer 3: 179-192. [DOI] [PubMed] [Google Scholar]
- Saucedo L.J. and Edgar, B.A. 2002. Why size matters: Altering cell size. Curr. Opin. Genet. Dev. 12: 565-571. [DOI] [PubMed] [Google Scholar]
- Shuster J.R. 1982. `Start' mutants of Saccharomyces cerevisiae are suppressed in carbon catabolite-derepressing medium. J. Bacteriol. 151: 1059-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M.S., Kahana, A., Wolf, A.J., Meisinger, L.L., Peterson, S.E., Goggin, C., Mahowald, M., and Gottschling, D.E. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150: 613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D. and Wittenberg, C. 1995. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes & Dev. 9: 2780-2794. [DOI] [PubMed] [Google Scholar]
- Sugimoto M., Kuo, M.-L., Roussel, M., and Sherr, C.J. 2003. Nucleolar arf tumour suppressor inhibits ribsomal RNA processing. Mol. Cell 11: 415-424. [DOI] [PubMed] [Google Scholar]
- Thomas G. 2000. An encore for ribosome biogenesis in the control of cell proliferation. Nat. Cell Biol. 2: E71-E72. [DOI] [PubMed] [Google Scholar]
- Thompson M., Haeusler, R.A., Good, P.D., and Engelke, D.R. 2003. Nucleolar clustering of dispersed tRNA genes. Science 302: 1399-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Cameron, S., Sass, P., and Wigler, M. 1988. SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes & Dev. 2: 517-527. [DOI] [PubMed] [Google Scholar]
- Tokiwa G. 1995. “Cell cycle control in Saccharomyces cerevisiae: G1 cyclin regulation and a connection with the Ras/cAMP signaling pathway.” Ph.D. thesis, State University of New York, Stony Brook, NY.
- Tokiwa G., Tyers, M., Volpe, T., and Futcher, B. 1994. Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature 371: 342-345. [DOI] [PubMed] [Google Scholar]
- Tsang C.K., Bertram, P.G., Ai, W., Drenan, R., and Zheng, X.F. 2003. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 22: 6045-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M., Tokiwa, G., and Futcher, B. 1993. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12: 1955-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson C.B., Lord, P.G., and Wheals, A.E. 1979. Dependency of size of Saccharomyces cerevisiae cells on growth rate. J. Bacteriol. 138: 92-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volarevic S., Stewart, M.J., Ledermann, B., Zilberman, F., Terracciano, L., Montini, E., Grompe, M., Kozma, S.C., and Thomas, G. 2000. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 288: 2045-2047. [DOI] [PubMed] [Google Scholar]
- Wade C., Shea, K.A., Jensen, R.V., and McAlear, M.A. 2001. EBP2 is a member of the yeast RRB regulon, a transcriptionally coregulated set of genes that are required for ribosome and rRNA biosynthesis. Mol. Cell. Biol. 21: 8638-8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Gari, E., Verges, E., Gallego, C., and Aldea, M. 2004. Recruitment of Cdc28 by Whi3 restricts nuclear accumulation of the G1 cyclin–CDK complex to late G1. EMBO J. 23: 180-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Pierce, M., Schneper, L., Guldal, C.G., Zhang, X., Tavazoie, S., and Broach, J.R. 2004. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLOS Biol. 2: 610-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J.R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437-440. [DOI] [PubMed] [Google Scholar]
- Wijnen H. and Futcher, B. 1999. Genetic analysis of the shared role of CLN3 and BCK2 at the G1–S transition in Saccharomyces cerevisiae. Genetics 153: 1131-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Schneider, C., Ottmers, L., Rodriguez, R., Day, A., Markwardt, J., and Schneider, B.L. 2002. Genomic scale mutant hunt identifies cell size homeostasis genes in S. cerevisiae. Curr. Biol. 12: 1992-2001. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Sohn, J.H., and Warner, J.R. 2003. Autoregulation in the biosynthesis of ribosomes. Mol. Cell. Biol. 23: 699-707. [DOI] [PMC free article] [PubMed] [Google Scholar]