Abstract
Glucocorticoid receptor (GR)-mediated transrepression of the transcription factors AP-1 and NF-κB, responsible for most of the anti-inflammatory effects of glucocorticoids, is initiated by the tethering of GR to the promoters of target genes. We report that this tethering is mediated by a nuclear isoform of the focal adhesion LIM domain protein Trip6. Trip6 functions as a coactivator for both AP-1 and NF-κB. As shown by chromatin immunoprecipitation, Trip6 is recruited to the promoters of target genes together with AP-1 or NF-κB. In the presence of glucocorticoids, GR joins the Trip6 complex. Reducing the level of Trip6 by RNA interference or abolishing its interaction with GR by dominant-negative mutation eliminates transrepression. We propose that GR tethering to the target promoter through Trip6 forms the basis of transrepression, and that Trip6 exerts its nuclear functions by acting as a molecular platform, enabling target promoters to integrate activating or repressing signals.
Keywords: Glucocorticoids, LIM domains, transactivation, repression, chromatin immunoprecipitation
Extracellular conditions trigger not only the activation of the multicomponent intracellular signaling network, but, at the same time, induce in-built mechanisms that limit the reaction. Such brake mechanisms are absolutely essential to maintain homeostasis. They can be cell-autonomous or involve organismic regulatory loops, and can be established at any level of the signaling network. For instance, an excessive inflammatory response is counteracted by the systemic release of glucocorticoids (Barnes 1998). Proinflammatory stimuli trigger, at the cellular level, specific gene expression. Glucocorticoids interfere with either the signal transduction pathways (Caelles et al. 1997; Kassel et al. 2001), with transcriptional (Jonat et al. 1990; Schüle et al. 1990; Yang-Yen et al. 1990) or even posttranscriptional processes (Boggaram et al. 1991; Amano et al. 1993). Control at the level of transcription is particularly intriguing. Members of the nuclear receptor family, upon hormone binding, inhibit the action of transcription factors involved in proliferative and pro-inflammatory responses, such as AP-1, NF-κB, CREB, and others (for reviews, see Beato et al. 1995; Göttlicher et al. 1998; Herrlich 2001; Karin and Chang 2001). Conversely, these latter transcription factors can interfere with the action of nuclear receptors, which are mostly involved in differentiation processes. The cross-talk is not necessarily mutually inhibitory. Yet ill-defined DNA–protein and protein–protein interaction conditions can lead to synergism (for review, see Lefstin and Yamamoto 1998). We focus here on a prototypical example of a negative cross-talk: the transrepression by the glucocorticoid receptor (GR) of AP-1 (Jonat et al. 1990; Schüle et al. 1990; Yang-Yen et al. 1990) and NF-κB activity (Ray and Prefontaine 1994; Scheinman et al. 1995).
The ability to modulate AP-1 is shared by most of the nuclear receptors, and requires their activation by lipophilic ligands, for example, steroid hormones, retinoic acid (RA), thyroid hormone (T3), or vitamin D. As transcription factors themselves, the liganded nuclear receptors select specific promoter elements where they assemble multiprotein complexes, including coactivators and corepressors (for reviews, see McKenna et al. 1999; Xu et al. 1999; Dilworth and Chambon 2001). Their ability to interfere with the action of AP-1 or NF-κB has been dissociated from their promoter-activating transcription factor function by mutation (Heck et al. 1994; Helmberg et al. 1995) or choice of (nonphysiologic) ligand (Heck et al. 1994; Vayssiere et al. 1997). In a prominent example, point mutations in the D-loop of GR prevent GR dimerization and DNA binding and thereby abolish most of the GR-dependent transcription of glucocorticoid-responsive genes. However, D-loop mutations do not affect the transrepression of AP-1 or NF-κB (Heck et al. 1994; Reichardt et al. 1998, 2001). DNA binding by the GR is thus dispensable for transrepression. Given that GR represses AP-1 or NF-κB while these factors are already bound to their promoter elements (König et al. 1992; Nissen and Yamamoto 2000; Rogatsky et al. 2001), a first step in transrepression may be the tethering of GR to AP-1 or NF-κB at the promoter of target genes. Several pieces of evidence suggest that the binding of GR to the transcription factors it represses is indirect, and that other factors are necessary for GR to repress AP-1 and NF-κB (discussed in Herrlich 2001). This idea prompted us to identify factors interacting with GR and potentially required for GR-mediated transrepression.
We have performed yeast two-hybrid screens using as a bait a GR mutant competent for transrepression but defective in transactivation, and have repeatedly isolated Trip6, an LIM domain protein belonging to the zyxin family involved in focal adhesion (Lee et al. 1995; Yi and Beckerle 1998; Wang et al. 1999). A nuclear isoform of Trip6, nTrip6, interacts in vitro and in vivo with both GR and the repressed transcription factors AP-1 and NF-κB. It acts as a coactivator for AP-1 and NF-κB, but, upon GR binding, it mediates transrepression. nTrip6 represents a novel class of chromatin-associated proteins that assemble multiple positive or negative factors to promoter-bound transcription factors.
Results
Identification by two-hybrid screen of putative cofactors for transrepression
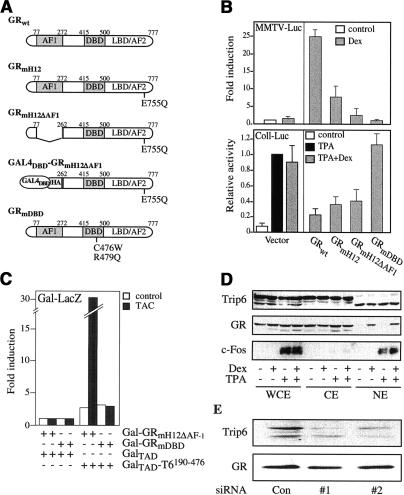
A two-hybrid screen was performed to isolate GR-interacting proteins that might be involved specifically in the transrepression by GR. For this purpose, the GR bait had to fulfill the following criteria: (1) the bait should be competent to repress AP-1; (2) it should be sensitive to hormone because transrepression is hormone-dependent; and (3) it should be defective in the transactivation function of GR to avoid isolation of GR-associating coactivators and direct activation of the reporter gene. Our preliminary studies showed that sequences N-terminal to the DNA-binding domain (DBD) are necessary, but that the transactivating domains (TADs) of the GR are not required for the repression of AP-1 (Jonat et al. 1990; E. Wade and S. Heck, unpubl.). To confirm this latter result, we disrupted the ligand-dependent TAD AF2 (Renaud et al. 1995; Moras and Gronemeyer 1998) by introducing a point mutation (E755Q) into helix 12 of the ligand-binding domain (LBD), termed GRmH12 (Fig. 1A), and by deleting the TAD AF1 from position 77 to 262 (Fig. 1A). The resulting mutant, GRmH12ΔAF1, was predicted to be defective in transactivation but still active in transrepression. In GR-negative Cos7 cells, GRmH12ΔAF1 could indeed not transactivate the prototypic GRE-containing MMTV promoter construct (MMTV-Luc), whereas it efficiently repressed in a hormone-dependent manner the AP-1-dependent collagenase I promoter (Coll-Luc) activated by phorbol ester (Fig. 1B, TPA). For use as a bait in the two-hybrid screen, the first 77 N-terminal residues of GRmH12ΔAF1, which are irrelevant for transrepression, were replaced by an HA tag and the GAL4-DBD (GAL4DBD–GRmH12ΔAF1) (Fig. 1A).
Figure 1.
Identification of a new GR-interacting protein: Trip6. (A) Schematic representation of wild-type (wt) GR and GR mutants. GAL4DBD–GRmH12ΔAF1 served as a bait in the two-hybrid screen. (AF) Activation function; (DBD) DNA-binding domain; (LBD) ligand-binding domain. (B) Transactivation and transrepression potential of the GR mutants shown in A. (Top) For transactivation, Cos7 cells were cotransfected with MMTV-luciferase reporter, the plasmids indicated, and treated as labeled. Normalized luciferase activities are plotted as fold induction (mean ± S.D. of three independent experiments). (Bottom) For transrepression, Cos7 cells were cotransfected with –517/+63-CollagenaseI-Luciferase reporter, the plasmids indicated, and treated as labeled. Normalized luciferase activities (mean ± S.D. of three independent experiments) are plotted relative to the value obtained in TPA-induced/Dex-untreated cells transfected with the empty expression vector. (C) Two-hybrid interaction of GR and Trip6. Y190 yeasts were transformed with the indicated plasmids. The β-galactosidase activity was measured after overnight incubation with vehicle alone or triamcinolone acetonide (TAC). (D) Subcellular localization of Trip6. HeLa cells were treated with TPA and dexamethasone (Dex) as indicated, and whole-cell extracts (WCE), cytoplasmic extracts (CE), and nuclear extracts (NE) were subjected to Western blotting using an anti-Trip6, an anti-c-Fos, and an anti-GR Ab. (E) Down-modulation of Trip6 by siRNA. HeLa cells were transfected with either a control siRNA (Con), or two different siRNAs targeting different sequences in Trip6 mRNA (#1 and #2). Whole-cell lysates were subjected to Western blotting using an anti-Trip6 and an anti-GR Ab.
To discriminate in our screen against interacting proteins that might have no function in transrepression, we counter-selected with the mutant GRmDBD (Fig. 1A, C476W, R479Q) defective in both transactivation and transrepression (Fig. 1B; Heck et al. 1994).
An HeLa cell cDNA library fused to Gal4–TAD was screened for hormone-dependent interaction with the fusion protein GAL4DBD–GRmH12ΔAF1 in Y190 yeast cells. Independent overlapping sequences of one clone were isolated more than 10 times. All isolates of this clone did not interact with the GAL4DBD fusion of GRmDBD, as illustrated for the longest isolate (Fig. 1C). This isolate was identical to part of a previously reported sequence, Trip6. Trip6 has first been isolated as a protein of unknown function interacting with both the thyroid hormone (TR) and the retinoid X (RXR) receptors (Lee et al. 1995). Trip6 is 476 amino acids long and contains three LIM domains in its C-terminal region. The longest isolate from the two-hybrid screen matches the Trip6 sequence from positions 190 to 476 (Trip6190–476).
Trip6 belongs to the zyxin family of LIM-domain proteins involved in focal adhesion (Lee et al. 1995; Yi and Beckerle 1998; Wang et al. 1999), and its localization is mainly cytosolic because of the presence of a nuclear export signal near the N terminus of the protein (Wang et al. 1999; Cuppen et al. 2000; Wang and Gilmore 2001). Trip6190–476 lacks this NES. If Trip6 were involved in GR-mediated transrepression, it should be nuclear, at least when transrepression occurs. To investigate its subcellular localization, we raised a rabbit antibody (Ab) against Trip6190–476. Western blotting analysis revealed the presence of three immunoreactive bands in whole-cell extracts from HeLa cells (Fig. 1D). The apparent size of the slower migrating band corresponded to the predicted size of full-length Trip6. We then studied the subcellular localization of these putative isoforms of Trip6. The two slower migrating bands were located exclusively in the cytosolic fraction, whereas the smallest isoform, called nTrip6 from here on, was exclusively nuclear (Fig. 1D). No modification of the expression pattern or the subcellular distribution of the three Trip6 isoforms was observed in cells treated with dexamethasone (Dex) or TPA. c-Fos and GR expression and localization confirmed the purity of the fractions (Fig. 1D).
According to RNA interference, the different bands observed by Western blotting were Trip6 isoforms (Fig. 1E). At 48 h after transfection into HeLa cells, two different siRNAs targeting Trip6 mRNA (siRNA #1 and #2) reduced the amount of the three immunoreactive bands, but not that of GR (half-life <24 h) (Fig. 1E; Dong et al. 1988; Hoeck et al. 1989), confirming that the two siRNAs are specific, and did not down-regulate translation at random or cause sequence-independent mRNA degradation. The relative effect of the two specific siRNAs on Trip6 isoforms differed. siRNA #1 efficiently reduced the expression of the two cytosolic isoforms and silenced nTrip6 only weakly (by 40%), whereas siRNA #2 was more efficient at reducing the expression of nTrip6 (by 80%) (Fig. 1E). The reason for this difference is not known.
The isoforms might arise from alternative splicing, because mRNA analysis revealed the existence of several splice variants (data not shown). Trip6 isoforms and their subcellular distribution were similar in all cell lines and primary cells tested (HEK-293, Cos7, A549, NIH3T3, mouse splenocytes, mouse thymocytes) (data not shown).
nTrip6 interacts with GR, c-Fos, and p65 (RelA)
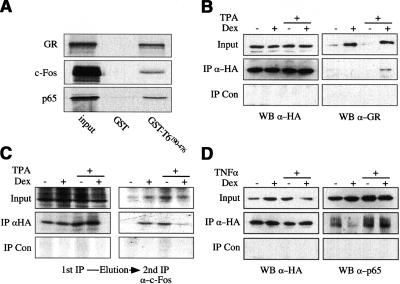
The nuclear localization of nTrip6 is consistent with its putative involvement in transrepression by the GR. If nTrip6 were to act on AP-1 and NF-κB, it would likely not only interact with GR, but also with the repressed transcription factors. To investigate the binding abilities of nTrip6, we performed GST-pull-down experiments using the longest isolate from the two-hybrid screen (GST–Trip6190–476). Specific pull-down interactions were observed with GR, c-Fos, and p65 (RelA), whereas no interaction was observed with GST alone (Fig. 2A).
Figure 2.
Trip6 interacts with GR, c-Fos, and NF-κB p65. (A) GST pull-downs. 35S-labeled, in vitro translated GR, c-Fos, and p65 were subjected to GST pull-down assays using either GST or GST-Trip6190–476. Input represents 10% of the in vitro translated material used in each assay. (B–D) CoIPs. (B,D) HeLa cells stably transfected with HA-Trip6 were treated with either TPA (B) or TNFα (D) in the presence or absence of dexamethasone (Dex) as indicated. Proteins from nuclear extract were immunoprecipitated using either a rat anti-HA or a rat isotype control Ab (Con). Input (1%) and immunoprecipitated proteins were subjected to Western blotting (WB) using a mouse anti-HA (B,D), an anti-GR (B), or an anti-p65 Ab (D). (C) Cells were metabolically labeled with [35S]methionine and [35S]cysteine, and treated as in B. Proteins immunoprecipitated with the anti-HA and the control Ab were eluted and subjected to a second IP with an anti-c-Fos Ab. Input (1%) and precipitated proteins were subjected to SDS-PAGE and autoradiography.
These in vitro interactions were confirmed by immunoprecipitation (IP) from nuclear extracts of cells under conditions in which transrepression of AP-1 and NF-κB occurs (Fig. 2). In a first series, HeLa cells stably expressing an HA-tagged version of Trip6 (HA-Trip6) were treated with TPA and Dex to induce AP-1 and GR activity, respectively. HA-Trip6 was present in nuclear extracts, and its level was not modified upon Dex and TPA treatment (Fig. 2B, input). GR was present in the nuclear extracts after treatment with Dex, irrespective of TPA activation (Fig. 2B, input). GR was found associated with HA-Trip6 only after treatment with Dex and TPA together (Fig. 2B). Neither HA-Trip6 nor GR was detected after IP with an isotype control Ab.
Because c-Fos comigrates with the IgG heavy chain, we labeled cells metabolically with [35S]-methionine and [35S]-cysteine to detect in vivo interactions with HA-Trip6. HA-Trip6 was immunoprecipitated from nuclear extracts, and associated proteins were eluted and subjected to a second IP using an anti-c-Fos Ab (Fig. 2C). A control IP with anti-c-Fos revealed a low basal level and induction by TPA. Interaction of HA-Trip6 with c-Fos occurred after treatment with Dex, with TPA, or with both. The level of c-Fos associated with Trip6 appeared to be lower in nuclear extracts from cells treated with both Dex and TPA (Fig. 2C), possibly because of the lower level of radiolabeled c-Fos immunoprecipitated in this condition. The interaction between c-Fos and Trip6 might thus be constitutive, and depend primarily on the level of c-Fos. No c-Fos was immunoprecipitated with an isotype control Ab (Fig. 2C).
In a third series of co-IPs, we aimed at detecting a Trip6–NF-κB interaction after TNFα activation. In the HeLa cells stably expressing HA-Trip6, NF-κB p65 appeared to be constitutively nuclear (Fig. 2D, input). Neither TNFα nor Dex treatment modified the levels of nuclear p65. Interaction between p65 and HA-Trip6 was observed under all conditions, more pronounced after TNFα activation (Fig. 2D). Thus, as observed with c-Fos, NF-κB p65 seems to be constitutively associated with Trip6 in the nucleus of HeLa cells.
nTrip6 is involved in the transrepression of AP-1 and NF-κB by GR
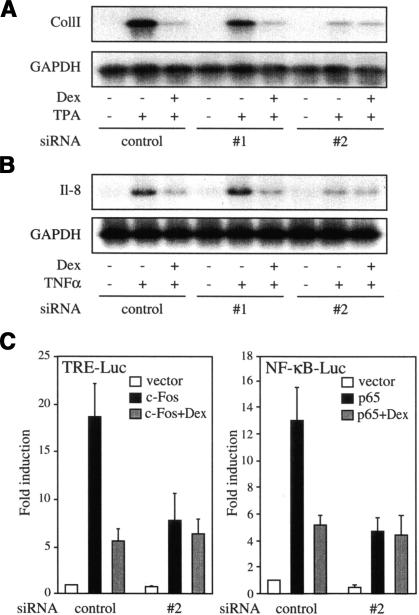
Knowing the association of nTrip6 with GR and with the two transcription factors repressed by GR, we aimed at proving a role for nTrip6 in the negative cross-talk. We used the Trip6-targeting siRNAs (Fig. 1E) to down-regulate the expression of the Trip6 isoforms and assayed for GR-mediated repression of the AP-1-driven collagenase I and of the NF-κB-dependent interleukin 8 (IL-8) genes. In cells transfected with the control siRNA, expression of collagenase I and IL-8 mRNAs induced by TPA and TNFα, respectively, was almost totally repressed by Dex (Fig. 3). Transfection of siRNA #1 affected neither their induction nor their repression by Dex. In cells transfected with siRNA #2, however, collagenase I and IL-8 mRNA induction was, to our surprise, severely reduced. Moreover, the ability of Dex to repress the remaining induction was abolished (Fig. 3). Expression of GAPDH mRNA was unaffected by either the Trip6-specific or the control siRNAs (Fig. 3). Because collagenase I and IL-8 promoters are complex, we studied the effect of Trip6 down-regulation in reporter gene assays using promoters containing only AP-1 and NF-κB response elements directly activated by cotransfection with c-Fos and p65, respectively (Fig. 3C). Both c-Fos- and p65-induced activation of the reporter genes and their repression by Dex were strongly reduced by siRNA #2 as compared with control siRNA (Fig. 3C). These results confirm that AP-1 and NF-κB, and not other transcription factors acting on collagenase I or IL-8 promoters, are direct targets of Trip6. Altogether, these results show that (1) Trip6 is necessary for AP-1- and NF-κB-dependent transcription; (2) Trip6 is required for GR-dependent repression of AP-1 and NF-κB; and (3) the shorter nuclear isoform of Trip6, nTrip6, is involved in these effects, because the two siRNAs exerted isoform-specific or preferential down-regulation (Fig. 1E).
Figure 3.
nTrip6 is required for glucocorticoid-mediated repression of collagenase I and interleukin 8 expression. HeLa cells were transfected with siRNAs as in Figure 1E and treated with either TPA (A) or TNFα (B), in the presence or absence of dexamethasone (Dex). Total RNA was prepared and subjected to Northern blotting using probes specific for collagenase I IL-8 or GAPDH. (C) HeLa cells were cotransfected with either the control siRNA or the siRNA #2 together with either an AP-1-driven reporter gene (TRE-Luc) and a c-Fos expression vector (left), or an NF-κB-driven reporter gene and a p65 expression vector, and treated with dexamethasone (dex) as indicated. Normalized luciferase activities are plotted as fold induction (mean ± S.D. of one representative experiment performed in triplicates).
nTrip6 is a coactivator for AP-1 and NF-κB
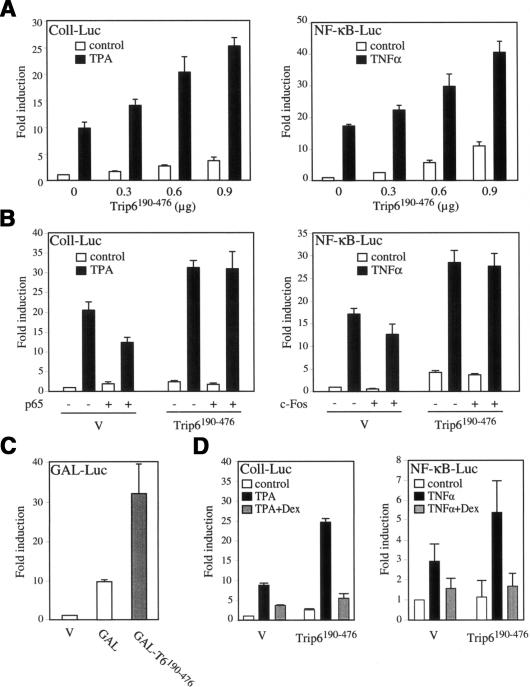
nTrip6 interacts with both AP-1 and NF-κB, and is necessary for transcriptional activation by both these transcription factors. These effects are reminiscent of a coactivator function. As described in the case of nuclear receptor coactivators (Horwitz et al. 1996; McKenna et al. 1999), a protein has to fulfill several criteria to qualify as a coactivator: (1) it should increase transactivation; (2) it should reverse the squelching between different transcription factors that compete for the same limiting coactivator; and (3) the coactivator function should be transferable. We thus studied the effect of nTrip6 on AP-1 and NF-κB transactivation in reporter gene assays. Overexpression of Trip6190–476 dose-dependently increased the responsiveness of the AP-1-dependent collagenase I promoter activity to TPA (Fig. 4A, left panel), as well as the responsiveness of an NF-κB-regulated promoter to TNFα (Fig. 4A, right panel). No effect of Trip6190–476 overexpression was observed on reporter genes driven by control promoters, the RSV promoter, or a minimal TATA-containing promoter (data not shown). Hence, nTrip6 is involved in AP-1- and NF-κB-mediated transactivation, and its expression level seems to be limiting for this action. If so, transactivation by AP-1 might be inhibited by concomitant activation of NF-κB if NF-κB competed for nTrip6, and vice versa (squelching). In reporter gene assays, AP-1-dependent transcription was, indeed, partially inhibited by overexpression of NF-κB p65. This inhibition was prevented by overexpression of Trip6190–476 (Fig. 4B, left panel). Similarly, NF-κB-dependent transcription was somewhat inhibited by overexpression of c-Fos, and the inhibition was reversed by increasing Trip6190–476 levels (Fig. 4B, right panel).
Figure 4.
nTrip6 acts as a coactivator for AP-1 and NF-κB. (A,B,D) HEK293 cells or A549 cells (D, right) were cotransfected with the luciferase reporter constructs indicated in the top left corner, together with expression vectors for Trip6190–476, p65 (B, left), or c-Fos (B, right), and treated with TPA (Coll-Luc) or TNFα (NF-κB-Luc) in the presence or absence of dexamethasone (Dex, D). (C) HEK293 cells were cotransfected with GAL-Luc reporter construct and expression vectors for either GALDBD or GALDBD–Trip6190–476. Normalized luciferase activities are plotted as fold induction (mean ± S.D. of one representative experiment performed in triplicates).
To explore whether nTrip6 coactivator function is transferable, we studied the effect of Trip6190–476 fused to GAL4–DBD on the expression of a reporter gene driven by GAL4–UAS. The fusions of Trip6190–476 strongly enhanced the transcription of the reporter gene, as compared with GAL4–DBD alone (Fig. 4C). These results strongly suggest that nTrip6 supplies a common coactivator function for both AP-1 and NF-κB.
Because nTrip6 levels in cells are limiting for its coactivator-like function (Fig. 4A), and because GR interacts with nTrip6 (Figs. 1, 2), one could hypothesize that GR represses by sequestering nTrip6 away from AP-1 and NF-κB. If this were the case, increasing the level of nTrip6 should rescue the repression. In reporter gene assays, Trip6190–476 overexpression increased the responsiveness of the promoter to TPA or TNFα, but it did not hamper GR-mediated repression (Fig. 4D). Thus repression of AP-1 and NF-κB by GR is not mediated by competition for limiting amounts of nTrip6.
nTrip6 forms a complex with GR and AP-1/NF-κB at target promoters
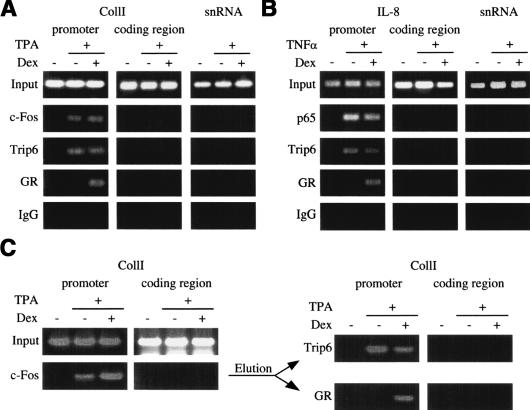
As a coactivator, nTrip6 should be associated with AP-1- and NF-κB-dependent promoters in the nucleus. To prove this, we performed chromatin immunoprecipitation (ChIP) experiments. Primers were chosen that amplify the region of the collagenase I promoter encompassing the AP-1-binding sites. Promoter occupancy by c-Fos was induced by TPA treatment of the cells, and was not inhibited by Dex (Fig. 5A). This confirms that GR does not influence AP-1 binding to its target promoter (König et al. 1992; Rogatsky et al. 2001). Like AP-1, nTrip6 was tethered to the collagenase I promoter upon TPA stimulation (Fig. 5A), consistent with nTrip6 being an AP-1 coactivator or adaptor for a coactivator. The promoter occupancy by Trip6 was not altered by Dex treatment (Fig. 5A). This confirms that GR-mediated AP-1 repression is not caused by a competition for limiting amounts of Trip6. GR was found associated with the collagenase I promoter in cells treated with TPA and Dex (Fig. 5A), confirming that during repression, GR is tethered to the AP-1-regulated promoter (Rogatsky et al. 2001). The collagenase I promoter was not amplified in ChIP performed with an isotype control Ab, and neither c-Fos nor nTrip6 or GR was found associated with an irrelevant promoter containing no AP-1 site, the U6 snRNA promoter (Fig. 5A). None of the proteins was found associated with a coding sequence of the collagenase I gene, showing that the interactions are specific for the promoter. Similar results were obtained using the promoter of an NF-κB target gene, IL-8 (Fig. 5B). nTrip6 was associated with the IL-8 promoter together with NF-κB p65 in cells activated with TNFα, and GR was tethered to the promoter in cells treated with TNFα and Dex (Fig. 5B).
Figure 5.
Occupancy of AP-1- and NF-κB-dependent promoters by nTrip6 and GR. Chromatin immunoprecipitation (ChIP) was performed in HeLa cells treated as in Figure 2B and D, using antibodies against c-Fos (A) or NF-κB p65 (B), and against Trip6 and GR, as well as an isotype control Ab. PCR was performed with primers that detect either the promoter or a coding region of collagenase I (CollI, A) or interleukin 8 (IL-8, B) genes, or the U6 snRNA promoter. (C) Chromatin double immunoprecipitation assay. A first ChIP was performed on HeLa cells treated as indicated using an anti-c-Fos Ab, and the immunoprecipitated material was eluted and subjected to a second ChIP using either an anti-Trip6 or an anti-GR Ab.
To ensure that the different proteins were recruited to the same promoter and not to different populations of the same promoter, we performed chromatin double IPs (ChDIP) on the collagenase I promoter (Fig. 5C). Chromatin was first immunoprecipitated with either the anti-c-Fos or an isotype control Ab, eluted from the beads, and subjected to a second IP using either the anti-Trip6 or the anti-GR Ab. PCR performed directly after the first IP with the anti-Fos Ab confirmed promoter occupancy by c-Fos after TPA induction in both Dex-untreated and -treated cells. After the second IP, nTrip6 was found associated with the c-Fos-occupied promoter in TPA-activated cells, and this association was not inhibited by Dex. GR was tethered to this promoter in cells treated with both TPA and Dex (Fig. 5C). The collagenase I promoter was not immunoprecipitated by the anti-Trip6 or the anti-GR Ab in eluates from IPs performed with an isotype control Ab (data not shown). These results confirm that c-Fos and Trip6 are recruited to the very same population of collagenase I promoters in TPA-activated cells, and that upon Dex treatment, GR is tethered to exactly these promoters, and not to a different population of promoters lacking nTrip6.
nTrip6 recruits GR to AP-1 and NF-κB at target promoters
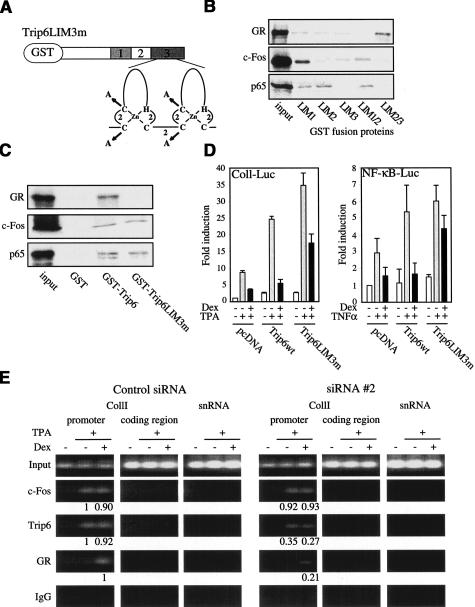
The C-terminal half of Trip6 carries three LIM domains (Fig. 6A). LIM domains are double zinc-finger structures, which serve as protein interaction modules (for review, see Dawid et al. 1998). Because nTrip6 interacted with both GR and AP-1 or NF-κB, interaction specificity may be mediated by different LIM domains, and destruction of individual LIM domains could act as dominant-negative mutations. We therefore mapped the GR, c-Fos, and p65 interaction surfaces on nTrip6 in pull-down experiments using GST fusions of either single LIM domains or combinations. GR interacted with the combination of LIM domains 2 and 3, but not with single LIM domains 2 or 3 (Fig. 6B; for nomenclature, see Fig. 6A). c-Fos was pulled down with LIM domain 1, and more weakly with the combination of LIM domains 1 and 2 (Fig. 6B). Finally, p65 interacted with the LIM domains 1 and 2, as well as with a combination of both (Fig. 6B).
Figure 6.
nTrip6 mediates GR tethering to the repressed promoter. (A) Schematic representation of Trip6LIM3m fused to GST. Trip6 harbors 3 LIM domains in its C-terminal patch, each consisting of two zinc fingers, as illustrated for the LIM domain 3 (LIM3). In Trip6LIM3m, the four coordinating cysteines in the two zinc fingers of LIM3 were mutated to alanines. (B) Mapping of Trip6 LIM domains interaction with GR, c-Fos, and NF-κB p65. 35S-labeled, in vitro translated GR, c-Fos, and p65 were subjected to GST pull-down assays using GST fusion of individual Trip6 LIM domains, or combinations of the LIM domains 1 and 2, or 2 and 3. Input represents 10% of the in vitro translated material used in each assay. (C) Trip6LIM3m does not interact with GR, but still interacts with c-Fos and NF-κB p65. GST pull-down assays were performed as in B using GST or GST fusions of Trip6 or Trip6LIM3m. (D) Trip6LIM3m has a dominant negative effect on AP-1 and NF-κB repression by GR. HEK-293 (left) or A549 cells (right) were cotransfected with the indicated luciferase reporter construct and expression vectors. Cells were treated with TPA (left) or TNFα (right), in the presence or absence of dexamethasone (Dex). Normalized luciferase activities (to renilla luciferase) are plotted as fold induction (mean ± S.D. of one representative experiment performed in triplicates). (E) nTrip6 mediates the tethering of GR to the repressed collagenase I promoter. Chromatin immunoprecipitation (ChIP) was performed as in Figure 5 in HeLa cells transfected with either the control siRNA or the siRNA #2, and treated with TPA in the presence or absence of Dex as indicated. The relative promoter occupancy by c-Fos, nTrip6, and GR (normalized to the input) is indicated below the bands.
To abolish the interaction between Trip6 and GR, we mutated in Trip6 the coordinating cysteines 399, 402 and 429, 432 to alanine in the two zinc fingers of LIM domain 3 (Trip6LIM3m), destroying its secondary structure without affecting LIM domains 1 and 2 (Fig. 6A). Whereas GST–Trip6, but not GST alone, interacted with GR, c-Fos, and p65, GST–Trip6LIM3m interacted with c-Fos and p65 but no longer with GR (Fig. 6C). Overexpression of Trip6LIM3m strongly increased TPA- or TNFα-induced expression of the appropriate reporter gene, as did wild-type Trip6 (Fig. 6D). This confirms that this Trip6 mutant is still tethered to the promoter and exerts a coactivator-like function. However, Trip6LIM3m severely reduced the Dex-mediated repression of both AP-1 and NF-κB, as compared with wild-type Trip6 (Fig. 6D). The dominant-negative effect on transrepression of this GR-noninteracting strongly suggests that GR is tethered to the repressed promoter through its interaction with nTrip6, and that this tethering is necessary for GR to repress AP-1 and NF-κB.
We expected that in the absence of Trip6, AP-1 would still be found associated with the collagenase promoter, but Dex-induced GR would not. To this end, we performed ChIPs in cells transfected with the Trip6 siRNA #2 (Fig. 6E). c-Fos recruitment to the collagenase I promoter was not affected by the siRNA. However, siRNA #2 reduced the promoter occupancy by Trip6 upon TPA activation, and reduced the tethering of GR to the promoter after treatment with TPA and Dex, as compared with the control siRNA (Fig. 6E). Altogether, these results demonstrate that Trip6 mediates the tethering of GR to the repressed promoter.
Discussion
In this paper we have identified an essential component of the cross-talk between GR and the proinflammatory transcription factors AP-1 and NF-κB. We discovered nTrip6 as a novel type of transcriptional cofactor, which assembles several regulatory proteins on specific promoters: the transcription factors AP-1, NF-κB, and GR. Recruited to AP-1 or NF-κB bound to target promoters, nTrip6 acts as or binds additional coactivator(s). GR binding to nTrip6 at the same time converts the complex into a repressed mode.
Trip6 is a ubiquitously expressed member of the zyxin family of focal adhesion proteins (Lee et al. 1995; Yi and Beckerle 1998; Wang et al. 1999) in which LIM domains serve as modules for protein–protein interactions. In overexpression studies, it has been proposed that Trip6 shuttles between cytoplasm and nucleus and its localization at the site of cell–cell contact and adhesion plaques depends on the presence of a strong nuclear export signal (NES) and nuclear export (Nix and Beckerle 1997; Cuppen et al. 2000; Wang and Gilmore 2001). We have not, however, been able to observe nuclear–cytoplasmic shuttling of endogenous Trip6 under the condition of AP-1, NF-κB, or GR activation (Fig. 1D). Here we show that various Trip6 proteins of at least three different sizes exist. The smallest protein, named nTrip6, is found in the nucleus and corresponds in size to our cDNA isolate that lacks the coding sequence for the NES. RNA interference experiments support the interpretation that the smallest protein is translated from a distinct mRNA rather than being generated by processing of the larger proteins. Preliminary comparison of several cDNA fragments from HeLa cells indicate variability in the part coding for the N terminus of the protein and suggests generation of several mRNAs by differential splicing (data not shown).
A prominent feature of nTrip6 discovered in this study is to serve as coactivator or adaptor for a coactivator for AP-1 and NF-κB. The composition of coactivating complexes is elaborate (for reviews, see McKenna et al. 1999; Xu et al. 1999; Dilworth and Chambon 2001). We do not yet know the detailed mechanism of transcriptional regulation by nTrip6. There are, however, precedents: two other LIM-domain proteins, paxillin and Hic-5/ARA55, serve as coactivators for steroid receptors (Yang et al. 2000; Kasai et al. 2003). nTrip6 shows many features of bona fide coactivators. The activation function can be transferred to GALDBD–Trip6 fusions (Fig. 4C; Zhao et al. 1999; Wang and Gilmore 2001). Reducing the level of nTrip6 by siRNA decreased and overexpression increased the transactivation by both AP-1 and NF-κB, suggesting that endogenous levels of nTrip6 can be limiting for AP-1 and NF-κB activity. This proposition is further supported by mutual squelching between AP-1 and NF-κB that was relieved by overexpression of Trip6 (Fig. 4B). And finally, nTrip6 was tethered to AP-1 and NF-κB at their target promoters (Fig. 5). Altogether, these data make nTrip6 a bona fide “coactivator” for AP-1 and NF-κB. However, apart from the three LIM domains in the C-terminal half, nTrip6 carries no other known functional domain, motif, or catalytic activity that could mediate transactivation. We favor, therefore, the hypothesis that the multi-LIM-domain protein nTrip6 recruits additional coactivators by protein–protein interaction.
In addition to the functions shared with many coactivators, nTrip6 has a novel and unique property. It serves as “the missing link” in the cross-talk between GR and AP-1 as well as NF-κB. Assembly of Jun:Fos or p65 complexes with GR through separate domains of nTrip6 appears to play a key role in this function. Simple competition of GR for limiting amounts of nTrip6 is ruled out because overexpression of Trip6 did not rescue GR-mediated repression (Fig. 4D), and glucocorticoid treatment did not reduce nTrip6 occupancy of AP-1 and NF-κB target promoters (Fig. 5). nTrip6, rather, tethers GR to these promoters, which results in a switch to repression. Therefore, an nTrip6 mutant only interacting with c-Fos and p65 but not with GR still increased transactivation by AP-1 and NF-κB, but exerted a dominant-negative effect on transrepression (Fig. 6). This view of the role of nTrip6 in transcriptional cross-talk is consistent with previous observations that the repressed transcription factors remain bound to the promoter elements (König et al. 1992; Nissen and Yamamoto 2000; Rogatsky et al. 2001) and that GR is associated with the promoters (Nissen and Yamamoto 2000; Rogatsky et al. 2001). nTrip6 may mediate transrepression also by other nuclear receptors (preliminary observations), explaining why Trip6 was first isolated as a protein of unknown function interacting with thyroid hormone and retinoid X receptors (Lee et al. 1995).
The GR DBD appears to be required for transrepression of both AP-1 and NF-κB (Jonat et al. 1990; Schüle et al. 1990; Heck et al. 1994; Scheinman et al. 1995; McKay and Cidlowski 1998), and point mutations in the DBD (C476W and R479Q) abolished GR-mediated repression of AP-1 (Heck et al. 1994; this paper). The finding that this mutant does not interact with Trip6 (Fig. 1C) further supports the key role of GR interaction with Trip6 in transrepression and points toward the DBD as an important interaction surface. Additional sequences neighboring the DBD may be required (E. Wade and S. Heck, unpubl.) given that replacing the MR DBD by the GR DBD could not fully convert the nonrepressing MR into an AP-1-repressing receptor (Pearce and Yamamoto 1993; Heck et al. 1994). The ligand-binding domain (LBD) is also needed, at least to establish hormone dependency of transrepression (Jonat et al. 1990; Schüle et al. 1990; McKay and Cidlowski 1998; Nissen and Yamamoto 2000). However, neither the transcriptional activation function in the LBD (AF-2) nor that in the N terminus (AF-1) appears to be required, because mutants of GR generated by us and others failed to transactivate from GRE-dependent promoters but were competent to repress AP-1 or NF-κB (Fig. 1B; Ray et al. 1999; Kucera et al. 2002). Therefore, competition of GR for coactivators as a mechanism of transrepression is ruled out.
Further questions on the mechanism of transrepression remain open. The possibility that GR blocks coactivator recruitment by nTrip6 is unlikely, because the repression of an NF-κB-dependent promoter appears to occur after binding of coactivators and formation of the preinitiation complex (Nissen and Yamamoto 2000). nTrip6 might, under the influence of GR, recruit an activity that blocks initiation or elongation of transcription from the preinitiation complex. This putative repressing activity would not likely involve a histone deacetylase (HDAC), because HDAC inhibitors did not affect GR-dependent transrepression (Vanden Berghe et al. 2002; our unpublished data).
In conclusion, we have defined a new class of transcriptional regulators represented by nTrip6, which by means of several interaction domains orchestrates the assembly of multiprotein complexes at the promoters of target genes. Depending on the availability of partner proteins such as nuclear GR, different complexes are formed that promote or prevent transcriptional activation.
Materials and methods
Plasmids
The expression vectors for GR are RSV-GRwt (A. Cato, Forschungszentrum Karlsruhe, Karlsruhe, Germany), RSV-GRmH12 (point mutation E755Q), RSV-GRmH12ΔAF-1 (deletion of the AF-1 in RSV-GRmH12 using BglII), and RSV-GRmDBD (C476W/R479Q) (Heck et al. 1994). For in vitro translation, GR was subcloned into pcDNA3.1(+). In pcDNA3.1HA, annealed oligonucleotides encoding the HA tag were ligated into the HindIII and EcoRI sites of pcDNA3.1(+) (Invitrogen). In pcDNA3.1HA-Trip6190–476, Trip6190–476 was PCR-amplified from HeLa cell cDNAs and subcloned into pcDNA3.1HA. In pcDNA3.1HA-Trip6LIM3m, the coordinating cysteine residues 399, 402, 429, and 432 in both zinc fingers of the LIM domain 3 (see Fig. 6B for nomenclature) were mutated to alanine using the QuickChange Site-Directed mutagenesis kit (Stratagene). In GST–Trip6190–476, Trip6190–476 was subcloned into pGEX-4T-3 (Amersham). GST-fusions of the LIM domains were generated by PCR-amplifying the individual LIM domains or combinations and subcloning into pGEX-4T-1 (Amersham). In GST–Trip6LIM3m, the SbfI/XhoI fragment of pcDNA3.1HA-Trip6LIM3m was subcloned into GST–Trip6. pcDNAGalDBD for mammalian expression was obtained by cloning the Gal4DBD (BamHI/XhoI fragment) of pAS-1CYH2 (Clontech) into pcDNA3.1(+). In pcDNAGalDBDTrip6190–476, Trip6190–476 was PCR-amplified and subcloned into pcDNAGalDBD. The sources of other plasmids are pcDNAc-Fos and pcDNAp65, F. Weih (Forschungszentrum Karlsruhe, Karlsruhe, Germany); pGEMc-Fos and pBATcJun, P. Angel (Deutsches Krebsforschungszentrum, Heidelberg, Germany); and pBlSK(+)p65, A. Cato.
The luciferase reporter constructs were as described: MMTV-Luc (Heck et al. 1997), Coll-Luc (Schneikert et al. 1996), TRE-Luc (5Xcoll-TRE-TATA-Luc) (van Dam et al. 1998), NF-κB-Luc (Lernbecher et al. 1993), and GAL-Luc (Weg-Remers et al. 2001). The Ubi-Renilla construct was made by replacing the growth hormone cDNA in pUbiGH (Schorpp et al. 1996) by Renilla reniformis luciferase cDNA (Promega).
For the two-hybrid screen, a Gal4DBD fusion of residues 262 to 777 of RSV-GRmH12ΔAF1 (Gal-GRmH12ΔAF-1), as well as the corresponding region of GRmDBD (Gal-GRmDBD), was constructed by subcloning into pAS2-1 (Clontech).
Cell culture and transfections
HeLa tk–, HEK-293T, Cos7, and A549 cells (ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum. HeLa Cos7 and A549 cells were transfected using FUGENE6 (Roche) or Oligofectamine (Invitrogen) in the case of siRNAs, HEK-293T using Lipofectamine 2000 (Invitrogen). HeLa cells stably expressing HA-Trip6 were established after transfection with pcDNA3.1HA-Trip6 and selection with G418.
All experiments were performed in serum-starved (24 h) cells. Unless otherwise stated, cells were treated with either TPA (50 ng/mL) for 3 h or TNFα (10 ng/mL, PromoCell) for 1 h, in the presence or absence of dexamethasone (10 nM). For reporter gene assays, cells were treated 24 h posttransfection and harvested 16 h posttreatment. Firefly luciferase activities were normalized to Renilla luciferase activity (Ubi-Renilla).
Two different synthetic siRNA duplexes targeting Trip6 sequence from position 853 to 873 (siRNA #1), and 1263 to 1283 (siRNA #2), as well as a control siRNA targeting luciferase, were purchased from MWG Biotech. For Western Blot analysis and reporter gene assays, cells were harvested 48 h posttransfection. For Northern Blot analysis, cells were activated 48 h posttransfection with either TPA for 5 h or TNFα for 3 h, in the presence or absence of dexamethasone. For chromatin immunoprecipitation, cells were activated 48 h posttransfection.
Yeast two-hybrid screen
All experiments were performed in the yeast reporter strain Y190 (Clontech). A clone expressing high levels of GAL4DBD–GRmH12ΔAF1 was used to screen a human HeLa MATCH-MAKER cDNA library (Clontech) by sequential transformation. Primary transformants (2.5 × 106) were selected for HIS-prototrophy in the presence of the synthetic glucocorticoid triamcinolone-acetonide at 100 μM (TAC; Sigma). Four-hundred colonies were analyzed for the second reporter gene, lacZ. The library plasmids of 150 positive colonies were retransformed into the yeast strain Y187, together with either GAL4DBD–GRmH12ΔAF1 to confirm the interaction, or with a plasmid encoding GAL4DBD to eliminate proteins interacting with the GAL4 moiety. Positive isolates were counterselected for the lack of interaction with the GAL4DBD fusion of GRmDBD that can neither transactivate nor transrepress. Inserts of 47 isolates were sequenced.
GST pull-down assays
pcDNAGR, pGEMc-Fos, pBATcJun, and pBlSK(+)p65 were transcribed and translated in vitro, using 1 μg of DNA, in the presence of 35S-methionine using the TNT Coupled Reticulocyte Lysate System (Promega) in a 50-μL reaction. Ten micrograms of purified GST-fusion protein or GST alone was attached to glutathione-Sepharose beads (Amersham) and incubated for 2 h at 4°C under agitation with 5 μL of in vitro translated proteins in pull-down buffer (40 mM HEPES-KOH at pH 7.9, 100 mM NaCl, 0.1 mM EDTA, 0.2% Triton X-100, 1 mM DTT, 1 mM PMSF). Bound proteins were washed three times with pull-down buffer, resolved by SDS-PAGE, and analyzed by autoradiography.
Cell fractionation and Western blotting
Half of the cell samples resuspended in lysis buffer (25 mM HEPES-KOH at pH 7.9, 1.5 mM EDTA, 50 mM NaCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, 5 μg/mL of each leupeptin, pepstatin A, and aprotinin) was sonicated and considered as whole-cell extract. The other half was lysed for 3 min on ice and centrifuged at 800g for 5 min. Cytosolic extracts (supernatants) were clarified by centrifugation at 15,000g for 20 min. Nuclei pellets were washed in lysis buffer without NP-40, resuspended in lysis buffer, and sonicated.
Western blot analyses were performed using a polyclonal Trip6 Ab raised against purified bacterially expressed Trip6190–476 (Genosys Biotechnologies), an anti-GR monoclonal Ab (clone 4H2; Novocastra), an anti-c-Fos polyclonal Ab (Upstate), an anti-HA monoclonal Ab (clone 12CA5; Roche), or an anti-NF-κB p65 polyclonal Ab (sc109; Santa Cruz).
Metabolic labeling and coimmunoprecipitation
When indicated, cells were metabolically labeled with L-[35S]methionine and L-[35S]cysteine (Redivue Promix; Amersham) for 1 h prior to treatment. Nuclei were prepared as described above, and extracted in lysis buffer under agitation for 1 h at 4°C. Nuclear extracts were precleared with Protein G Plus/Protein A-agarose (Calbiochem) for 1 h, and incubated overnight with either a rat monoclonal anti-HA (clone 3F10; Roche), a rabbit polyclonal anti-c-Fos (sc-52; Santa Cruz), or an isotype control Ab (normal rat IgG1; Santa Cruz). Immune complexes were then precipitated by a 1-h incubation with Protein G Plus/Protein A-agarose. Immobilized proteins were washed four times with lysis buffer, and either eluted in SDS-sample buffer and subjected to Western blot analysis, or eluted in elution buffer (50 mM Tris-HCl at pH 7.5, 1% SDS, 5 mM DTT) by boiling for 5 min, diluted in lysis buffer, subjected to a second immunoprecipitation using the anti-c-Fos Ab as above, resolved by SDS-PAGE, and analyzed by autoradiography.
Northern blotting
Total RNA extraction and Northern blotting was performed as described (Kassel et al. 2001), using as probes cDNA fragments for collagenase I (Jonat et al. 1990), interleukin 8 (IMAGE Consortium, Resource Center of the German Human Genome Project), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Fort et al. 1985).
Chromatin immunoprecipitation
ChIP was performed essentially as described (Da Silva et al. 2003). Immunoprecipitations were performed using either an anti-GR (sc-1002; Santa Cruz), an anti-c-Fos (sc52; Santa Cruz), the anti-p65, the anti-Trip6, or an isotype control Ab (nonimmune rabbit IgG; Santa Cruz). The PCR primer pairs used were 5′-GCAGAGTGTGTCTCTTTCGCACAC-3′ and 5′-GCCC TTCCAGAAAGCCAGAGGCTG-3′ for the collagenase I promoter region –219/–41; 5′-GATGTGGCTCAGTTTGTCCT-3′ and 5′-CTTGGCAAATCTGGCGTGTA-3′ for the collagenase I coding region +837/+1024; 5′-GGGCCATCAGTTGCAAATC-3′ and 5′-TTCCTTCCGGTGGTTTCTTC-3′ for the IL-8 promoter region –121/+61; 5′-GCCTTCCATAGTCTCCAAAT-3′ and 5′-TGGTCCACTCTCAATCACTC-3′ for the IL-8 coding region +879/+1095; and 5′-GGCCTATTTCCCATGATTCC-3′ and 5′-ATTTGCGTGTCATCCTTGC-3′ for the U6 snRNA promoter region –245/+85. For chromatin double immunoprecipitation (ChDIP), complexes immunoprecipitated with either the anti-c-Fos or the isotype control Ab were eluted by incubation with 10 mMDTT for 30 min at 37°C and diluted 1:500 in RIPA buffer (Da Silva et al. 2003), followed by reprecipitation with either the anti-Trip6 or the anti-GR Ab.
Semiquantification was performed in the exponential phase of PCR amplification by densitometric analysis using the NIH-Image program, and the data were normalized to the DNA input used for each individual immunoprecipitation.
Acknowledgments
This work was supported by the German Science Foundation (DFG grant no. HE 551/13-1) and by the European Commission (HPRN-CT-2002-00256).
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.322404.
References
- Amano Y., Lee, S.W., and Allison, A.C. 1993. Inhibition by glucocorticoids of the formation of interleukin-1 α, interleukin-1 β, and interleukin-6: Mediation by decreased mRNA stability. Mol. Pharmacol. 43: 176-182. [PubMed] [Google Scholar]
- Barnes P.J. 1998. Anti-inflammatory actions of glucocorticoids: Molecular mechanisms. Clin. Sci. (Lond) 94: 557-572. [DOI] [PubMed] [Google Scholar]
- Beato M., Herrlich, P., and Schütz, G. 1995. Steroid hormone receptors: Many actors in search of a plot. Cell 83: 851-857. [DOI] [PubMed] [Google Scholar]
- Boggaram V., Smith, M.E., and Mendelson, C.R. 1991. Posttranscriptional regulation of surfactant protein-A messenger RNA in human fetal lung in vitro by glucocorticoids. Mol. Endocrinol. 5: 414-423. [DOI] [PubMed] [Google Scholar]
- Caelles C., Gonzalez-Sancho, J.M., and Munoz, A. 1997. Nuclear hormone receptor antagonism with AP-1 by inhibition of the JNK pathway. Genes & Dev. 11: 3351-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppen E., van Ham, M., Wansink, D.G., de Leeuw, A., Wieringa, B., and Hendriks, W. 2000. The zyxin-related protein TRIP6 interacts with PDZ motifs in the adaptor protein RIL and the protein tyrosine phosphatase PTP-BL. Eur. J. Cell Biol. 79: 283-293. [DOI] [PubMed] [Google Scholar]
- Da Silva C.A., Heilbock, C., Kassel, O., and Frossard, N. 2003. Transcription of stem cell factor (SCF) is potentiated by glucocorticoids and interleukin-1β through concerted regulation of a GRE-like and an NF-κB response element. FASEB J. 17: 2334-2336. [DOI] [PubMed] [Google Scholar]
- Dawid I.B., Breen, J.J., and Toyama, R. 1998. LIM domains: Multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 14: 156-162. [DOI] [PubMed] [Google Scholar]
- Dilworth F.J. and Chambon, P. 2001. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene 20: 3047-3054. [DOI] [PubMed] [Google Scholar]
- Dong Y., Poellinger, L., Gustafsson, J.A., and Okret, S. 1988. Regulation of glucocorticoid receptor expression: Evidence for transcriptional and posttranslational mechanisms. Mol. Endocrinol. 2: 1256-1264. [DOI] [PubMed] [Google Scholar]
- Fort P., Marty, L., Piechaczyk, M., el Sabrouty, S., Dani, C., Jeanteur, P., and Blanchard, J.M. 1985. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 13: 1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttlicher M., Heck, S., and Herrlich, P. 1998. Transcriptional cross-talk, the second mode of steroid hormone receptor action. J. Mol. Med. 76: 480-489. [DOI] [PubMed] [Google Scholar]
- Heck S., Kullmann, M., Gast, A., Ponta, H., Rahmsdorf, H.J., Herrlich, P., and Cato, A.C. 1994. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J. 13: 4087-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck S., Bender, K., Kullmann, M., Göttlicher, M., Herrlich, P., and Cato, A.C. 1997. I κB α-independent downregulation of NF-κB activity by glucocorticoid receptor. EMBO J. 16: 4698-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmberg A., Auphan, N., Caelles, C., and Karin, M. 1995. Glucocorticoid-induced apoptosis of human leukemic cells is caused by the repressive function of the glucocorticoid receptor. EMBO J. 14: 452-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrlich P. 2001. Cross-talk between glucocorticoid receptor and AP-1. Oncogene 20: 2465-2475. [DOI] [PubMed] [Google Scholar]
- Hoeck W., Rusconi, S., and Groner, B. 1989. Down-regulation and phosphorylation of glucocorticoid receptors in cultured cells. Investigations with a monospecific antiserum against a bacterially expressed receptor fragment. J. Biol. Chem. 264: 14396-14402. [PubMed] [Google Scholar]
- Horwitz K.B., Jackson, T.A., Bain, D.L., Richer, J.K., Takimoto, G.S., and Tung, L. 1996. Nuclear receptor coactivators and corepressors. Mol. Endocrinol. 10: 1167-1177. [DOI] [PubMed] [Google Scholar]
- Jonat C., Rahmsdorf, H.J., Park, K.K., Cato, A.C., Gebel, S., Ponta, H., and Herrlich, P. 1990. Antitumor promotion and antiinflammation: Down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell 62: 1189-1204. [DOI] [PubMed] [Google Scholar]
- Karin M. and Chang, L. 2001. AP-1–glucocorticoid receptor crosstalk taken to a higher level. J. Endocrinol. 169: 447-451. [DOI] [PubMed] [Google Scholar]
- Kasai M., Guerrero-Santoro, J., Friedman, R., Leman, E.S., Getzenberg, R.H., and DeFranco, D.B. 2003. The Group 3 LIM domain protein paxillin potentiates androgen receptor transactivation in prostate cancer cell lines. Cancer Res. 63: 4927-4935. [PubMed] [Google Scholar]
- Kassel O., Sancono, A., Kratzschmar, J., Kreft, B., Stassen, M., and Cato, A.C. 2001. Glucocorticoids inhibit MAP kinase via increased expression and decreased degradation of MKP-1. EMBO J. 20: 7108-7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König H., Ponta, H., Rahmsdorf, H.J., and Herrlich, P. 1992. Interference between pathway-specific transcription factors: Glucocorticoids antagonize phorbol ester-induced AP-1 activity without altering AP-1 site occupation in vivo. EMBO J. 11: 2241-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera T., Waltner-Law, M., Scott, D.K., Prasad, R., and Granner, D.K. 2002. A point mutation of the AF2 transactivation domain of the glucocorticoid receptor disrupts its interaction with steroid receptor coactivator 1. J. Biol. Chem. 277: 26098-26102. [DOI] [PubMed] [Google Scholar]
- Lee J.W., Choi, H.S., Gyuris, J., Brent, R., and Moore, D.D. 1995. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol. Endocrinol. 9: 243-254. [DOI] [PubMed] [Google Scholar]
- Lefstin J.A. and Yamamoto, K.R. 1998. Allosteric effects of DNA on transcriptional regulators. Nature 392: 885-888. [DOI] [PubMed] [Google Scholar]
- Lernbecher T., Muller, U., and Wirth, T. 1993. Distinct NF-κ B/Rel transcription factors are responsible for tissue-specific and inducible gene activation. Nature 365: 767-770. [DOI] [PubMed] [Google Scholar]
- McKay L.I. and Cidlowski, J.A. 1998. Cross-talk between nuclear factor-κ B and the steroid hormone receptors: Mechanisms of mutual antagonism. Mol. Endocrinol. 12: 45-56. [DOI] [PubMed] [Google Scholar]
- McKenna N.J., Xu, J., Nawaz, Z., Tsai, S.Y., Tsai, M.J., and O'Malley, B.W. 1999. Nuclear receptor coactivators: Multiple enzymes, multiple complexes, multiple functions. J. Steroid Biochem. Mol. Biol. 69: 3-12. [DOI] [PubMed] [Google Scholar]
- Moras D. and Gronemeyer, H. 1998. The nuclear receptor ligand-binding domain: Structure and function. Curr. Opin. Cell Biol. 10: 384-391. [DOI] [PubMed] [Google Scholar]
- Nissen R.M. and Yamamoto, K.R. 2000. The glucocorticoid receptor inhibits NFκB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 14: 2314-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix D.A. and Beckerle, M.C. 1997. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: A potential mechanism for communication between sites of cell adhesion and the nucleus. J. Cell Biol. 138: 1139-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce D. and Yamamoto, K.R. 1993. Mineralocorticoid and glucocorticoid receptor activities distinguished by nonreceptor factors at a composite response element. Science 259: 1161-1165. [DOI] [PubMed] [Google Scholar]
- Ray A. and Prefontaine, K.E. 1994. Physical association and functional antagonism between the p65 subunit of transcription factor NF-κ B and the glucocorticoid receptor. Proc. Natl. Acad. Sci. 91: 752-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D.W., Suen, C.S., Brass, A., Soden, J., and White, A. 1999. Structure/function of the human glucocorticoid receptor: Tyrosine 735 is important for transactivation. Mol. Endocrinol. 13: 1855-1863. [DOI] [PubMed] [Google Scholar]
- Reichardt H.M., Kaestner, K.H., Tuckermann, J., Kretz, O., Wessely, O., Bock, R., Gass, P., Schmid, W., Herrlich, P., Angel, P., et al. 1998. DNA binding of the glucocorticoid receptor is not essential for survival. Cell 93: 531-541. [DOI] [PubMed] [Google Scholar]
- Reichardt H.M., Tuckermann, J.P., Göttlicher, M., Vujic, M., Weih, F., Angel, P., Herrlich, P., and Schütz, G. 2001. Repression of inflammatory responses in the absence of DNA binding by the glucocorticoid receptor. EMBO J. 20: 7168-7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud J.P., Rochel, N., Ruff, M., Vivat, V., Chambon, P., Gronemeyer, H., and Moras, D. 1995. Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature 378: 681-689. [DOI] [PubMed] [Google Scholar]
- Rogatsky I., Zarember, K.A., and Yamamoto, K.R. 2001. Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase-3 response element that mediates regulation by phorbol esters and hormones. EMBO J. 20: 6071-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinman R.I., Gualberto, A., Jewell, C.M., Cidlowski, J.A., and Baldwin Jr., A.S. 1995. Characterization of mechanisms involved in transrepression of NF-κ B by activated glucocorticoid receptors. Mol. Cell. Biol. 15: 943-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneikert J., Peterziel, H., Defossez, P.A., Klocker, H., Launoit, Y., and Cato, A.C. 1996. Androgen receptor-Ets protein interaction is a novel mechanism for steroid hormone-mediated down-modulation of matrix metalloproteinase expression. J. Biol. Chem. 271: 23907-23913. [DOI] [PubMed] [Google Scholar]
- Schorpp M., Jager, R., Schellander, K., Schenkel, J., Wagner, E.F., Weiher, H., and Angel, P. 1996. The human ubiquitin C promoter directs high ubiquitous expression of transgenes in mice. Nucleic Acids Res. 24: 1787-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüle R., Rangarajan, P., Kliewer, S., Ransone, L.J., Bolado, J., Yang, N., Verma, I.M., and Evans, R.M. 1990. Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell 62: 1217-1226. [DOI] [PubMed] [Google Scholar]
- van Dam H., Huguier, S., Kooistra, K., Baguet, J., Vial, E., van der Eb, A.J., Herrlich, P., Angel, P., and Castellazzi, M. 1998. Autocrine growth and anchorage independence: Two complementing Jun-controlled genetic programs of cellular transformation. Genes & Dev. 12: 1227-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Berghe W., De Bosscher, K., Vermeulen, L., De Wilde, G., and Haegeman, G. 2002. Induction and repression of NF-κ B-driven inflammatory genes. Ernst Schering Res. Found. Workshop 40: 233-278. [DOI] [PubMed] [Google Scholar]
- Vayssiere B.M., Dupont, S., Choquart, A., Petit, F., Garcia, T., Marchandeau, C., Gronemeyer, H., and Resche-Rigon, M. 1997. Synthetic glucocorticoids that dissociate transactivation and AP-1 transrepression exhibit antiinflammatory activity in vivo. Mol. Endocrinol. 11: 1245-1255. [DOI] [PubMed] [Google Scholar]
- Wang Y. and Gilmore, T.D. 2001. LIM domain protein Trip6 has a conserved nuclear export signal, nuclear targeting sequences, and multiple transactivation domains. Biochim. Biophys. Acta 1538: 260-272. [DOI] [PubMed] [Google Scholar]
- Wang Y., Dooher, J.E., Koedood Zhao, M., and Gilmore, T.D. 1999. Characterization of mouse Trip6: A putative intracellular signaling protein. Gene 234: 403-409. [DOI] [PubMed] [Google Scholar]
- Weg-Remers S., Ponta, H., Herrlich, P., and König, H. 2001. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J. 20: 4194-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Glass, C.K., and Rosenfeld, M.G. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9: 140-147. [DOI] [PubMed] [Google Scholar]
- Yang L., Guerrero, J., Hong, H., DeFranco, D.B., and Stallcup, M.R. 2000. Interaction of the τ2 transcriptional activation domain of glucocorticoid receptor with a novel steroid receptor coactivator, Hic-5, which localizes to both focal adhesions and the nuclear matrix. Mol. Biol. Cell 11: 2007-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang-Yen H.F., Chambard, J.C., Sun, Y.L., Smeal, T., Schmidt, T.J., Drouin, J., and Karin, M. 1990. Transcriptional interference between c-Jun and the glucocorticoid receptor: Mutual inhibition of DNA binding due to direct protein–protein interaction. Cell 62: 1205-1215. [DOI] [PubMed] [Google Scholar]
- Yi J. and Beckerle, M.C. 1998. The human TRIP6 gene encodes a LIM domain protein and maps to chromosome 7q22, a region associated with tumorigenesis. Genomics 49: 314-316. [DOI] [PubMed] [Google Scholar]
- Zhao M.K., Wang, Y., Murphy, K., Yi, J., Beckerle, M.C., and Gilmore, T.D. 1999. LIM domain-containing protein trip6 can act as a coactivator for the v-Rel transcription factor. Gene Expr. 8: 207-217. [PMC free article] [PubMed] [Google Scholar]