Abstract
MSH4, a meiosis-specific member of the MutS-homolog family of genes, is required for normal levels of recombination and fertility in budding yeast, mouse, and Caenorhabditis elegans. In this paper, we report the identification and characterization of the Arabidopsis homolog of MSH4 (AtMSH4). We demonstrate that AtMSH4 expression can only be detected in floral tissues, consistent with a role in reproduction. Immunofluorescence studies indicate that its expression is limited to early meiotic prophase I, preceding the synapsis of homologous chromosomes. A T-DNA insertional mutant (Atmsh4) exhibited normal vegetative growth but a severe reduction in fertility, consistent with a meiotic defect; this was confirmed by cytological analysis of meiosis. RNAi-induced down-regulation of the MSH4 gene resulted in a similar fertility and meiotic phenotype. We demonstrate that prophase I chromosome synapsis is delayed and may be incomplete in Atmsh4, and metaphase I chiasma frequency is greatly reduced to ∼15% of wild type, leading to univalence and nondisjunction. We show that these residual chiasmata are randomly distributed among cells and chromosomes. These features of chiasma frequency and distribution in Atmsh4 show close parallels to MSH4-independent crossovers in budding yeast that have been proposed to originate by a separate pathway. Furthermore, the characteristics of the MSH4-independent chiasmata in the Atmsh4 mutant closely parallel those of second-pathway crossovers that have been postulated from Arabidopsis crossover analysis and mathematical modeling. Taken together, this evidence strongly indicates that Arabidopsis possesses two crossover pathways.
Keywords: Arabidopsis, meiosis, recombination, chiasmata, MSH4
Understanding of the genetic regulation of meiosis has advanced rapidly in recent years because of the application of molecular genetics, in combination with genetic analysis and cytology (Roeder 1997; Zickler and Kleckner 1999). Much of this effort has been focused on the simple unicellular fungus Saccharomyces cerevisiae (budding yeast), but parallel studies have been conducted in a range of other eukaryotic model species such as the filamentous fungus Sordaria macrospora, the invertebrate animals Caenorhabditis elegans and Drosophila melanogaster, the mouse Mus musculus, and the flowering plant Arabidopsis thaliana. Part of the motivation for these parallel studies is to establish the similarities and differences of meiotic regulation between different eukaryote groups (Loidl 2000) that should further our understanding of the fundamentals of meiosis.
The interrelated events of chromosome pairing, synapsis, and recombination are central to meiotic function and progression (Zickler and Kleckner 1999). The association of homologous chromosome pairs and their subsequent separation in division I of meiosis is a uniquely meiotic phenomenon that underpins both chromosome reduction and the regular disjunction of chromosomes to the meiotic products (Hawley 1988). Any disturbance of these events can lead to nondisjunction and aneuploidy, or in extreme cases to nuclear nonreduction. The double-strand break repair (DSBR) model (Szostak et al. 1983; Sun et al. 1991) provides a robust and well-characterized explanation of the molecular mechanism of meiotic recombination. According to the current version of this model, recombination is initiated by the formation of programmed double-strand breaks that are catalyzed by the SPO11 protein (Keeney 2001). Several other proteins are known to be required for DSB formation, including the RAD50, MRE11, XRS2 complex that is also required for the processing of DSBs to give 3′-single-stranded tails. Subsequently, RAD51 and DMC1 coat the 3′-single-stranded tails and promote single end invasions (SEIs) of homologous intact duplexes. Further molecular changes consequent upon SEI lead to capture of the 3′-end on the other side of the break (second end capture or SEC) resulting in recombination intermediates termed double Holliday junctions (dHjs). It was originally proposed that differential resolution of dHjs gave either crossover (CO) or noncrossover (NCO) products (Holliday 1964). The MutL homologs MLH1 and MLH3 occur as foci that correlate in number and position with crossovers, and they were therefore initially thought to have important roles in this transition (Roeder 1997). More recent studies in budding yeast have concluded that the crossover/noncrossover decision is made much earlier than originally thought, probably during leptotene, not long after DSB formation (Allers and Lichten 2001; Börner et al. 2004).
It has long been recognized that the MutS homologs MSH4 and MSH5 play a key role in promoting crossover formation in eukaryotes (Ross-Macdonald and Roeder 1994; Zalevsky et al. 1999). In Escherichia coli, the MutS protein recognizes and binds to mismatched nucleotides and forms part of the MutHLS system for mismatch repair (MMR) of DNA damage. Budding yeast possesses six homologs of MutS (MSH1–6) that are widely conserved among other eukaryotes. Three of these (MSH2, MSH3, and MSH6) have been shown to participate in somatic mismatch repair. MSH4 and MSH5, on the other hand, are not involved in mismatch repair, but instead have meiosis specific roles in recombination. Mutants of MSH4 in yeast and mouse show defects in chromosome synapsis, and the yeast mutant exhibits a severe reduction in crossing over (Ross-Macdonald and Roeder 1994; Kneitz et al. 2000). The HIM14 gene of C. elegans encodes a germ-line-specific member of the MutS family that is apparently the nematode ortholog of the yeast MSH4 gene. Interestingly, the him14 mutant displays apparently normal meiotic chromosome synapsis (Zalevsky et al. 1999), but recombination is virtually eliminated.
Börner et al. (2004) found that yeast mutants lacking the meiotic proteins Zip1, Zip2, Zip3, Mer3, and/or Msh5 (ZMM proteins) were coordinately defective in SEIs, dHjs, and crossover products, whereas DSBs and noncrossovers formed normally, implying that these proteins act coordinately to promote COs very early in the recombination process. By implication, this places MSH4, which acts in concert with MSH5 as a heterodimer, in the same time frame. This timing is partly supported by Colaiacovo et al. (2003), who found that MSH4 and MSH5 are required for the timely disappearance of RAD51 foci in C. elegans. They concluded that MSH4 and MSH5 act at a relatively early step in recombination, after loading of the RAD51 protein but before dHj resolution, and that they act locally at the sites of nascent recombination events.
An important recent development is the suggestion that budding yeast possesses an alternative pathway for CO formation, based on the processing of non-dHj intermediates. This pathway requires the activity of an endo-nuclease, MUS81, that acts as a heterodimer with another protein MMS4 (de los Santos et al. 2003; Hollingsworth and Brill 2004). This realization led to the proposal that there are two classes of crossovers in budding yeast originating from two distinct biochemical pathways. Most crossovers belong to class I, exhibit crossover interference, and are dependent on MSH4/MSH5. Class II crossovers are a minority, do not exhibit interference, and are dependent on MUS81/MMS4 (de los Santos et al. 2003). However, evidence from other studies indicates that the contribution of these two classes to total crossovers can vary between different organisms. In the fission yeast, Schizosaccharomyces pombe, all crossovers appear to be class II events, whereas in C. elegans, msh4 and msh5 mutants completely eliminate crossing over, indicating that all crossovers in this organism belong to class I. The situation in mice is currently unknown, although it is known that MSH4 and MSH5 are required for the proper execution of meiosis and that MUS81 activity has been detected in somatic cells (Hollingsworth and Brill 2004).
A. thaliana has emerged as an important flowering plant model for the molecular analysis of many genetic and developmental processes, including meiosis. Because meiotic genes catalyzing recombination are highly conserved, it is not surprising to find that homologs of many of these genes have been identified in Arabidopsis (Caryl et al. 2003). As part of this wider investigation, the Arabidopsis homolog of MSH4 (AtMSH4) has been identified, cloned, and characterized. In addition to the availability of genomic tools and resources for genetic analysis, Arabidopsis possesses excellent meiotic cytology at light and electron microscope levels (Albini 1994; Ross et al. 1996). This feature has greatly facilitated the analysis of meiotic mutant phenotypes (e.g., Ross et al. 1997; Mercier et al. 2003), including detailed recombination assessment by chiasma analysis (Sanchez-Moran et al. 2001). Furthermore, Arabidopsis lacks meiotic check-point controls that are a feature of some organisms. This means that meiosis proceeds to its end point (tetrads) despite whatever defects are present. This has the great advantage that aspects of the mutant phenotype are not confused with abnormalities resulting from meiotic arrest and the onset of apoptosis. In this paper, we present a detailed molecular characterization of the AtMSH4 gene and its expression and assess its role in meiosis by a cytological and cytogenetical analysis of a T-DNA insertional mutant. Evidence is presented that two classes of crossovers exist in Arabidopsis, supporting an earlier suggestion based on the analysis and modeling of Arabidopsis genetic crossover data (Copenhaver et al. 2002).
Results
Identification and structure of AtMSH4
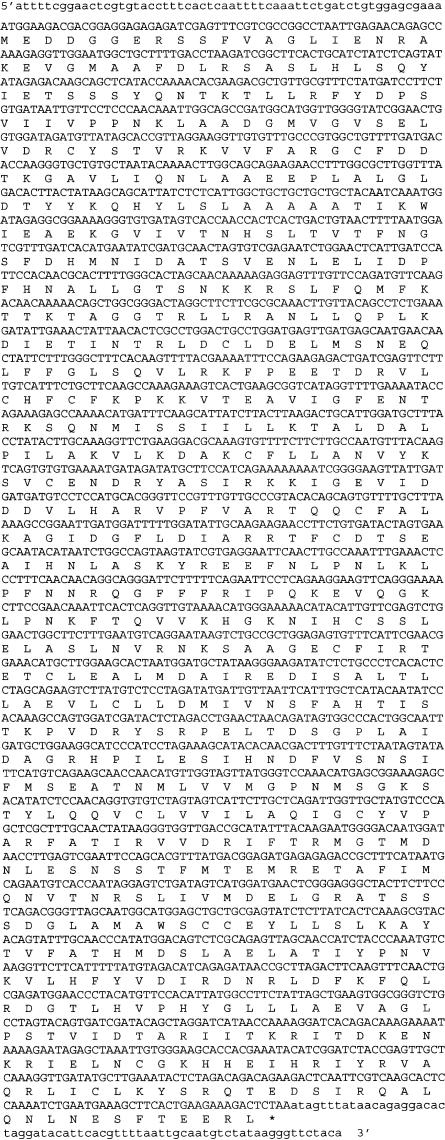
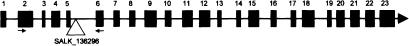
To identify a putative MSH4 gene in Arabidopsis, a homology search was performed using BLAST (NCBI) with the human, mouse, and yeast MSH4 amino acid sequences against the Arabidopsis genome sequence. The BLAST search produced one clear candidate with significant homology located at locus At4g17380. Primers designed against part of the coding region of this gene were used to amplify the corresponding cDNA using RT–PCR, which was cloned and sequenced. A full-length cDNA clone was then obtained using 5′- and 3′-RACE PCR with gene-specific primers based on the RT–PCR-derived sequence. Nucleotide sequence analysis of the cDNA sequence revealed that it comprised 2503 bp with an open reading frame of 2376 bp (Fig. 1). Comparison of the AtMSH4 cDNA sequence and the published genomic sequence of the At4g17380 locus (TAIR) allowed the intron/exon structure of the gene to be determined. AtMSH4 is composed of 23 exons and 22 introns (Fig. 2), which is different from the predicted gene structure (NM_117842). Sequence comparison revealed that the predicted sequence, at 1725 bp, is significantly shorter because of the omission of exons 7–10, 13, 22, and 23. Furthermore, the experimentally derived sequence exhibits a higher degree of homology than the predicted sequence with the MSH4 homologs from yeast and human.
Figure 1.
Nucleotide sequence of the Arabidopsis thaliana AtMSH4 cDNA including 5′- and 3′-untranslated regions. The deduced amino acid sequence of the AtMSH4 protein is presented below the cDNA.
Figure 2.
Map of the 5.6-kb At4g17380 locus showing the exon/intron organization of AtMSH4. The exons are represented by black boxes, the triangle shows where the SALK-136296 T-DNA has inserted, and the arrows indicate the positions of the primers used for the RT–PCR expression analysis (Fig. 3).
The AtMSH4 protein sequence
The AtMSH4 open reading frame encodes a predicted protein of 792 residues with a molecular mass of 89 kDa and pI of 7.26. Comparison of this amino acid sequence in computer databases indicated that it was most highly related to human MSH4 (35% identity and 57% similarity). It also had significant homology to the S. cerevisiae MSH4 (26% identity and 47% similarity), leading us to propose that we have identified the Arabidopsis MSH4 homolog. The protein contains several sequence motifs that are of potential functional significance. It has a putative nuclear localization signal (FKPKK, residues 266–269). AtMSH4 also contains highly conserved MutS domains including MutS II, III, IV, V (NCBI, conserved domain search). These domains are present in the meiosis-specific MutS homologs from yeast, humans, and C. elegans. The meiosis-specific MutS homologs do not contain domain I, which is essential to mismatch repair activity.
Domain I comprises the mismatch-recognition site, and in its absence the MutS dimer is unlikely to recognize mismatched DNA bases (Lamers et al. 2000). When present, this domain also forms part of the core of the MutS dimer, and its absence is predicted to leave a hole, large enough to encompass two DNA duplexes side by side (Obmolova et al. 2000). The fold of domain II that is present in AtMSH4 (residues 10–142) forms the other part of the core of the MutS dimer. Interestingly, this resembles the prokaryotic Holliday junction resolvase (RuvC) (Ariyoshi et al. 1994). Domain III contains a leucine zipper (residues 292–314) and is likely to connect domains II, IV, and V by peptide bonds to give the MutS structure (Obmolova et al. 2000). Domain IV is structurally similar to DNA gyrase and topoisomerase II in its dimeric form, and it has been proposed that it contains a suitable geometry for clamping DNA crossovers or Holliday junctions (Timsit 2001). Domain V contains the helix–turn–helix motif (residues 710–728) involved in dimerization of the MutS partners, which in the case of AtMSH4 is likely, by analogy with other systems, to be a homolog of MSH5. AtMSH4 also contains an ATPase domain, with the ATP-binding site (GPNMSGKS, residues 553–560). The ATPase is part of the ABC transporter superfamily, which includes RAD50 and may be involved in recruiting MutL homologs when the MutS heterodimer is clamped to DNA.
Expression of the AtMSH4 gene
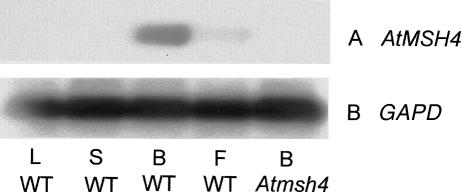
We examined the expression of AtMSH4 transcripts in different tissues using RT–PCR with gene-specific primers. Authenticity of the RT–PCR products was checked by hybridization to a radiolabeled AtMSH4 cDNA probe. There was no detectable AtMSH4 expression in somatic tissue (leaf and stem). The highest level of expression was found in bud and at a lower level in flower (Fig. 3). Bud tissue contains both male and female meiocytes at a range of meiotic stages, and although flowers represent a slightly later developmental stage than bud, this material also contains some embryo mother sac cells that are still in meiosis. This is caused by the asynchrony of meiosis in Arabidopsis, whereby meiosis in female reproductive tissues occurs later than in male tissues (Armstrong and Jones 2001). Thus these results are consistent with a meiotic role for AtMSH4 and are in agreement with results in other species such as S. cerevisiae, where expression of the gene is only detectable during meiosis (Ross-Macdonald and Roeder 1994), and humans, where expression is limited to the testes and ovaries (Paquis-Flucklinger et al. 1997).
Figure 3.
Expression analysis shows tissue-specific expression of AtMSH4. (A) RT–PCR analysis of expression in wild-type leaf (L), stem (S), flower bud (B), and open flower (F), and Atmsh4 mutant flower bud (lane 5). (B) Control for A using housekeeping gene GAPD.
AtMSH4 protein localizes to meiotic chromosomes in early prophase I
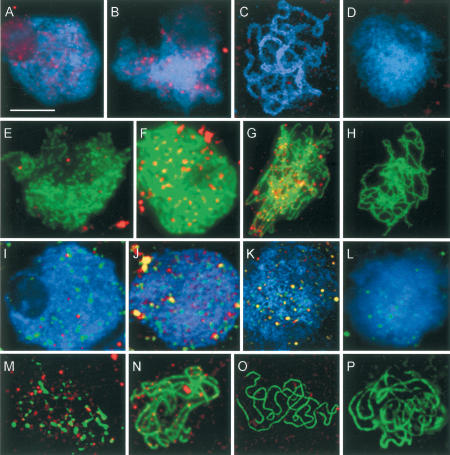
The intracellular distribution and chronology of AtMSH4 protein in meiocytes was investigated by fluorescence immunolocalization on DAPI-stained spread preparations of anthers from immature flower buds, using a polyclonal antibody raised in rabbit against AtMSH4 fusion protein (see Materials and Methods). The results indicated that AtMSH4 protein is restricted to meiocytes at prophase I of meiosis. Fluorescent foci first appeared in leptotene nuclei, persisting through zygotene and into very early pachytene (Fig. 4A–C). The foci were most numerous (80–100) over mid-leptotene nuclei, gradually diminishing in abundance through later leptotene and zygotene. By early zygotene, the number of foci reduced to ∼40 per nucleus, and by early pachytene, very few foci could be detected; later pachytenes were entirely devoid of foci, apart from prominent signals at the ends of each pachytene bivalent. These could indicate a specific association of AtMSH4 MSH4 protein with telomeric regions.
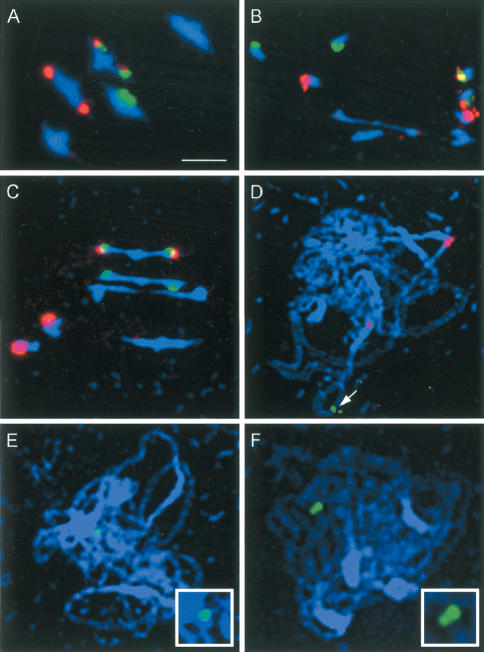
Figure 4.
(A–C) Immunolocalization of AtMSH4 protein (red) to early (A), mid (B), and late (C) wild-type prophase I nuclei, showing that AtMSH4 protein is present from mid-leptotene to late zygotene. Dual immunolocalization of AtMSH4 protein (red) with AtASY1 (E–G; green), AtRAD51 (I–K; green), and AtZYP1 (M–O; green), to early (E,I,M), mid (F,J,N), and late (G,K,O) wild-type prophase I nuclei. The patterns of colocalization with these three proteins allow a more precise description of the expression pattern of AtMSH4 (see text for details). (D,H,L,P) The corresponding immunolocalization patterns of these proteins in Atmsh4 mutant prophase I cells, showing expression of AtASY1, AtRAD51, and ATZYP1 but nonexpression of AtMSH4. Bar, 10 μm.
To determine more precisely the temporal and spatial distribution of AtMSH4 in meiotic nuclei, dual immunolocalization was performed using anti-AtMSH4 in combination with antibodies to other meiotic proteins (ASY1, AtRAD51, ZYP1) whose chronology had been previously established. ASY1, an axis-associated protein that is required for chromosome synapsis and normal level of recombination (Caryl et al. 2000), is first detected in G2 meiotic interphase/early leptotene, as discrete foci, ∼5–9 h post-S phase, well before the beginning of leptotene (Armstrong et al. 2003). By leptotene, ASY1 extends along the entire lengths of chromosome axes, giving continuous linear signals that conveniently mark the unsynapsed axes. In contrast, AtRAD51 protein first appears during leptotene, considerably later than ASY1 (Mercier et al. 2003). Dual immunolocalization experiments indicate that AtMSH4 appears after the loading of ASY1 onto meiotic chromosomes, when ASY1 labeling is continuous, that is, at leptotene. At this stage, AtMSH4 appears as numerous discrete foci that colocalize with ASY1-labeled axes (Fig. 4E–G), although at these early stages there are additionally many foci that are not axis-associated surrounding the spread chromosomes; these reduce in number by zygotene. In contrast, AtMSH4 and AtRAD51 foci appear almost simultaneously at leptotene and show strong colocalization in late leptotene/early zygotene nuclei (Fig. 4I–K). There are, however, some indications that AtRAD51 may precede AtMSH4 by a short interval, because we occasionally observed AtRAD51 signals over leptotene nuclei lacking AtMSH4 signals. These events can be shown to occur before zygotene and the onset of chromosome synapsis because in dual immunolocalization experiments, both AtRAD51 and AtMSH4 loading preceded the appearance of ZYP1, an Arabidopsis protein that immunolocalizes only to synapsed chromosome regions and is homologous to the yeast SC central region protein ZIP1 (J.D. Higgins and S.J. Armstrong, unpubl.). Furthermore, during zygotene the AtMSH4 foci were almost entirely limited to unsynapsed axes, that is, regions devoid of ZYP1 staining (Fig. 4M–O). The relatively few instances of colocalization of ZYP1 and AtMSH4 were confined to the extremities of some ZYP1 stretches, that is, to the boundaries of synapsed and unsynapsed regions. Evidently AtMSH4 protein does not persist on chromosome axes in Arabidopsis once synapsis is achieved.
Isolation and characterization of a T-DNA insertional Atmsh4 mutation
The SALK T-DNA lines database was screened for potential insertions in the AtMSH4 gene. A single putative insertional mutant line was identified, and a seed sample was obtained. Plants of this line were grown to maturity, and seed were collected and resown. Homozygous progeny plants, identified by PCR using gene-specific primers and the T-DNA left-border primer Lba1, showed normal vegetative growth and development, but they exhibited moderate to severe reduction in fertility, indicative of a probable meiotic defect. Mean silique length was reduced to 5.72 mm, compared with 12.75 mm in wild type, and mean seed-set was 3.53 per silique, representing 7.15% of normal wild-type seed set for the Col accession.
Fluorescence in situ hybridization (FISH) analysis showed that this line had just one T-DNA insertion site located in a mid-arm position on chromosome 4 (Fig. 5D). This was confirmed by Southern analysis, which gave a single band when genomic DNA was probed with a diagnostic T-DNA fragment (data not shown). PCR using gene-specific primers and a primer designed to the T-DNA left-border sequence confirmed the insertion and precisely mapped the insertion site within locus At4g17380 to position 97097026 in intron 5 of the AtMSH4 gene. Because the insert is located near the beginning of the gene, a null phenotype is likely to result. This was confirmed by failure to detect AtMSH4 expression in the mutant either by immunofluorescence (Fig. 4) or by RT–PCR (Fig. 3).
Figure 5.
(A–C) Representative metaphase I nuclei of wild-type (A) and Atmsh4 (B,C) cells after FISH to mark the locations of 5S rDNA (red) and 45S rDNA (green). (D–F) FISH of DNA probes to pachytene-stage nuclei of wild-type Arabidopsis. (D) The localization of the T-DNA insert (green) to chromosome 4 (arrowed); red signals show the locations of 5S rRNA loci on chromosomes 4 and 5. E and F illustrate single (E) and twin (F) BAC FISH signals (green) of the types recorded in the BAC synapsis assay. Bar, 5 μm.
Cytological investigation of DAPI-stained spread preparations confirmed that meiosis was, indeed, defective in the Atmsh4 mutant (Fig. 6). The most obvious phenotypic expression of this defect was the presence of numerous univalents at metaphase I, associated with a marked reduction in chiasma frequency (Figs. 5B,C, 6J). The mean chiasma frequency of Atmsh4, based on counts from 60 pollen mother cells, was 1.55 per cell (0.31 per chromosome pair), compared with 9.86 chiasmata per cell in wild type (1.97 per chromosome pair). In fact, 72% of chromosome pairs in Atmsh4 lacked chiasmata and were present as univalents at metaphase I. As expected, this resulted in frequent errors of chromosome segregation and the production of first and second division products having variable chromosome numbers (Fig. 6K,L).
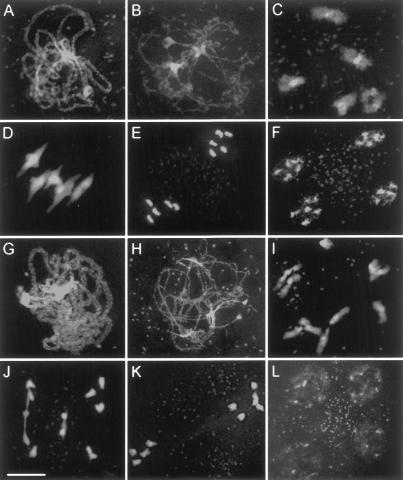
Figure 6.
Representative meiotic stages from wild-type (A–F) and Atmsh4 (G–L) pollen mother cells. (A,G) Pachytene. (B,H) Early diplotene. (C,I) Diakinesis. (D,J) Metaphase I. (E,K) Metaphase II. (F,L) Tetrads. Pachytene and early diplotene are generally similar in appearance in wild type and Atmsh4 (but see text). At diakinesis (I) and metaphase I (J), some univalents are present in Atmsh4 that can lead to uneven chromosome distribution to the second division (K) and tetrad (L) nuclei. Bar, 10 μm.
RNA interference confirms the meiotic function of AtMSH4
Formally, in the absence of a second mutant allele, we cannot exclude the remote possibility that a second-site untagged mutation is responsible for the meiotic phenotype. Therefore, to confirm the meiotic function of the AtMSH4 gene, we carried out an RNA interference (RNAi) experiment (Hamilton and Baulcombe 1999) to show that silencing or down-regulation of the AtMSH4 gene results in reduced fertility and reduced chiasma frequency. A 687-bp fragment of AtMSH4 cDNA was cloned into the pHannibal vector in sense and antisense orientations for generation of RNAi lines (see Materials and Methods). After transformation, 55 T1 transformants were identified based on seedling resistance to kanamycin, transferred to soil, and grown to maturity. T1 plants were initially screened for reduced silique length, as an indication of reduced fertility, and 10 candidate plants were then investigated cytologically, of which five showed reduced chiasma frequency to varying degrees (Fig. 7). Three of these plants (numbers 22, 25, and 28) were selected for further analysis. The mean cell chiasma frequencies of these plants were 1.90, 1.80, and 1.75, values that are similar to the chiasma frequency of the T-DNA mutant (1.55). Confirmation that the meiotic phenotypes observed resulted from RNAi-induced silencing or down-regulation of AtMSH4 was obtained by finding that these plants showed decreased levels of AtMSH4 transcript (see Supplementary Figs. 1, 2). Furthermore, immunolocalization using the anti-AtMSH4 antibody detected greatly reduced levels of AtMSH4 protein compared with wild-type controls in prophase I pollen mother cells of these plants (see Supplementary Fig. 3).
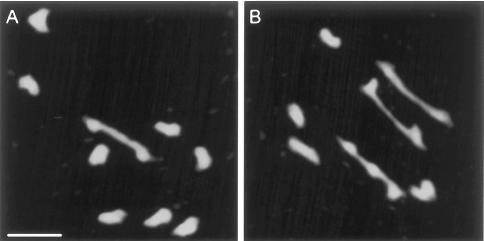
Figure 7.
DAPI-stained metaphase I cells from RNAi plant 25 showing severely reduced chiasma frequencies. Cell in A has a single bivalent and eight univalents (1 chiasma); cell in B has three bivalents and four univalents (3 chiasmata). Bar, 10 μm.
Early steps of meiotic chromosome morphogenesis, pairing, and recombination occur in the Atmsh4 mutant
Despite the disruption of later meiotic stages, the early development of meiotic nuclei appeared to proceed normally in Atmsh4 mutants. DAPI-stained meiotic interphase, leptotene, and zygotene were superficially normal in appearance. This semblance of normality was reinforced by finding that several key meiotic proteins that are required for chromosome morphogenesis, and for some essential steps in chromosome synapsis and recombination, were present in the mutant and appeared in the same sequence as in wild type during these early stages. SWI1 is a protein required for meiotic axis development and sister-chromatid cohesion that is present from late G1 interphase to the end of the meiotic S phase (data not shown). ASY1 is an axis-associated protein required for chromosome synapsis that appears during G2 interphase, whereas AtRAD51 catalyzes a key early step in meiotic recombination and appears during leptotene. Antibodies to these proteins and ZYP1 were applied to preparations of early meiotic stages of Atmsh4, and in each case their pattern of immunolocalization was indistinguishable from wild type (Fig. 4H,L,P). In contrast, the normal expression of AtMlh3, a protein associated with later stages of recombination, is AtMSH4-dependent because distinct AtMLH3 foci cannot be detected by immunolocalization in the Arabidopsis msh4 mutant (Fig. 4D).
Synapsis of homologous chromosomes in the Atmsh4 mutant is delayed and may be incomplete
Light microscopical (LM) analysis of DAPI- and silver-stained spreads initially indicated that synapsis was essentially complete in the Atmsh4 mutant. However, the higher resolution of electron microscopy (EM) revealed that fully and normally synapsed pachytenes were relatively uncommon in Atmsh4. Most nuclei at this stage showed some, usually limited, regions of asynapsis (Fig. 8B). However, in synapsed regions, the SCs appeared quite normal with normal dimensions and properly developed central elements. This cytological phenotype is suggestive of delayed synapsis in the Atmsh4 mutant, which would lead to a high proportion of cells showing partial synapsis and could result in a proportion of cells failing to achieve full synapsis. To investigate this possibility, a time-course experiment was conducted, based on BrdU labeling, as recently described by Armstrong et al. (2003). This experiment showed that the progress of cells through prophase I and the time course of synapsis are, indeed, delayed in Atmsh4, compared with wild type. The interval from S phase to the end of prophase I (diakinesis/metaphase I) lasts 30 h in the wild-type control, whereas the same interval lasts a minimum of 38 h in Atmsh4, which is a delay of ∼8 h.
Figure 8.
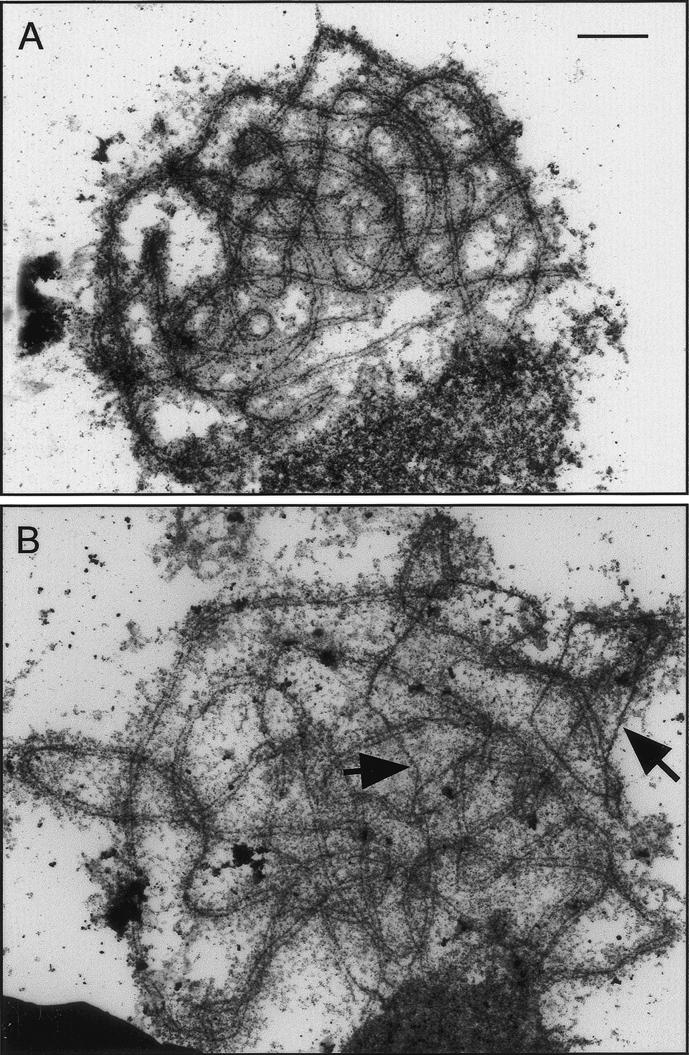
Electron micrographs of spread and silver-stained pachytene nuclei of wild-type (A) and Atmsh4 (B) cells. Synapsis is complete in wild type (A), but desynapsed regions are present in the Atmsh4 (B) example (arrowed). Bar, 2 μm.
In addition to direct LM and EM observations, synapsis and chromosome organization were investigated by fluorescence in situ hybridization (FISH) of BAC probes to prophase I nuclei. It was reasoned that if synapsis is locally incomplete in Atmsh4, this might be seen as a greater proportion of doublet as opposed to single FISH signals. A single Arabidopsis BAC (F24J1), located interstitially on the long arm of chromosome 1, was selected for analysis. FISH signals were scored as doublets if two dots could be clearly resolved, even if they were contiguous; otherwise they were scored as singles (Fig. 5E,F). In wild-type meiocytes, there were almost three times as many single signals as doublets (61:22), whereas in Atmsh4, we observed the opposite, that is, almost three times as many doublets as singles (46:16). This difference is highly significant (χ[1]2 = 32.72, P ≤ 0.001). This may reflect incomplete synapsis in Atmsh4, but could also be revealing an altered chromatin state in the mutant.
The overall conclusion from the cytological analysis of pairing/synapsis is that synapsis is often locally incomplete in Atmsh4, but that SC, where it forms, appears to exhibit normal structure and dimensions. Fully synapsed pachytenes were rather uncommon in Atmsh4, leading to the suggestion that these synaptic defects may be associated with delayed synapsis, a hypothesis that was confirmed by a time-course experiment.
Residual chiasmata in the Atmsh4 mutant are randomly distributed
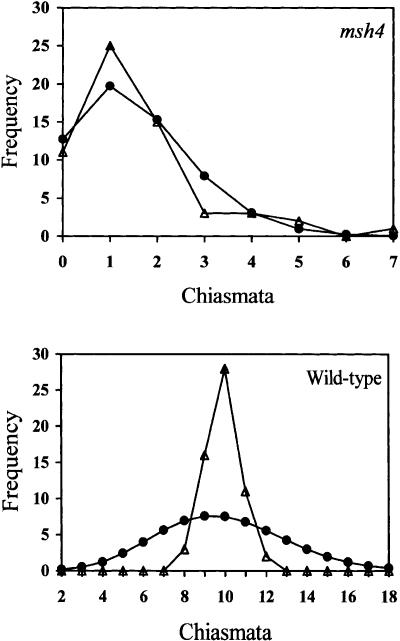
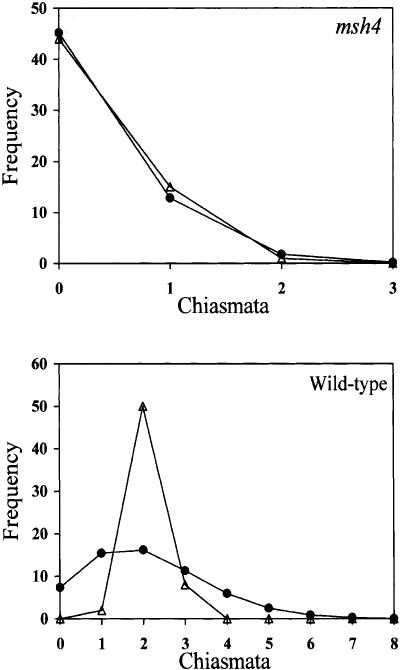
The Atmsh4 mutant is not entirely recombination-deficient, as it forms an average of 1.55 chiasmata and 1.40 bivalents per cell. The distributions of chiasma numbers per cell in Atmsh4 and in wild type (Col) are presented graphically in Figure 9. It can be seen that, despite the low mean chiasma frequency of Atmsh4, its chiasma numbers per cell are quite variable, ranging from 0 to 7, although the distribution is markedly skewed with its mode in the one-chiasma class. In fact, this distribution does not differ significantly from a Poisson distribution (χ(3)2 = 5.348; P > 0.1), indicating that the distribution of these residual chiasmata among cells is random. In contrast, wild type has many more chiasmata (9.86 per cell) and their distribution among cells deviates significantly from a Poisson distribution (χ(11)2 = 80.60, P < 0.001). Wild-type meiocytes exhibit a large excess of chiasma numbers around the mean, and no cases of fewer than eight or more than 12 chiasmata per cell were observed in our sample of 60 cells.
Figure 9.
Observed (open triangles) and Poisson-predicted (solid circles) distributions of chiasma numbers per cell for Atmsh4 mutant (top) and wild type (bottom).
In a similar way, the distributions of chiasmata were analyzed for each chromosome separately, in both Atmsh4 and wild type, and with the same conclusion. Whereas the distributions of chiasma numbers per chromosome for Atmsh4 fitted the Poisson distribution, in wild type the equivalent distributions were strongly non-Poissonian. Figure 10 shows the observed and Poisson-predicted frequencies for chromosome 3. Very similar results were obtained for the other four chromosomes (see Supplementary Table 1). These results provide additional evidence that the residual chiasmata that are formed by Atmsh4 are randomly distributed.
Figure 10.
Observed (open triangles) and Poisson-predicted (solid circles) distributions of chiasma numbers per chromosome 3 for Atmsh4 mutant (top) and wild type (bottom).
The detailed analysis of chiasma frequency and distribution in Atmsh4 was conducted on data collected from pollen mother cells. Cytological analysis of embryo-sac mother cells from ovules show that female meiosis is also defective in Atmsh4, based on the observation of numerous univalents and reduced chiasma frequency at metaphase I (data not shown).
Discussion
The meiotic phenotype of msh4 mutants in diverse organisms includes a variable chromosome pairing/synapsis defect, a severe reduction in crossing over, and (in yeast) the abolition or reduction of crossover interference. In this paper, we present details of the Atmsh4 mutant phenotype, based on detailed cytological and immunocytological observations combined with quantitative cytogenetical analysis of chiasma frequencies and distributions.
AtMSH4 expression and chromosome localization
It has been reported, based on immunofluorescence studies, that MSH4 protein localizes to discrete foci on meiotic chromosomes of yeast and mouse from early zygotene to pachytene (Kneitz et al. 2000; Novak et al. 2001). However, a later study including both immunofluorescence and EM immunogold localization concluded that MSH4 protein is loaded onto mouse chromosomes somewhat later in zygotene, at the time that early nodules are losing the RAD51/DMC1 component and undergoing a transformation to transitional nodules (Moens et al. 2002). The present study, based on dual localization of AtMSH4 with a panel of antibodies to other meiotic proteins, shows that AtMSH4 protein appears during leptotene. It localizes to unsynapsed axes during leptotene and zygotene, but is not present on synapsed regions of zygotene nuclei. Accordingly, as synapsis proceeds AtMSH4 decreases in abundance and, apart from a few residual foci, has disappeared by early pachytene. Interpretation of immunofluorescent MSH4 foci hinges on two factors. First, we have very limited knowledge or understanding of how the appearance and persistence of foci relate to the time of action of the molecules being detected. Second, the relationship of SC nucleation (at recombination initiation sites) to SC polymerization to give continuous SCs is likely to vary depending on organism, sex, and other factors. Therefore, the interspecific variation that has been reported regarding MSH4 loading onto chromosomes is not too surprising. The main conclusion to be drawn from our observations in Arabidopsis is that AtMSH4 protein is loaded onto chromosome axes very early in prophase I, probably soon after the appearance of DSBs and almost concurrently with the loading of AtRAD51. This is consistent with a role for AtMSH4 in an early step of crossover formation.
The absolute numbers of AtMSH4 foci associated with chromosome axes are likely significantly less than the total numbers detected by immunofluorescence because at early stages some of the foci are apparently not axis-associated. The best estimate of axis-associated AtMSH4 foci, corresponding to sites of synaptic initiation/recombination intermediates, comes from counts of coimmunolocalized AtMSH4 and AtRAD51, that gave ∼22 per nucleus in late leptotene/early zygotene. This number is significantly greater than the number of chiasmata (9.86), in contrast to the situation in budding yeast, where the number of crossovers (∼90) exceeds the number of ZMM-Synaptic Initiation Complexes (ZMM-SICs; ∼60) (Fung et al. 2004). It is, however, well known from earlier EM-based studies that the numbers of early SICs (early recombination nodules) exceed the eventual numbers of chiasmata in Angiosperm plants (e.g., Albini and Jones 1987). One frequently cited explanation for this excess is that plants require additional early recombinational interactions, over and above those destined to be crossovers, to ensure efficient homologous pairing of their relatively large chromosomes (Zickler and Kleckner 1999).
Residual MSH4 immunolocalization signals on pachytene nuclei are confined to the telomeric regions of each bivalent, indicating a specific association of MSH4 protein with telomeric regions and possibly with telomeric DNA. This is an intriguing parallel with the demonstration that the recombination protein RAD51D localizes to telomeres in meiotic and somatic cells of mouse, where it has been shown to be required for telomere maintenance (Tarsounas et al. 2004).
AtMSH4 is required for normal chromosome synapsis
One of the most variable aspects of the msh4 mutant phenotype in different species is its effect on chromosome synapsis during zygotene/pachytene. At one extreme, the him14 mutant of C. elegans shows apparently normal synapsis, although recombination in this mutant is virtually eliminated (Zalevsky et al. 1999). This finding is entirely consistent with the knowledge that, unlike budding yeast, mice, and Arabidopsis, synapsis in C. elegans is independent of recombination and depends instead on the existence of cis-acting pairing centers (Zetka and Rose 1995; Dernburg et al. 1998). At the other extreme, Kneitz et al. (2000) reported severe abnormalities of synapsis in msh4 knockout male mice; partial synapsis occurred in 70% of spermatocyte nuclei but mostly between nonhomologous chromosomes.
The Arabidopsis Atmsh4 phenotype most closely resembles that seen in budding yeast, in that superficially synapsis appears to proceed normally but on closer inspection is seen to be imperfect. In budding yeast, synapsis is delayed and is incomplete in 50% of meiocytes. In Arabidopsis, there is also some evidence of incomplete synapsis and evidence is presented that this is associated with delayed meiotic progression and delayed synapsis. It is not surprising, however, to find that SC polymerization is affected in this mutant as in other msh4 mutants. Much of the variation in phenotype that has been described is likely a reflection of interspecies differences in the relative timing and duration of SC nucleation versus SC polymerization.
AtMSH4 function; two classes of crossovers in Arabidopsis?
It was originally proposed that MSH4 interacts with recombination intermediates to influence their resolution (Ross-Macdonald and Roeder 1994) by binding to Holliday junctions and promoting their resolution in favor of crossing over (Novak et al. 2001). The more complex phenotype that is now apparent, combined with other factors, require a reassessment of the time and mode of action of MSH4. Another proposal was that the MSH4 protein may have dual or multiple functions in early and late events of chromosome pairing/synapsis/recombination, in which case it may participate in several different steps of the recombination pathway and be involved in interactions with different proteins at different stages (Kneitz et al. 2000). Alternatively, the primary defect may be at an early step in the recombination pathway, implying that the various facets of the meiotic phenotype are all consequences of this initial defect (Zickler and Kleckner 1999). Colaiacovo et al. (2003) also argue for a relatively early role for MSH4/MSH5 in the recombination process in C. elegans, based on the finding that these proteins are required for the timely disappearance of RAD51 foci. Our immunolocalization data are certainly consistent with this proposition. AtMSH4 protein is abundantly present as early as leptotene and is confined to unsynapsed axes during zygotene.
A possible early role for AtMSH4, based on the Atmsh4 mutant phenotype, could be to designate sites that are destined to become crossovers, after further downstream processing, and thereby establish interference. However, evidence from budding yeast indicates that the crossover/noncrossover decision precedes MSH4 action because msh4 mutants exhibit normal (not elevated) levels of noncrossovers despite a marked reduction in crossover frequencies (Ross-Macdonald and Roeder 1994, discussed by Bishop and Zickler 2004). Börner et al. (2004) also argue that recombinational interactions in budding yeast are differentiated into crossover and noncrossover types very early, “prior to SEI formation, and therefore prior to stable strand exchange.” They suggest that crossover control, including the imposition of interference, might be imposed at a point where DSBs are engaged in nascent undetected interactions with homologs, prior to SEI. The role of MSH4, and other associated proteins in the ZMM complex, must therefore be downstream of the crossover/noncrossover decision, to promote the crossover process (SEI and/or dHj formation) at sites that have been preselected for crossing over (Bishop and Zickler 2004; Börner et al. 2004). Current opinion holds that only a proportion of recombinational interactions are selected for processing into crossovers via SEIs and dHjs in budding yeast. The majority, it is thought, proceed by another pathway to noncrossovers, possibly by a synthesis-dependent strand-annealing mechanism (Allers and Lichten 2001) that does not involve long-lived SEIs and dHjs.
The position of the T-DNA insert in the Atmsh4 mutant and the absence of detectable MSH4 protein make it highly likely that the gene is completely disrupted, thereby giving a null phenotype. Therefore, the retention of a few chiasmata in Atmsh4 suggests that these represent a subset of chiasmata that are AtMSH4-independent. This suggestion is given added credence by the finding that these residual chiasmata are distributed randomly among cells and among chromosomes, implying, among other things, absence of crossover interference. This situation has strong parallels with recent proposals for the existence of two distinct classes of crossovers in S. cerevisiae as suggested by Zalevsky et al. (1999) and Novak et al. (2001), only one of which exhibits interference. de los Santos et al. (2003) proposed that these two classes are promoted by biochemically distinct pathways in S. cerevisiae. Class I events exhibit interference and are promoted by an MSH4/5-based complex, whereas class II events do not exhibit interference and are promoted by an MMS4/MUS81-based complex. The decrease of crossing over in budding yeast mms4 and mus81 mutants is modest, indicating that the majority of crossovers in wild-type yeast belong to class I.
Fung et al. (2004) have recently reported that synaptic initiation complexes (SICs), containing ZMM proteins, show interference in early prophase I nuclei of wild-type budding yeast, but interestingly also in zip1 and msh4 mutants that do not exhibit genetically detected crossover interference. The importance of this observation is that it demonstrates that interference is established at an early stage in the crossover process, well in advance of synapsis. The apparent conflict between the cytological and genetic interference observations in these mutants is most readily explained by the proposal that the crossovers that are abolished in msh4 and zip1 mutants are precisely those that exhibit genetic interference in wild type.
Börner et al. (2004) have estimated that ∼15% of crossovers in yeast arise independently of the main class I (ZMM) pathway, and suggest that these could be products of the noncrossover branch of the recombination pathway. This proportion is significant in view of the discussion that follows. In addition, de los Santos et al. (2003) indicate that class II crossovers appear to be more prominent between shorter chromosomes of budding yeast, suggesting that recombination responds to chromosome size by modulating the relative numbers of class I and class II crossovers. Interestingly in view of our findings, Copenhaver et al. (2002) have argued that the genetically determined crossover distribution in Arabidopsis is compatible with the existence of two pathways for crossing over. The quartet mutation of Arabidopsis was exploited to analyze crossover distribution in meiotic tetrads. They found that the fit to a theoretical “chi-square” distribution, which has been widely applied to modeling crossover interference, was substantially improved by assuming an additional set of crossovers sprinkled at random among those distributed as per chi-square. Based on this analysis they argued for the existence for two pathways for crossing over in Arabidopsis, as in yeast but unlike Drosophila and C. elegans, only one of which exhibits interference. Furthermore, they estimated that the proportion of crossovers without interference (p) was generally close to 0.20, although it occasionally fell below 0.10. Our finding that the residual chiasmata in the Atmsh4 mutant constitute 0.157 (1.55/9.86) of the wild-type value is remarkably consistent with this prediction. The proposal by Börner et al. (2004) that ∼15% of crossovers in yeast arise as minority products of the noncrossover recombination pathway is similarly consistent. It is, of course, an inference that the AtMSH4-independent chiasmata in our Atmsh4 mutant are also present in wild type as part of its normal complement of chiasmata. However, the analysis conducted by Copenhaver et al. (2002) supports this contention. Taken together, the various sources of evidence indicate strongly that Arabidopsis possesses two crossover pathways, exactly as predicted for budding yeast.
Copenhaver et al. (2002) found that chromosomes 1, 3, and 5 gave comparable estimates of p, with p = 0 ruled out, but for the short chromosomes, 2 and 4, there were insufficient data to rule out p = 0. They concluded that although the short chromosomes may not differ from the others with respect to p, it remained possible that crossing over on chromosomes 2 and 4 occurs only or primarily by the interference pathway. Our cytological observations on the Atmsh4 mutant indicate that there is no fundamental difference regarding the proportion of chiasmata without interference (class II) between the shorter chromosomes, 2 and 4 (0.162), and the longer chromosomes, 1, 3, and 5 (0.154). It would appear, therefore, that the smaller size, acrocentric structure, and nucleolus-organizing regions of chromosomes 2 and 4 have no effect on the balance of crossovers originating by interference and noninterference pathways, unlike the situation proposed to exist in budding yeast (de los Santos et al. 2003).
As class II crossovers are randomly distributed, they clearly do not ensure the occurrence of an obligate chiasma per bivalent that is required for efficient chromosome disjunction. It would be interesting, therefore, to know whether class I crossovers alone could satisfy this requirement. We infer that the great majority (85%) of cells in wild-type Arabidopsis have only 0, 1, or 2 class II chiasmata, whereas 98.3% have from 0 to 5 chiasmata belonging to this class. It is therefore reasonable to suppose that class I crossovers could satisfy the “obligate chiasma rule,” but to be sure we need to analyze meiosis in a mutant that is defective in the class II pathway.
The proposed existence of a minority subclass of chiasmata that do not exhibit interference has implications for current estimates of recombination frequency in Arabidopsis based on chiasma analysis. We infer that ∼80% of cells in wild-type Arabidopsis have at least one (one to seven) interference-free class II crossover. A proportion of these are therefore likely to occur nearby other crossovers and, as such, will be difficult or impossible to detect cytologically. This may explain the small but significant deficit of metaphase I chiasmata compared with genetic estimates of recombination frequency in Arabidopsis (Sanchez-Moran et al. 2001).
Materials and methods
The A. thaliana ecotype Columbia (0) was used in this study for wild-type analysis. The T-DNA insertion line SALK_136296 was obtained from the SALK institute via NASC for mutant analysis.
Plants were grown in a soil-based compost in a greenhouse under supplementary light (16 h light, 8 h dark cycle). Bud material consisted of the top of the inflorescence, cut above the first open flower, harvested throughout the plants' flowering period. Flowers comprised flowers with open petals, including mature stamens and carpels. Leaf material was a mixture of rosette and cauline leaves. Stem material consisted of internodal stem sections.
Nucleic acid extraction
Plant genomic DNA was isolated using the Nucleon Phytopure Kit (Amersham Life Sciences) and with the extract'n'amp plant PCR kit (Sigma-Aldrich). Bacterial plasmid DNA was isolated with the Wizard Prep kit (Promega). Total RNA was isolated using the RNeasy miniprep kit (QIAGEN) and then treated with DNase I (Promega) to remove residual genomic DNA.
Cloning the full-length AtMSH4 cDNA
The Superscript II RNase- reverse transcriptase (Invitrogen) was used with an Oligo-dT(16) primer to synthesize cDNA from 10 μg of Columbia (0) total bud RNA. The cDNA was subjected to PCR using REDTAQ (ABgene) with gene-specific primers F1, 5′-CGCATATGGCGCTTGGTTTAGACACTTAC-3′, and R1, 5′-CTCTCGAGTTCATCCATGACTATCAGACTC-3′, designed to amplify an internal cDNA fragment based on the TAIR prediction of At4g17380. A 1833-bp fragment was cloned into pCR2.1 (Invitrogen) and sequenced. This sequence was used to design specific primers for RACE–PCR. 3′-RACE was carried out using the 17AP, 5′-GACTCGAGTCGACATCGATTTT TTTTTTTTTTTTT-3′ and 3′RACE 5′-GAGTCGAATTCCA GCACGTT-3′, and 3′-RACE nested 5′-GATAGTCATGGA TGAACTCG-3′ primers. The 5′-RACE was carried out as follows: cDNA was synthesized from 20 μg of total bud RNA using the Superscript II RNase- reverse transcriptase (Invitrogen) with the R2 primer 5′-GCGTTGTGGAATGGATCAATG-3′. The cDNA was tailed with dCTP using Terminal Transferase (Roche). Nested PCR was then performed with the following gene-specific primers: 5′-RACE, 5′-AGAGATAATGCTGCTT ATA-3′ and 5′-RACE nested 5′-GTAAGTGTCTAAACCAA GCG-3′ together with the 5′-RACE Abridged Anchor primer 5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′ (Invitrogen). The PCR products were cloned into pDrive (QIAGEN) and sequenced. The annotated sequence is lodged with GenBank, accession number AY646927.
Semiquantitative RT–PCR for transcript expression
The Onestep RT–PCR kit (QIAGEN) was used to amplify gene-specific products from DNase I-treated total RNA extracted from Arabidopsis wild-type Col(0) leaf, stem, bud, flower, and bud tissue from Salk_136296 and RNAi plants 15, 22, and 28. Primers were designed to amplify a housekeeping gene GAPD to equalize RNA loading into the RT–PCR reaction. These were GAPDF1, 5′-CTTGAAGGGTGGTGCCAAGAAG G-3′, and GAPDR1, 5′-CCTGTTGTCGCCAACGAAGTCAG-3′. The AtMSH4 gene-specific primers were MSH4EXPF1, 5′-CATATGGAGGTTGGAATGGCTGC-3′, and MSH4EXPR1, 5′-GCGTTGTGGAATGGATCAATG-3′. DNA was transferred from an agarose gel onto Hybond N+ membrane (Amersham) by capillary blotting. The blot was hybridized in modified Church and Gilbert (1984) buffer and probed using radiolabeled F1/R1 PCR product (Feinberg and Vogelstein 1983, 1984). Blots were washed at high stringency (0.2× SSC, 0.1% SDS). Radioactivity was detected by autoradiography.
T-DNA insertion site mapping
The T-DNA insertion site of SALK_136296 was mapped with primers LBa1 and R2. The PCR products were cloned into pDrive (QIAGEN) and sequenced. Pairs of primers were used to determine if the plants were homozygous or heterozygous for the T-DNA insertion. For SALK_136296, primers F1 and MSH4EXPR1 were used to amplify the wild-type genomic region and primers LBa1 and T-DNAR2 to amplify the region where the T-DNA had inserted.
Construction of AtMSH4 RNA interference cassette
A 687-bp fragment of AtMSH4 (between 508 and 1195 bp relative to the ATG+1) was amplified from Arabidopsis bud cDNA with the following primers showing restriction sites in bold: RNAiF, 5′-GGATCCTCGGAGAATCTGGAACTCATGGATC C-3′ (BamHI, XhoI), and RNAiR, 5′-GAAGCTTGAATTCCT CACGATACTTACTGGCCAG-3′ (HindIII, EcoRI). The PCR fragment was cloned into pDrive (QIAGEN) and sequenced. An EcoRI/XhoI digest produced a sense fragment when ligated into the same sites of pHannibal (Wesley et al. 2001) and a BamHI/HindIII digest produced an antisense fragment when ligated into the same sites of pHannibal. An NotI digest of pHannibal containing the CaMV35S promoter, sense-intron-antisense AtMSH4 sequences, and OCS terminator produced a 4.3-kb fragment that was ligated into the binary vector pART27 (Gleave 1992).
Plant transformation
The binary plasmid pART27 containing the AtMSH4 RNAi construct was introduced into Agrobacterium tumefaciens (LBA4404) by electroporation. Arabidopsis plants were transformed using the floral-dip method (Clough and Bent 1998). The harvested seeds were selected on Murashige and Skoog (1962) medium containing 50 mg/mL kanamycin. Kanamycin-resistant plantlets were transferred to soil in the greenhouse. PCR was performed on these plants to test for the presence of the pHannibal intron with primers pHanF, 5′-TCCCAACTGTA ATCAATCC-3′, and pHanR, 5′-GACAAGTGATGTGTAAG ACG-3′.
Antibody production
An NdeI site was designed into primer F1 and an XhoI site into primer R1. The 1833-bp cloned fragment containing primers F1/R1 was digested with NdeI/XhoI and ligated in-frame into the NdeI/XhoI restriction sites of expression vector pET21b (Novagen). The expression vector was transferred to E. coli Bl21 (Novagen) cells. Upon induction, the 611-amino acid recombinant protein accumulated as insoluble inclusion bodies. Purified, refolded recombinant protein was prepared as described previously (Kakeda et al. 1998). Rabbit and rat polyclonal anti-sera were produced against the recombinant protein (ISL).
Nucleic acid sequencing
Automated nucleotide sequencing was carried out by the Genomics Laboratory, Biosciences, University of Birmingham, UK.
Cytological procedures
Mutant and wild-type pollen mother cells (PMCs) were examined by light microscopy in DAPI- and silver-stained spreads as described by Ross et al. (1996) and Armstrong et al. (2001). Female meiosis was examined in DAPI-stained spreads of embryo sac mother cells as described by Armstrong and Jones (2001).
Fluorescence immunolocalization was carried out as described by Armstrong et al. (2002) and Mercier et al. (2003). The following antibodies were used: anti-AtMSH4 (rabbit + rat), anti-AtSWl1 (rabbit), anti-ASY1 (rat), anti-AtRAD51 (rabbit), anti-ZYP1 (rat), and anti-AtMLH3 (rabbit).
Chiasmata were recorded by light microscopy from spread PMCs after fluorescence in situ hybridization (FISH) to identify individual bivalents (Sanchez-Moran et al. 2001).
FISH experiments to confirm the location of the T-DNA insertion and to detect the BAC used in the synapsis assay were carried out as described by Caryl et al. (2000) and Armstrong et al. (2001).
The meiotic time-course experiment was conducted as described by Armstrong et al. (2003).
Electron microscopical examination of wild-type and mutant PMCs was performed on spread preparations by a modification of the method described by Albini (1994). Individual flower buds from five inflorescences were separated, kept on moist filter paper, and graded by size. Only buds smaller than 0.2 mm, containing leptotene-to-pachytene stages, were used. Individual anthers were dissected out on damp filter paper, producing a pool of ∼30 anthers. The anthers were placed together in a cavity slide containing 10 μL of digestion medium (Albini 1994; Armstrong et al. 2003). The anthers were gently tapped with a brass rod to release meiocytes and left for 5 min in a moisture chamber at 33°C. Two-microliter aliquots of this suspension were placed onto 10-μL drops of 1% Lipsol (1% Lipsol detergent in freshly distilled water buffered to pH 9.0 with borate buffer) on glow-discharged plastic-coated slides. This suspension was left in a moisture chamber at 33°C for a further 5 min. The meiocyte suspension was gently dispersed over the slide using a glass spreader and finally fixed by adding 4% paraformaldehyde (pH 8.0) to the slide, and placed in a fume cupboard until dry. The slides were silver-stained as described by Albini (1994). The stained slides were examined by light microscopy, and well-spread cells were marked with black ink before floating the plastic supporting film onto a clean water surface. Copper “finder” grids were placed over the marked areas, and the plastic film was picked up on clean paper and dried. Grids were examined in a JEOL 1200 EX electron microscope.
Statistical procedures
The observed and Poisson-expected numbers of chiasmata per bivalent and per cell were tested for agreement by means of chi-squared (χ2) tests. For the purposes of these tests, the “chiasma number” classes were grouped such that none of the expected values fell below 1 and not more than a third were in the range 1–5 (Lancaster 1966). The degrees of freedom in each case were given by the number of classes (after any grouping) minus 2 (because it was necessary to estimate a parameter, μ, the mean).
Acknowledgments
We are grateful to the Biotechnology and Biological Science Research Council for financial support. Nancy Kleckner read an early draft of the paper and made several helpful comments and suggestions. Valuable technical support was provided by Stephen Price, Karen Staples, and the University of Birmingham EM Unit.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.317504.
References
- Albini S.M. 1994. A karyotype of the Arabidopsis thaliana genome derived from synaptonemal complex analysis at prophase I of meiosis. Plant J. 5: 665-672. [Google Scholar]
- Albini S.M. and Jones, G.H. 1987. Synaptonemal complex spreading in Allium cepa and A. fistulosum. I. The initiation and sequence of pairing. Chromosoma 95: 324-338. [Google Scholar]
- Allers T. and Lichten, M. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47-57. [DOI] [PubMed] [Google Scholar]
- Ariyoshi M., Vassylyev, D.G., Iwasaki, H., Nakamura, H., Shinagawa, H., and Morikawa, K. 1994. Atomic structure of the RuvC resolvase: A Holliday junction-specific endonuclease from E. coli. Cell 78: 1063-1072. [DOI] [PubMed] [Google Scholar]
- Armstrong S.J. and Jones, G.H. 2001. Female meiosis in wildtype Arabidopsis thaliana and in two meiotic mutants. Sex Plant Reprod. 13: 177-183. [Google Scholar]
- Armstrong S.J., Franklin, F.C.H., and Jones, G.H. 2001. Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J. Cell Sci. 114: 4207-4217. [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Caryl, A.P., Jones, G.H., and Franklin, F.C.H. 2002. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115: 3645-3655. [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Franklin, F.C.H., and Jones, G.H. 2003. A meiotic time-course for Arabidopsis thaliana. Sex. Plant Reprod. 16: 141-149. [Google Scholar]
- Bishop D.K. and Zickler, D. 2004. Early decision: Meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117: 9-15. [DOI] [PubMed] [Google Scholar]
- Börner G.V., Kleckner, N., and Hunter, N. 2004. Crossover/noncrossover differentiation, synaptonemal complex formation and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29-45. [DOI] [PubMed] [Google Scholar]
- Caryl A.P., Armstrong, S.J., Jones, G.H., and Franklin, F.C.H. 2000. A homologue of the yeast gene HOP1 is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109: 62-71. [DOI] [PubMed] [Google Scholar]
- Caryl A.P., Jones, G.H., and Franklin, F.C.H. 2003. Dissecting plant meiosis using Arabidopsis thaliana mutants. J. Exp. Bot. 54: 25-38. [DOI] [PubMed] [Google Scholar]
- Church G.M. and Gilbert, W. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. 81: 1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J. and Bent, A.F. 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735-743. [DOI] [PubMed] [Google Scholar]
- Colaiacovo M.P., MacQueen, A.J., Martinez-Perez, E., McDonald, K., Adamo, A., La Volpe, A., and Villeneuve, A.M. 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but not for proper completion of recombination. Dev. Cell 5: 463-474. [DOI] [PubMed] [Google Scholar]
- Copenhaver G.P., Housworth, E.A., and Stahl, F.W. 2002. Crossover interference in Arabidopsis. Genetics 160: 1631-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T., Hunter, N., Lee, C., Larkin, B., Loidl, J., and Hollingsworth, N.M. 2003. The Mus81/Mms81 endonuclease acts independently of double Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A.F., McDonald, K., Moulder, G., Barstead, R., Dresser, M., and Villeneuve, A.M. 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387-398. [DOI] [PubMed] [Google Scholar]
- Feinberg A.P. and Vogelstein, B. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132: 6-13. [DOI] [PubMed] [Google Scholar]
- ____. 1984. Addendum: A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137: 266-267. [DOI] [PubMed] [Google Scholar]
- Fung J.C., Rockmill, B., Odell, M., and Roeder, G.S. 2004. Imposition of crossover interference through nonrandom distribution of synapsis initiation complexes. Cell 116: 795-802. [DOI] [PubMed] [Google Scholar]
- Gleave A.P. 1992. A versatile binary vector system with a T-DNA organisational structure conductive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203-1207. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe, D.C. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950-952. [DOI] [PubMed] [Google Scholar]
- Hawley R.S. 1988. Exchange and chromosome segregation in eukaryotes. In Genetic recombination (eds. R. Kucherlapati and G.R. Smith), pp. 497-528. American Society for Microbiology, Washington, DC.
- Holliday R. 1964. A mechanism for gene conversion in fungi. Genet. Res. 5: 282-304. [DOI] [PubMed] [Google Scholar]
- Hollingsworth N.M. and Brill, S.J. 2004. The Mus81 solution to resolution: Generating meiotic crossovers without Holliday junctions. Genes & Dev. 18: 117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakeda K., Jordan, N.D., Conner, A., Ride, J.P., Franklin-Tong, V.E., and Franklin, F.C.H. 1998. Identification of residues in a hydrophilic loop of the Papaver rhoeas S protein that play a crucial role in recognition of incompatible pollen. Plant Cell 10: 1723-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1-53. [DOI] [PubMed] [Google Scholar]
- Kneitz B., Cohen, P.E., Avdievich, E., Zhu, L., Kane, M.F., Hou Jr., H., Kolodner, R.D., Kucherlapati, R., Pollard, J.W., and Edelmann, W. 2000. MutS homolog 4 localization is required for chromosome pairing during meiosis in male and female mice. Genes & Dev. 14: 1085-1097. [PMC free article] [PubMed] [Google Scholar]
- Lamers M.H., Perrakis, A., Enzlin, J.H., Winterwerp, H.H.K., de Wind, N., and Sixma, T.K. 2000. The crystal structure of DNA mismatch repair protein MutS binding to a G-T mismatch. Nature 407: 711-717. [DOI] [PubMed] [Google Scholar]
- Lancaster H.O. 1966. The chi-squared distribution. John Wiley and Sons, New York.
- Loidl J. 2000. Meiosis in budding yeast and in multicellular eukaryotes—Similarities and differences. In Chromosomes today (eds. E. Olmo and C.A. Redi), Vol. 13, pp. 123-137. Birkhauser Verlag, Switzerland. [Google Scholar]
- Mercier R., Armstrong, S.J., Horlow, C., Jackson, N.P., Makaroff, C.A., Vezon, D., Pelletier, G., Jones, G.H., and Franklin, F.C.H. 2003. The meiotic protein Swi1 is required for axial element formation and recombination initiation in Arabidopsis thaliana. Development 130: 3309-3318. [DOI] [PubMed] [Google Scholar]
- Moens P.B., Kolas, N.K., Tarsounas, M., Marcon, E., Cohen, P., and Spyrapoulos, B. 2002. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA–DNA interactions without reciprocal recombination. J. Cell Sci. 115: 1611-1622. [DOI] [PubMed] [Google Scholar]
- Murashige T. and Skoog, F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant 15: 473-497. [Google Scholar]
- Novak J.E., Ross-Macdonald, P.B., and Roeder, G.S. 2001. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158: 1013-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obmolova G., Ban, C., Hsieh, P., and Yang, W. 2000. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407: 703-710. [DOI] [PubMed] [Google Scholar]
- Paquis-Flucklinger V., Santucci-Darmanin, S., Paul, R., Saunieres, A., Turc-Carel, C., and Desnuelle, C. 1997. Cloning and expression analysis of a meiosis-specific MutS homolog: The human MSH4 gene. Genomics 44: 188-194. [DOI] [PubMed] [Google Scholar]
- Roeder G.S. 1997. Meiotic chromosomes: It takes two to tango. Genes & Dev. 11: 2600-2621. [DOI] [PubMed] [Google Scholar]
- Ross K.J., Fransz, P., and Jones, G.H. 1996. A light microscope atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4: 507-516. [DOI] [PubMed] [Google Scholar]
- Ross K.J., Fransz, P., Armstrong, S.J., Vizir, I., Mulligan, B., Franklin, F.C.H., and Jones, G.H. 1997. Cytological characterisation of four meiotic mutants of Arabidopsis isolated from T-DNA transformed lines. Chromosome Res. 5: 551-559. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P. and Roeder, G.S. 1994. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79: 190-195. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E., Armstrong, S.J., Santos, J.L., Franklin, F.C.H., and Jones, G.H. 2001. Chiasma formation in Arabidopsis thaliana accession Wassileskjia and in two meiotic mutants. Chromosome Res. 9: 121-128. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco, D., and Szostak, J.W. 1991. Extensive 3′-over-hanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64: 1155-1161. [DOI] [PubMed] [Google Scholar]
- Szostak J.W., Orr-Weaver, T.L., Rothstein, R.J., and Stahl, F.W. 1983. The double-strand-break repair model for recombination. Cell 33: 25-35. [DOI] [PubMed] [Google Scholar]
- Tarsounas M., Munoz, P., Claas, A., Smiraldo, P.G., Pittman, D.L., Blasco, M.A., and West, S.C. 2004. Telomere maintenance requires the RAD51D recombination/repair protein. Cell 117: 337-347. [DOI] [PubMed] [Google Scholar]
- Timsit Y. 2001. Convergent evolution of MutS and topoisomerase II for clamping DNA crossovers and stacked Holliday junctions. J. Biomol. Struct. Dynamics 19: 215-218. [DOI] [PubMed] [Google Scholar]
- Wesley S.V., Helliwell, C.A., Smith, N.A., Wang, M.B., Rouse, D.T., Liu, Q., Gooding, P.S., Singh, S.P., Abbott, D., Stoutjesdijk, P.A., et al. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581-590. [DOI] [PubMed] [Google Scholar]
- Zalevsky J., MacQueen, A., Duffy, J.B., Kemphues, K.J., and Villeneuve, A.M. 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka M. and Rose, A. 1995. The genetics of meiosis in Caenorhabditis elegans. Trends Genet. 11: 27-31. [DOI] [PubMed] [Google Scholar]
- Zickler D. and Kleckner, N. 1999. Meiotic chromosomes: Integrating structure and function. Ann. Rev. Genet. 33: 603-754. [DOI] [PubMed] [Google Scholar]