Abstract
Diploidization, which follows whole genome duplication events, does not occur evenly across the genome. In salmonid fishes, certain pairs of homeologous chromosomes preserve tetraploid loci in higher frequencies toward the telomeres due to residual tetrasomic inheritance. Research suggests this occurs only in homeologous pairs where one chromosome arm has undergone a fusion event. We present a linkage map for Arctic charr (Salvelinus alpinus), a salmonid species with relatively fewer chromosome fusions. Genotype by sequencing identified 19,418 SNPs, and a linkage map consisting of 4508 markers was constructed from a subset of high quality SNPs and microsatellite markers that were used to anchor the new map to previous versions. Both male- and female-specific linkage maps contained the expected number of 39 linkage groups. The chromosome type associated with each linkage group was determined, and 10 stable metacentric chromosomes were identified, along with a chromosome polymorphism involving the sex chromosome AC04. Two instances of a weak form of pseudolinkage were detected in the telomeric regions of homeologous chromosome arms in both female and male linkage maps. Chromosome arm homologies within the Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss) genomes were determined. Paralogous sequence variants (PSVs) were identified, and their comparative BLASTn hit locations showed that duplicate markers exist in higher numbers on seven pairs of homeologous arms, previously identified as preserving tetrasomy in salmonid species. Homeologous arm pairs where neither arm has been part of a fusion event in Arctic charr had fewer PSVs, suggesting faster diploidization rates in these regions.
Keywords: diploidization, duplicated genes, epigenetic modification, linkage map, salmonid fishes, transmission genetics, transposition
Whole genome duplications (WGDs) are rare evolutionary events that drastically alter genomic architecture by producing duplicate copies of every chromosome. The doubling of all loci can be a major driving force of evolution, as there is the potential to produce loci with novel functions through the accumulation of mutations that were formerly selected against (Ohno 1970). WGDs provide a surfeit of genetic information that can be associated with adaptive innovation and evolutionary change (Taylor and Raes 2004; Moghadam et al. 2005; Nakatani et al. 2007; Berthelot et al. 2014). WGDs have been important events in the evolutionary history of vertebrate animals (Smith et al. 2013). Two WGDs occurred in the common ancestor of all vertebrates, and these (referred to as the 1R and 2R WGDs) yielded a karyotype of between 40 and 52 chromosomes from the protovertebrate karyotype of 10–13 chromosomes (Nakatani et al. 2007). A third (3R) WGD occurred ∼400 million yr ago (MYA) in the ancestor of teleost fish, while a fourth salmonid specific WGD (Ss4R) took place ∼96 MYA in the ancestor of salmonid fishes (Allendorf and Thorgaard 1984; Berthelot et al. 2014; Macqueen and Johnston 2014; Lien et al. 2016). Subsequent chromosome fusions and fissions caused changes in chromosome numbers and genomic architecture in specific vertebrate lineages (Kasahara et al. 2007; Nakatani et al. 2007).
Following WGD, the genome undergoes the process of diploidization, where it reverts from a tetraploid (4n) to a diploid (2n) state. Specifically, chromosome pairs that shared a common genetic ancestor prior to WGD (termed homeologs), diverge from one another due to genomic rearrangements, gene deletion, pseudogenization, and mutation (Comai 2005; Bergthorsson et al. 2007). Salmonids are in the midst of the diploidization process, in that some regions of the genome have diverged into two pairs of diploid loci, while in other regions, residual tetrasomy occurs as the result of multivalent formation and recombination among homeologues during meiosis (Berthelot et al. 2014; Allendorf et al. 2015; May and Delany 2015; Lien et al. 2016). For example, 52% of genes in the rainbow trout (Oncorhynchus mykiss) genome have diploidized since the Ss4R WGD, while the other 48% of genes have retained both copies, and have yet to revert to a diploid state (Berthelot et al. 2014). Similarly, work on the Atlantic salmon (Salmo salar) genome has found that 55% of genes have been retained as two functional copies since the Ss4R WGD (Lien et al. 2016). Chromosomal architecture appears to play a role in determining which genomic regions undergo homeologous recombination during meiosis in these fishes (Brieuc et al. 2014; Lien et al. 2011; Kodama et al. 2014; Waples et al. 2015). All recombining pairs of homeologs appear to include one large chromosome, either a metacentric produced through the Robertsonian fusion of two chromosome arms, or a fused acrocentric resulting from the tandem fusion of two smaller acrocentric chromosomes. This suggests that the large size of these chromosomes may provide the stability necessary for homeologous recombination (Brieuc et al. 2014; Kodama et al. 2014; Lien et al. 2016). Furthermore, the telomeric regions of homeologs show the slowest rates of diploidization within several salmonid species, as illustrated by a relatively high number of paralogous loci (Brieuc et al. 2014; Waples et al. 2015; Lien et al. 2016).
The development of detailed linkage maps has shown that lineage specific changes in chromosomal architecture have taken place in salmonids following WGD. Maps are available for species of Salmo and Oncorhynchus (Rexroad et al. 2008; Gonen et al. 2014; Limborg et al. 2015; McKinney et al. 2015; Waples et al. 2015; Tsai et al. 2016). In the karyotypes of certain species such as Atlantic salmon, a large number of chromosomes are the product of fusions (e.g., 42/58) (Phillips and Ráb 2001). Variation in genomic architecture may cause the rate of diploidization to vary across the genomic landscape of the different taxa, and which regions of the genome undergo residual tetrasomy (Lien et al. 2011; Brieuc et al. 2014; Kodama et al. 2014; Lien et al. 2016). Similar studies of taxa with more basal karyotypes, such as the Salvelinus species, would improve our understanding of the role that genomic rearrangements have in the diploidization process. For instance, karyotype data suggests that only 20 of the 78 chromosomes (2n = 78) in Arctic charr (Salvelinus alpinus) are metacentric, suggesting that fewer Robertsonian fusion events have occurred in their evolutionary history (Hartley 1989; Phillips et al. 2002). The study of genomic evolution in Arctic charr is currently limited by the low resolution of available genetic linkage maps, and the paucity of known molecular markers (Woram et al. 2004; Norman et al. 2011; Timusk et al. 2011). Expanding the genomic resources of Arctic charr through the addition of several thousand SNPs will make it possible to track the genomic rearrangements that have shaped the modern Arctic charr karyotype.
The initial aims of this study were to (1) increase the number of known genetic markers in the Arctic charr genome using genotype by sequencing (GBS); (2) create a second generation genetic linkage map of the Arctic charr genome using newly identified SNP markers, and integrate the revised map to previous versions primarily based on microsatellite loci; and (3) identify the chromosome type associated with each linkage group, and use this information to test for the existence of acrocentric-acrocentric homeologous pairs in the genome. These are of interest because a lack of homeologous recombination could cause diploidization to occur faster in homeologous pairs of this type. Using the data produced to meet the above goals, we also aimed to (4) compare genomic data from Arctic charr to the genomes of rainbow trout and Atlantic salmon to identify chromosome arm homologies across these three species, allowing the characterization of conserved genomic rearrangements and fusion/fission events unique to the Arctic charr lineage; and, also, (5) identify putative duplicate loci, and assess their distribution across the Arctic charr genome. As we analyzed sequence data, it became apparent that there were signatures of significant transposon activity within the Arctic charr genome. We therefore used this opportunity to characterize transposable element (TE) activity in the Arctic charr genome, and see how TE distribution is affected by residual tetrasomy, chromosome architecture, and the uneven distribution of duplicate loci throughout the genome.
Transposon activity is associated with important evolutionary transitions, adaptation to novel environments, and extensive changes in genome evolution (de Boer et al. 2007; Schrader et al. 2014; Staton and Burke 2015). In fact, the estimated time of radiation of Salmoninae into Salmo, Oncorhynchus, and Salvelinus (14–23 MYA) (Macqueen and Johnston 2014) coincides with a known spike in TE activity (de Boer et al. 2007). TEs cause sequence deletion or duplication due to unequal homologous recombination and segmental duplication, and they facilitate genomic rearrangements (Kazazian 2004). Genomic regions with an accumulation of TEs appear to evolve faster than the rest of the genome (Schrader et al. 2014). Therefore, TE activity might be influenced by the rate of residual tetrasomy in particular chromosomal regions, or vice versa. Reduced presence of TEs has been observed in the duplicated regions of Atlantic salmon, chum salmon (O. keta), and chinook salmon (O. tshawytscha) that lag behind in the diploidization process (Larson et al. 2015; McKinney et al. 2015; Waples et al. 2015; Lien et al. 2016). Therefore, our final goal was to characterize TE activity in the Arctic charr genome, and determine if there is a relationship between residual tetrasomy and TE activity in the telomeric regions of homeologs that undergo homeologous recombination.
Materials and Methods
Source mapping panel
The analysis utilized 85 full-siblings, and their parents, from a single family of Fraser strain Arctic charr obtained from the Coastal Zones Research Institute (CZRI), Shippagan, NB Canada. The Fraser strain originated from collections of fish from the Fraser River, Labrador, Canada, in the 1980s. The family was produced on November 6, 2012, and reared communally until March 11, 2014, at which time each fish was PIT tagged, weighed (to the nearest gram), measured (fork length), and samples of adipose fin were removed for DNA analysis. DNA was extracted using a commercial kit (Qiagen DNeasy Blood & Tissue), as per manufacturer’s instructions, and treated with RNase A to remove any RNA. The samples were then quantified using a Qubit Fluorometer to ensure that all DNA concentrations exceeded 50 ng/µl.
Sequencing analysis
The DNA samples were submitted for GBS (Elshire et al. 2011) at the Cornell Institute of Biotechnology. DNA from each progeny was added to a single well of a 96-well plate, while the parents were analyzed in triplicate, to increase sequencing depth and provide the information necessary for linkage mapping based on the inheritance of SNP alleles. The four grandparents of the family were also added to a single well each. Samples were digested with the restriction enzyme EcoT22I, and unique barcode sequence adapters (4–8 bp in length) were ligated to each of the DNA samples (three for each parent) such that the DNA sequence data could be assigned to a specific individual or parental subsample. After the barcodes were added, sequencing primers, and the samples from all 95 wells containing DNA samples (and a single blank control well) were pooled. Paired end sequencing primers with oligonucleotides that allow binding to the sequencing flowcell were then added to the pooled samples. Polymerase chain reaction (PCR) was then used to amplify the DNA fragment pool, and the resulting DNA products were analyzed for fragment size. The DNA samples were then sequenced on an Illumina Hisequation 2000 high-throughput sequencing instrument, and replicated across two flowcells. The sequencing process produced 100 bp single-end reads. GBS sequence reads are available in the NCBI sequence read archive (www.ncbi.nlm.nih.gov/sra) under the BioProject accession number #SRP026259 and BioSample accession numbers #SAMN06165956 and #SAMN06165957.
SNP identification from raw sequence data
Raw sequence reads were analyzed using the UNEAK pipeline, part of the Tassel 3.0 software package produced by the maize genetics laboratory at Cornell University (Glaubitz et al. 2014). The UNEAK pipeline allows for the identification of SNPs in species where a reference genome is not available. The 5′ sequence barcodes were used to define individual-specific reads, but were trimmed prior to sequence analysis. Alignment of all the sequence data was conducted to produce a master “tag” list for the dataset. Tags are unique sequences of up to 64 bp in length observed across multiple reads. Further alignment of the master tag list identified tag pairs with a single base pair mismatch, and these were considered SNPs (with a sequencing error tolerance rate parameter of 0.03) (for more information, see Glaubitz et al. 2014). Note the program only considered tag pairs with a 1 bp mismatch to be SNPs, so any 64 bp read with 2+ SNPs would be excluded. The number of times each tag is observed in the sequencing data from each individual is used to determine the individual’s genotype for a particular SNP.
SNP filtering
The inventory list of SNP genotypes identified within the UNEAK pipeline was analyzed manually to remove markers where: (1) data were available for <75/85 progeny; (2) the genotype for one or both of the parents was missing. Markers where SNP inheritance displayed significant segregation distortion (i.e., SNPs with G values >6.693, P < 0.01) were identified and removed using LINKMFEX (Danzmann 2016).
Microsatellite genotyping
To anchor the newly identified SNP markers to previous Arctic charr linkage maps, genotypes at 102 microsatellite loci (Supplemental Material, File S1) from known locations across the Arctic charr genome were determined for all progeny and parents using established genotyping methods (Moghadam et al. 2007; Timusk et al. 2011; Norman et al. 2014).
Linkage mapping
The high quality SNPs selected for mapping, and the microsatellite marker anchors, were assessed for genetic linkage using LINKMFEX (Danzmann 2016). The SNPs were split into three categories: heterozygous male, heterozygous female, and double heterozygote (DH) SNPs where both parents were heterozygotes. The SNPs where only a single parent was heterozygous had high information content, given that all progeny genotypes are informative for linkage mapping. Double heterozygous SNP markers are informative in only about half of the progeny, given that linkage phases cannot be assigned in heterozygous progeny. Markers that are heterozygous in only one parent are problematic in that SNP mapping locations cannot be compared between parents. Data from additional mapping panels will be required to compare map orders between the sexes for these markers. Double heterozygous SNP markers were assigned to specific linkage groups, but were not added to specific map locations.
Linkage groups were identified using a logarithm of odds (LOD) threshold of 10 for linkage group assessment. We first created male-parent- and female-parent-specific linkage maps using the SNPs that were heterozygous in only one parent using LINKMFEX. In order to integrate the male and female linkage maps with one another, SNPs where both parents were heterozygous were added to each of the two datasets, and analyzed in LINKMFEX for genetic linkage with a LOD=10 threshold. It was then possible to identify overlapping sets of male- and female-specific linkage groups.
To determine the location of unlinked markers, repeated analysis was performed using descending LOD scores (LOD = 6 down to LOD = 3) using LINKMFEX, and LOD = 3 additions were accepted if they created joinings of two or more linkage groups that are known to be homeologous to one another. For some markers, a pseudolinkage (see below) of known homeologous chromosome arms was detected within the range of LOD = 3.0 to LOD = 5.0, and were therefore accepted. Linkage groups were named according to historical designations based on previous microsatellite marker assignments (Woram et al. 2004; Norman et al. 2011; Timusk et al. 2011). For each linkage group, marker order was determined in OneMap using the record algorithm (Margarido et al. 2007). The record algorithm was selected for ordering because it consistently gave the shortest map distances [i.e., smallest number of adjacent double cross-over (DCO) alignment points] out of the three possible OneMap ordering functions (ug, rcd, and record). This option also gave shorter map lengths than those produced by LINKMFEX. Male and female marker orderings were determined separately for each linkage group. As mentioned above, markers heterozygous in both parents were not ordered. OneMap marker ordering was further refined using the LINKMFEX program Adjacent-DCO-Count_Ripple-Check. This program was used to identify and reorder marker placements causing adjacent DCOs in the ordering. Adjacent double cross-overs are biologically unlikely in salmonids, given the high levels of chromatid interference detected during meiosis (Sakamoto et al. 2000; Danzmann and Gharbi 2001; Allendorf et al. 2015) in these species. The revised marker ordering minimized the number of adjacent DCOs in the dataset. Final map distances were calculated using the MAPDIS-V program in LINKMFEX, and selecting the option to ignore adjacent DCO events. We chose this option, as we considered that remaining adjacent DCOs may be due to errors in genotyping calls.
sdY marker
The progeny were genotyped for the sexually dimorphic gene (sdY) located on the Y-chromosome, using the PCR and agarose gel visualization methods described in Yano et al. (2012) with two modifications. First, we substituted insulin-like growth factor binding protein 5 (IGFBP5) as a positive control, using primers we developed: DQ206713-F3 (CCACCAGCTAATTACTGCAA) and DQ206713-R3 (GTAGAATTTGGCTGGCCCTA). Second, the following PCR temperature cycling conditions was used: denaturation for 5 min at 95°, followed by five cycles of 95° for 1 min, 58° for 30 sec, and 72° for 30 sec, then 30 cycles of 95° for 30 sec, 58° for 30 sec, and 72° for 30 sec, followed by a final 10 min at 72°. The sdY marker was validated based on conformity of the sdY genotypes to phenotypic assessment of the individual’s sex in this population.
Comparison with rainbow trout and Atlantic salmon genomes
SNP sequences were compared to the rainbow trout draft genome, and the Atlantic salmon genome (Berthelot et al. 2014; Lien et al. 2016), using the following BLASTn parameter settings (-word_size 11 -gapopen 5 -evalue 0.00001 -gapextend 2 -reward 2 -penalty -3) (Altschul et al. 1990). Blast hits were filtered, and the hits with the lowest e-value were used for subsequent homology identification. In the case of equivalent e-values, all Blast hits were retained, and considered equal “top hits” for the given SNP.
Moveable genetic elements and homeologies
Following Blast comparison of the Arctic charr markers to the rainbow trout and Atlantic salmon genomes, SNPs that aligned to adjacent regions of a single rainbow trout, and Atlantic salmon chromosome arm, allowed for the identification of arm homologies across the species. Within the SNP clusters that displayed consistent homology to a given Atlantic salmon, or rainbow trout, chromosome arm, individual markers sometimes showed homologies to disparate regions of the genome. Two hypotheses about the cause of these disparate SNPs were tested: (1) the SNPs lie within moveable, and/or highly repetitive DNA sequences; (2) the SNPs may be aligning to a homeologous chromosome arm with highly similar sequences. To test for the existence of repetitive sequences and moveable DNA elements, BLASTn (same parameters as above) was used to compare the linkage map SNPs to Repbase Update’s database of known vertebrate moveable and repetitive DNA elements (Jurka et al. 2005). The distribution of TE Blast hits in the linkage map was used to assess TE activity in the Arctic charr genome. TE activity in each linkage group was determined by assessing the proportion of markers in each linkage group that displayed significant Blast alignments to TEs.
Genomic architecture and residual tetrasomy
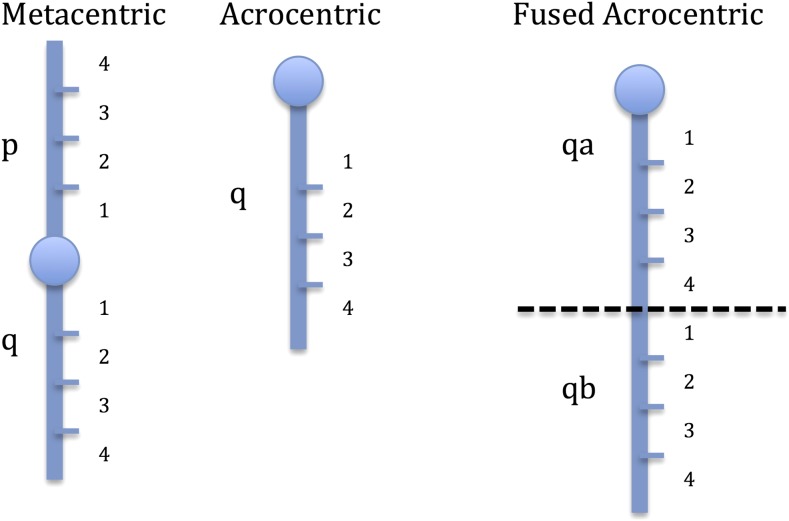
Paralogous sequence variants (PSVs) were identified to assess the distribution of duplicate loci through the Arctic charr genome. To do this, fixed heterozygote SNPs were used as PSV markers. These SNPs were heterozygous in both parents, and 100% of the progeny. The lack of homozygous progeny suggests that these are duplicate, monomorphic, loci with a single base-pair difference, causing them to appear heterozygous in all individuals. Since these PSVs lack recombinant progeny, their location relative to the linkage map SNPs cannot be determined. However, their linkage group affinities can be inferred based upon comparative homologies. We tentatively assigned these PSVs to Arctic charr linkage groups by aligning the PSVs and linkage map SNPs to the Atlantic salmon genome using BLASTn, and comparing their top hit locations. Two characteristics were assessed: (1) the distribution of PSVs between, and within, chromosomes of different genomic architectures, including Acrocentric Homeolog Pairs (AHPs), and High Residual Tetrasomy Arms (HRTAs); (2) the distribution of PSVs along chromosome arms relative to the centromere or telomere (see Figure 1). AHPs were defined as homeologous pairs of Arctic charr chromosome arms where neither arm in the pair is fused with another chromosome arm. HRTAs are defined as pairs of homeologous chromosomes that have higher levels of duplicate loci than the rest of the genome in multiple salmonid species (Danzmann et al. 2008; Lien et al. 2011, 2016; Brieuc et al. 2014), and likely form multivalents during meiosis due to crossing-over between their homeologous arms (Sakamoto et al. 2000)
Figure 1.
Visual representation of how Atlantic salmon chromosome arms were divided into quarters to assess Arctic charr PSV and map marker BLASTn hit distributions. Circles represent centromeres.
The distribution of duplicate loci in the genome was assessed using the Blast TopHit dataset, which consisted of markers in the Arctic charr linkage map and PSVs. The linkage map markers represent diploid loci, and the PSVs represent tetraploid loci. These markers were assigned to Atlantic salmon chromosome arm based on their top BLASTn hit locations (determined by lowest observed e-value). For markers with BLASTn hits of equal e-values to a single chromosome arm, only a single hit per chromosome arm was retained in the dataset. In the case of markers (both PSVs and linkage map markers) with equal BLASTn hit locations on two Atlantic salmon chromosome arms, the top Blast hit to each chromosome arm was retained in the dataset. Markers with equal top Blast hits to three or more chromosome arms, and markers with no Blast hits in the Atlantic salmon genome, were removed from the dataset.
To further assess whether any of the apparent single copy SNP markers may be duplicate copies of one another, we performed a BLASTn analysis of all SNPs against all SNPs (non-PSVs) in the database (see parameter settings above). Duplicate pairs exceeding 95% identity, and a 95% overlap in length, and occurring on separate chromosome arms, were considered potential homeologs of one another. Duplicates mapping to the same linkage groups were considered regional marker duplicates, unless they mapped adjacent to one another, indicating some type of tandem duplication.
We tested if duplicate loci are preserved in a higher frequency on HRTAs. Using Atlantic salmon as a reference, the HRTA homeolog pairs are represented by: Ssa02p/Ssa05q, Ssa11qa/Ssa26, Ssa16qa/Ssa17qa, Ssa03q/Ssa06p, Ssa12qa/Ssa02q, Ssa07q/Ssa17qb, and Ssa04p/Ssa08q. Using a contingency chi-square test, the number of linkage map and PSV top BLASTn hits on the HRTAs was compared to the number of hits on all other chromosome arms.
An additional contingency chi-square test was performed to test if Arctic charr AHPs preserve duplicate loci in the same manner as the rest of the genome. Putative AHPs were identified based on homologies in the Arctic charr genome, and their respective Atlantic salmon (Lien et al. 2016), and rainbow trout (Danzmann et al. 2008; Berthelot et al. 2014), homeologies. Based on this information, the following AHPs were identified in Arctic charr: AC02/AC36, AC05/AC29, AC19/AC32, and AC30/AC31. A contingency chi-square test compared the number of linkage map and PSV top BLASTn hits on acrocentric homeolog chromosome arms to the number of hits on all other chromosome arms.
The base pair hit location (s.start) for each marker in the Blast TopHit dataset was used to assess the PSV and linkage map markers’ distributions along Atlantic salmon chromosome arms. Each Atlantic salmon chromosome arm was divided equally into four quarters based on the known base-pair start and end locations (Lien et al. 2016). Quarter 1 was closest to the centromere, and quarter 4 was telomeric (Figure 1). All marker hits were then assigned to a chromosome arm quarter, and data from all the chromosome arms were merged. A chi-square test (goodness-of-fit) was performed to see if the frequency of linkage map SNPs and PSVs varied across different chromosome arm quarters. In addition to the above tests, BLASTn was used to compare the PSVs to Repbase update’s list of vertebrate TEs. The number of TE hits in PSVs was then compared to the number of TE hits in the linkage map SNPs using a contingency chi-square test.
We tested if PSVs in Arctic charr aligned more toward the telomeres of Atlantic salmon chromosome arms compared to linkage map SNPs. Tetrasomy is more readily preserved near the telomeres of chromosomes and most cross-overs in multivalent chromosome formations occur toward the telomeres (Sakamoto et al. 2000). Therefore, residual tetrasomic inheritance would likely persist in these regions for a longer period of time following the Ss4R WGD, and highly similar duplicate loci are expected to be found in high frequency closer to the telomeres of chromosome arms (Brieuc et al. 2014; Allendorf et al. 2015).
Data availability
Details on data used in this study can be found in Materials and Methods under the Sequencing Analysis section, and in the supplementary files.
Results and Discussion
SNP identification and linkage mapping
GBS of the Arctic charr family produced 4 × 108 sequence reads (roughly 4.7 million reads per progeny). Using the UNEAK pipeline (Tassel 3.0), 19,418 SNPs were then identified. Following the SNP filtering process, and the addition of microsatellite markers, the mapping dataset consisted of 4536 markers (see File S1 for the DNA sequence corresponding to each SNP marker).
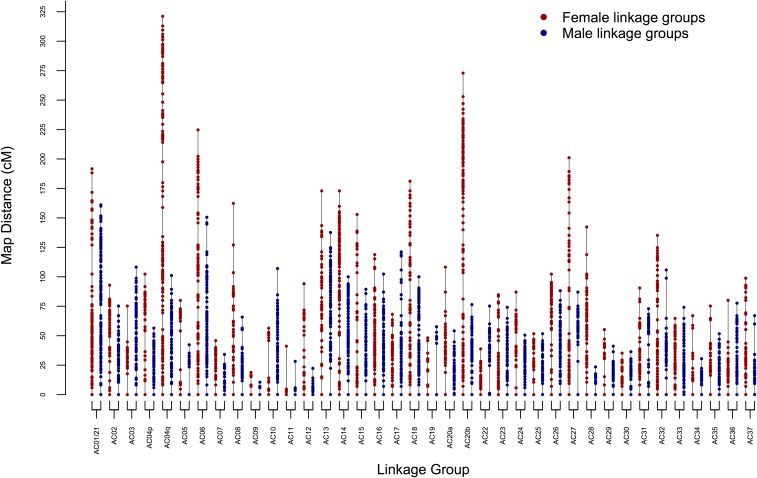
The linkage mapping process produced 39 linkage groups containing 4508 markers (4405 SNPs, 1 sdY, and 102 SSR), while 28 markers (24 SNP and 4 SSR) remained unlinked. Separate male and female maps were produced (File S2). A total of 1538 markers was ordered in the male map, spanning a distance of 2808.5 cM, while 1709 markers were ordered in the female map, covering 4302.7 cM (Figure 2 and Table 1). Markers that were heterozygous in both parents were not ordered, as mentioned previously, but 1283 of these were assigned to linkage groups (linkage group assignments are found in File S2).
Figure 2.
Visual representation of Arctic charr linkage groups. Female linkage groups are shown in red, and male linkage groups are shown in blue. Each point along the length of the line represents a single marker, or zero recombination cluster of several markers.
Table 1. List of Arctic charr linkage groups; the number of markers in each linkage group and the map distance covered (centiMorgans) by each male and female linkage group.
| Linkage Group | Male Marker Number | Male Distance (cM) | Female Marker Number | Female Distance (cM) | Unordered Markers | Chromosome Type |
|---|---|---|---|---|---|---|
| AC01/21 | 116 | 161.1 | 94 | 268.1 | 92 | AC01 is M/AC21 is A |
| AC02 | 52 | 75.2 | 39 | 92.9 | 47 | A |
| AC03 | 55 | 108.2 | 24 | 75.2 | 34 | M |
| AC04p | 39 | 56.4 | 41 | 102.3 | 22 | A/split M |
| AC04q | 55 | 101.1 | 132 | 321.1 | 43 | Fused A/split M |
| AC05 | 12 | 42.3 | 22 | 80 | 85 | A |
| AC06 | 74 | 150.5 | 102 | 224.7 | 33 | M |
| AC07 | 23 | 34.1 | 24 | 45.8 | 29 | A |
| AC08 | 38 | 65.8 | 46 | 162.3 | 24 | M |
| AC09 | 4 | 10.5 | 9 | 18.8 | 76 | A |
| AC10 | 50 | 107 | 15 | 56.4 | 13 | A |
| AC11 | 5 | 28.2 | 8 | 41.1 | 10 | A |
| AC12 | 12 | 22.3 | 24 | 94.1 | 10 | A |
| AC13 | 73 | 137.6 | 55 | 172.9 | 18 | M |
| AC14 | 74 | 100 | 87 | 172.9 | 47 | M |
| AC15 | 49 | 89.4 | 46 | 152.9 | 76 | M |
| AC16 | 51 | 102.3 | 65 | 118.8 | 27 | A |
| AC17 | 46 | 121.1 | 40 | 68.2 | 19 | Fused A |
| AC18 | 68 | 100 | 62 | 181.1 | 34 | M |
| AC19 | 9 | 57.6 | 17 | 48.2 | 78 | A |
| AC20a | 37 | 54.1 | 37 | 108.2 | 7 | A |
| AC20b | 47 | 76.4 | 128 | 272.9 | 38 | M |
| AC22 | 31 | 75.2 | 31 | 38.8 | 20 | A |
| AC23 | 45 | 74.1 | 38 | 84.7 | 25 | A |
| AC24 | 35 | 50.5 | 33 | 87 | 6 | A |
| AC25 | 31 | 51.7 | 24 | 51.7 | 18 | A |
| AC26 | 50 | 88.2 | 43 | 102.3 | 57 | A |
| AC27 | 34 | 87 | 61 | 201.1 | 34 | M |
| AC28 | 16 | 23.5 | 48 | 142.3 | 9 | A |
| AC29 | 28 | 41.1 | 17 | 55.2 | 37 | A |
| AC30 | 27 | 36.4 | 26 | 35.2 | 3 | A |
| AC31 | 35 | 72.9 | 40 | 90.5 | 33 | A |
| AC32 | 37 | 105.8 | 67 | 135.2 | 30 | A |
| AC33 | 48 | 74.1 | 44 | 64.7 | 62 | A |
| AC34 | 22 | 30.5 | 18 | 81.1 | 8 | A |
| AC35 | 36 | 51.7 | 24 | 75.2 | 29 | A |
| AC36 | 47 | 77.6 | 32 | 80 | 30 | A |
| AC37 | 27 | 67 | 46 | 98.8 | 20 | A |
| Totals | 1538 | 2808.5 | 1709 | 4302.7 | 1283a | Metacentric: 10b |
| Acrocentric: 27–29 | ||||||
| Split meta: 1c |
The chromosome type [metacentric (M) or acrocentric (A)] is also shown for each linkage group.
Note that there are 4508 markers in the linkage map, but the total marker numbers here sum to 4530. This is because a small number of markers were successfully ordered in both the male and female maps, and are therefore counted twice in this row.
10 metacentric assumes AC20b is metacentric in structure, and in karyotypes where AC04p/q are joined, 11 metacentrics would be observed.
27 acrocentrics would be observed if AC20b is metacentric, and AC04 was metacentric in the karyotype, while 29 acrocentrics would be present in the configuration where AC04p and AC04q form separate arms. “Fused A” designations indicate acrocentric arms that appear to be composed to two ancestral teleost chromosome arms.
Microsatellite anchors
Of the 102 microsatellite markers genotyped, 98 were successfully added to the linkage map (four remained unlinked). Five markers were duplicated and mapped to two linkage groups. Of 39 linkage groups, 36 contained one or more microsatellite markers, allowing the new SNP-based linkage groups to be aligned with the microsatellite-based linkage maps (Woram et al. 2004; Timusk et al. 2011; Norman et al. 2012). The identity of three linkage groups without microsatellite markers (AC25, AC30, and AC31) was determined based on BLASTn search results, and previously identified arm homologies among the salmonid species compared.
Chromosome type and salmonid arm homologies
A comparison of the Arctic charr linkage map to the Atlantic salmon (Lien et al. 2016) and rainbow trout genomes (Berthelot et al. 2014) based on the BLASTn analysis is presented in Table 2. The top Blast hit data for each marker are available in File S2 and File S3. Nine linkage groups (AC01, -03, -06, -08, -13, -14, -15, -18, and -27) were identified as metacentric chromosomes based on homologies to two salmonid chromosome arms. Two potential split metacentric groupings were identified (AC04 and AC20). One set involves the sex-linkage group AC04, and has been identified as possessing a fusion polymorphism in Arctic charr (Moghadam et al. 2007). Our data also suggest that a two acrocentric vs. one metacentric polymorphism exists for AC04 (see below). The q-arm in this chromosome set involves a fusion of two ancestral salmonid chromosome arms that are homologous to Omy02q and Omy25, while the p-arm is homologous to Omy24. None of these arms are homeologous to one another, and are therefore unlikely to show pseudolinkage affinities. The second set of chromosomes (AC20 group) involves a small (AC20a) and a large (AC20b) acrocentric arm, where the large arm appears to be composed of two fused ancestral salmonid arms that are homologous to Omy12p/q and Ots09/q. Therefore, it is more likely that AC20b represents an entire metacentric chromosome, and AC20a represents a separate acrocentric arm, rather than AC20a/20b representing a large metacentric chromosome comprised of joined homeologous arms (Woram et al. 2004). This would support the suggestion that there are 10 stable metacentrics in North American Arctic charr (Hartley 1989; Phillips et al. 2002), with the AC04 polymorphism generating an additional metacentric in some individuals. The homologous chromosome arm of AC20a in rainbow trout is Omy13q, and, since Omy12q/Omy13q are homeologs, they may show pseudolinkage affinities. Twenty-seven linkage groups appear to be acrocentric, including two (AC04 and AC17) that result from a tandem fusion of qa and qb chromosome arms. AC17 appears to include segments homologous to both Omy16q and 20q. The total chromosome arm number (NF) observed was 100, containing 52 haploid ancestral arm segments.
Table 2. Salmonid chromosome arm homologies referenced to Arctic charr linkage group homologies.
| Ssa Homeolog Paira | Atlantic Salmon | Rainbow Troutb | Arctic Charr | Chinook Salmonc | Ssa Homeolog Pair | Atlantic Salmon | Rainbow Trout | Arctic Charr | Chinook Salmon |
|---|---|---|---|---|---|---|---|---|---|
| 10qa/b | Ssa16qa | Omy01p15 | AC26 | Ots06p | 27 | Ssa14qb | Omy14p | AC30 | Ots31 |
| 01qa/b | Ssa18qa14 | Omy01q14 | AC25 | Ots06q14 | 09qb | Ssa05p | Omy14q | AC06q | Ots21 |
| 02p1 | Ssa05q1 | Omy02p1 | AC06p | Ots231 | 19qa | Ssa298 | Omy15p | AC27q | Ots298 |
| 16qa&23 | Ssa10qb | Omy02q15 | AC04qb | Ots19 | 07q7 | Ssa17qb7 | Omy15q7 | AC24 | Ots177 |
| 05q1 | Ssa02p1 | Omy03p1 | AC35 | Ots03p1 | 28&29 | SSa19qb8 | Omy16p16 | AC18p | Ots248 |
| 21 | Ssa25 | Omy03q9 | AC02 | Ots03q | 15qb | Ssa13qa1 | Omy16q1 | AC17qa | Ots221 |
| 10qa/b | Ssa23 | Omy04p10 | AC13p | Ots01p | 22 | Ssa12qb | Omy17p11 | AC01p | Ots02p |
| 15qa | Ssa06q | Omy04q | AC14q | Ots18 | 12qa6 | Ssa02q6 | Omy17q6 | AC01q | Ots02q6 |
| 11qb&13qb | Ssa01qb | Omy05p | AC29 | Ots20 | 17qa3 | Ssa16qb3 | Omy18p3 | AC27p | Ots14p3 |
| 23 | Ssa10qa | Omy05q10 | AC16 | Ots05q | 14qb | Ssa27 | Omy18q | AC31 | Ots13p |
| 20qa | Ssa24 | Omy06p | AC15p | Ots04p | 08q/04p4 | Ssa04p/08q4 | Omy19p4 | AC34 | Ots11p4 |
| 11qa2 | Ssa262 | Omy06q2 | AC15q | Ots04q2 | 09qa | Ssa01p | Omy19q | AC09 | Ots11q |
| 16qb3 | Ssa17qa3 | Omy07p3 | AC12 | Ots07p3 | 05p/19qa/b&01qa | Ssa09qb/28 | Omy20p16 | AC08q | Ots25 |
| 12qb | Ssa22 | Omy07q11 | AC11 | Ots07q | 29 | Ssa19qa | Omy20q | AC17qb | Ots25 |
| 06q | Ssa15qa | Omy08p | AC28 | Ots05p | 18qb | Ssa07p7 | Omy21p7 | AC03p | Ots15p7 |
| 03p | Ssa14qa | Omy08q12 | AC32 | Ots10q | 17qb7 | Ssa07q | Omy21q13 | AC03q | Ots15q |
| 07p | Ssa18qb | Omy09p13 | AC37 | Ots10p | 25 | Ssa21 | Omy22p9 | AC36 | Ots26 |
| 13qa | Ssa15qb1 | Omy09q1 | AC07 | Ots16q1 | Omy22q | ||||
| 13qb | Ssa04q | Omy10p | AC23 | Ots30 | 28&29 | Ssa01qa14 | Omy2314 | AC18q | Ots01q14 |
| 04p/08q4 | Ssa08q/04p4 | Omy10q4 | AC13q | Ots344 | 20qb | Ssa09qc | Omy24 | AC04pd | Ots14q |
| 24/29 | 20qa/19qa | Omy11p | AC14p | Ots16p/12p | 01p | Ssa09qa | Omy25 | AC04qa | Ots08p |
| 24 | Ssa20qa | Omy11q | AC33 | Ots12p | 262 | Ssa11qa2 | Omy262 | AC10 | Ots12q2 |
| 04q | Ssa13qb | Omy12p | AC20b-2 | Ots09p | 09qc | Ssa20qb | Omy27 | AC22 | Ots13q |
| 06q/01qb&04q5 | 03q/13qb5 | Omy12q5 | AC20b-1 | Ots09q5 | 14qa | Ssa03p | Omy2812 | AC19 | Ots28 |
| 02q6 | Ssa12qa6 | Omy13p6 | AC21 | Ots326 | 05p/19qa/b&01qa | Ssa09qb/28 | Omy29 | Ac08p | Ots08q |
| 03q5 | Ssa06p5 | Omy13q5 | AC20a | Ots275 | 01qb&4p | Ssa11qb | Sex | AC05 | Ots33 |
These were determined based on the most common BLASTn hit locations of a linkage group’s markers when compared to the Atlantic salmon genome and the rainbow trout draft genome. Additionally, homologies with Chinook salmon chromosome arms are presented based on known homologies in Atlantic salmon and rainbow trout, though direct BLASTn comparison of the Arctic charr linkage map and Chinook salmon genome was not performed. The column “Ssa Homeolog pair” shows each chromosome arm’s homeolog partner derived from a common pre-Ss4R ancestor.
Cells in columns 1 and 6 with matching superscript numbers represent HRTA identified in Atlantic Salmon [data from Lien et al. (2016)].
Homeolog pairs identified in rainbow trout based on high numbers of duplicate markers have matching superscript numbers in columns 3 and 8 [data from Danzmann et al. (2008) and Berthelot et al. (2014)].
Homeolog pairs identified in Chinook salmon based on high numbers of duplicate markers have matching superscript numbers in columns 5 and 10 [data from Brieuc et al. (2014)].
Indicates sex the linkage group of Arctic charr.
Sex-linkage group polymorphism in AC04
Previous work has shown AC04 to be polymorphic, and taking the form of either a single linkage group (type 1) or two unlinked linkage groups (type 2) (Moghadam et al. 2007). The mapping parents in this study are both type 2 individuals; microsatellite markers associated with AC04 are found on both AC04p and AC04q (Woram et al. 2004; Moghadam et al. 2007; Timusk et al. 2011). Previous research identified three salmonid chromosome arms homologous with AC04 (Timusk et al. 2011). Two of these arms are homologous with AC04q, and the third is homologous with AC04p (Table 2). AC04p also contains the sex-determining gene, sdY (Yano et al. 2013). Although two separate, type 2 AC04 linkage groups are observed in the current linkage map, the polymorphic nature of this linkage group, and the evidence presented here, is indicative of a fusion/fission polymorphism for the sex-determining chromosome of Arctic charr. Affinity of the sdY marker to the Slml-family of TEs supports the suggestion that the reported sex-linkage difference between North American (AC04) and European (AC01/21) Arctic charr may be the result of a translocation through TE movements (Woram et al. 2003; Küttner et al. 2011).
Pseudolinkage
Two pairs of homeologous linkage groups (AC01q/21 and AC13q/34) were detected as possessing a weak pseudolinkage to one another (LOD ≥ 3–5). Interestingly, pseudolinkage was detected between both homeologous pairs of linkage groups in both the male and female parents (Table 3). Previously, pseudolinkage was thought to occur only within male meiosis, with rare reports of female pseudolinkage (Ostberg et al. 2013; Allendorf et al. 2015). Our identification of pseudolinkage in a female confirms that multivalents are also likely formed during female meioses.
Table 3. Allele counts of the two homeolog pairs displaying pseudolinkage within females.
| Marker Pair | Genotypes | Linkage Group | Parental Phases (Marker A/Marker B) | Recombinant Phases (Marker A/Marker B) | Chi-Squared | P-value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker A | Marker B | Marker A | Marker B | Marker A | Marker B | Alleles | Count | Alleles | Count | ||
| TP47181 | TP21253 | G,C | A,G | AC13 | Pseudolink | G/A | 32 | C/A | 23 | 18.7 | 3.20E−04 |
| C/G | 25 | G/G | 5 | ||||||||
| TP21253 | TP30908 | A,G | T,C | Pseudolink | AC34 | A/T | 43 | A/C | 12 | 36 | 7.50E−08 |
| G/C | 23 | G/T | 7 | ||||||||
| TP47181 | TP30908 | G,C | T,C | AC13 | AC34 | G/T | 23 | C/T | 27 | 4.2 | 0.243 |
| C/C | 21 | G/C | 14 | ||||||||
| TP10591 | TP15996 | T,G | G,A | AC21 | Pseudolink | T/G | 14 | G/G | 4 | 15.8 | 0.0013 |
| G/A | 13 | T/A | 1 | ||||||||
| TP15996 | TP32826 | G,A | A,T | Pseudolink | AC01 | G/A | 18 | A/A | 0 | 33 | 3.20E−07 |
| A/T | 14 | G/T | 0 | ||||||||
| TP10591 | TP32826 | T,G | A,T | AC21 | AC01 | T/A | 22 | G/A | 23 | 7.9 | 0.049 |
| G/T | 27 | T/T | 10 | ||||||||
These instances appear to result from an excess of parental phase genotypes. Flanking markers from both linkage groups with the most complete genotypes, along with the principle marker causing pseudolinkage, are displayed. Note, for AC21/AC01, the marker causing pseudolinkage (TP15996) was heterozygous in both parents (ab X ab cross). Therefore, the phases for half of the progeny could not be ascertained. Chi-squared goodness of fit tests were performed for each pair of alleles, comparing the observed genotype frequencies to a null hypothesis of a 1:1:1:1 genotype distribution.
Pseudolinkage is a phenomenon arising due to segregation of gametes following preferential pairing of homeologous chromosome arms during meiosis I (Allendorf et al. 2015; May and Delany 2015). Preferential as opposed to random pairing typically occurs in cases of hybridization, where the homeolog pairs provided by one parent may be more closely related to one another due to their species-specific ancestry. Therefore, within the hybrid, the homeologs from one parent may therefore be more likely to pair with one another and recombine. Crossing-over tends not to occur between homeologous markers located very close to the centromere. If preferential pairing occurs at meiosis I, this leads to a significant excess of nonparental genotypes being produced following meiosis II, because alternate disjunction occurs as the multivalents separate at meiosis I [see Allendorf et al. (2015) and May and Delany (2015) for a more detailed explanation of these models]. This produces a statistical linkage between the two homeologous chromosome arms characterized by a significant excess of nonparental gametes, and is a characteristic feature of hybrid salmonids.
Random pairing of homeologous pairs may, however, still occur as the genome undergoes the process of diploidization within species. This may involve the random formation of both bivalents, and multivalents, between the homeologous pairs, such that gametic expectation models cannot be precisely defined. This may result in varying levels of exchange among alleles for loci that are located proximal and distal to chiasmata junctions along the length of randomly paired homeologous chromosomes [see Sakamoto et al. (2000), for an explanation]. We have developed models to explain this weaker form of pseudolinkage, and will present these in a subsequent publication.
Homeologies
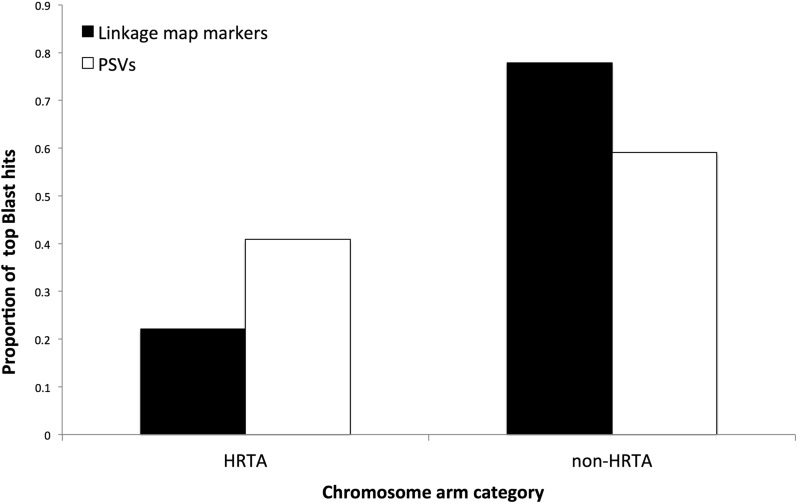
Arm homologies identified in Atlantic salmon, rainbow trout, and chinook salmon (Table 2) with Arctic charr, were used to identify homeologous arm pairings within Arctic charr (Table 4). Lower levels of duplicate marker regions were detected for certain arms, but can nonetheless be inferred based upon the recent extensive survey of duplicate gene copies in Atlantic salmon (Hermansen et al. 2016) (Table 4). All seven homeologous pairs in Arctic charr identified as HRTAs in other species contain one chromosome arm that has undergone a chromosome fusion. One pair of HRTA identified in Atlantic salmon (Ssa09qc/20qb) (Lien et al. 2016) did not show a high number of duplicates in either rainbow trout or chinook salmon. Similarly a HRTA region in both rainbow trout and chinook salmon (Omy01q and Ots06q/Omy23 and Ots01q), was not identified as an HRTA in Atlantic salmon (Table 2). Four AHPs, where neither homeolog has undergone a fusion event since the Ss4R, were identified in Arctic charr (Table 5). AHP likely undergo minimal residual tetrasomic inheritance, given that AHP multivalent pairings are unlikely due to structural instability (May and Delany 2015). We found support for this prediction in that the number of PSVs aligning to AHPs was significantly lower compared to the number aligning to the other chromosome arms (P < 0.0001). Of the linkage map markers that successfully aligned to the Atlantic salmon genome, 20% (861/4290) aligned to HRTA chromosome arms, while only 14.0% of PSVs (158/1130) had top hit locations on AHPs. These results suggest that fewer duplicate loci exist on AHPs, which might be indicative of more rapid diploidization relative to the rest of the genome (Figure 3).
Table 4. Reference table of homeologous chromosome pairs in Arctic charr.
| Homeolog 1 | Homeolog 2 |
|---|---|
| AC01p | AC11 |
| AC01q | AC21 |
| AC02 | AC36 |
| AC03p | AC24 |
| AC03q | AC37 |
| AC04p | AC14p/AC33a |
| AC04p | AC22 |
| AC04qa | AC09 |
| AC04qb | AC26 |
| AC06 | AC08p/AC08q |
| AC06p | AC35 |
| AC07 | AC17qa |
| AC08p | AC18p?a |
| AC10 | AC15q |
| AC12 | AC27p |
| AC13p | AC16 |
| AC13q | AC34 |
| AC14q | AC28a |
| AC15p | AC14p/AC14q/AC33a |
| AC17qb | AC27q |
| AC18p | AC27q |
| AC18q | AC25 |
| AC19 | AC32 |
| AC20b-1 | AC20a |
| AC23 | AC04qb/AC05/AC20b-1/AC-20b-2a |
| AC29 | AC05/AC20b-2a |
| AC30 | AC31 |
Identified through their homologies in Atlantic salmon, and homeologies identified in Hermansen et al. (2016).
Table 5. Reference table for the analysis of Arctic charr duplicate loci distribution.
| Atlantic Salmon Chromosome Arm | Arctic Charr Linkage Group | High Residual Tetrasomy Arms (HRTA) | Acrocentric Homeolog Pairs (AHP) | Atlantic Salmon Chromosome Arm | Arctic Charr Linkage Group | High Residual Tetrasomy Arms (HRTA) | Acrocentric Homeolog Pairs (AHP) | |
|---|---|---|---|---|---|---|---|---|
| Ssa01p | AC09 | Ssa13qa | AC17qa | |||||
| Ssa01qa | AC18q | Ssa13qb | AC20b-2 | |||||
| Ssa01qb | AC29 | AHP | Ssa14qa | AC32 | AHP | |||
| Ssa02p | AC35 | HRTA | Ssa14qb | AC30 | AHP | |||
| Ssa02q | AC01q | HRTA | Ssa15qa | AC28 | ||||
| Ssa03p | AC19 | AHP | Ssa15qb | AC07 | ||||
| Ssa03q | AC20b-1/AC20b-2 | HRTA | Ssa16qa | AC26 | ||||
| Ssa04p | AC13q/AC-34 | HRTA | Ssa16qb | AC27p | HRTA | |||
| Ssa04q | AC23 | Ssa17qa | AC12 | HRTA | ||||
| Ssa05p | AC06q | Ssa17qb | AC24 | HRTA | ||||
| Ssa05q | AC06p | HRTA | Ssa18qa | AC25 | ||||
| Ssa06p | AC20a | HRTA | Ssa18qb | AC37 | ||||
| Ssa06q | AC14q | Ssa19qa | AC17qb/AC14p | |||||
| Ssa07p | AC03p | Ssa19qb | AC18p | |||||
| Ssa07q | AC03q | HRTA | Ssa20qa | AC14p/AC33 | ||||
| Ssa08q | AC13q/AC34 | HRTA | Ssa20qb | AC22 | ||||
| Ssa09qa | AC04qa | Ssa21 | AC36 | AHP | ||||
| Ssa09qb | AC08p/AC08q | Ssa22 | AC11 | |||||
| Ssa09qc | AC04p | Ssa23 | AC13p | |||||
| Ssa10qa | AC16 | Ssa24 | AC15p | |||||
| Ssa10qb | AC04qb | Ssa25 | AC02 | AHP | ||||
| Ssa11qa | AC10 | HRTA | Ssa26 | AC15q | HRTA | |||
| Ssa11qb | AC05 | AHP | Ssa27 | AC31 | AHP | |||
| Ssa12qa | AC21 | HRTA | Ssa28 | AC08p/AC08q | ||||
| Ssa12qb | AC01p | Ssa29 | AC27q | |||||
The Atlantic salmon chromosome arms are listed alongside their Arctic charr homologs. The table lists all chromosome arms classified as belonging to the HRTA or AHP categories.
Figure 3.
BLASTn hit locations in the Atlantic salmon genome for Arctic charr PSVs, and linkage map markers; 40.8% (462/1130) of PSVs had their top hit locations on HRTAs, while only 22.1% (950/4290) of the linkage map SNPs had their top Blast hit locations on HRTAs.
Duplicate loci
The distribution of PSVs throughout the Arctic charr genome appeared to be nonrandom, with certain regions preserving higher numbers of duplicate loci. BLASTn alignment of PSVs, and linkage map SNPs to the Atlantic salmon genome, was performed to compare the locations of linkage map markers and PSVs. The top BLASTn hit locations (based on lowest e-values) were used to assess the distribution of SNPs throughout the genome. Direct comparison of the markers through linkage mapping was not possible, given that PSVs lack any type of segregation pattern, and therefore cannot be mapped. For markers with BLASTn hits of equal e-values on a single chromosome arm, only a single hit was used per chromosome arm in the dataset. In the case of markers (both linkage map markers and PSVs) with BLASTn hit locations of equal e-values on two Atlantic salmon chromosomes, a BLASTn hit to each chromosome arm was retained in the dataset, as these markers represented potential duplicate loci. Markers with equal BLASTn hits to three or more locations were excluded from the dataset.
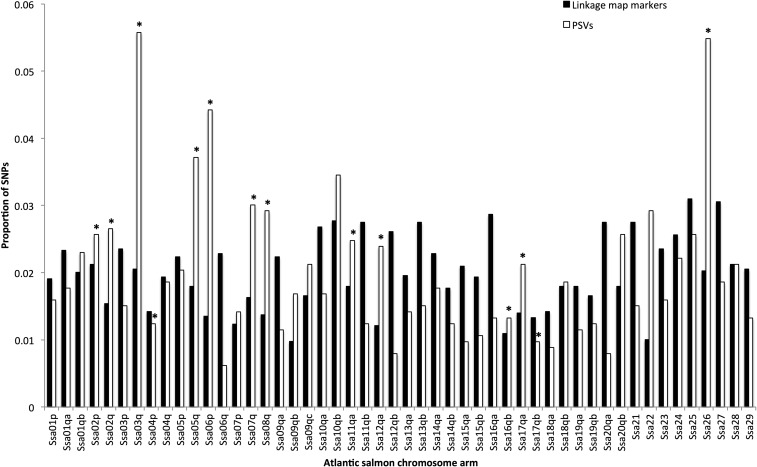
The analysis identified 1130 PSV Blast hits in the Atlantic salmon genome (Figure 4), in addition to the 4429 SNPs assigned to the linkage map. If the PSV distribution throughout the genome is random, and chromosome arm size was the only factor influencing their distribution, then the proportion of PSVs aligning to each chromosome would be similar to the proportion of linkage map SNPs aligning to each chromosome arm. To assess the effect of genomic architecture on the preservation of duplicate loci, chromosome arms were binned into two categories: HRTAs (listed in Table 5) and non-HRTA (other) salmonid chromosome arms. A contingency test showed higher numbers of PSVs aligned to the HRTAs than expected (P < 0.0001) (Figure 3). Of the 14 chromosome arms in the HRTA category, 12 had higher proportions of PSV Blast hits than linkage map marker Blast hits (Figure 4). The two HRTA arms with lower numbers of PSV BLASTn hits (Ssa04p and Ssa17qb) had homeologs with high numbers of PSV hits (Ssa08q and Ssa07q, respectively). This suggests that the lower number of PSV hits on the HRTA arms may be due to more PSVs aligning to their homeologs because of small sequence differences. High numbers of PSVs are observed in regions that preserved residual tetrasomy longer after the Ss4R than other parts of the genome, which may be indicative of historically reduced diploidization rates (Brieuc et al. 2014).
Figure 4.
The top BLASTn hit locations of Arctic charr linkage map SNPs, and Arctic charr PSVs, across Atlantic salmon chromosome arms. The data are shown in the proportion of hits from a category (Map SNPs or PSVs) in order to account for the bias of Atlantic salmon chromosome size. Arms with a * are HRTAs identified in Lien et al. (2016).
Several other Atlantic salmon arms displayed high numbers of PSV BLASTn hits, notably Ssa09qb, Ssa09qc, Ssa10qb, and Ssa22. The Arctic charr homologs of these arms are AC08p/q, AC04p, AC04qb, and AC11, respectively. All of these arms, or their homeologs in Arctic charr, have undergone a fusion. Interestingly, these arms also share homology to rainbow trout chromosome arms with high numbers of duplicates (Table 2). Previously, it had been thought that one metacentric chromosome must be present in a homeolog pair to provide the stability necessary for homeologous pairing, and multivalent formation (Kodama et al. 2014). However, Lien et al. (2016) have recently shown that metacentric structures are not a requirement for homeologous pairing, given that two HRTA pairs in Atlantic salmon (Ssa11qa/Ssa26 and Ssa16qb/Ssa17qa) preserve high sequence similarity without the presence of a metacentric, but, in both cases, one of the arms has undergone a fusion event. This suggests that fused acrocentrics, as well as metacentric chromosomes, provide the structural stability necessary for homeologous recombination.
The conservation of duplicate loci on HRTAs does not appear to be due to current chromosome structure, because the HRTAs do not display homologous chromosome arm fusions across species (with the exception of AC03/Ssa07/Omy21/Ots15). Therefore, the slower diploidization in these regions may be attributed to some aspect of their evolutionary past. For instance, certain fusions may have arisen in the common ancestor of all these salmonid lineages following the Ss4R. This could have provided the HRTA homeolog pairs with the ability to form multivalents during meiosis, and undergo residual tetrasomy, thereby slowing diploidization rates. A large number of species-specific fusion/fission events since the more recent divergence of salmonids (Macqueen and Johnston 2014) could disjoin previous chromosome structures, and also explain why the seven HRTA homeolog pairs preserve duplicate loci in multiple species, despite the large variation in karyotypic structures across current salmonid species.
To search for possible duplicate SNP positions that could have arisen from the Ss4R event, we reciprocally BLASTn aligned all linkage map SNP markers against each other, and retained those duplicate pairs that shared ≥95% identity to one another, as well as retaining ≥95% of an overlap in their length distributions. Surprisingly, of the 362 duplicate pairs identified, only 18 pairs were interchromosomal duplicate pairs. Of these 18 pairs, only three were considered to be possible WGD paralogs, as the other 15 pairs involved either one or both SNPs with TE-specific sequence (see File S4). One of the potential paralogs that was not associated with TE, involved markers from identified homeolog pair of AC03q/37. Two of the potential paralogs not associated with TEs involved markers from the linkage groups AC06p and AC32, which is not an identified homeolog region in the Arctic charr map. However, these two paralog sets show homology to the Ssa02p/05q paralogs, and therefore may indicate a small region of previously unidentified homeology.
The vast majority of duplicate pair markers appear to be intrachromosomal duplicates (95.1%) (File S4). To assess whether chromosome structure may have had an influence on the distribution of these duplicates, we compared the proportion of duplicated SNP markers between metacentric vs. acrocentric type linkage groups. Duplicates within AC04p and AC04q were included in the metacentric grouping, as well as duplicates within AC17, given that this acrocentric appears to be composed of fused chromosome arms. No differences were detected in the distribution of duplicates between the chromosome types (P > 0.05) (Table 6).
Table 6. Arctic charr linkage map location of duplicate pair SNPs that shared ≥95% identity with one another.
| Linkage Group (LG) | Inter-LG Pair Membersa | Intra-LG Duplicates | Proportion Marker Duplicatesb | Linkage Group (LG) | Inter-LG Pair Members | Intra-LG Duplicates | Proportion Marker Duplicates |
|---|---|---|---|---|---|---|---|
| AC01 | 4 | 30 | 0.192 | AC20a | 2 | 12 | 0.15 |
| AC02 | 0 | 20 | 0.149 | AC20b | 0 | 28 | 0.136 |
| AC03 | 3 | 16 | 0.149 | AC21 | 0 | 18 | 0.125 |
| AC04p | 1 | 22 | 0.224 | AC22 | 0 | 10 | 0.125 |
| AC04q | 3 | 30 | 0.133 | AC23 | 1 | 22 | 0.206 |
| AC05 | 0 | 16 | 0.136 | AC24 | 0 | 8 | 0.114 |
| AC06 | 4 | 26 | 0.129 | AC25 | 2 | 16 | 0.219 |
| AC07 | 0 | 8 | 0.108 | AC26 | 3 | 32 | 0.219 |
| AC08 | 1 | 12 | 0.115 | AC27 | 0 | 24 | 0.192 |
| AC09 | 0 | 12 | 0.136 | AC28 | 0 | 6 | 0.086 |
| AC10 | 0 | 6 | 0.08 | AC29 | 1 | 16 | 0.2 |
| AC11 | 0 | 0 | 0 | AC30 | 1 | 8 | 0.143 |
| AC12 | 0 | 2 | 0.044 | AC31 | 0 | 22 | 0.204 |
| AC13 | 0 | 30 | 0.211 | AC32 | 3 | 16 | 0.121 |
| AC14 | 0 | 20 | 0.099 | AC33 | 1 | 30 | 0.2 |
| AC15 | 0 | 20 | 0.12 | AC34 | 0 | 2 | 0.043 |
| AC16 | 3 | 26 | 0.183 | AC35 | 0 | 22 | 0.256 |
| AC17 | 0 | 16 | 0.16 | AC36 | 0 | 8 | 0.075 |
| AC18 | 0 | 28 | 0.175 | AC37 | 3 | 30 | 0.133 |
| AC19 | 0 | 24 | 0.233 |
Number of SNPs where the duplicate member maps to a different linkage group.
Number of duplicates/total number of SNPs mapped to the LG.
Duplicate marker pairs do not map next to one another within linkage groups, suggesting that they are not tandem repeats. These marker duplicates may be part of larger segmental duplicate blocks that can vary in size from 1 to 400 kb (Mendivil Ramos and Ferrier 2012), but alignment of these regions to more complete scaffold contigs would be needed to test this idea. The reason so few apparent WGD paralog regions were detected is likely due to the high stringency in which duplicated regions were identified. Interchromosomal 4R paralogs have average identity levels ranging from 86 to 90% (Moghadam et al. 2011; Berthelot et al. 2014), with average levels up to 96% for protein-coding duplicate regions (Berthelot et al. 2014). Hence, an analysis using lowered stringency cut-offs will likely reveal higher frequencies of WGD paralog regions than those reported here.
Many of the duplicate markers appear to align to identical locations within both the rainbow trout and Atlantic salmon genomes, but in opposite strand orientations (see File S4). Upon closer inspection of these duplicates, we observed that several reads aligning to each contig cluster were longer in either the 5′ or 3′ direction, and these reads spanned an EcoT22I cut-site on either side. Both the 3′ side of the forward strand, and 5′ side of the reverse strand were bordered by the 5 bp signature of a DNA strand cut by EcoT22I. This suggests that an internal cut-site may have been skipped due to an epigenetic modification, and that an additional EcoT22I cut-site, 20–30 bp away, flanked the uncut site.
We queried all 425,391,431 reads obtained from the fish used in this study, and determined that the presence of uncut EcoT22I sites averaged 2.8%, while those found in the paired duplicates having extended reads ranged from 4 to 6% supporting the suggestion that these regions may be more epigenetically modified. However, many of these reads also appeared as chimeric religations, highlighting that enzymes susceptible to epigenetic modification may lead to genotyping errors (Jiang et al. 2016).
Distribution of Arctic charr duplicated markers in relation to the Atlantic salmon genome
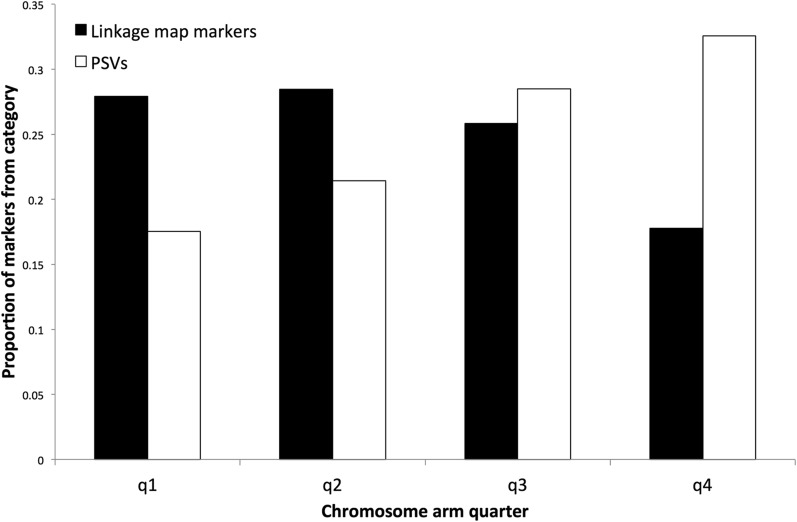
BLASTn alignments of the Arctic charr duplicated markers were not uniformly distributed along the length of Atlantic salmon chromosomes from the Lien et al. (2016) assembly. Significantly more Arctic charr PSVs aligned to the 4th quarter (telomeric ends) of chromosome arms (P < 0.0001) (Figure 5). This suggests that PSVs are preserved near the telomeres in the regions of chromosomes that undergo residual tetrasomy (Wright et al. 1983; Allendorf et al. 2015; May and Delany 2015). Previous studies in salmonids have also observed that duplicate loci are present in higher frequencies near the telomeres (Sakamoto et al. 2000; Brieuc et al. 2014; Kodama et al. 2014; Larson et al. 2015; McKinney et al. 2015; Waples et al. 2015). There was also a significant reduction of linkage map markers with top Blast hits in the 4th quarter of chromosomes, which suggests that residual tetrasomy may generate high numbers of duplicate loci that cause telomeric regions to be under-represented with markers (Allendorf et al. 2015).
Figure 5.
Distribution of the top Blast hits of Arctic charr linkage map SNPs and PSVs across Atlantic salmon chromosome arms. q1 is the quarter closest to the centromere, q4 is the quarter closest to the telomere.
Transposon activity
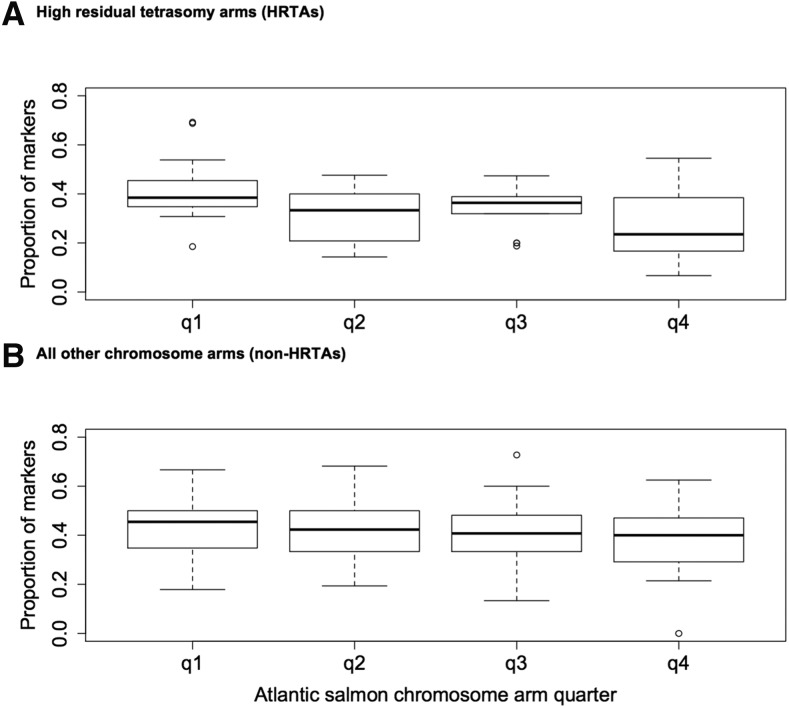
Recent studies of salmonid genomes have observed lower instances of sequence homology to TEs within genomic regions characterized as preserving tetrasomy (McKinney et al. 2015; Waples et al. 2015; Lien et al. 2016). We assessed TE distributions through BLASTn alignments to the Atlantic salmon genome to determine if chromosomes homologous to HRTA had lower levels of TE alignments. Of the 4405 linkage map SNPs, 1608 (36.5%) had homology with transposable elements in Repbase Update’s list of known vertebrate TEs (Jurka et al. 2005). This was a lower proportion of TE hits than expected as repetitive elements comprise 58–60% of the Atlantic salmon genome, and suggests that our dataset underrepresents the proportion of repetitive elements in Arctic charr. We detected a small but significant reduction in TE activity between HRTAs (30.9% of SNPs on these arms had significant TE hits), and non-HRTA chromosome arms (35.8%) (P = 0.0009) (Table 7). Using the Atlantic salmon genome as a scaffold, we detected no significant difference in the frequency of TE hits between the telomeric and centromeric regions of chromosomes (P = 0.1322) (Figure 6).
Table 7. TE distribution of the SNPs aligning to the Atlantic salmon chromosome arms, and PSVs markers not found in the linkage map.
| No TE Hits | TE Hits | |
|---|---|---|
| Linkage groups containing HRTAs | 976 | 436 |
| Linkage Groups with no HRTAs | 2572 | 1436 |
| Total | 3548 | 1872 |
There was a slight but significant reduction in TE activity seen on HRTA (30.9% of SNPs with significant TE hits) relative to all other chromosome arms (35.8% with significant TE hits) (P = 0.0009).
Figure 6.
Proportion of Arctic charr Map SNPs and PSVs with TE hits across the length of chromosome arms. SNPs were grouped based on their alignment to Atlantic salmon chromosome arms. q1 is the quarter closest to the centromere, q4 is the quarter closest to the telomere. Atlantic salmon chromosome arms were grouped according to whether they were HRTAs (A) or non-HRTAs (B).
Conclusion
We have presented a SNP-based linkage map of the Arctic charr genome, which is comprised of 4508 markers spanning 39 linkage groups. The map was used to identify the chromosome type of each linkage group, and the homologous chromosome arms in other salmonid species. Using data from the Atlantic salmon genome, we have identified putative homeologous arm pairs in Arctic charr. Based on the distribution of PSV, we suggest that genomic architecture is influencing diploidization rate in the Arctic charr genome, with higher levels of duplicate loci being preserved on HRTAs and lower numbers of duplicate loci preserved on AHPs. Transposon activity was also quantified, but we failed to detect a strong influence of genomic architecture on TE distribution. Pseudolinkage was also detected in both the male and female parents, and this involved two HRTA homeolog pairs (AC01q/21 and AC13q/34). This map also characterized the genome of a salmonid species with a more basal karyotype, and shows how these differences in genomic architecture have influenced diploidization.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.038026/-/DC1.
Acknowledgments
We thank Claude Pelletier from the Coastal Zones Research Institute, Shippagan, New Brunswick, for providing the mapping family. The project was overseen and coordinated by the Coastal Zones Research Institute and we thank Andre Dumas and Rodrigue Yossa for their important role in this regard. This work was funded by Natural Sciences and Engineering Research Council of Canada (NSERC) (Discovery Grant Program), and the Atlantic Innovation Fund/Atlantic Canada Opportunities Agency for the project “Aquaculture development and profitable commercialization of Arctic charr in Canada.”
Footnotes
Communicating editor: R. Houston
Literature Cited
- Allendorf F. W., Thorgaard G. H., 1984. Tetraploidy and the evolution of salmonid fishes, pp. 1–46 in Evolutionary Genetics of Fishes, edited by Turner B. J. Plenum Press, New York. [Google Scholar]
- Allendorf F. W., Bassham S., Cresko W. A., Limborg M. T., Seeb L. W., et al. , 2015. Effects of crossovers between homeologs on inheritance and population genomics in polyploid-derived salmonid fishes. J. Hered. 106: 217–227. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bergthorsson U., Andersson D. I., Roth J. R., 2007. Ohno’s dilemma: evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. USA 104: 17004–17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot C., Brunet F., Chalopin D., Juanchich A., Bernard M., et al. , 2014. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 5: 3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieuc M. S. O., Waters C. D., Seeb J. E., Naish K. A., 2014. A dense linkage map for chinook salmon (Oncorhynchus tshawytscha) reveals variable chromosomal divergence after an ancestral whole genome duplication event. G3 (Bethesda) 4: 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Danzmann, R. G., 2016 LINKMFEX: linkage analysis package for outcrossed families with male female exchange of the mapping parent. (Click on links to the ‘Software’ and ‘LINKMFEX’ folders.) Available at: http://www.uoguelph.ca/∼rdanzman/.
- Danzmann R. G., Gharbi K., 2001. Gene mapping in fishes: a means to an end. Genetica 111: 3–23. [DOI] [PubMed] [Google Scholar]
- Danzmann R. G., Davidson E. A., Ferguson M. M., Gharbi K., Koop B. F., et al. , 2008. Distribution of ancestral proto-Actinopterygian chromosome arms within the genomes of 4R-derivative salmonid fishes (Rainbow trout and Atlantic salmon). BMC Genomics 9: 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J. G., Yazawa R., Davidson W. S., Koop B. F., 2007. Bursts and horizontal evolution of DNA transposons in the speciation of pseudotetraploid salmonids. BMC Genomics 8: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K., et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubitz J. C., Casstevens T. M., Lu F., Harriman J., Elshire R. J., et al. , 2014. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9: e90346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen S., Lowe N. R., Cezard T., Gharbi K., Bishop S. C., et al. , 2014. Linkage maps of the Atlantic salmon (Salmo salar) genome derived from RAD sequencing. BMC Genomics 15: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley S. E., 1989. Chromosomes and constitutive heterochromatin distribution in Arctic charr, Salvelinus alpinus (L.) (Pisces: Salmonidae). Genetica 79: 161–166. [Google Scholar]
- Hermansen R. A., Hvidsten T. R., Sandve S. R., Liberles D. A., 2016. Extracting functional trends from whole genome duplication events using comparative genomics. Biol. Proced. Online 18: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Wang H., Michal J. J., Zhou X., Liu B., et al. , 2016. Genome wide sampling sequencing for SNP genotyping: methods, challenges and future development. Int. J. Biol. Sci. 12: 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V. V., Pavlicek A., Klonowski P., Kohany O., et al. , 2005. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110: 462–467. [DOI] [PubMed] [Google Scholar]
- Kasahara M., Naruse K., Sasaki S., Nakatani Y., Qu W., et al. , 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447: 714–719. [DOI] [PubMed] [Google Scholar]
- Kazazian H. H., Jr, 2004. Mobile elements: drivers of genome evolution. Science 303: 1626–1632. [DOI] [PubMed] [Google Scholar]
- Kodama M., Brieuc M. S. O., Devlin R. H., Hard J. J., Naish K. A., 2014. Comparative mapping between coho salmon (Oncorhynchus kisutch) and three other salmonids suggests a role for chromosomal rearrangements in the retention of duplicated regions following a whole genome duplication event. G3 (Bethesda) 4: 1717–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küttner E., Nilsson J., Skulason S., Gunnarsson S., Ferguson M. M., et al. , 2011. Sex chromosome polymorphisms in Arctic charr and their evolutionary origins. Genome 54: 852–861. [DOI] [PubMed] [Google Scholar]
- Larson W. A., McKinney G. J., Limborg M. T., Everett M. V., Seeb L. W., et al. , 2015. Identification of multiple QTL hotspots in sockeye salmon (Oncorhynchus nerka) using genotyping-by-sequencing and a dense linkage map. J. Hered. 107: 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S., Gidskehaug L., Moen T., Hayes B. J., Berg P. R., et al. , 2011. A dense SNP-based linkage map for Atlantic salmon (Salmo salar) reveals extended chromosome homeologies and striking differences in sex-specific recombination patterns. BMC Genomics 12: 615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S., Koop B. F., Sandve S. R., Miller J. R., Matthew P., et al. , 2016. The Atlantic salmon genome provides insights into rediploidization. Nature 533: 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limborg M. T., McKinney G., Seeb L. W., Seeb J. E., 2015. Recombination patterns reveal information about centromere location on linkage maps. Mol. Ecol. Resour. 16: 655–661. [DOI] [PubMed] [Google Scholar]
- Macqueen D. J., Johnston I. A., 2014. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc. Biol. Sci. 281: 20132881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margarido G. R. A., Souza A. P., Garcia A. A. F., 2007. Onemap: software for genetic mapping in outcrossing species. Hereditas 144: 78–79. [DOI] [PubMed] [Google Scholar]
- May B., Delany M. E., 2015. Meiotic models to explain classical linkage, pseudolinkage, and chromosomal pairing in tetraploid derivative salmonid genomes: II. Wright is still right. J. Hered. 106: 762–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney G., Seeb L., Larson W., Gomez-Uchida D., Limborg M., et al. , 2015. An integrated linkage map reveals candidate genes underlying adaptive variation in chinook salmon (Oncorhynchus tshawytscha). Mol. Ecol. Resour. 16: 769–783. [DOI] [PubMed] [Google Scholar]
- Mendivil Ramos O., Ferrier D. E., 2012. Mechanisms of gene duplication and translocation and progress towards understanding their relative contributions to animal genome evolution. Int. J. Evol. Biol. 2012: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadam H. K., Ferguson M. M., Danzmann R. G., 2005. Evidence for Hox gene duplication in rainbow trout (Oncorhynchus mykiss): a tetraploid model species. J. Mol. Evol. 61: 804–818. [DOI] [PubMed] [Google Scholar]
- Moghadam H. K., Ferguson M. M., Danzmann R. G., 2007. Linkage variation at the sex-determining locus within Fraser strain Arctic charr (Salvelinus alpinus). J. Fish Biol. 71: 294–301. [Google Scholar]
- Moghadam H. K., Ferguson M. M., Danzmann R. G., 2011. Whole genome duplication: challenges and considerations associated with sequence orthology assignment in Salmoninae. J. Fish Biol. 79: 561–574. [DOI] [PubMed] [Google Scholar]
- Nakatani Y., Takeda H., Kohara Y., Morishita S., 2007. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 17: 1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman J. D., Danzmann R. G., Glebe B., Ferguson M. M., 2011. The genetic basis of salinity tolerance traits in Arctic charr (Salvelinus alpinus). BMC Genet. 12: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman J. D., Robinson M., Glebe B., Ferguson M. M., Danzmann R. G., 2012. Genomic arrangement of salinity tolerance QTLs in salmonids: a comparative analysis of Atlantic salmon (Salmo salar) with Arctic charr (Salvelinus alpinus) and rainbow trout (Oncorhynchus mykiss). BMC Genomics 13: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman J. D., Ferguson M. M., Danzmann R. G., 2014. Transcriptomics of salinity tolerance capacity in Arctic charr (Salvelinus alpinus): a comparison of gene expression profiles between divergent QTL genotypes. Physiol. Genomics 46: 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., 1970. Evolution by Gene Duplication. Springer, New York. [Google Scholar]
- Ostberg C. O., Hauser L., Pritchard V. L., Garza J. C., Naish K. A., 2013. Chromosome rearrangements, recombination suppression, and limited segregation distortion in hybrids between Yellowstone cutthroat trout (Oncorhynchus clarkii bouvieri) and rainbow trout (O. mykiss). BMC Genomics 14: 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R., Ráb P., 2001. Chromosome evolution in the Salmonidae (Pisces): an update. Biol. Rev. Camb. Philos. Soc. 76: 1–25. [DOI] [PubMed] [Google Scholar]
- Phillips R. B., Matsuoka M. P., Reed K. M., 2002. Characterization of charr chromosomes using fluorescence in situ hybridization. Environ. Biol. Fishes 64: 223–228. [Google Scholar]
- Rexroad C. E., III, Palti Y., Gahr S. A., Vallejo R. L., 2008. A second generation genetic map for rainbow trout (Oncorhynchus mykiss). BMC Genet. 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Danzmann R. G., Gharbi K., Howard P., Ozaki A., et al. , 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155: 1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader L., Kim J. W., Ence D., Zimin A., Klein A., et al. , 2014. Transposable element islands facilitate adaptation to novel environments in an invasive species. Nat. Commun. 5: 1717–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. J., Kuraku S., Holt C., Sauka-Spengler T., Jiang N., et al. , 2013. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat. Genet. 45: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staton S. E., Burke J. M., 2015. Evolutionary transitions in the Asteraceae coincide with marked shifts in transposable element abundance. BMC Genomics 16: 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. S., Raes J., 2004. Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 38: 615–643. [DOI] [PubMed] [Google Scholar]
- Timusk E. R., Ferguson M. M., Moghadam H. K., Norman J. D., Wilson C. C., et al. , 2011. Genome evolution in the fish family Salmonidae: generation of a brook charr genetic map and comparisons among charrs (Arctic charr and brook charr) with rainbow trout. BMC Genomics 12: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H. Y., Robledo D., Lowe N. R., Bekaert M., Taggart J. B., et al. , 2016. Construction and annotation of a high density SNP linkage map of the Atlantic salmon (Salmo salar) genome. G3 (Bethesda) 6: 2173–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples R. K., Seeb L. W., Seeb J. E., 2015. Linkage mapping with paralogs exposes regions of residual tetrasomic inheritance in chum salmon (Oncorhynchus keta). Mol. Ecol. Resour. 16: 17–28. [DOI] [PubMed] [Google Scholar]
- Woram R. A., Gharbi K., Sakamoto T., Hoyheim B., Holm L., et al. , 2003. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 13: 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woram R. A., McGowan C., Stout J. A., Gharbi K., Ferguson M. M., et al. , 2004. A genetic linkage map for Arctic char (Salvelinus alpinus): evidence for higher recombination rates and segregation distortion in hybrid vs. pure strain mapping parents. Genome 47: 304–315. [DOI] [PubMed] [Google Scholar]
- Wright J. E., Johnson K., Hollister A., May B., 1983. Meiotic models to explain classical linkage, pseudolinkage and chromosome pairing in tetraploid derivative salmonid genomes. Isozymes 10: 239–260. [PubMed] [Google Scholar]
- Yano A., Guyomard R., Nicol B., Jouanno E., Quillet E., et al. , 2012. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 22: 1423–1428. [DOI] [PubMed] [Google Scholar]
- Yano A., Nicol B., Jouanno E., Quillet E., Fostier A., et al. , 2013. The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y- chromosome sequence in many salmonids. Evol. Appl. 6: 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Details on data used in this study can be found in Materials and Methods under the Sequencing Analysis section, and in the supplementary files.