Abstract
By combining 7077 SNPs and 61 microsatellites, we present the first linkage map for some of the early diverged lineages of the common frog, Rana temporaria, and the densest linkage map to date for this species. We found high homology with the published linkage maps of the Eastern and Western lineages but with differences in the order of some markers. Homology was also strong with the genome of the Tibetan frog Nanorana parkeri and we found high synteny with the clawed frog Xenopus tropicalis. We confirmed marked heterochiasmy between sexes and detected nonrecombining regions in several groups of the male linkage map. Contrary to the expectations set by the male heterogamety of the common frog, we did not find male heterozygosity excess in the chromosome previously shown to be linked to sex determination. Finally, we found blocks of loci showing strong transmission ratio distortion. These distorted genomic regions might be related to genetic incompatibilities between the parental populations, and are promising candidates for further investigation into the genetic basis of speciation and adaptation in the common frog.
Keywords: linkage map, Rana temporaria, Spanish lineages, recombination cold spots, heterochiasmy
Intra- and interspecific comparative genomic studies shed light on basic evolutionary processes such as sexual differentiation, adaptation, and speciation (Koonin et al. 2000; Kocher 2004; Nadeau and Jiggins 2010; Bachtrog 2013). While comparative genomic studies have mostly focused on model organisms, such as humans and mice (e.g., Bernstein et al. 2005), the advances in next-generation sequencing technology have facilitated genomic analysis in species lacking a reference genome (Kocher 2004; Miller et al. 2007; Gagnaire et al. 2013; Brelsford et al. 2016a). Amphibians are good models for comparative genomics due to their extraordinary biodiversity, worldwide distribution, and wide range of variation in genome size and karyotype (Duellman and Trueb 1986; Green and Sessions 1991). In particular, the common frog, Rana temporaria, has a wide distribution range across contrasting environmental conditions and very well reported cases of local adaptation (Miaud et al. 1999; Laurila et al. 2002; Laugen et al. 2003; Cano et al. 2004; Phillimore et al. 2010).
Phylogenetic analyses of the common frog have revealed multiple, deeply diverged evolutionary lineages or clades across Europe (Palo et al. 2004; Vences et al. 2013). There are two widely distributed Western and Eastern clades, which separated ∼0.7 MYA (million years ago) (Palo et al. 2004), and whose contact zone is in Central Europe (Schmeller et al. 2008; Teacher et al. 2009; Vences et al. 2013). In northwest Spain, another divergent evolutionary lineage, frequently referred to as subspecies R. t. parvipalmata (Seoane 1885), has been identified. This lineage is basal (i.e., divergence 1.12 MYA) to the Western and Eastern clades (Veith et al. 2002, 2003; Palo et al. 2004). In addition, there is evidence for at least one other lineage, a sister group of R. t. parvipalmata, also located in northern Spain (Vences et al. 2013).
Earlier linkage maps of R. temporaria have been constructed using individuals from the Western and Eastern clades (Cano et al. 2011; Rodrigues et al. 2013, 2016). The Western and Eastern lineage maps showed different locus order that might indicate potential genomic rearrangements (e.g., in linkage groups Xt3, Xt5, Xt6, and Xt7B), which might be linked to adaptive processes (Lee 2002). However, we still lack a comprehensive understanding of the genomic differences among lineages of the common frog, and especially about the genome structure of the early diverged lineages in the north of the Iberian Peninsula.
The aim of the present study is to compare recombination patterns, synteny, and putative sex-linked chromosomes of the Iberian lineages with more recently diverged lineages of the common frog. To accomplish these objectives, we used Restriction site-Associated DNA sequencing (RAD-seq) and microsatellite markers to construct a high-density consensus linkage map of R. temporaria, based on a cross between parents from the southwestern end of the species range in the north of the Iberian Peninsula.
Materials and Methods
Mapping family and DNA extraction
This study is based on a large full-sib family from a single R. temporaria cross. Adults were sampled in two localities from the North of Spain (Bárcena Mayor, Cantabria, N 43.13103 W 4.17879; and Vega de Candioches, León, N 42.99910 W 5.92124). We crossed a male from Candioches [high altitude: 1687 m.a.s.l. (metres above sea level)] and a female from Bárcena (low altitude: 551 m.a.s.l.) belonging to two different mtDNA clades (Vences et al. 2013; A. G. Nicieza, unpublished data). Frogs from these localities are exposed to contrasting environmental conditions (i.e., highland with short growing season and cold winters vs. lowland with long growing season and mild winters). The controlled cross was carried out in the laboratory of the Amphibian Facilities at the University of Oviedo. Female and male abdomens were pressed gently by hand to obtain eggs and sperm without harming the animals. Tadpoles were individualized at Gosner stage 25 and were kept at 12 hr light:12 hr dark photoperiod and 14° until they reached Gosner stage 42. Then, they were killed with an overdose of benzocaine and frozen. DNA from the brains of parents and 184 F1 offspring was extracted with the DNeasy Blood and Tissue Kit (QIAGEN). DNA quality was assessed via 0.7% agarose gel electrophoresis and DNA concentration was determined with a Qubit fluorometer. DNA extractions were diluted to 100 ng/μl for subsequent library preparation.
RAD-seq
Two libraries were prepared according to the slightly modified protocol of Elshire et al. (2011) (Supplemental Material, File S1). Briefly, DNA was digested with restriction enzymes PstI, and BamHI and the fragments of each individual were ligated to one of the 94 modified Illumina adapters, which contain barcode sequences, with T4 DNA ligase. Adapter-ligated DNA was combined to two library pools consisting of 92 offspring and the two parents in each pool. After purification, DNA fragments of a certain size (300–600 bp range) were selected in each library using an E-Gel iBase Power System as in Pukk et al. (2015). Size-selected fragments were amplified by a PCR using 18 cycles and purified with a QIAquick PCR purification kit (QIAGEN). An Agilent 2100 Bioanalyzer system was used to check the quality and quantity of size-selected and amplified libraries. At the end, fragments of average 400 bp length (80% of fragments with size range 300–550 bp) from the two libraries were sequenced on two paired-end lanes (2 × 100) with the Illumina HiSequation 2000 in the Illumina Genome Analyzer platform at the Center for Genomic Regulation in Barcelona, Spain.
Genotyping
Raw Illumina reads with low quality were discarded as well as reads with ambiguous barcode sequences. As the forward and reverse reads did not overlap, forward reads were used for the first part of the analysis. We demultiplexed the sequences and barcodes were trimmed as a result of this process. Low quality ends were discarded resulting in 91 bp reads. Because of the lack of a reference genome for R. temporaria, the reads from the presumably heterogametic parent (i.e., sire) were used to generate the reference sequence. To that end, identical reads were first collapsed with FASTQ/A Collapser of the FASTX-toolkit (Gordon and Hannon 2010). Second, the remaining sire’s sequences were clustered with CD-HIT EST (Li 2015) at 90% of similarity. An assembly de novo was performed with the resulting contigs in the MIRA assembler (Chevreux 2005) and a sire-based reference sequence was obtained for each contig. Third, the reads of the dam and sire were aligned to this reference sequence using the software Bowtie2 (Langmead and Salzberg 2012). The dataset of forward read contigs was obtained after discarding contigs mapped with < 10 reads or > 1000 reads for each parent, as well as those with > 10% of mismatch. We used the information from the forward read to select the corresponding reverse read and create a dataset for both forward and reverse read contigs. Fourthly, to select informative loci, we conducted parental SNP calling with the software Samtools (Li et al. 2009). A heterozygotic genotype was called when the minor allele frequency (MAF) among reads was > 0.1. Uninformative SNPs (i.e., both parents with alternative homozygous genotypes) were discarded. Finally, the selected SNPs were used to map the progeny reads. Following DaCosta and Sorenson (2014) and minimizing the mismatching, contigs mapped with < 200 or > 20,000 total reads were rejected.
SNP-calling was performed again with a quality threshold of 100 and a maximum of four SNPs allowed per contig. Parents and progeny were deemed heterozygotes if the MAF was > 0.1 and SNPs were excluded if > 25% of progeny genotypes were missing and > 6% of progeny showed noncompatible genotypes. SNPs showing segregation distortion were not included in the initial linkage maps (X2 test, p < 0.05). However, to later determine potentially incompatible regions between the two lineages, we retained loci showing relatively mild segregation distortion (X2 test, p-value 0.05–0.005) for construction of new linkage maps and permutation analysis (see Transmission ratio distortion). Genotyping error was calculated for both sire and dam by comparing replicated genotype calls obtained from two separate sequencing lanes.
Microsatellites
In addition to the SNPs, microsatellite markers were also included to enable direct comparison with the previous maps (Cano et al. 2011; Rodrigues et al. 2013). From 116 tested markers (File S3), 113 microsatellites were successfully amplified within 19 multiplex reactions following the protocol of the QIAGEN Multiplex PCR Master Mix (2 ×) at half volume (25 μl). Amplification reactions started with an initial polymerase activation step at 95° for 15 min, followed by 40 cycles of: 94° for 30 sec, 55° (for the markers: Rib01, Rib 06, Rib 08, and Rib15) or 60° (for the rest) for 90 sec, and 72° for 60 sec; and finally, an extension stage of 60° for 30 min. Microsatellite analysis was performed using an ABI 3100 automatic DNA Sequencer.
Genotypes were determined using the software GENEMARKER v 2.4 (Soft Genetics, State College, PA). Uninformative microsatellites, as well as those with null alleles, were excluded from subsequent analysis (File S3).
As for the SNPs from the RAD-seq, microsatellites showing Mendelian segregation violations (X2 test, p < 0.05) were removed before the linkage analysis.
Map construction
Markers, both SNPs and microsatellites, were assigned according to their segregation pattern to five categories: nnxnp, lmxll, efxeg, abxcd, and hkxhk. Sex-specific maps were constructed from informative markers of each sex (nnxnp and lmxll) using the cross type “doubled haploid” (DH) and Maximum Likelihood algorithm in MSTmap (Wu et al. 2008). This algorithm has been shown to be more successful at ordering the loci compared to other methods, such as weighted least squares and minimum sum of adjacent recombination fractions (Hackett and Broadfoot 2003). The Kosambi mapping function was used to calculate the genetic distance between markers and a p-value of 1 × 10−8 was used as the threshold for clustering the markers into the linkage groups. Due to the lack of linkage phase information, the dataset was duplicated, changing the phase of each marker (Gadau et al. 2001; Brelsford et al. 2016a). Duplicated linkage groups were eliminated manually in the output. As a conservative approach, we used the option “detect bad data” in the software. In addition, possible double crossovers were transformed to unknown (U) iteratively followed by an additional round of linkage map reconstruction, until no possible double crossover was found.
The average linkage map was created using the option appropriate for an outbred full-sib family (cross pollinator, CP) in Joinmap 4.1 (Van Ooijen 2011). Identical loci were excluded to decrease computational time and added again after map construction. Linkage groups were identified with a LOD threshold of eight. Small linkage groups (i.e., < 4 markers) were excluded from further analysis. The order of the markers within each linkage group was determined with the Maximum Likelihood mapping algorithm, which assumes no crossover interference. Therefore, the distance between markers was calculated using the Haldane mapping function. Spatial sampling was used with five thresholds: 0.1, 0.05, 0.03, 0.02, and 0.01. Three map optimization rounds were run in each spatial sample, using the following parameters: chain length was 10,000, cooling control parameter was 0.0001, and the rounds were stopped after 100,000 chains without improvement. Finally, for the multipoint estimation of recombination frequencies, we used a burn-in of 100,000, five cycles of Monte Carlo Expectation Maximization (chain length per cycle: 100,000), and six sampling periods for recombination frequency. Stabilization of the recombination frequencies was monitored with the sum of recombination frequencies of adjacent fragments and the mean number of recombination.
The name of the linkage groups follows the Xenopus tropicalis homology. The expected genome length and observed genome coverage were calculated according to Chakravarti et al. (1991) and Cervera et al. (2001), respectively. The observed length (GO) was the sum of the observed length of the linkage groups. The expected length (GE) was the sum of the expected length of the linkage groups, which was calculated multiplying the observed length of each linkage group by the factor (m + 1) / (m − 1), m being the number of markers of the linkage group. The observed genome coverage, understood as the proportion of the genome comprised in our recombination map, was the ratio GO/GE.
Transmission ratio distortion markers
Earlier studies have demonstrated that excluding markers that show segregation distortion from linkage mapping may result in the exclusion of certain chromosome regions from the map (Cervera et al. 2001; Doucleff et al. 2004). However, a large number of distorted markers may also increase the chance of type I errors and may result in inaccurate estimation of genetic distances (Cervera et al. 2001). Therefore, we included only markers with moderate levels of segregation distortion (X2, p-values from 0.05 to 0.005) in the linkage analysis. None of the markers caused big gaps (i.e., >50 cM) of recombination distance. To identify distorted regions, instead of only distorted markers, we used a kernel smoothing and permutation test (for details see Bruneaux et al. (2013) and Ozerov et al. (2016)). By taking into account differences in marker density, this strategy increases the statistical power for detecting regions where several adjacent markers show high distortion. For this analysis, we ran one million permutations in the R software v. 3.0.2 (R Core Team 2013).
Analysis of the homology
The genomes from X. tropicalis (version 9.0, xenbase.org) and Nanorana parkeri (version 2, gigadb.org) were used to evaluate the homology and the synteny among amphibian genomes. References from both forward and reverse sire contigs containing SNPs of the linkage map were aligned to a draft genome of R. temporaria (File S2) using Bowtie2 (Langmead and Salzberg 2012). The draft scaffolds were selected when forward and reverse references aligned within 600 bp from each other because, during library construction, the size of the fragments was <550 bp. This allowed the use of longer sequences for interspecific comparisons, increasing the power of the homology searches. We searched for homology in NCBI Nucleotide (http://ncbi.nlm.nih.gov) and Swissprot (http://uniprot.org) databases as well as in X. tropicalis and N. parkeri genomes using blast. Homologous sequences of other species were retained if the best hit e-value was five orders of magnitude higher than the second-best hit e-value (Brelsford et al. 2016a,b). To visualize the synteny between R. temporaria and X. tropicalis, we used the software Circos plot v 0.69-2 (Krzywinski et al. 2009) as in Yang et al. (2014).
Data availability
File S1 contains a detailed protocol of the library preparation. File S2 contains the pipeline for the construction of the draft genome. File S3 contains detailed information about the microsatellites used (i.e., accession numbers, core motifs, and references). File S4 contains sex-specific and average maps and a comparison with previous microsatellite maps. File S5 contains supplementary figures and tables being: Figure S1, linear model that fits the relation between male and female number of markers; Figure S2, linkage group specific male vs. female recombination lengths; Figure S3, map length and number of markers compared to Swiss map; Figure S4, kernel smoothing results; Table S1, summary of distorted maps, linkage group lengths, number of markers, and sex-specific recombination rates. File S6 contains a figure of the distorted sex-specific map. File S7 contains sex-specific distorted maps and the results of the kernel analysis. File S8 contains the genotypic information of the individuals. The draft genome is available at https://figshare.com/s/c0ff6bacbfc4572e1ff1 and marker sequences are available at https://figshare.com/s/24fa6c7cd2f133467207.
Results
Microsatellite and RAD analysis
Out of the 113 amplified microsatellites, 77 were heterozygous in at least one parent. Twelve of them possessed null alleles and four loci showed Mendelian segregation violation (p-value < 0.0001), resulting in a final set of 61 informative microsatellites (File S3). In the whole dataset, 19 offspring exhibited three alleles in at least one microsatellite locus, suggesting potential partial trisomy, and were excluded from subsequent analysis.
A total of 162 F1 offspring were used to construct the linkage map, after discarding three individuals with < 5000 reads. Approximately 634 million reads were retained after the quality filtering. Over 10 million reads were obtained from each parent, while on average 3.8 million of reads were sequenced per offspring (ranging from 2 to 6.8 million). The assembly from the parental read alignment resulted in 88,497 contigs. From those, 13,203 contigs were polymorphic and were used for mapping the reads from the progeny and SNP calling. Over 13,600 SNPs were initially identified, from which 7217 markers passed a X2 test for Mendelian segregation (p-value > 0.05). The estimated genotyping error inferred based on independent technical replicates was ∼5% (5.5% sire and 5.3% dam).
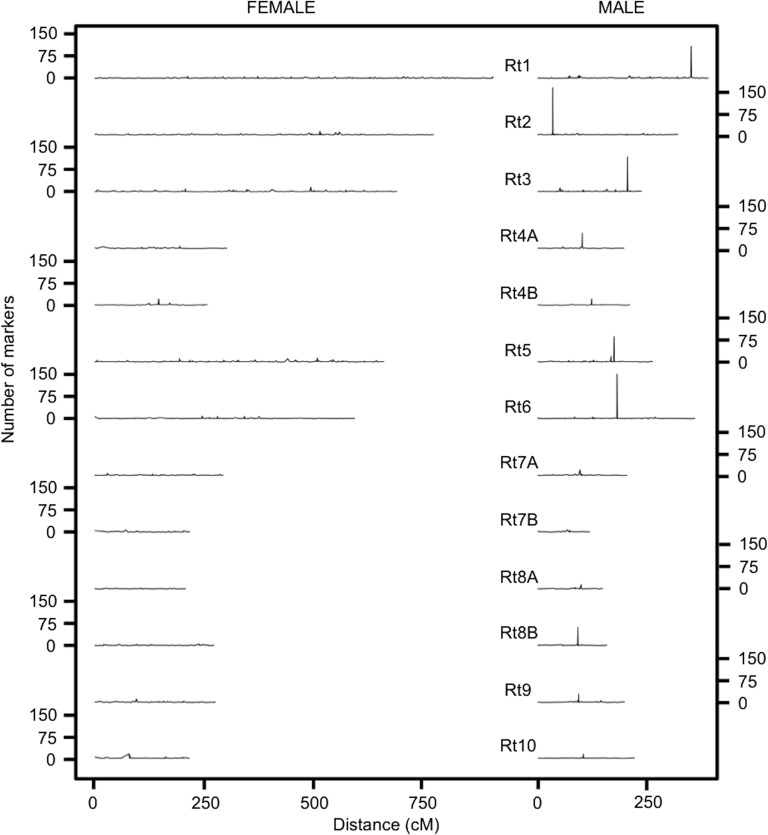
Sex-specific linkage maps were constructed using 3644 markers (51 microsatellites and 3593 SNPs) and 3180 markers (51 microsatellites and 3129 SNPs), for female and male respectively. After discarding linkage groups formed by <4 markers, 13 linkage groups were recovered in both maps (Table 1). A total of 3596 markers in the female map and 3105 markers in the male map were assigned to the 13 linkage groups (File S4). Our map presented an observed genome coverage of 99% for both sexes. The average recombination rate was 1.35 times higher and the total length was roughly twofold (1.87 times) larger in the female map than in the male map. Interestingly, all male linkage groups contained a cluster of markers with zero recombination, henceforth referred to as recombination cold spot (Figure 1). The number of clustered nonrecombining markers was higher in the largest linkage groups (i.e., Rt1, Rt2, Rt3, Rt5, and Rt6) ranging from 86 to 159 markers, while 9–62 nonrecombining markers were detected in smaller linkage groups in the male map. The nonrecombining regions in the female map were generally smaller (<23 markers). However, the linkage groups Rt4B, Rt7B, and Rt10 showed nonrecombining regions of similar length in both sexes (Table 1).
Table 1. Linkage group length, total number of markers, recombination rate, and number of markers in the nonrecombining region by sex.
| LG | Length (cM) | No. Markers | Recombination Rate | No. Markers Within Cold Spot | ||||
|---|---|---|---|---|---|---|---|---|
| Male | Female | Male | Female | Male | Female | Male | Female | |
| Rt1 | 392.838 | 914.948 | 551 | 631 | 0.71 | 1.45 | 108 | 8 |
| Rt2 | 323.074 | 778.775 | 435 | 497 | 0.74 | 1.57 | 159 | 13 |
| Rt3 | 239.262 | 694.948 | 383 | 453 | 0.62 | 1.53 | 117 | 16 |
| Rt4A | 199.211 | 304.537 | 178 | 160 | 1.12 | 1.9 | 52 | 9 |
| Rt4B | 212.494 | 259.109 | 114 | 161 | 1.86 | 1.61 | 22 | 22 |
| Rt5 | 265.076 | 664.933 | 323 | 481 | 0.82 | 1.38 | 86 | 7 |
| Rt6 | 362.237 | 597.954 | 422 | 339 | 0.86 | 1.76 | 150 | 10 |
| Rt7A | 205.762 | 295.579 | 144 | 177 | 1.43 | 1.67 | 19 | 8 |
| Rt7B | 119.879 | 218.716 | 70 | 125 | 1.71 | 1.75 | 9 | 9 |
| Rt8A | 149.881 | 209.416 | 85 | 96 | 1.76 | 2.18 | 15 | 4 |
| Rt8B | 159.626 | 273.802 | 135 | 158 | 1.18 | 1.73 | 62 | 6 |
| Rt9 | 200.646 | 277.64 | 133 | 173 | 1.51 | 1.6 | 28 | 12 |
| Rt10 | 222.21 | 217.664 | 132 | 145 | 1.68 | 1.5 | 16 | 17 |
| Total | 3052.196 | 5708.021 | 3105 | 3596 | 1.23 | 1.67 | ||
LG, linkage group; No., number.
Figure 1.
Difference in recombination between sexes. While the female map shows a more uniform recombination rate across all linkage groups, the male map exhibits nonrecombining regions as peaks of high marker density.
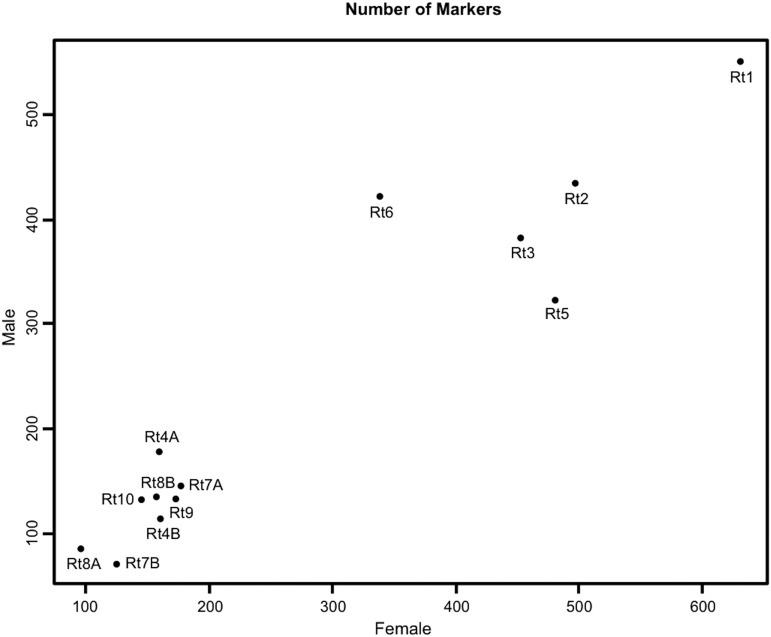
Our results showed that the linkage group Rt1 is the longest and contains the largest number of markers in both sexes (Figure 1 and Figure 2). The female map presented a slightly higher number of markers per linkage group, except for -Rt4A and Rt6 (Table 1). Rt6 deviated from the general trend due to a larger number of markers in the male than in the female map (Figure 2 and Figure S1). Furthermore, female map showed higher recombination length than the male map but some linkage groups deviated above or below from the general trend (Figure S2). For instance, Rt6 was longer in the male map and Rt10 was shorter in the female map compared to other linkage groups. Rt3 exhibited the lowest recombination rate in the male map. Finally, Rt7B was the shortest linkage group in the male map with the smallest number of markers (Table 1).
Figure 2.
Female vs. male number of markers for each linkage group.
Compared with the recent RAD-based linkage map in the common frog (Brelsford et al. 2016b), the number of markers and recombination length of the linkage groups in our study was similar (coefficient of determination, female map length R2 = 0.83, number of markers in female map R2 = 0.92, number of markers in male map R2 = 0.90, all p-values < 0.05) except for the length of the male linkage groups (coefficient of determination, R2 = 0.09, p-value > 0.05) (Figure S3). The lack of correlation between length of male linkage groups and the number of markers supports the independence between number of crossovers and chromosome size in males.
Transmission ratio distortion
In order to evaluate the segregation distortion patterns along the chromosomes, 521 and 401 distorted markers of dam and sire, respectively, were added to construct new sex-specific maps. Among the distorted markers, a total of 500 SNPs segregating in the dam (12.2% of total number of markers) and 375 SNPs segregating in the sire (10.8% of total number of markers) were successfully assigned within the 13 linkage groups (File S7). The resulting map was 663 cM longer in the female and 398 cM longer in the male compared to the maps without including distorted markers. Average recombination rate was similar in distorted and nondistorted maps (Table S1). The distribution of the distorted markers exhibited a nonuniform pattern along and among linkage groups (File S6). For example, a large number of distorted markers was observed on Rt1 in contrast with the low number on Rt4B and Rt7A. Furthermore, there were differences between sexes. Kernel smoothing results showed a higher distortion in the female than in the male map (Figure S4). Only Rt2, Rt5, Rt7B, Rt8B, and Rt9 from the male map exhibited regions with signals of high distortion based on the kernel analysis, while all female linkage groups except Rt9 contained at least one area with an excess of distorted markers. Interestingly, Rt6 and Rt9 from the male map and Rt7B from the female map showed clusters of distorted markers within the recombination cold spots (Figure 3).
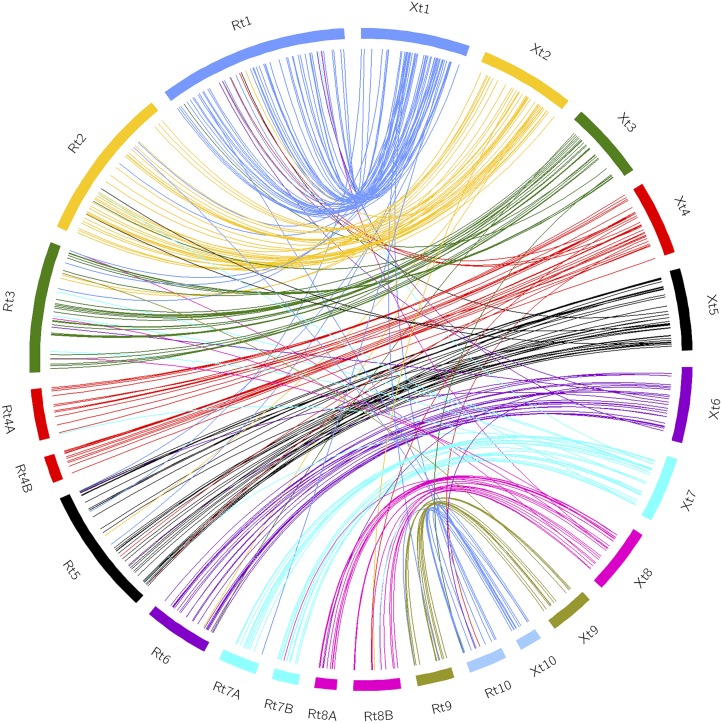
Figure 3.
Circos plot showing the strong synteny between X. tropicalis and R. temporaria. To obtain better visualization, the number of base pairs of the R. temporaria linkage groups was multiplied by 100.
Average map
We used 7278 markers (61 microsatellites and 7217 SNPs) to construct the average linkage map and 7138 markers were successfully assigned to 13 linkage groups. All the linkage groups reached stabilization after five Monte Carlo Expectation Maximization cycles. All markers were located within the same linkage groups as in sex-separated maps. However, as expected, the average map showed less accurate marker order than the female map.
Similarity analysis
A total of 4161 forward and reverse reference sequences from the sire were aligned to the R. temporaria draft genome. In 2359 cases (56.7%), the pair of forward and reverse reference sequences aligned onto the same scaffold within 600 bp (range from 190 to 473 bp). From a total of 2331 scaffolds selected, 1184 aligned to the N. parkeri genome (50.8%) and 305 aligned to the X. tropicalis genome (13%). In addition, 81% of homologous sequences between R. temporaria and X. tropicalis were located in the same chromosome and the synteny was also strong, as is shown in the Circos plot (Figure 3).
Discussion
We have generated the densest linkage map for the common frog to date. The combined use of SNPs and microsatellites allowed in-depth comparison with the existing linkage maps for the species allowing, for the first time, a detailed analysis of the early diverged Iberian lineages in relation to the more recently diverged Western and Eastern lineages.
Both the number and relative length of our linkage groups matched the karyotype described for the species, with five large and eight short chromosomes (Spasić-Bošković et al. 1997). Furthermore, we observed a strong synteny between the R. temporaria linkage map and the genome of X. tropicalis. Heterochiasmy was pronounced, with a lower recombination rate in the male map. Finally, some regions exhibited a strong transmission ratio distortion, which might be indicative of genetic incompatibilities between the lineages used in our experimental cross. Below, we discuss in detail the recombination patterns and evolutionary processes that may have shaped the differences found among the lineages of R. temporaria.
Comparison with previous microsatellite maps
The obtained consensus linkage map was consistent with previous microsatellite maps based on the Western and Eastern lineages (Cano et al. 2011; Rodrigues et al. 2013, 2016). However, compared to the first linkage map based on Swedish populations from the Eastern lineage (Cano et al. 2011), our map revealed several differences. For example, markers that showed significant linkage in groups 1, 4, and 12 from Cano et al. (2011) were unlinked in our map and located in other linkage groups (File S4). On the other hand, linkage groups 15 and 2 from Cano et al. (2011) were joined within the largest group, Rt1, in our map (File S4). Separation of these two linkage groups in Cano et al. (2011) was likely due to lower coverage. Since Rodrigues et al. (2016) also found similar inconsistencies in their map based on Swedish populations, the observed discrepancies may be caused by differences in coverage and methodology. Nevertheless, our results revealed frequent change in marker order compared to the published maps (File S4), hence, further research is needed to determine if some of these changes can reflect the occurrence of independent chromosomal rearrangements, such as inversions, among the different lineages.
Recombination rate
Similar to earlier studies, we detected large differences in recombination rate between sexes (Berset-Brändli et al. 2008; Cano et al. 2011; Rodrigues et al. 2013; Brelsford et al. 2016b). The average recombination rate was 1.36 times larger in the female than in the male map. This heterochiasmy is smaller than that observed in maps from the Eastern (1.76 times; Cano et al. 2011) and Western lineages, both based solely on microsatellite markers (82.5 times; Rodrigues et al. 2013) and SNPs (3.15 times; Brelsford et al. 2016b). In relation to the total length of the maps, our female map was 1.87 times larger than the male map, similar to the first Eastern clade map (1.52 times, Cano et al. 2011) but more discordant with the second Eastern clade map (17.4 times, Rodrigues et al. 2016) and the Western clade maps, which ranged from 3.37 to 72 times (Rodrigues et al. 2013; Brelsford et al. 2016b). All in all, the heterochiasmy pattern observed in this study was similar to the first Eastern lineage map (Cano et al. 2011), which was also based on an outcross. More families from the North Iberian lineages would be needed to confirm these findings.
Concerning the sex-specific recombination patterns, female recombination rates were fairly constant overall while male recombination rates were reduced in the majority of the linkage groups (Figure 1). Similar to Brelsford et al. (2016b), a cluster of markers with no recombination occurred in every chromosome, forming an extensive suppressed-recombination region in the largest linkage groups. In amphibians, male recombination is usually restricted to telomeric areas and the nonrecombining region is near the center of the linkage group (Morescalchi and Galgano 1973), coinciding with the centromere and paracentromere area (King 1991). This pattern was found in Hyla arborea (Brelsford et al. 2016a), and it is in accordance with the metacentric/submetacentric chromosomes described in the karyotype of R. temporaria based on populations from the Eastern lineage (Spasić-Bošković et al. 1997). However, the position of the nonrecombining regions in our map was shifted to one side in Rt1, Rt2, and Rt3 (Figure 1). Furthermore, subtelomeric/telomeric recombination cold spots were also observed in some linkage groups of the Western clade [see Figure 1 in Brelsford et al. (2016b)]. The authors suggested that the shifted position in their map could be due to a lack of coverage at the chromosome ends (A. Brelsford, personal communication). However, because of the higher coverage of the current map and the good homology with the terminal regions of the chromosomes in X. tropicalis, the map presented here reflects the actual position of the recombination cold spots in the chromosomes well. Hence, our results call for further karyotype and whole-genome sequencing work between the Iberian and more recently diverged lineages to establish whether centromere repositioning has occurred in the species.
The Iberian Peninsula has been suggested as the place where R. temporaria originated (Vences et al. 2013) and the Northwest Iberian lineages diverged 1.12 MYA from the lineage for which the karyotype of the species was described (i.e., Balkans), being isolated in different refugia during the last glaciation. Centromere repositioning events have been observed at similar evolutionary time scales but only at the interspecific level. For example, five centromere repositioning events have occurred between donkeys and zebras, which diverged from each other 1–2.78 MYA (Carbone et al. 2006; Vilstrup et al. 2013).
Putative sex linkage group
R. temporaria, as is the case in other Rana species, exhibits male heterogamety (XY). If sex determination is strictly genetic and there is no recombination between sex-chromosomes, X and Y are expected to accumulate differences as a result of large rearrangements and/or degeneration of the Y chromosome (Charlesworth and Charlesworth 2000; Charlesworth et al. 2005). Thus, over time, heterogamety is expected to result in an excess of heterozygotes in the linkage group(s) containing the sex-determining gene(s). Based on this expectation, and evidence in H. arborea frogs, Brelsford et al. (2016a) proposed a method to detect sex chromosomes based solely on heterozygosity differences among linkage groups.
Nevertheless, X and Y chromosomes are homomorphic in the common frog and recombination between them still seems to occur as reported for other amphibian species (Stöck et al. 2013; Dufresnes et al. 2014). Perrin (2009) suggested that occasional sex-reversal events could maintain X–Y recombination because XY females would prevent the decay of the Y chromosome. In fact, spontaneous sex-reversed individuals, including observed XX males and possibly XY females, have been documented in some populations (Matsuba et al. 2008; Perrin 2009; Alho et al. 2010). Previous studies found several sex-linked markers in the linkage group Rt1 and it has been considered as the putative sex chromosome for the species (e.g., Matsuba et al. 2008; Alho et al. 2010; Cano et al. 2011; Rodrigues et al. 2013). However, both Brelsford et al. (2016b) and our study failed to find the expected heterozygosity excess in Rt1 as a putative sex chromosome. Since we lacked phenotypic sex information for our family, we cannot establish whether nongenetic sex determination plays a role in our results, although this was the case in Brelsford et al. (2016b).
On the other hand, Rt6 showed markedly higher male heterozygosity in our cross (Figure 2 and Figure S1) and harbored two sex-linked markers in the Finnish population of Kilpisjärvi (BFG267 and BFG239; C. Matsuba, unpublished data). Therefore, this linkage group warrants further research to determine its potential involvement in sex determination or whether another source of differentiation among the lineages crossed is causing this large male heterozygosity. The possibility of multiple chromosomes participating in the sex determination of the common frog has also been discussed in previous work (Cano et al. 2011; Rodrigues et al. 2013, 2014) and demonstrated recently by Rodrigues et al. (2016). Alternatively, a large autosomal supergene with two differentiated haplogroups in the crossed lineages could also produce higher male heterozygosity, if the male were heterozygous and the female were homozygous (Brelsford et al. 2016a).
Segregation distortion
Many biological processes acting before or after fertilization can cause transmission ratio distortion (Fishman et al. 2001; Kuittinen et al. 2004). We found clusters of distorted markers in the nonrecombining regions of the linkage groups Rt6 and Rt9 in the male and Rt7B in the female, based on the randomization and kernel smoothing approach. There are nongenetic sources of segregation distortion such as genotyping errors, sampling biases, and comigration (Rogers et al. 2007; Zhou et al. 2015). Our genotyping-by-sequencing dataset contained ∼5% of genotyping errors based on analysis of replicated samples, despite the rather strict quality control. However, such technical artifacts are not expected to systematically cluster together in relation to other loci. Thus, the distorted genomic regions with blocks of distorted markers found in this study are likely related to biological processes such as meiotic drive, lineage incompatibilities, or outbreeding depression (Zhou et al. 2015). Since we observed high mortality and deformity rates in our experiment, these genomic regions are good candidates to investigate potential genetic incompatibilities. Furthermore, loci with non-Mendelian inheritance could have greater evolutionary importance than our current knowledge suggests (Lyttle 1991; Taylor and Ingvarsson 2003). However, analysis of gametes and replicate families generated preferably using backcross breeding design would be needed to further understand the relationship between genome structure and segregation distortion.
Homology analysis
The comparative genomic analysis between R. temporaria, X. tropicalis, and N. parkeri supported the strong homology among amphibian genomes (Brelsford et al. 2013, 2016a). As expected from their close phylogenetic relationship (Pyron and Wiens 2011), N. parkeri showed higher homology with R. temporaria. These species diverged 90 MYA while X. tropicalis and R. temporaria split ∼208 MYA (divergence times retrieved from timetree.org). The estimated homology between R. temporaria and X. tropicalis in this study (12.8%) is slightly higher than that observed (10%) for the Western lineage of this species (Brelsford et al. 2016b). Our study confirmed the finding of Brelsford et al. (2016b), that X. tropicalis chromosomes 4, 7, and 8 were split into two pairs in R. temporaria.
Conclusions
The constructed high-density consensus linkage map provides an important resource for further research in the evolutionary biology of R. temporaria, facilitating the search for genes of adaptive relevance. In addition, due to the conserved synteny among amphibians, this linkage map represents a valuable tool for further comparative genomic studies. Our work indicates that the genome structure is generally conserved between common frog lineages while the position of the recombination cold spots and marker order can vary. Finally, genomic regions showing strong transmission distortion found here are promising candidates for studying incipient speciation processes.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.036459/-/DC1.
Acknowledgments
We thank Cristina García for help in fieldwork and tadpole care and Leticia Viesca for help with laboratory tasks. We are indebted to Luming Yang who kindly shared his Circos plot script with us. We also thank Alan Brelsford for assistance with data analysis and interpretation and Nicolas Perrin for providing additional details about his published work. We are grateful to John Loehr for improving English grammar. Our research was supported by the following grants: Spanish Ministry of Education (reference CGL2011-23443), Ministry of Economy and Competitiveness (reference BES-2012-055220), National Parks Autonomous Agency (reference MARM 428/211), Estonian Ministry of Education and Research (institutional research funding project IUT8-2), and the Academy of Finland (grant no. 266321).
Footnotes
Communicating editor: J. A. Birchler
Literature Cited
- Alho J. S., Matsuba C., Merilä J., 2010. Sex reversal and primary sex ratios in the common frog (Rana temporaria). Mol. Ecol. 19(9): 1763–1773. [DOI] [PubMed] [Google Scholar]
- Bachtrog D., 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14(2): 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D. K., et al. , 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120(2): 169–181. [DOI] [PubMed] [Google Scholar]
- Berset-Brändli L., Jaquiéry J., Broquet T., Ulrich Y., Perrin N., 2008. Extreme heterochiasmy and nascent sex chromosomes in European tree frogs. Proc. R. Soc. Lond. B Biol. Sci. 275(1642): 1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelsford A., Stöck M., Betto‐Colliard C., Dubey S., Dufresnes C., et al. , 2013. Homologous sex chromosomes in three deeply divergent anuran species. Evolution 67(8): 2434–2440. [DOI] [PubMed] [Google Scholar]
- Brelsford A., Dufresnes C., Perrin N., 2016a High-density sex-specific linkage maps of a European tree frog (Hyla arborea) identify the sex chromosome without information on offspring sex. Heredity 116(2): 177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelsford A., Rodrigues N., Perrin N., 2016b High‐density linkage maps fail to detect any genetic component to sex determination in a Rana temporaria family. J. Evol. Biol. 29(1): 220–225. [DOI] [PubMed] [Google Scholar]
- Bruneaux M., Johnston S. E., Herczeg G., Merilä J., Primmer C. R., et al. , 2013. Molecular evolutionary and population genomic analysis of the nine‐spined stickleback using a modified restriction‐site‐associated DNA tag approach. Mol. Ecol. 22(3): 565–582. [DOI] [PubMed] [Google Scholar]
- Cano J. M., Laurila A., Palo J., Merilä J., 2004. Population differentiation in G matrix structure due to natural selection in Rana temporaria. Evolution 58(9): 2013–2020. [DOI] [PubMed] [Google Scholar]
- Cano J. M., Li M.-H., Laurila A., Vilkki J., Merilä J., 2011. First-generation linkage map for the common frog Rana temporaria reveals sex-linkage group. Heredity 107(6): 530–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L., Nergadze S. G., Magnani E., Misceo D., Cardone M. F., et al. , 2006. Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics 87(6): 777–782. [DOI] [PubMed] [Google Scholar]
- Cervera M.-T., Storme V., Ivens B., Gusmao J., Liu B. H., et al. , 2001. Dense genetic linkage maps of three Populus species (Populus deltoides, P. nigra and P. trichocarpa) based on AFLP and microsatellite markers. Genetics 158(2): 787–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A., Lasher L. K., Reefer J. E., 1991. A maximum likelihood method for estimating genome length using genetic linkage data. Genetics 128(1): 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 2000. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355(1403): 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Marais G., 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95(2): 118–128. [DOI] [PubMed] [Google Scholar]
- Chevreux, B., 2005 MIRA: an automated genome and EST assembler. Ph.D. Thesis, German Cancer Research Center Heidelberg, Heidelberg.
- DaCosta J. M., Sorenson M. D., 2014. Amplification biases and consistent recovery of loci in a double-digest RAD-seq protocol. PLoS One 9(9): e106713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucleff M., Jin Y., Gao F., Riaz S., Krivanek A. F., et al. , 2004. A genetic linkage map of grape, utilizing Vitis rupestris and Vitis arizonica. Theor. Appl. Genet. 109(6): 1178–1187. [DOI] [PubMed] [Google Scholar]
- Duellman W. E., Trueb L., 1986. Biology of Amphibians. JHU Press, Baltimore, MD. [Google Scholar]
- Dufresnes C., Stöck M., Brelsford A., Perrin N., 2014. Range-wide sex-chromosome sequence similarity supports occasional XY recombination in European tree frogs (Hyla arborea). PLoS One 9(6): e97959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K., et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6(5): e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L., Kelly A. J., Morgan E., Willis J. H., 2001. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159(4): 1701–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadau J., Gerloff C. U., Krüger N., Chan H., Schmid‐Hempel P., et al. , 2001. A linkage analysis of sex determination in Bombus terrestris (L.) (Hymenoptera: Apidae). Heredity 87(2): 234–242. [DOI] [PubMed] [Google Scholar]
- Gagnaire P. A., Normandeau E., Pavey S. A., Bernatchez L., 2013. Mapping phenotypic, expression and transmission ratio distortion QTL using RAD markers in the Lake Whitefish (Coregonus clupeaformis). Mol. Ecol. 22(11): 3036–3048. [DOI] [PubMed] [Google Scholar]
- Gordon A., Hannon G. J., 2010. Fastx-toolkit. FASTQ/A short-reads pre-processing tools. Available online at: http://hannonlab.cshl.edu/fastx_toolkit/
- Green D. M., Sessions S. K., 1991. Amphibian Cytogenetics and Evolution. Academic Press, San Diego, CA. [Google Scholar]
- Hackett C. A., Broadfoot L. B., 2003. Effects of genotyping errors, missing values and segregation distortion in molecular marker data on the construction of linkage maps. Heredity 90(1): 33–38. [DOI] [PubMed] [Google Scholar]
- King M., 1991. The evolution of heterochromatin in the amphibian genome, pp. 359–391 in Amphibian Cytogenetics and Evolution, edited by Green D. M., Sessions S. K. Academic Press, San Diego, CA. [Google Scholar]
- Kocher T. D., 2004. Adaptive evolution and explosive speciation: the cichlid fish model. Nat. Rev. Genet. 5(4): 288–298. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Aravind L., Kondrashov A. S., 2000. The impact of comparative genomics on our understanding of evolution. Cell 101(6): 573–576. [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., et al. , 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9): 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuittinen H., de Haan A. A., Vogl C., Oikarinen S., Leppälä J., et al. , 2004. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168(3): 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9(4): 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugen A. T., Laurila A., Räsänen K., Merilä J., 2003. Latitudinal countergradient variation in the common frog (Rana temporaria) development rates – evidence for local adaptation. J. Evol. Biol. 16(5): 996–1005. [DOI] [PubMed] [Google Scholar]
- Laurila A., Karttunen S., Merilä J., 2002. Adaptive phenotypic plasticity and genetics of larval life histories in two Rana temporaria populations. Evolution 56(3): 617–627. [DOI] [PubMed] [Google Scholar]
- Lee C. E., 2002. Evolutionary genetics of invasive species. Trends Ecol. Evol. 17(8): 386–391. [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16): 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., 2015. Fast program for clustering and comparing large sets of protein or nucleotide sequences, pp. 173–177 in Encyclopedia of Metagenomics, edited by Nelson K. E. Springer, New York, NY. [Google Scholar]
- Lyttle T. W., 1991. Segregation distorters. Annu. Rev. Genet. 25(1): 511–581. [DOI] [PubMed] [Google Scholar]
- Matsuba C., Miura I., Merilä J., 2008. Disentangling genetic vs. environmental causes of sex determination in the common frog, Rana temporaria. BMC Genet. 9(1): 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaud C., Guyétant R., Elmberg J., 1999. Variations in life-history traits in the common frog Rana temporaria (Amphibia: Anura): a literature review and new data from the French Alps. J. Zool. (Lond.) 249(01): 61–73. [Google Scholar]
- Miller M. R., Dunham J. P., Amores A., Cresko W. A., Johnson E. A., 2007. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 17(2): 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morescalchi A., Galgano M., 1973. Meiotic chromosomes and their taxonomic value in amphibia Anura. Caldasia 11: 41–50. [Google Scholar]
- Nadeau N. J., Jiggins C. D., 2010. A golden age for evolutionary genetics? Genomic studies of adaptation in natural populations. Trends Genet. 26(11): 484–492. [DOI] [PubMed] [Google Scholar]
- Ozerov M. Y., Gross R., Bruneaux M., Vähä J. P., Burimski O., et al. , 2016. Genome‐wide introgressive hybridization patterns in wild Atlantic salmon influenced by inadvertent gene flow from hatchery releases. Mol. Ecol. 25: 1275–1290. [DOI] [PubMed] [Google Scholar]
- Palo J. U., Schmeller D. S., Laurila A., Primmer C. R., Kuzmin S. L., et al. , 2004. High degree of population subdivision in a widespread amphibian. Mol. Ecol. 13(9): 2631–2644. [DOI] [PubMed] [Google Scholar]
- Perrin N., 2009. Sex reversal: a fountain of youth for sex chromosomes? Evolution 63(12): 3043–3049. [DOI] [PubMed] [Google Scholar]
- Phillimore A. B., Hadfield J. D., Jones O. R., Smithers R. J., 2010. Differences in spawning date between populations of common frog reveal local adaptation. Proc. Natl. Acad. Sci. USA 107(18): 8292–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukk L., Ahmad F., Hasan S., Kisand V., Gross R., et al. , 2015. Less is more: extreme genome complexity reduction with ddRAD using Ion Torrent semiconductor technology. Mol. Ecol. Resour. 15(5): 1145–1152. [DOI] [PubMed] [Google Scholar]
- Pyron R. A., Wiens J. J., 2011. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 61(2): 543–583. [DOI] [PubMed] [Google Scholar]
- R Core Team , 2013. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rodrigues N., Betto‐Colliard C., Jourdan‐Pineau H., Perrin N., 2013. Within‐population polymorphism of sex‐determination systems in the common frog (Rana temporaria). J. Evol. Biol. 26(7): 1569–1577. [DOI] [PubMed] [Google Scholar]
- Rodrigues N., Merilä J., Patrelle C., Perrin N., 2014. Geographic variation in sex‐chromosome differentiation in the common frog (Rana temporaria). Mol. Ecol. 23(14): 3409–3418. [DOI] [PubMed] [Google Scholar]
- Rodrigues N., Vuille Y., Brelsford A., Merilä J., Perrin N., 2016. The genetic contribution to sex determination and number of sex chromosomes vary among populations of common frogs (Rana temporaria). Heredity 117(1): 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S. M., Isabel N., Bernatchez L., 2007. Linkage maps of the dwarf and normal lake whitefish (Coregonus clupeaformis) species complex and their hybrids reveal the genetic architecture of population divergence. Genetics 175(1): 375–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeller D. S., Palo J. U., Merilä J., 2008. A contact zone between two distinct Rana temporaria lineages in northern Germany. Alytes 25(3–4): 93–98. [Google Scholar]
- Seoane V. L., 1885. On two forms of Rana from NW Spain. The Zoologist 9: 169–172. [Google Scholar]
- Spasić-Bošković O., Tanić N., Blagojević J., Vujošević M., 1997. Comparative cytogenetic analysis of European brown frogs: Rana temporaria, R. dalmatina and R. graeca. Caryologia 50(2): 139–149. [Google Scholar]
- Stöck M., Savary R., Betto‐Colliard C., Biollay S., Jourdan‐Pineau H., et al. , 2013. Low rates of X‐Y recombination, not turnovers, account for homomorphic sex chromosomes in several diploid species of Palearctic green toads (Bufo viridis subgroup). J. Evol. Biol. 26(3): 674–682. [DOI] [PubMed] [Google Scholar]
- Taylor D. R., Ingvarsson P. K., 2003. Common features of segregation distortion in plants and animals. Genetica 117(1): 27–35. [DOI] [PubMed] [Google Scholar]
- Teacher A. G. F., Garner T. W. J., Nichols R. A., 2009. European phylogeography of the common frog (Rana temporaria): routes of postglacial colonization into the British Isles, and evidence for an Irish glacial refugium. Heredity 102(5): 490–496. [DOI] [PubMed] [Google Scholar]
- Van Ooijen J. W., 2011. Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet. Res. 93(05): 343–349. [DOI] [PubMed] [Google Scholar]
- Veith M., Vences M., Vieites D. R., Nieto-Roman S., Palanca A., 2002. Genetic differentiation and population structure within Spanish common frogs (Rana temporaria complex; Ranidae, Amphibia). Folia Zool. 51(4): 307–318. [Google Scholar]
- Veith M., Kosuch J., Vences M., 2003. Climatic oscillations triggered post-Messinian speciation of Western Palearctic brown frogs (Amphibia, Ranidae). Mol. Phylogenet. Evol. 26(2): 310–327. [DOI] [PubMed] [Google Scholar]
- Vences M., Hauswaldt J. S., Steinfartz S., Rupp O., Goesmann A., et al. , 2013. Radically different phylogeographies and patterns of genetic variation in two European brown frogs, genus Rana. Mol. Phylogenet. Evol. 68(3): 657–670. [DOI] [PubMed] [Google Scholar]
- Vilstrup J. T., Seguin-Orlando A., Stiller M., Ginolhac A., Raghavan M., et al. , 2013. Mitochondrial phylogenomics of modern and ancient equids. PLoS One 8(2): e55950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Bhat P. R., Close T. J., Lonardi S., 2008. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 4(10): e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Koo D. H., Li D., Zhang T., Jiang J., et al. , 2014. Next‐generation sequencing, FISH mapping and synteny‐based modeling reveal mechanisms of decreasing dysploidy in Cucumis. Plant J. 77(1): 16–30. [DOI] [PubMed] [Google Scholar]
- Zhou W., Tang Z., Hou J., Hu N., Yin T., 2015. Genetic map construction and detection of genetic loci underlying segregation distortion in an intraspecific cross of Populus deltoides. PLoS One 10(5): e0126077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
File S1 contains a detailed protocol of the library preparation. File S2 contains the pipeline for the construction of the draft genome. File S3 contains detailed information about the microsatellites used (i.e., accession numbers, core motifs, and references). File S4 contains sex-specific and average maps and a comparison with previous microsatellite maps. File S5 contains supplementary figures and tables being: Figure S1, linear model that fits the relation between male and female number of markers; Figure S2, linkage group specific male vs. female recombination lengths; Figure S3, map length and number of markers compared to Swiss map; Figure S4, kernel smoothing results; Table S1, summary of distorted maps, linkage group lengths, number of markers, and sex-specific recombination rates. File S6 contains a figure of the distorted sex-specific map. File S7 contains sex-specific distorted maps and the results of the kernel analysis. File S8 contains the genotypic information of the individuals. The draft genome is available at https://figshare.com/s/c0ff6bacbfc4572e1ff1 and marker sequences are available at https://figshare.com/s/24fa6c7cd2f133467207.